Introduction
NOVA alternative splicing regulator 1 (NOVA1) is a
member of the NOVA family (1–6). The
role of NOVA1 was first reported in neurological diseases (6). For example, NOVA1 regulates neuronal
microRNA (miRNA/miR) expression via a variety of mechanisms
(6). Decreased NOVA1 expression
was observed in the gastric cancer microenvironment, and in tumor
cells was correlated with the progression and poor prognosis of
gastric cancer (4); however, the
role of NOVA1 in regulating the epithelial to mesenchymal
transition (EMT) in gastric cancer remains unknown.
miRs represent a class of small non-coding RNA
molecules and are important post-transcriptional regulators that
target key genes, such as NOVA1, which has been associated with the
progression of gastric cancer (7).
Numerous miRs are dysregulated in gastric cancer (8). For example, the expression of
miR-27a-3p was increased in gastric cancer tissues and cell lines
(8). Experimental evidence has
demonstrated that miR-27a-3p is a potential oncogene (9–15).
It was reported that miR-27a-3p can contribute to progression of
gastric cancer by regulating different genes, such as BTG
antiproliferation factor 2 (9,10).
Circulating miR-27a-3p has been proposed as a biomarker for the
early diagnosis and prognostic evaluation of patients with gastric
cancer (11); high expression
levels of miR-27a-3p were associated with poor overall survival
(12). Additionally, miR-27a-3p
expression within tumors has been proposed as a potential biomarker
for predicting chemosensitivity and chemotherapy resistance in
metastatic gastric cancer (12–15).
EMT serves critical roles in the regulation of chemosensitivity
(16); however, the mechanism
underlying the effects of miR-27a-3p on EMT in gastric cancer
requires further investigation.
Materials and methods
Clinical specimens
The present study was approved by the Ethics
Committees of Renmin Hospital of Wuhan University (Wuhan, China)
and Cixi People's Hospital (Cixi, China). Written informed consent
was obtained from patients prior to the collection of tissues.
Gastric cancer samples were obtained from those diagnosed with
primary gastric cancer. Patients did not receive preoperative
chemotherapy or radiation at Renmin Hospital of Wuhan University
and Cixi People's Hospital between January 1990 and January 2002.
In total, 108 gastric cancer samples were immediately frozen in
liquid nitrogen following surgical resection and were then stored
at −80°C. The patients, (75 males and 33 females; age range, 35–90
years). Samples of the corresponding adjacent normal tissue were
obtained >3 cm away from the site at which the primary tumor was
sampled (17). All tumor tissues
and adjacent normal tissues were reviewed in a blind manner by two
pathologists. The duration of the follow-up was 9 years. The AJCC
Cancer Staging system was used for tumor classification (18).
Cell culture
The AGS gastric cancer cell line obtained from
Changhai Hospital, Second Military Medical University (Shanghai,
China) (19) were cultured with
RPMI-1640 medium (Sigma-Aldrich; Merck KGaA) containing 10% fetal
bovine serum (Shanghai ExCell Biology, Inc.) and 100 mg/ml
penicillin and 100 mg/ml streptomycin (Gibco; Thermo Fisher
Scientific, Inc.) in a humidified atmosphere with 5% CO2
at 37°C. Cellular morphologies were observed using an optical
microscopy (Leica Microsystems, Inc.).
Scramble- and NOVA1-short hairpin
RNA-expressing plasmids, control miR (mock)/Pre-miR-27a-3p
Mock and NOVA1-shRNA expressing plasmids (Sequence,
5′-CCAGATAAGCTTGCCACCATGGAAGGGGCCC-3′) were obtained from Tiangen
Biotech Co., Ltd. Mock (5′-CACUCGGACCCUGAGCUGCCG-3′) and
Pre-miR-27a-3p (5′-UUCACAGUGGCUAAGUUCCGC-3′) were obtained from
Ambion, Inc. (Ambion; Thermo Fisher Scientific, Inc.).
Transfection experiments
Transfection experiments were performed as described
previously (20). Cells
(8×102 cells/well) were seeded in six-well plates prior
to transfection for each experiment. For transfection experiments,
cells were cultured in serum-free medium without antibiotics at 60%
confluence for 24 h. Subsequently, cells were using FuGENE HD
transfection reagent (Promega Corporation) according to the
manufacturer's protocol. In total, 5 µg Pre-miR-27a-3p or control
miR mimics and 10 µg NOVA1 or mock-shRNA expressing plasmids were
used. After incubation for 6 h, the medium was removed and replaced
with serum-free medium without antibiotics, and cells were
incubated for 24 h. Then, the western blot analysis,
Immunofluorescence assay and luciferase reporter assay and reverse
transcription-quantitative polymerase chain reaction (RT-qPCR) were
performed as described below.
Western blot analysis
Western blot analysis was performed as previously
described (20). Total protein was
prepared using extraction buffer comprising NaCl/Pi
containing 0.5% Triton X-100, 1 mM EDTA, 1 mM PMSF and 1X complete
protease inhibitors (Roche Diagnostics). The concentration of each
protein lysate was determined using a bicinchoninic acid protein
assay kit (Thermo Fisher Scientific, Inc.). Total protein (50
µg/lane) was separated by 12% SDS-PAGE. Subsequently, samples were
transferred to nitrocellulose membranes and blocked for 60 min at
room temperature in 5% skimmed milk. The membranes were
immunoblotted using the following primary antibodies:
Anti-fibronectin (cat. no. ab2413; 1:500; Abcam), anti-N-cadherin
(cat. no. ab18203;1:500; Abcam), anti-E-cadherin (cat. no. ab40772;
1:500; Abcam), anti-keratin 8 (cat. no. ab53280; 1:500; Abcam),
anti-NOVA1 (cat. no. ab183024; 1:500; Abcam) and anti-β-actin (cat.
no. ab5694; 1:500; Abcam) overnight at 4°C. Subsequently,
anti-rabbit secondary antibody (cat. no. ab6940; 1:10,000; Abcam)
was used to incubate membranes for 30 min at room temperature. The
specific protein bands were visualized by Odyssey Infrared Imaging
system (LI-COR Biosciences). β-actin expression was used as an
internal control to confirm equal loading of the protein
samples.
Bioinformatics analysis
Bioinformatics analysis was performed as described
previously (21). miRanda (release
date, August 2010; http://www.microrna.org/microrna/home.do) was used to
identify the target genes of miR-27a-3p.Immunofluorescence
assay. Immunofluorescence assay was performed as described
previously (20). After
transfection with plasmids, the cells were fixed in 4%
paraformaldehyde for 15 min at room temperature, and then blocked
with 20% goat serum (Gibco; Thermo Fisher Scientific, Inc.)
blocking solution for 20 min at room temperature.
Antibody staining
Rabbit antibody against NOVA1 (cat. no. ab183024;
1:200 dilution; Abcam) were added, and the mixtures were incubated
overnight at 4°C. After washing three times with
NaCl/Pi, cells were incubated with appropriate
Cy2-conjugated secondary antibodies (cat. no. ab6940; 1:10,000;
Abcam) for 30 min at 37°C. After washing with NaCl/Pi, the samples
were observed under a laser scanning confocal microscope (Leica
Microsystems). DAPI staining was used to stain nuclei for 30 min at
37°C. Representative images (magnifications, ×100) are presented.
Fluorescence intensities were analyzed using ImageJ software
(version 1.37v; http://rsb.info.nih.gov/ij/index.html).
Immunocytochemistry analysis
Immunocytochemistry was performed as described
previously (22).
Immunohistochemistry was performed on 10% formalin-fixed for 24 h,
paraffin-embedded tissue sections using the envision peroxidase
detection method. Sections (thickness, 4-µm) were deparaffinized in
xylene for 10 min at 4°C, dehydrated through graded ethanol series
and subsequently treated with 0.3% hydrogen peroxide in methanol
for 30 min at room temperature to eliminate the endogenous
peroxidase activity. For antigen retrieval, the sections were
microwaved in 10 mM citrate buffer (pH 6.0) for 10 min. The primary
antibody anti-CRP (cat. no. ab211631; 1:200, Abcam) was incubated
for 12 h at 4°C after blocking non-specific binding with 10% normal
goat serum (Gibco; Thermo Fisher Scientific, Inc.) in PBS for 30
min at room temperature. Anti-rabbit secondary antibody (cat. no.
ab211631; ab150077; 1:10,000, Abcam) was used for 60 min at room
temperature. Sections were incubated with envision peroxidase
complex (Dako; Agilent Technologies, Inc.) for 20 min at room
temperature. Sections were counterstained with Mayer's hematoxylin
for 10 min at room temperature after immunostaining prior to
mounting. If ≥10% cells were stained, the expression levels of CRP
were defined as ‘High’ expression. Otherwise, the expression levels
of CRP were defined as ‘low’ expression.
Luciferase reporter assay
Luciferase reporter plasmids containing the
3′-untranslated region (3′-UTR) of human NOVA1 mRNA were obtained
from Tiangen Biotech Co., Ltd. Luciferase reporter assay was
performed as described previously (20). For reporter assays, cells was
transiently transfected with reporter plasmids (Tiangen Biotech
Co.) and miRNA using Lipofectamine 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). Reporter assays were performed 36 h
post-transfection using the Dual-luciferase assay-system (Promega
Corporation), normalized for transfection efficiency by
cotransfected Renilla-luciferase.
Real-time PCR for miRNA
expression
The analysis of miR expression via Real-time PCR was
performed as described (23).
Total RNA was isolated from the cells or tissues using the mirVana
miRNA Isolation kit (cat. no. AM1561; Ambion; Thermo Fisher
Scientific, Inc.). The detection of the mature form of miRNAs was
performed using the mirVana RT-qPCR miRNA Detection kit (SYBR
Green) and RT-qPCR Primer Sets, according to the manufacturer's
instructions (Ambion; Thermo Fisher Scientific, Inc.). For the qPCR
of miR-27a-3p, the forward primer was: 5′-CAGTTCACAGTGGCTAAGA-3′;
and the reverse primer was: 5′-CAGTTTTTTTTTTTTTTTGCGGAA-3′. The
quantification was performed using the 2−∆∆Cq method
(24). The U6 small nuclear RNA
was used as an internal control. The thermocycling conditions were
as follows: One cycle at 50°C for 2 min, one cycle at 95°C 10 min
and 40 cycles of 95°C for 15 sec and 60°C for 30 sec. The primers
used were as follows: U6 forward, 5′-GCTTCGGCAGCACATATACTAAAAT-3′;
reverse, 5′-CGCTTCACGAATTTGCGTGTCAT-3′.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
The analysis of mRNA via RT-qPCR was performed as
described previously (20).
Briefly, total cellular RNA was extracted from cultured cells using
TRIzol reagent (Invitrogen, Thermo Fisher Scientific, Inc.) and 2
µg total RNA was reverse-transcribed using M-MLV reverse
transcriptase (Promega Corporation) according to the manufacturer's
protocol. The thermocycling conditions were as follows:
Denaturation for 30 sec at 95°C, annealing for 45 sec at 52–58°C
depending on the primers used, and extension for 45 sec at 72°C.
Each PCR reaction was performed for 28–32 cycles. qRT-PCR was
performed using a StepOne™ real-time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Fast SYBR Green Master
Mix was obtained from Applied Biosystems (Thermo Fisher Scientific,
Inc.). Data are shown as a relative expression level after
normalization to GAPDH. The primers employed were as follows: NOVA1
forward, 5′-TACTGAGCGAGTGTGCTTGAT-3′, reverse,
5′-GTCTGGGGTTGTAGAATGCTG-3′; E-cadherin forward,
5′-TCAACGATCCTGACCAGCAGTTCG-3′, reverse,
5′-GGTGAACCATCATCTGTGGCGATG-3′; keratin 8 forward,
5′-GAAGACCACCAGCGGCTATG-3′, reverse, 5′-GTCAGAGGACTCAGACACCAG-3′;
fibronectin forward, 5′-TTTTGACAACGGGAAGCATTATCAGATAA-3′; reverse,
5′-TGATCAAAACATTTCTCAGCTATTGG-3′; N-cadherin forward,
5′-CATCCCTCCAATCAACTTGC-3′, reverse 5′-ATGTGCCCTCAAATGAAACC-3′; and
GAPDH forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse,
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. The quantification of PCR performed
was performed using the 2−ΔΔCq method (24).
Statistical analysis
The results were analyzed using SAS software
(version 9.4; SAS Institute, Inc.). Samples were analyzed with a
Student's t-test for the comparison of two groups (two-tailed),
unless otherwise indicated, such as a χ2 test. Data are
presented as the mean ± SEM from experiments performed in
triplicate. Overall survival was analyzed via a Kaplan-Meier test.
The log-rank test was used to examine the statistical significance
of the difference between groups. Survival was investigated based
on NOVA1 expression (two-tailed). P<0.05 was considered to
indicate a statistically significant difference.
Results
miR-27a-3p expression is inversely
associated with overall survival and overexpression of miR-27a-3p
promotes EMT in AGS cells
To detect miR-27a-3p expression in adjacent normal
tissues and gastric cancer tissues, we performed RT-qPCR. We
isolated microRNA from 10 pairs of normal tissues and gastric
cancer tissues. The results revealed that miR-27a-3p expression
levels were significantly upregulated in gastric cancer tissues
compared with normal tissues (Fig.
1A). To determine whether miR-27a-3p expression was associated
with the overall survival of patients with gastric cancer, we
analyzed tumor miR-27a-3p expression in patients with an overall
survival ≥40 months and those with an overall survival <40
months. We observed that miR-27a-3p expression was significantly
reduced in those with an overall survival ≥40 months than those
with an overall survival <40 months (Fig. 1B). The results indicated that
miR-27a-3p could be associated with progression of gastric
cancer.
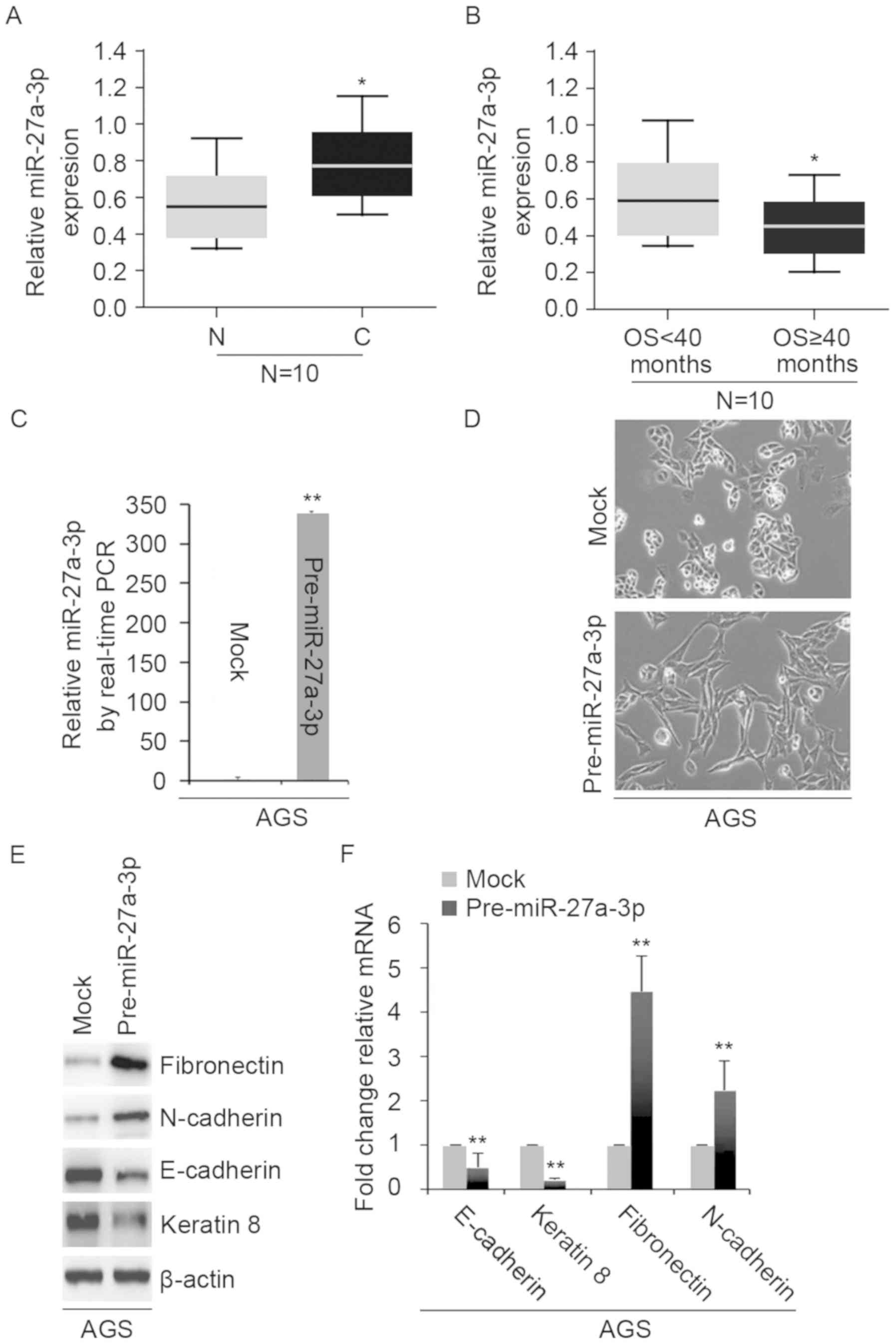 | Figure 1.miR-27a-3p expression is inversely
associated with overall survival and overexpression of miR-27a-3p
promotes epithelial-mesenchymal transition in AGS cells. (A)
RT-qPCR for the analysis of miR-27a-3p expression in 10 pairs of
gastric cancer tissues and adjacent normal tissues. *P<0.05 vs.
N. (B) RT-qPCR for the analysis of miR-27a-3p expression in gastric
cancer tissues and adjacent normal tissues. *P<0.05 vs. OS<40
months. (C) RT-qPCR for the analysis of miR-27a-3p expression in
AGS cells transfected as indicated. **P<0.01 vs. mock. (D) AGS
cells were transfected as indicated and analyzed via microscopy
(n=3), Scale bars, 50 µm. (E) Western blot analysis for
fibronectin, N-cadherin, E-cadherin and keratin 8 expression in AGS
cells transfected as indicated (n=3). (F) RT-qPCR for fibronectin,
N-cadherin, E-cadherin and keratin 8 in AGS cells transfected as
indicated. **P<0.01 vs. mock. C, cancer tissues; miR, microRNA;
mock, control miR; N, adjacent normal tissues; OS, overall
survival; RT-qPCR, reverse transcription-quantitative polymerase
chain reaction. |
To investigate the role of miR-27a-3p in
vitro, we transfected AGS cells with control miR and
pre-miR-27a-3p, we performed RT-qPCR. We observed that miR-27a-3p
expression was significantly increased in the cells transfected
with pre-miR-27a-3p than in the control group (Fig. 1C); overexpression of miR-27a-3p
revealed visible changes in the morphology of AGS cells (Fig. 1D). The phenotype was changed from a
cobblestone-like to a spindle-like morphology. To confirm that
these morphological changes in AGS cells were promoted by EMT, we
performed western blotting and RT-qPCR to examine the expression of
epithelial and mesenchymal markers in AGS cells. We observed that
the expression of E-cadherin and keratin 8 (epithelial markers) was
markedly suppressed, whereas that of N-cadherin and fibronectin
(mesenchymal markers) were promoted by pre-miR-27a-3p compared with
in the control group (Fig. 1E and
F).
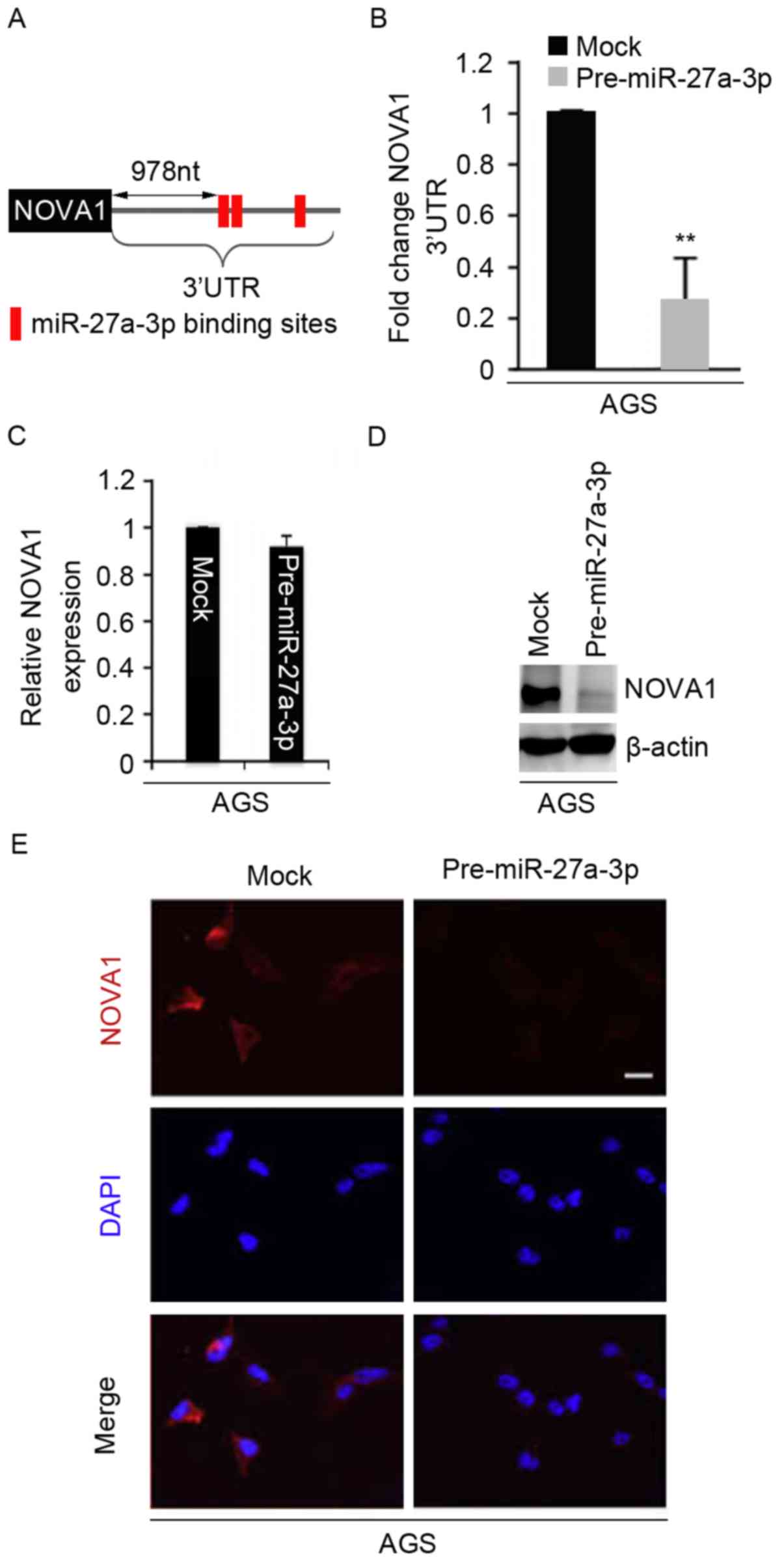
miR-27a-3p inhibits NOVA1 expression
in AGS cells
miR-27a-3p expression was determined to be inversely
associated with overall survival and overexpression of miR-27a-3p
promoted EMT in AGS cells. We sought to determine the mechanism
underlying the effects of miR-27a-3p. miRNAs are a novel class of
small (~22 nucleotides) noncoding RNAs, and negatively regulate
protein-coding gene expression by targeting mRNA degradation or
inhibit translation (9–11). Thus, we suggested that miR-27a-3p
might promote EMT by regulating the expression of its target genes.
To identify the target genes of miR-27a-3p, we used miRanda, a
commonly used prediction algorithm (http://www.microrna.org/microrna/home.do). Numerous
target genes, including NOVA1, were identified. Decreased NOVA1
expression has been observed in gastric cancer, and correlated with
the progression and poor prognosis of this disease (4). Thus, we reasoned that miR-27a-3p may
promote EMT by regulating NOVA1 expression. The target sites on the
3′UTR of NOVA1 are presented in Fig.
2A. To examine whether miR-27a-3p regulates the 3′UTR of NOVA1,
we performed a luciferase reporter assay. We observed that the
luciferase activities of luciferase reporter plasmids were
significantly inhibited by pre-miR-27a-3p compared with the
control. To identify whether miR-27a-3p could regulate NOVA1
expression in AGS cells, we used RT-qPCR to determine NOVA1 mRNA
expression. The results demonstrated that miR-27a-3p did not
inhibit NOVA1 mRNA expression (Fig.
2C). Furthermore, we performed western blotting and an
immunofluorescence assay to examine NOVA1 protein expression in AGS
cells. We observed that NOVA1 protein expression was notably
inhibited by pre-miR-27a-3p (Fig. 2D
and E).
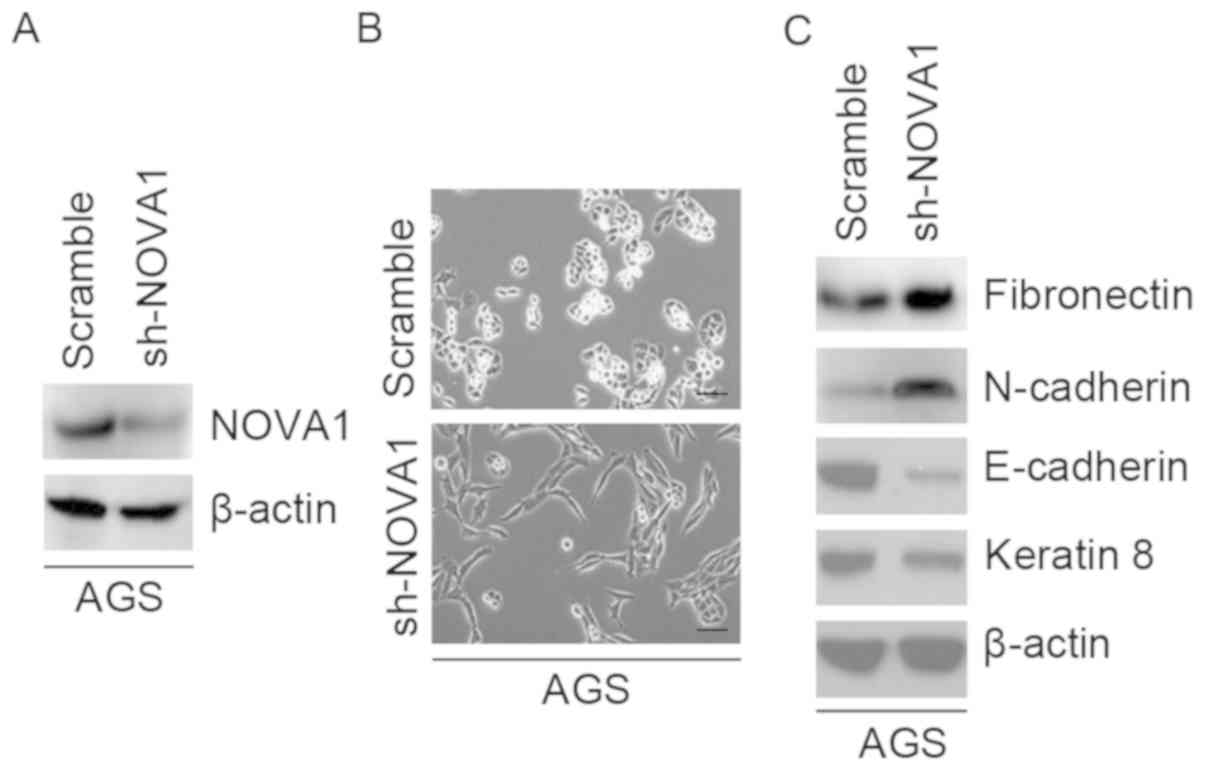
Silencing of NOVA1 promotes EMT in AGS
cells
To study the role of NOVA1 in gastric cancer, we
transfected AGS cells with shNOVA1- or scramble-expression
plasmids. We observed that NOVA1 protein expression was inhibited
by shNOVA1 (Fig. 3A). Silencing of
NOVA1 promoted visible changes in the morphology of AGS cells
(Fig. 3B). The phenotype was
changed from a spindle-like to a cobblestone-like morphology. In
order to confirm whether the morphological changes of AGS cells
were promoted by EMT, we performed western blotting to examine the
expression of epithelial and mesenchymal markers in AGS cells. We
observed that E-cadherin and keratin 8 (epithelial markers) were
downregulated, while that of N-cadherin and fibronectin
(mesenchymal markers) were promoted by shNOVA1 (Fig. 3C).
Association between NOVA1 expression
and the clinicopathological features of gastric cancer
We observed that the expression of NOVA1 was
associated with the majority of the clinicopathological variables
(age, sex, tumor size and histological classification); however,
decreased NOVA1 expression was linked to lymph node metastasis and
tumor stages (Table I).
 | Table I.NOVA1 expression in relation to the
clinicopathological characteristics of gastric cancer. |
Table I.
NOVA1 expression in relation to the
clinicopathological characteristics of gastric cancer.
|
|
| NOVA1
expression |
|
|---|
|
|
|
|
|
|---|
| Clinicopathological
features | n | Positive | Negative
(%)a |
P-valuesb |
|---|
| Age (median) |
| 65 | 64 | >0.05 |
| Sex |
|
|
|
|
| M | 75 | 21 | 54 (73) | >0.05 |
| F | 33 | 7 | 26 (80) |
|
| Tumor size
(cm) |
| 7.1 | 7.6 | >0.05 |
| Histology |
|
|
| >0.05 |
|
Undifferentiated
carcinoma | 50 | 11 | 39 (77) |
|
|
Differentiated carcinoma | 58 | 16 | 42 (73) |
|
| Lymph node
metastasis |
|
|
| <0.05 |
| − | 34 | 10 | 24 (72) |
|
| + | 74 | 18 | 56 (76) |
|
| Stage |
|
|
| <0.05 |
| 1 | 21 | 6 | 15 (70) |
|
| 2 | 20 | 6 | 14 (69) |
|
| 3 | 29 | 8 | 21 (72) |
|
| 4 | 38 | 7 | 31 (81) |
|
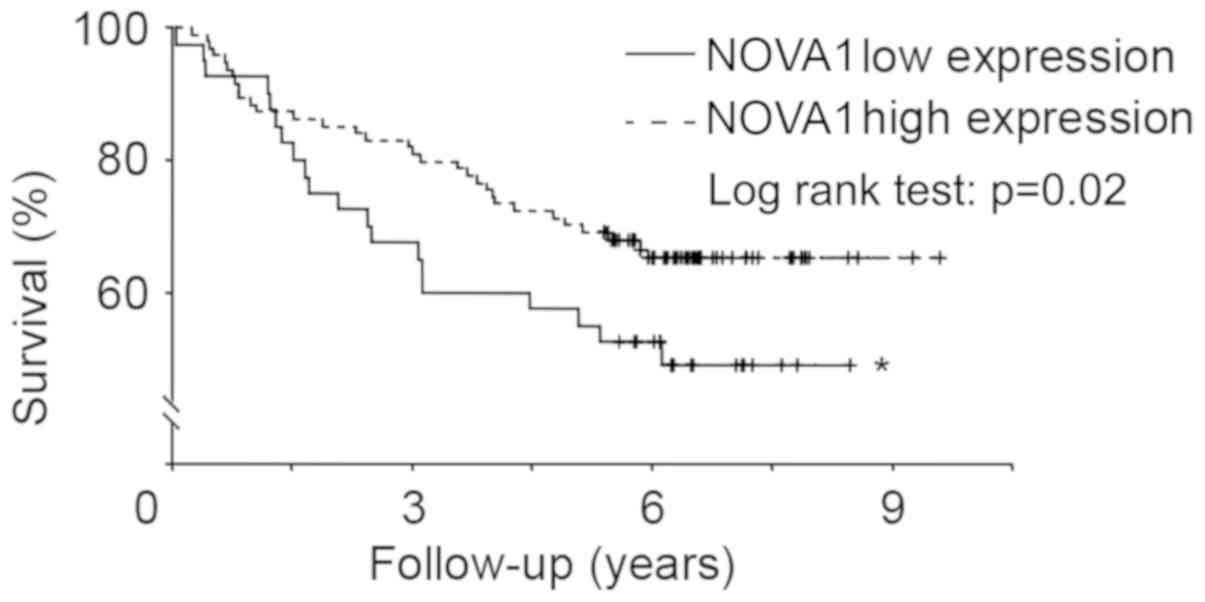
Expression of NOVA1 protein and
overall survival
Kaplan-Meier curves were generated to investigate
overall survival; patients were stratified based on the expression
of tumor NOVA1 protein. Using a log-rank test, we observed that the
two overall survival curves were significantly different; survival
among gastric cancer patients with low NOVA1 protein expression was
significantly poorer than those with high NOVA1 protein expression
(Fig. 4).
Discussion
Decreased NOVA1 expression has been observed in the
tumor microenvironment, and correlates with progression and poor
prognosis of gastric cancer (2,4). EMT
has been proposed as one of main causes for cancer progression and
poor prognosis (25,26). additionally, >4,000 genes are
involved in EMT (27). In the
present study, we showed that silencing of NOVA1 promoted EMT in
gastric cancer cells. For example, silencing of NOVA1 promoted
visible changes in the morphologies of gastric cancer cells;
cellular morphologies were changed from a cobblestone-like
phenotype to a spindle-like phenotype. Decreased E-cadherin and
keratin 8 expression, or increased fibronectin and N-cadherin
expression have been proposed as hallmarks of EMT (28,29).
We observed that E-cadherin and keratin 8 expression were
decreased, while that of fibronectin and N-cadherin were increased
in response to sh-NOVA1. These results suggested that decreased
NOVA1 expression in tumors may be associated with the progression
and poor prognosis of patients with gastric cancer (2,4).
Consistent with a recent report (4), we observed that NOVA1 expression was
associated with lymph node metastasis and tumor stage in Chinese
patients with gastric cancer.
Accumulating evidence indicates that miRNAs are
involved in tumorigenesis and tumor progression of various cancers,
via EMT and the formation of cancer-initiating cells (30). In esophageal squamous cell
carcinoma, miR-27a-3p has been proposed as a tumor suppressive
miRNA (31). However, an
experimental model demonstrated that it could be an oncogene in
osteosarcoma (32), laryngeal
cancer (33), breast cancer
(34), colorectal cancer (35) and gastric cancer (9,36,).
Several genes (MAX-interacting protein 1, F-box and F-box WD-repeat
containing protein 7) have been proposed as target genes of
miR-27a-3p (37,38).
Bioinformatics analysis indicated that NOVA1 could
be a potential target gene of miR-27a-3p, and three putative
binding sites of miR-27a-3p were found in the 3′UTR of NOVA1 mRNA.
We observed that overexpression of miR-27a-3p inhibited NOVA1
protein expression in AGS cells. The results suggested that NOVA1
was a target gene of miR-27a-3p. The levels of circulating
miR-27a-3p are increased in patients with gastric cancer (11); miR-27a-3p expression was determined
to be upregulated in gastric cancer tissues (9). Consistent with a recent report
(9), we observed a statistically
significant difference in the expression of miR-27a-3p between
adjacent normal tissues and gastric cancer tissues. In
additionally, miR-27a-3p expression is correlated with distant
metastasis, lymph node metastasis and advanced clinical stage
(9). This is in line with our
results, in which upregulated miR-27a-3p levels were associated
with shorter survival. EMT participates in the tumorigenesis and is
involved in the progression of cancer (39). Our results suggested that
miR-27a-3p promoted EMT in gastric cancer cells. These findings
indicate a possible molecular mechanism underlying the clinical
observations of gastric cancer, in which miR-27a-3p may participate
in the progression of gastric cancer.
Acknowledgements
Not applicable.
Funding
The present study was supported by Cixi People's
Hospital.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
KL and XZ conceived the study, collected the
experimental data and drafted the manuscript. XC and XW contributed
to the experimental work and data analysis. All authors edited and
approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of Renmin Hospital of Wuhan University and Cixi People's
Hospital. All subjects gave explicit written informed consent at
the time of enrollment.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Villate O, Turatsinze JV, Mascali LG,
Grieco FA, Nogueira TC, Cunha DA, Nardelli TR, Sammeth M, Salunkhe
VA, Esguerra JL, et al: Nova1 is a master regulator of alternative
splicing in pancreatic beta cells. Nucleic Acids Res.
42:11818–11830. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Yoon SO, Kim EK, Lee M, Jung WY, Lee H,
Kang Y, Jang YJ, Hong SW, Choi SH and Yang WI: NOVA1 inhibition by
miR-146b-5p in the remnant tissue microenvironment defines occult
residual disease after gastric cancer removal. Oncotarget.
7:2475–2495. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhi F, Wang Q, Deng D, Shao N, Wang R, Xue
L, Wang S, Xia X and Yang Y: miR-181b-5p downregulates NOVA1 to
suppress proliferation, migration and invasion and promote
apoptosis in astrocytoma. PLoS One. 9:e1091242014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kim EK, Yoon SO, Jung WY, Lee H, Kang Y,
Jang YJ, Hong SW, Choi SH and Yang WI: Implications of NOVA1
suppression within the microenvironment of gastric cancer:
Association with immune cell dysregulation. Gastric Cancer.
20:438–447. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Shen B, Zhang Y, Yu S, Yuan Y, Zhong Y, Lu
J and Feng J: MicroRNA-339, an epigenetic modulating target is
involved in human gastric carcinogenesis through targeting NOVA1.
FEBS Lett. 589:3205–3211. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Störchel PH, Thümmler J, Siegel G,
Aksoy-Aksel A, Zampa F, Sumer S and Schratt G: A large-scale
functional screen identifies Nova1 and Ncoa3 as regulators of
neuronal miRNA function. EMBO J. 34:2237–2254. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Browne G, Taipaleenmäki H, Stein GS, Stein
JL and Lian JB: MicroRNAs in the control of metastatic bone
disease. Trends Endocrinol Metab. 25:320–327. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Vimalraj S, Miranda P, Ramyakrishna B and
Selvamurugan N: Regulation of breast cancer and bone metastasis by
microRNAs. Dis Markers. 35:369–387. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ding L, Zhang S, Xu M, Zhang R, Sui P and
Yang Q: MicroRNA-27a contributes to the malignant behavior of
gastric cancer cells by directly targeting PH domain and
leucine-rich repeat protein phosphatase 2. J Exp Clin Cancer Res.
36:452017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Liu T, Tang H, Lang Y, Liu M and Li X:
MicroRNA-27a functions as an oncogene in gastric adenocarcinoma by
targeting prohibitin. Cancer Lett. 273:233–242. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Park JL, Kim M, Song KS, Kim SY and Kim
YS: Cell-free miR-27a, a potential diagnostic and prognostic
biomarker for gastric cancer. Genomics Inform. 13:70–75. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang D, Wang H, Liu R, Li H, Ge S, Bai M,
Deng T, Yao G and Ba Y: miRNA27a is a biomarker for predicting
chemosensitivity and prognosis in metastatic or recurrent gastric
cancer. J Cell Biochem. 115:549–556. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Danza K, Silvestris N, Simone G, Signorile
M, Saragoni L, Brunetti O, Monti M, Mazzotta A, De Summa S, Mangia
A and Tommasi S: Role of miR-27a, miR-181a and miR-20b in gastric
cancer hypoxia-induced chemoresistance. Cancer Biol Ther.
17:400–406. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhao Q, Li Y, Tan BB, Fan LQ, Yang PG and
Tian Y: HIF-1α induces multidrug resistance in gastric cancer cells
by inducing miR-27a. PLoS One. 10:e01327462015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhao X, Yang L and Hu J: Down-regulation
of miR-27a might inhibit proliferation and drug resistance of
gastric cancer cells. J Exp Clin Cancer Res. 30:552011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Z, Li Y, Ahmad A, Azmi AS, Kong D,
Banerjee S and Sarkar FH: Targeting miRNAs involved in cancer stem
cell and EMT regulation: An emerging concept in overcoming drug
resistance. Drug Resist Updat. 13:109–118. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Fan Y, Shen B, Tan M, Mu X, Qin Y, Zhang F
and Liu Y: TGF-β-induced upregulation of malat1 promotes bladder
cancer metastasis by associating with suz12. Clin Cancer Res.
20:1531–1541. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Washington K: 7th edition of the AJCC
cancer staging manual: Stomach. Ann Surg Oncol. 17:3077–3079. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang F, Bi J, Xue X, Zheng L, Zhi K, Hua J
and Fang G: Up-regulated long non-coding RNA H19 contributes to
proliferation of gastric cancer cells. FEBS J. 279:3159–3165. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Liao XH, Lu DL, Wang N, Liu LY, Wang Y, Li
YQ, Yan TB, Sun XG, Hu P and Zhang TC: Estrogen receptor α mediates
proliferation of breast cancer MCF-7 cells via a
p21/PCNA/E2F1-dependent pathway. FEBS J. 281:927–942. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36((Database Issue)): D149–D153. 2008.PubMed/NCBI
|
|
22
|
Riemenschneider MJ, Hirblinger M,
Vollmann-Zwerenz A, Hau P, Proescholdt MA, Jaschinski F,
Rothhammer-Hampl T, Wosikowski K, Janicot M and Leo E: TGF-β
isoforms in cancer: Immunohistochemical expression and
Smad-pathway-activity-analysis in thirteen major tumor types with a
critical appraisal of antibody specificity and immunohistochemistry
assay validity. Oncotarget. 6:26770–26781. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Liu L, Yu X, Guo X, Tian Z, Su M, Long Y,
Huang C, Zhou F, Liu M, Wu X and Wang X: miR-143 is downregulated
in cervical cancer and promotes apoptosis and inhibits tumor
formation by targeting Bcl-2. Mol Med Rep. 5:753–760.
2012.PubMed/NCBI
|
|
24
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tsuji T, Ibaragi S and Hu GF:
Epithelial-mesenchymal transition and cell cooperativity in
metastasis. Cancer Res. 69:7135–7139. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Gavert N and Ben-Ze'ev A:
Epithelial-mesenchymal transition and the invasive potential of
tumors. Trends Mol Med. 14:199–209. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zavadil J, Bitzer M, Liang D, Yang YC,
Massimi A, Kneitz S, Piek E and Bottinger EP: Genetic programs of
epithelial cell plasticity directed by transforming growth
factor-beta. Proc Natl Acad Sci USA. 98:6686–6691. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mani SA, Guo W, Liao MJ, Eaton EN, Ayyanan
A, Zhou AY, Brooks M, Reinhard F, Zhang CC, Shipitsin M, et al: The
epithelial-mesenchymal transition generates cells with properties
of stem cells. Cell. 133:704–715. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Busch T, Armacki M, Eiseler T, Joodi G,
Temme C, Jansen J, von Wichert G, Omary MB, Spatz J and Seufferlein
T: Keratin 8 phosphorylation regulates keratin reorganization and
migration of epithelial tumor cells. J Cell Sci. 125:2148–2159.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hur K, Toiyama Y, Takahashi M, Balaguer F,
Nagasaka T, Koike J, Hemmi H, Koi M, Boland CR and Goel A:
MicroRNA-200c modulates epithelial-to-mesenchymal transition (EMT)
in human colorectal cancer metastasis. Gut. 62:1315–1326. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhu L, Wang Z, Fan Q, Wang R and Sun Y:
microRNA-27a functions as a tumor suppressor in esophageal squamous
cell carcinoma by targeting KRAS. Oncol Rep. 31:280–286. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Salah Z, Arafeh R, Maximov V, Galasso M,
Khawaled S, Abou-Sharieha S, Volinia S, Jones KB, Croce CM and
Aqeilan RI: miR-27a and miR-27a* contribute to metastatic
properties of osteosarcoma cells. Oncotarget. 6:4920–4935. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Tian Y, Fu S, Qiu GB, Xu ZM, Liu N, Zhang
XW, Chen S, Wang Y, Sun KL and Fu WN: MicroRNA-27a promotes
proliferation and suppresses apoptosis by targeting PLK2 in
laryngeal carcinoma. BMC Cancer. 14:6782014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou S, Huang Q, Zheng S, Lin K, You J and
Zhang X: miR-27a regulates the sensitivity of breast cancer cells
to cisplatin treatment via BAK-SMAC/DIABLO-XIAP axis. Tumour Biol.
37:6837–6845. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Liang J, Tang J, Shi H, Li H, Zhen T, Duan
J, Kang L, Zhang F, Dong Y and Han A: miR-27a-3p targeting RXRα
promotes colorectal cancer progression by activating Wnt/β-catenin
pathway. Oncotarget. 8:82991–83008. 2017.PubMed/NCBI
|
|
36
|
Zhou L, Liang X, Zhang L, Yang L, Nagao N,
Wu H, Liu C, Lin S, Cai G and Liu J: miR-27a-3p functions as an
oncogene in gastric cancer by targeting BTG2. Oncotarget.
7:51943–51954. 2016.PubMed/NCBI
|
|
37
|
Xu W, Liu M, Peng X, Zhou P, Zhou J, Xu K,
Xu H and Jiang S: miR-24-3p and miR-27a-3p promote cell
proliferation in glioma cells via cooperative regulation of MXI1.
Int J Oncol. 42:757–766. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wu XZ, Wang KP, Song HJ, Xia JH, Jiang Y
and Wang YL: miR-27a-3p promotes esophageal cancer cell
proliferation via F-box and WD repeat domain-containing 7 (FBXW7)
suppression. Int J Clin Exp Med. 8:15556–15562. 2015.PubMed/NCBI
|
|
39
|
Peng Z, Wang CX, Fang EH, Wang GB and Tong
Q: Role of epithelial-mesenchymal transition in gastric cancer
initiation and progression. World J Gastroenterol. 20:5403–5410.
2014. View Article : Google Scholar : PubMed/NCBI
|