Introduction
Glioma is the most common brain tumor type in
humans, results in a high mortality rate and is divided into either
a low grade (I and II) or high grade (II and IV) type, based on its
histological features (1).
Glioblastoma multiforme (GBM) is classified as a grade IV glioma
according to the World Health Organization (WHO) grading system
(2). Despite advancement in
treatment of GBM, the majority of patients continue to succumb to
the disease, which exhibits an overall survival rate of <3 years
(3). Following surgery,
chemotherapy, including DNA alkylating antineoplastic drug
temozolomide (TMZ) chemotherapy, is used as the first-line
treatment for patients with GBM and successfully improves overall
survival time (4). However,
acquired de novo resistance limits the efficacy of TMZ for
patients with GBM (5,6). The lack of knowledge regarding the
initiation and development of GBM results in difficulty in treating
patients with GBM. Therefore, an investigation of the molecular
mechanism regulating GBM is urgently required.
MicroRNAs (miRNAs/miRs) are small, non-coding,
single stranded RNA molecules that are ubiquitously expressed in
human cells (7). miRNAs function
as negative regulators of gene expression through binding to the
complementary sites on the 3′-untranslated region (UTR) of target
mRNAs, and decrease target gene expression via the degradation of
mRNA or the inhibition of translation (8). The expression of miRNA is controlled
by DNA histone modification and other epigenetic factors, and
miRNAs serve an important function in a number of biological
processes, including cell differentiation, cell proliferation, the
cell cycle and cell motility (9–11).
The initiation and development of human cancer is frequently
accompanied by miRNA deregulation (12,13).
In GBM, accumulating evidence has demonstrated that the aberrant
expression of miRNAs contributes to cancer progression (12,14).
The analysis of gene expression and the matched miRNA profile in
patients with GBM has revealed a RNA-RNA interaction network that
regulates GBM cell proliferation (14). miR-296 expression has been revealed
to be increased in the primary tumor endothelial cells compared
with normal brain endothelial cells (15). Furthermore, the expression of
miR-296 has been indicated to be associated with cell invasion and
the multi-drug resistance of glioma cells (16,17).
Further investigation is necessary to determine the complexity of
the miRNA network in GBM.
Inhibitor of β-catenin and T cell factor (TCF)
(ICAT) is a well-characterized negative regulator of Wnt signaling
activity, which functions by blocking the binding of TCF to
β-catenin (18). ICAT is reported
to be deregulated in a number of human tumor types, while its
function in carcinogenesis remains yet to be determined (19,20).
In hepatocellular carcinoma, ICAT promotes the
epithelial-to-mesenchymal transition, and is targeted and inhibited
by miR-424-5p (21). In GBM, ICAT
is downregulated and has been indicated to inhibit cell
proliferation, migration and invasion, and induce cell apoptosis in
GBM cells (22). ICAT expression
is regulated by miRNAs in a number of different cancer types,
including hepatocellular carcinoma and breast cancer (21,23).
The mechanisms by which ICAT is regulated by miRNAs has, to the
best of our knowledge, not yet been determined in GBM.
Materials and methods
Patients
Glioma tissues and normal brain tissues were
collected from the Affiliated Hospital of North Sichuan Medical
College (Sichuan, China) between June 2014 and July 2018. GBM
tissues from patients with WHO grade II, III and IV tumor types
were obtained during standard surgery, and 10 patients were
included for each grade. The 10 normal brain tissues were obtained
during surgery in patients with intractable epilepsy. All
participants provided written informed consent prior to tissue
sampling. The present study was ethically approved and conducted
under the supervision of the Ethics Committee of North Sichuan
Medical College (approval no. NSREC20140622H). Patients were
enrolled in the study if their diagnosis was histologically
confirmed by two neuropathologists based on the 2007 WHO
classification guidelines (24).
Cell lines
293 cells and human GBM cell lines U251 and U138MG
were purchased from the Type Culture Collection of the Chinese
Academy of Sciences (Shanghai, China). Cell lines were maintained
in Dulbecco's modified Eagle's medium (Gibco; Thermo Fisher
Scientific, Inc., Waltham, MA, USA) containing 10% fetal bovine
serum (Gibco; Thermo Fisher Scientific, Inc.) and 1%
penicillin/streptomycin (Invitrogen; Thermo Fisher Scientific,
Inc.) and incubated at 37°C in a humidified incubator with 5%
CO2.
miR-296-3p mimics and miR-296-3p
antagonist transfection
miR-negative control (NC) mimics
(5′-UUCUCCGAACGUGUCACGUTT-3′), miR-296-3p mimics
(5′-AGGGCCCCCCCUCAAUCCUGU-3′), miR-NC antagonist
(5′-CAGUACUUUUGUGUAGUACAA-3′) and miR-296-3p antagonist
(5′-ACAGGAUUGAGGGGGGGCCCU-3′) were purchased from Guangzhou RiboBio
Co., Ltd. (Guangzhou, China). For miR-296-3p antagonization or
miR-296-3p overexpression, 200 nM miR-296-3p antagonist or
miR-296-3p mimics were mixed with Lipofectamine 2000®
(Invitrogen; Thermo Fisher Scientific, Inc.), sustained for 15 min
at room temperature then added to 1×106 cells which were
seeded into six-well plates. The efficiency of the miR-296-3p
antagonist and miR-296-3p mimics was detected at 48 h following
transfection.
Silencing of ICAT in cells
ICAT siRNA (5′-GAUGGGAUCAAACCUGACA-3′) and control
siRNA (5′-AAUUCUCCGAACGUGUCACGU-3′) were purchased from GenePharma
(Suzhou, China). In total, 50 nM ICAT siRNA or control siRNA was
transfected into 2×106 U251 cells in six-well plates
with Lipofectamine RNAiMax (Invitrogen; Thermo Fisher Scientific,
Inc.) according to the manufacturer's protocol. The transfection
efficiency was detected by western blotting 72 h after
transfection.
Cell proliferation assay
Cell proliferation was detected using a Cell
Counting Kit-8 (CCK-8) assay (Dojindo Molecular Technologies,
Inc.). A total of 2×104 cells were plated into 96-well
plates. At 0, 24, 48 and 96 h, a total of 10 µl CCK-8 solution was
added into the culture medium and maintained for 2 h. Culture
medium containing CCK-8 solution was subsequently transferred to a
96-well plate. Absorbance was measured at 450 nm and each well was
analyzed using a microplate reader (Bio-Rad Laboratories, Inc.) to
determine cell number.
Cell cycle analysis
Cell cycle distribution was analyzed using flow
cytometry. A cell cycle assay was performed according to the
manufacturers protocol of the Cell Cycle Analysis kit (Beyotime
Institute of Biotechnology, Haimen, China). A total of
1×106 cells were collected, washed with cold phosphate
buffered saline (PBS; Invitrogen; Thermo Fisher Scientific, Inc.)
and fixed in 70% ethanol at 4°C for 24 h. Cells were subsequently
washed with cold PBS and stained in a propidium iodide/RNase A
mixture at room temperature for 30 min. Subsequent to incubation
for 30 min at room temperature, the cells were analyzed using a
fluorescence-activated cell sorting Caliber system (BD
Biosciences). The data were analyzed with the FlowJo software V
10.0.7 (BD Biosciences).
Western blotting
Lysates were prepared from 1×106 cells
using RIPA lysis buffer (Sigma Aldrich; Merck KGaA). The
concentration of each sample was determined with a bicinchoninic
acid Protein Assay Kit (Thermo Fisher Scientific, Inc.). Antibodies
for GAPDH (mouse; G8795; 1:10,000) were purchased from Sigma
Aldrich (Merck KGaA). P21 (rabbit; 2947; 1:2,000), cyclin D1
(rabbit; 2978; 1:2,000), c-Myc (rabbit; 5605; 1:2,000), AKT
(rabbit; 4691; 1:2,000), phosphorylated (p-)AKT (rabbit; 4060;
1:2,000), ERK1/2 (rabbit; 4695; 1:2,000), p-ERK1/2 (rabbit; 9101;
1:2,000) antibodies were bought from Cell Signaling Technology.
ICAT (rabbit; ab129011; 1:1,000) and β-catenin (rabbit, ab16051,
1:1,000) antibody was obtained from Abcam. Horseradish
peroxidase-conjugated secondary antibodies against mouse
(SA00001-1; 1:100,000) and rabbit (SA00001-2; 1:100,000) were
obtained from Proteintech Group. Western blot was conducted
following a standard procedure. 20 µg protein lysate was loaded
into an 8% SDS gel and separated via electrophoresis. Proteins on
gel were transferred into a PVDF membrane. The membrane was blocked
using 5% non-fat milk at room temperature for 1 h and then
incubated with the indicated primary antibody overnight at 4°C. On
the next day, the membrane was washed and incubated with the
appropriate secondary antibody for 1 h at room temperature.
Finally, membrane was developed using ECL western blotting
substrate (Thermo Fisher Scientific, Inc.) and images were obtained
using ImageQuant LAS 4000 (GE Healthcare Life Sciences). Western
blotting data were quantified with Image J software Version.1.6.0
(National Institute of Science).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA from tissues and 1×106 cells
was extracted using TRIzol Reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. The
quality and concentration of RNA was detected with NanoDrop 2000
(Thermo Fisher Scientific, Inc.). For detection of miR-296-3p
expression, RNA was reverse transcribed to cDNA using a stem-loop
primer with RevertAid First Strand cDNA kit (Thermo Fisher
Scientific, Inc.). For mRNA expression analysis, RNA was reverse
transcribed to first-strand cDNA using PrimeScript RT Master Mix
(Takara Bio, Inc.). qPCR was performed using SYBR Premix Ex Taq kit
(Takara Bio, Inc.). U6 and GAPDH served as internal controls for
the semi-quantification of miR-296-3p and genes expression,
respectively. The relative miR-296-3p and genes expression was
calculated by the 2−ΔΔCq method (25). qPCR thermocycling conditions were
as follows: Initial denaturation at 95°C for 30 sec, followed by 35
cycles of 95°C for 15 sec and 60°C for 30 sec. The primer sequences
were as follows: Stem-loop primer,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGGGAGAG-3′;
miR-296-3p-forward, 5′-GCCGAGGAGGGTTGGGTGGA-3′; miR-296-3p-reverse,
5′-CTCAACTGGTGTCGTGGA-3′; U6-forward, 5′-CTCGCTTCGGCAGCACA-3′;
U6-reverse, 5′-AACGCTTCACGAATTTGCGT-3′; ICAT-forward,
5′-CCTATGCAGGGGTGGTCAAC-3′; ICAT-reverse,
5′-CGACCTGGAAAACGCCATCA-3′; GAPDH-forward,
5′-AACGTGTCAGTGGTGGACCTG-3′; GAPDH-reverse,
5′-AGTGGGTGTCGCTGTTGAAGT-3′.
Bioinformatic analysis
The potential target genes of miR-296-3p were
predicted using the online software miRanda V 2010 (http://www.microrna.org/microrna/home.do) (26). The target site prediction of all
conserved miRNAs was downloaded from the software. Then, the
Database for Annotation, Visualization and Integrated Discovery
(https://david.ncifcrf.gov/) was used to
screen for Wnt signaling related genes (27,28).
Dual luciferase reporter assay
The 3′UTR of ICAT mRNA was amplified from cDNA of
293 cells followed by insertion into pmirGLO plasmid (Promega
Corporation) to construct pmirGLO-ICAT 3′UTR-wild-type (WT).
pmirGLO-ICAT 3′UTR-Mutant (Mut) with mutation of predicted
miR-296-3p binding sites was created by introducing site mutations
of pmirGLO-ICAT 3′UTR-WT using Quick site-directed mutagenesis kit
(Agilent Technologies, Inc.). For dual luciferase assay,
1×106 U251 cells were transfected with 2 µg pmirGLO-ICAT
3′UTR-WT or pmirGLO-ICAT 3′UTR-Mut accompanied with miR-296-3p
mimics or miR-NC mimics and an internal control Renilla
plasmid (hRluc-neo). After 24 h, the relative luciferase activity
of each well was measured using a Dual-Glo luciferase Assay System
(Promega Corporation) following the manufacture's protocol. The
firefly luciferase activity was normalized to Renilla
luciferase activity.
Immunofluorescence
In total, 1×105 U251 cells were grown on
glass slides in 24-well plates, transfected with miR-NC antagonist
or miR-296-3p antagonist and incubated for 48 h at 37°C. Wells were
then washed with PBS and treated with 4% paraformaldehyde for 30
min at room temperature. Cells were subsequently permeabilized and
blocked using PBS containing 0.1% Triton X-100, in 1% bovine serum
albumin, for 1 h at room temperature. A β-catenin (rabbit; 1:100;
cat. no. ab16051; Abcam) antibody was used as the primary antibody
to incubate the glass slides for 2 h at room temperature, and Alexa
Fluor 594-conjugated secondary antibody (1:50,000; cat. no. R37117;
Invitrogen; Thermo Fisher Scientific, Inc.) was applied to detect
fluorescence (1 h at room temperature). DAPI (Vector Laboratories,
Inc.) was finally added into the glass slides to stain cell nuclei.
The slides were observed and the representative images were
captured using the Leica DM5000 B microscope (Leica Microsystems,
Inc.).
Statistical analysis
All data were statistically analyzed using GraphPad
Prism 7 (GraphPad Software, Inc.) and presented as the mean ±
standard deviation. Differences between two groups were compared
using a paired Student's t-test. All experiments were repeated at
least three times. For comparison among three groups, the data were
firstly analyzed using a one-way analysis of variance, followed by
a Newman Keul's test. Pearson's correlation analysis was used for
the analysis of the correlation between miR-296-3p expression and
ICAT mRNA levels in GBM tissues. P<0.05 was considered to
indicate a statistically significant result.
Results
miR-296-3p is upregulated in GBM
tissues
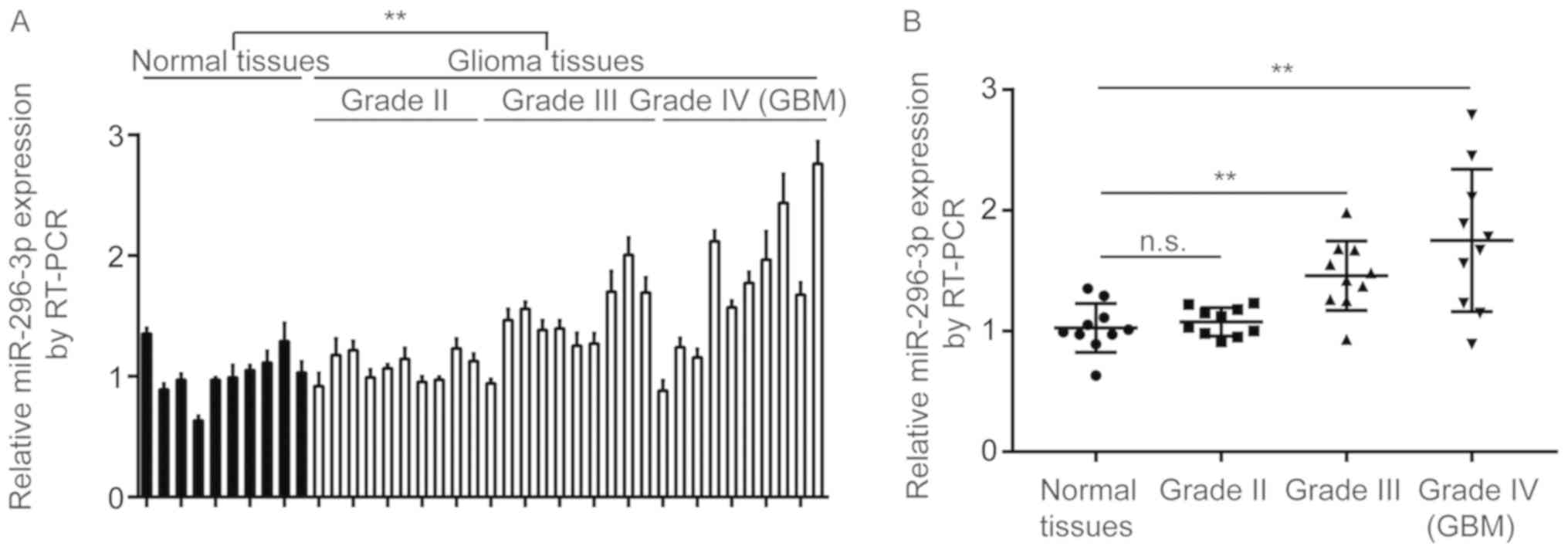
To determine the expression of miR-296-3p in GBM
tissues, a total of 10 normal brain tissues and 40 GBM tissues (10
grade I, 10 grade II, 10 grade III and 10 grade IV) were collected,
and miR-296-3p expression was determined using a reverse
transcription-quantitative polymerase chain reaction (RT-qPCR).
miR-296-3p expression was demonstrated to be significantly
increased in GBM tissues compared with normal tissues (P<0.01;
Fig. 1A). Additionally, miR-296-3p
was indicated not to be overexpressed in GBM tissues from patients
at grade II, but significantly increased in GBM tissues from
patients at grade III and grade IV compared with normal tissues
(P<0.01; Fig. 1B).
Antagonization of miR-296-3p
suppresses cell proliferation and alters cell cycle distribution in
U251 cells
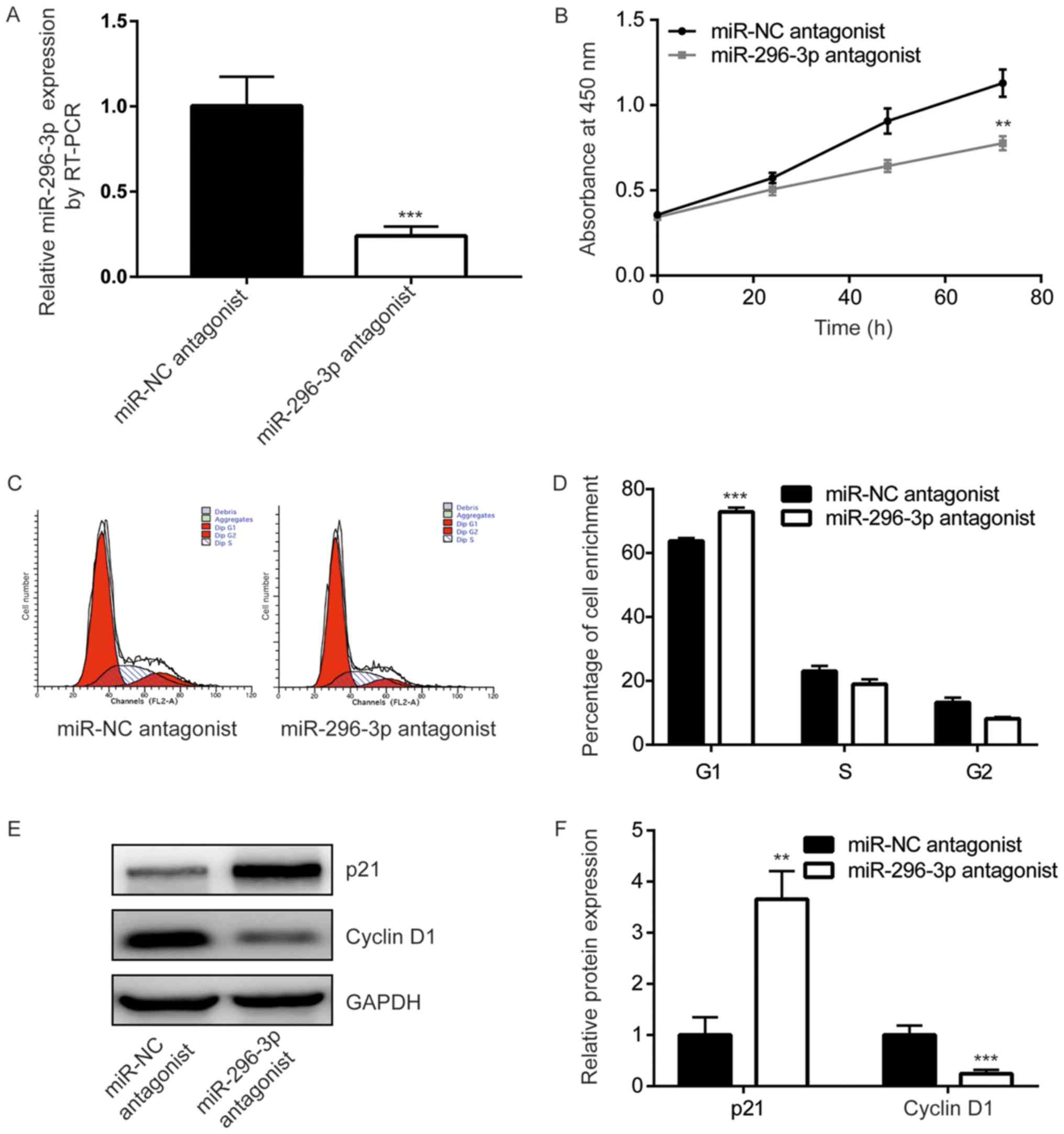
To study the function of miR-296-3p in GBM cells,
miR-296-3p expression was downregulated, using transfection with a
miR-296-3p antagonist, in U251 cells. Cells transfected with the
miR-296-3p antagonist demonstrated a significant 4-fold decrease in
miR-296-3p expression compared with those transfected with the
negative control (P<0.001; Fig.
2A). The results of the CCK-8 assay indicated that miR-296-3p
downregulation significantly inhibited U251 cell growth (P<0.01;
Fig. 2B). Additionally, flow
cytometry revealed that the miR-296-3p antagonist induced a
significant increase of cells enriched in the G0/G1 phase compared
with the control cells, indicating the redistribution of the cell
cycle (P<0.001; Fig. 2C and D).
Western blot analysis was performed to detect a number of key genes
that have been indicated to be associated with the G1/S checkpoint.
The expression levels of p21, an inhibitor of cell cycle
progression, were demonstrated to be significantly increased
following transfection with the miR-296-3p antagonist compared with
the negative control (P<0.01). Furthermore, cyclin D1 expression
levels were revealed to be significantly decreased following
transfection with miR-296-3p antagonist compared with the negative
control (P<0.001; Fig. 2E and
F). These results suggested that miR-296-3p may promote cell
cycle progression to facilitate GBM cell proliferation.
miR-296-3p negatively regulates ICAT
expression in GBM cells
To investigate the mechanism of miR-296-3p in GBM
cells, miRanda was used to predict the potential target genes of
miR-296-3p. Among these target genes, Database for Annotation,
Visualization and Integrated Discovery software was used and the
results demonstrated that the 3′UTR of ICAT, a key regulator of Wnt
signaling, could complementary bind to miR-296-3p (data not shown).
ICAT is a negative regulator of the Wnt signaling pathway and is
considered to be a tumor suppressor in GBM cells (20). With similar results observed in
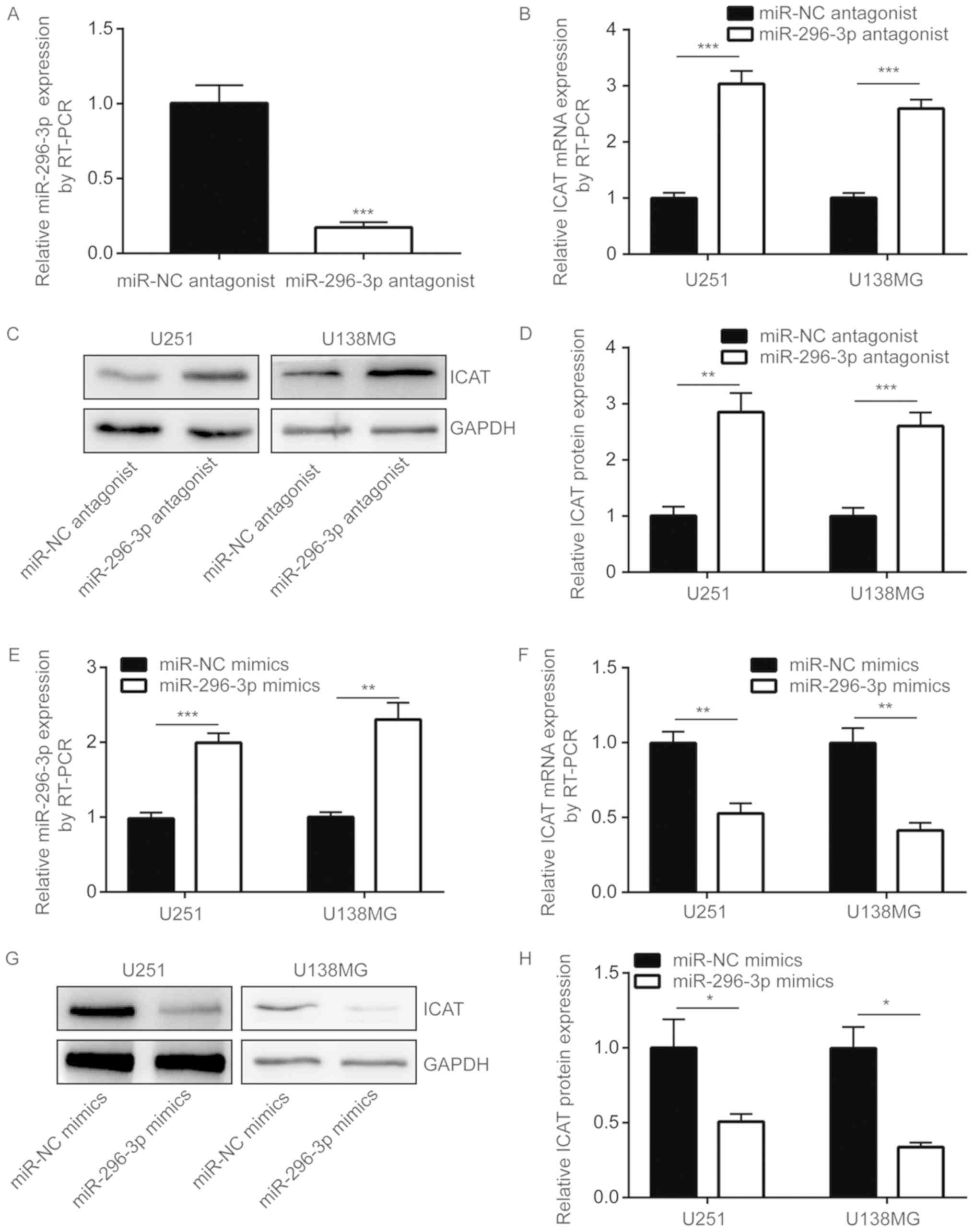
U251 cells, the transfection of miR-296-3p antagonist also
significantly decreased miR-296-3p expression levels in U138MG
cells (P<0.001; Fig. 3A).
miR-296-3p downregulation was also indicated to significantly
increase ICAT mRNA expression levels in U251 and U138MG cells
(P<0.001; Fig. 3B).
Additionally, ICAT protein expression levels were also
significantly increased following transfection with an miR-296-3p
antagonist (P<0.01; Fig. 3C and
D). miRNA mimics are chemically synthesized miRNAs, and
following transfection into cells, miRNA mimics have been revealed
to mimic the function of a miRNA (12). Therefore, miR-296-3p mimics were
used to overexpress miR-296-3p in U251 and U138MG cells.
Transfection of miR-296-3p mimics resulted in a 2-fold increase in
miR-296-3p expression levels in U251 and U138MG cells (P<0.01;
Fig. 3E). In contrast, the
overexpression of miR-296-3p significantly decreased ICAT
expression at the mRNA and protein levels (P<0.05; Fig. 3F-H).
miR-296-3p regulates the
ICAT-regulated signaling network in GBM cells
The overactivation of phosphoinositide-3-kinase
(PI3K)-protein kinase B (AKT), mitogen-activated protein
kinase-extracellular signal-regulated kinase (ERK) and Wnt
signaling pathways serves an important function in cell
proliferation and cell cycle regulation in GBM cells (29–31).
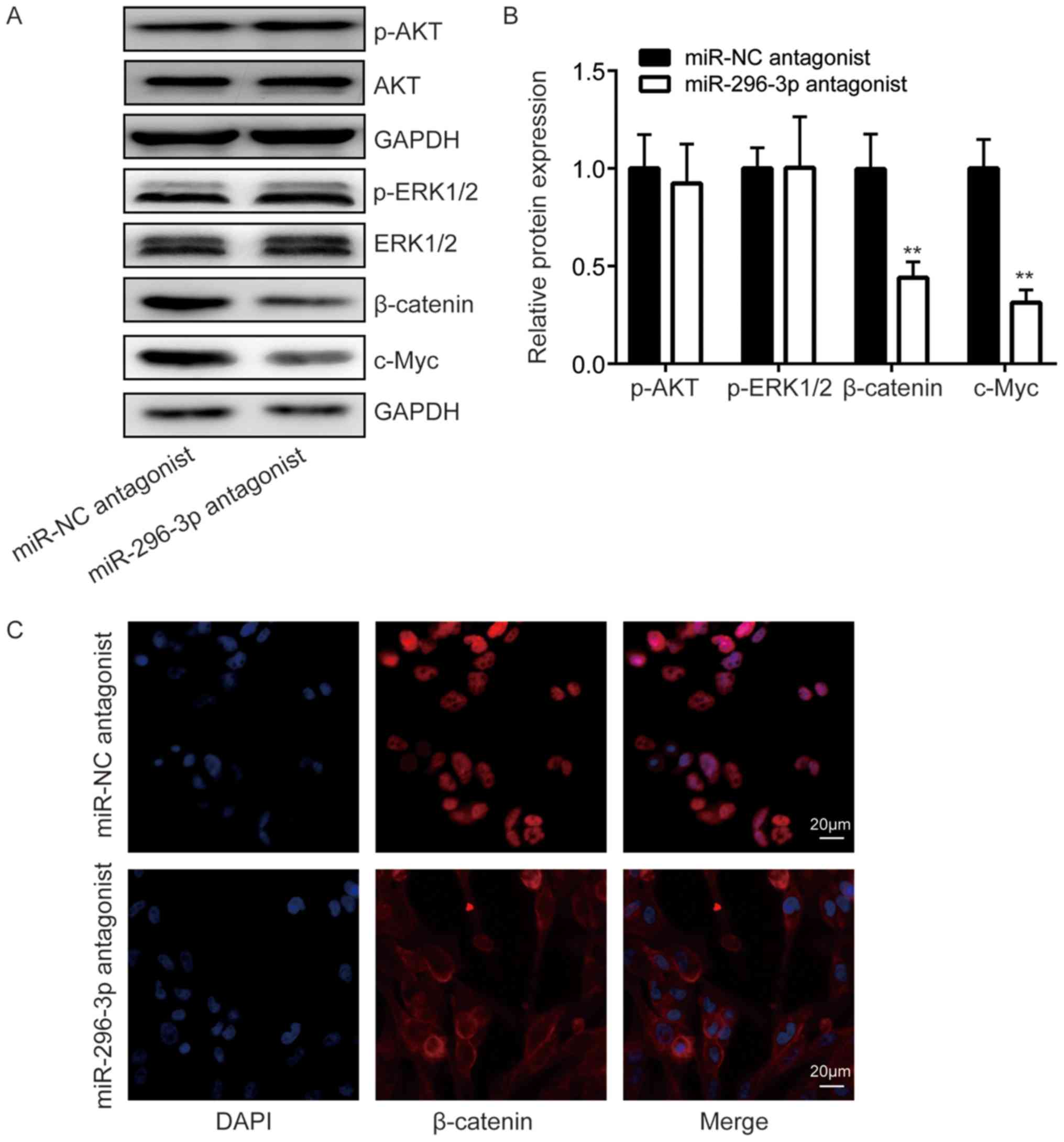
Using western blot analysis, β-catenin, a signal transducer of the
Wnt signaling pathway, was indicated to be significantly decreased
following the downregulation of miR-296-3p compared with the
negative control (P<0.01), while p-AKT and p-ERK1/2 expression
did not differ (Fig. 4A and B).
Additionally, the expression of Wnt signaling target gene c-Myc was
also significantly decreased in cells transfected with miR-296-3p
antagonist compared with the negative control (P<0.01; Fig. 4A and B). Furthermore, using
immunofluorescence, it was observed that β-catenin was located in
the nucleus of U251 cells (Fig.
4C). Transfection of a miR-296-3p antagonist resulted in the
translocation of β-catenin from the nucleus to the cytoplasm of
cells (Fig. 4C). These results
suggested that miR-296-3p may activate the Wnt signaling pathway
via the regulation of ICAT.
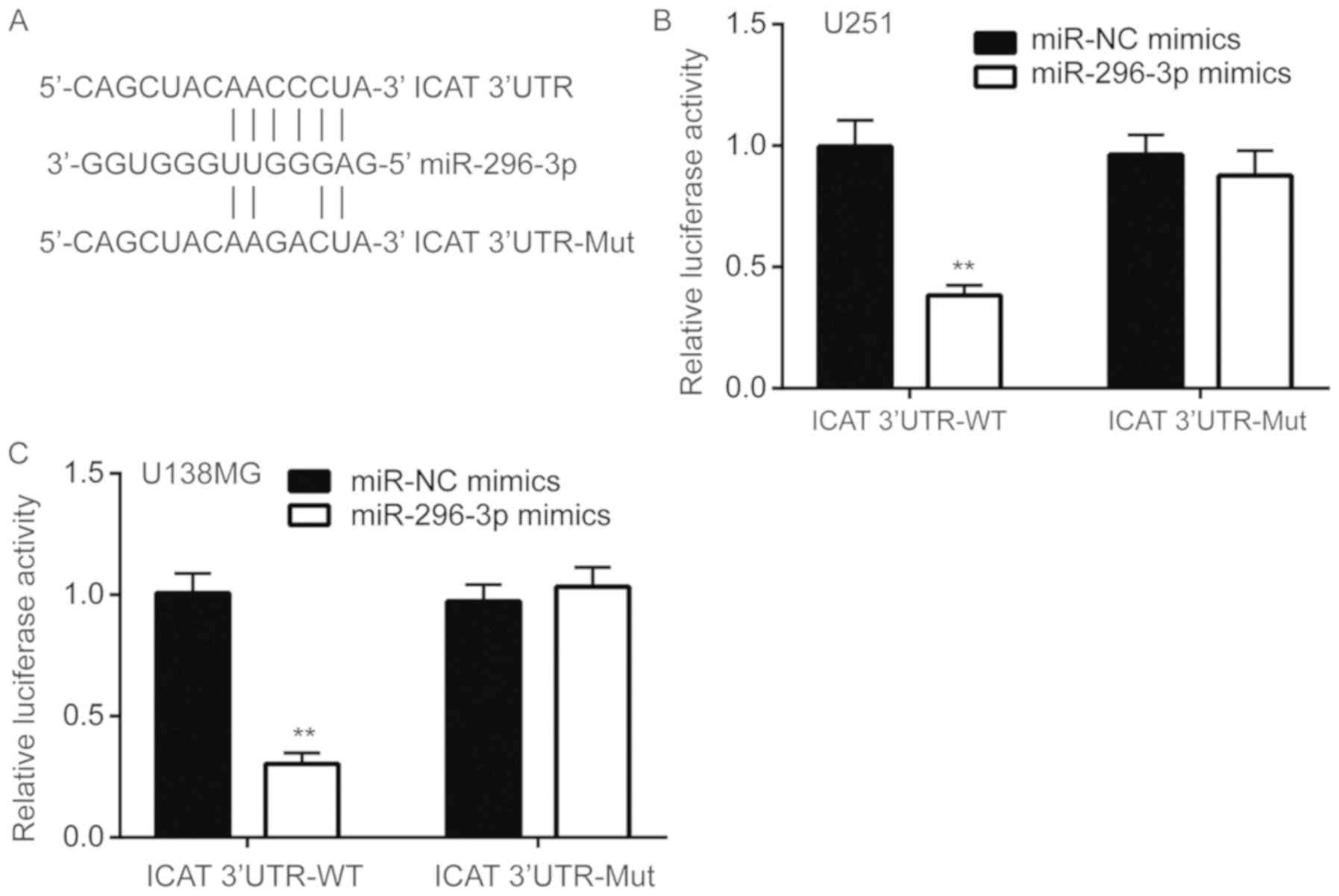
miR-296-3p directly binds to the 3′UTR
of ICAT mRNA
To assess whether miR-296-3p directly regulated ICAT
expression, miRanda was used to align the sequences of miR-296-3p
and ICAT 3′UTR. A complementary site was observed between
miR-296-3p and ICAT 3′UTR (Fig.
5A). Luciferase reporter assays were performed to functionally
verify whether miR-296-3p directly targets ICAT in U251 cells. Wild
type ICAT-3′UTR resulted in significantly decreased luciferase
activity relative to mutated ICAT-3′UTR in U251 cells
co-transfected with miR-296-3p mimics compared with the negative
control (P<0.01; Fig. 5B),
indicating that ICAT was a target gene of miR-296-3p in U251 cells.
Similar results were also observed in U138MG cells (P<0.01;
Fig. 5C).
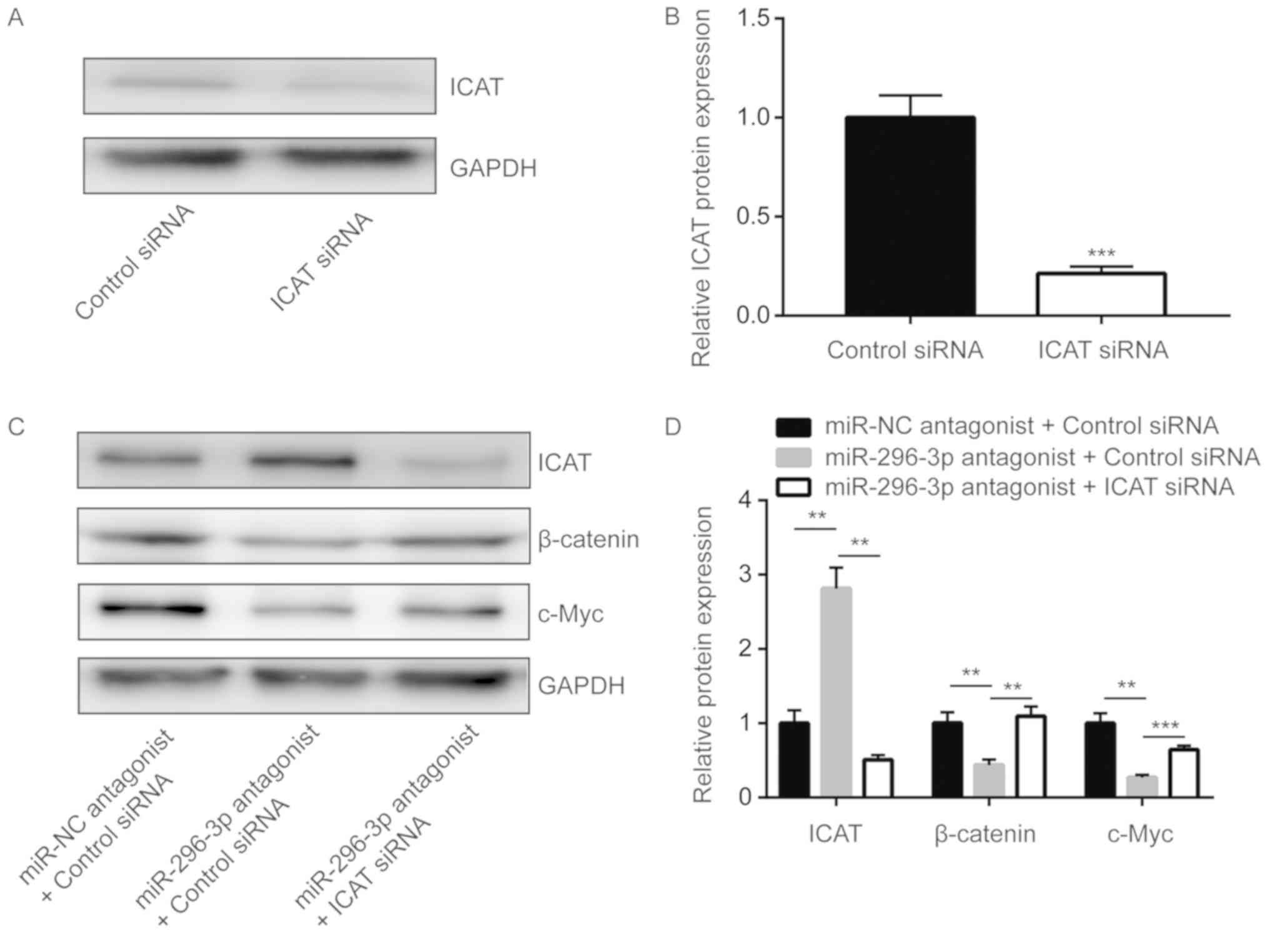
miR-296-3p regulates Wnt signaling via
ICAT to control glioma cell proliferation
ICAT small-interfering RNA was transfected into U251
cells to significantly downregulate ICAT expression compared with
the control (P<0.0001; Fig. 6A and
B). Silencing ICAT significantly reversed the downregulation of
β-catenin and c-Myc that was induced by miR-296-3p antagonist
(P<0.01; Fig. 6C and D).
Furthermore, miR-296-3p downregulation-induced cell growth arrest
was also demonstrated to be significantly reversed upon ICAT
silencing in U251 cells (P<0.0001; Fig. 6E).
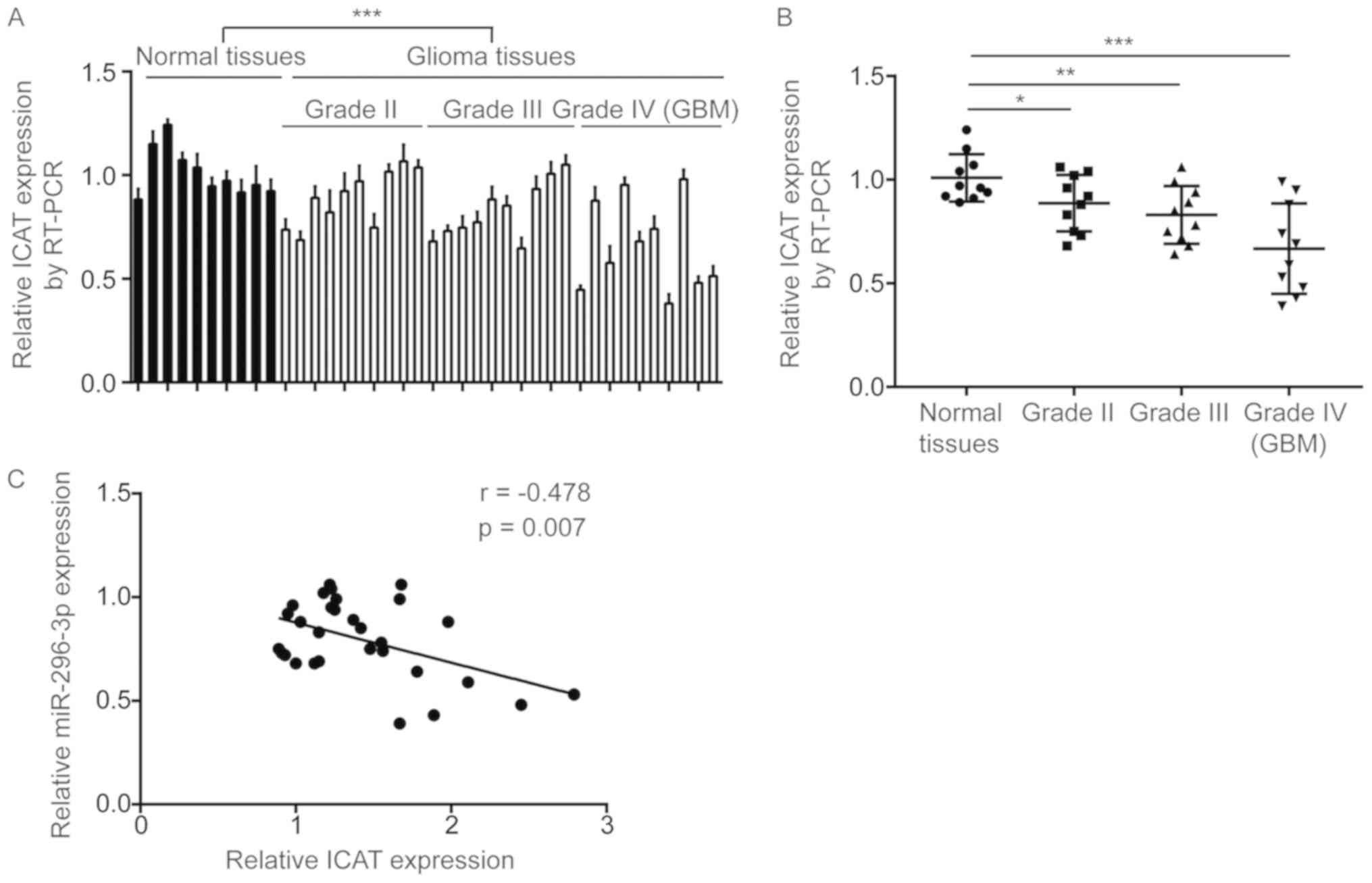
miR-296-3p level is negatively
correlated with ICAT levels in GBM tissues
Subsequently, the association between miR-296-3p and
ICAT expression was assessed in the present study. RT-qPCR
indicated that ICAT mRNA levels were significantly increased in GBM
tissues compared with normal brain tissues (P<0.0001; Fig. 7A). Additionally, significantly
decreased ICAT expression was observed in GBM tissues from patients
at all grades in comparison with normal brain tissues (P<0.05;
Fig. 7B). Pearson correlation
analysis indicated a strong and significant negative correlation
between miR-296-3p expression and ICAT mRNA levels in GBM tissues
(r=−0.478, P=0.007; Fig. 7C).
Discussion
Accumulating evidence has suggested that miRNAs are
associated with the initiation and development of GBM (32–34).
A number of miRNAs have been demonstrated to be aberrantly
expressed, and are indicated to be promising biomarkers for
patients with GBM (35,36). miR-296-3p was reported to be
downregulated in GBM tissues from 30 patients compared with 12
normal brains, and the downregulation of miR-296-3p was indicated
to promote the development of multi-drug resistance (17). However, one other study indicated
that miR-296-3p is negatively regulated by neurofibromatosis 2 in
GBM cells, and analysis of The Cancer Genome Atlas GBM dataset
demonstrated that a high expression of miR-296-3p was associated
with the short overall survival time of patients with GBM, in a
large cohort (16). In the present
study, an elevation of miR-296-3p was observed in GBM tissues,
particularly in GBM tissues from patients at late grades compared
with normal brain tissues, which supports the oncogenic function of
miR-296-3p, as a high expression of miR-296-3p predicts poor
survival in GBM. The downregulation of miR-296-3p levels in U251
cells was also indicated to inhibit cell proliferation and alter
the cell cycle distribution. Additionally, the expression of p21
and cyclin D1 differed following the downregulation of miR-296-3p.
The results of the present study suggested that miR-296-3p may
promote the proliferation of GBM cells via the regulation of cell
cycle progression.
The Wnt/β-catenin signaling pathway is an important
pathway in the maintenance of stem cells, and has been indicated to
be aberrantly activated in a number of cancer types, including in
GBM (37–39). Through blocking the binding of TCF
to β-catenin, ICAT has been observed to function as an inhibitor of
the Wnt signaling pathway (18).
However, the function of ICAT in carcinogenesis remains to be
controversial. A previous study demonstrated decreased ICAT
expression in GBM tissues and a tumor suppressor function of ICAT
in GBM cells (22). In the present
study, through the analysis of ICAT mRNA expression in GBM tissues
and normal tissues, the downregulation of ICAT in GBM tissues was
also observed, particularly in tissues from patients at grade III
and IV. The expression of ICAT was negatively correlated with
miR-296-3p in GBM tissues. Inconsistently, ICAT expression was
decreased in grade II glioma, whereas miR-296-3p levels were not
increased, which may reflect the complexity of ICAT regulation in
GBM, suggesting that ICAT may be regulated by other mechanisms in
patients with grade II GBM. In cells, the deregulation of a number
of miRNAs was reported to promote the aberrant expression of ICAT
(21,23,40).
miR-296-3p was revealed to negatively regulate ICAT expression in
U251 cells. Using miRanda, ICAT was predicted as a potential target
gene of miR-296-3p, which was further verified using a dual
luciferase reporter assay in two glioblastoma cell lines.
Additionally, miR-296-3p downregulation was observed to result in
the decreased expression of β-catenin and its target genes (cyclin
D1 and c-Myc) in U251 cells, suggesting that miR-296-3p may
activate the Wnt signaling pathway via the repression of ICAT
expression. The cyclin D1-cyclin-dependent kinase 4/6 complex is
important for cell cycle progression in the G1 phase (41). The overexpression of cyclin D1 has
been observed in a number of cancer types and has been indicated to
promote cell proliferation (42).
Cyclin D1 protein expression decreases as cells enter the S phase,
but the expression of cyclin D1 is still important for passing the
G0/G1 phase, and the low expression of cyclin D1 will result in
cell accumulation in the G1 phase (41). The results of the present study
suggested that miR-296-3p may positively regulate cyclin D1
expression via Wnt signaling to control GBM cell cycle progression
and promote GBM cell growth. miR-296-3p has been suggested to
target MK2 and PRKCA in nasopharyngeal carcinoma and lung
adenocarcinoma, respectively (43,44).
The downregulation of MAPK activated protein kinase 2 and protein
kinase Cα was observed to inhibit PI3K/AKT and Ras signaling,
resulting in the downregulation of c-Myc (43,44).
In the present study, the expression of key proteins of PI3K/AKT,
Ras and Wnt signaling was determined following the downregulation
of miR-296-3p in GBM. The downregulation of Wnt signaling was only
observed in GBM cells (43,44),
but the observation excluded the previously reported regulatory
association between miR-296-3p and c-Myc. The results of the
present study indicated that miR-296-3p promoted c-Myc expression
via activating the Wnt signaling. However, the association between
miR-296-3p and Wnt, PI3K/AKT and Ras signaling was only assessed in
U251 cells. Due to the complexity of the signaling network in
glioblastoma, further studies should investigate the potential
function of miR-296-3p on the signaling network in two glioblastoma
cell lines.
In conclusion, the present study revealed that
miR-296-3p was elevated in GBM tissues and promoted cell
proliferation in GBM cells by suppressing ICAT. Therefore, the
results of the present study identified a novel molecular mechanism
in which miR-296-3p was overexpressed in GBM tissues to activate
the Wnt signaling pathway and indicated that miR-296-3p may be a
novel biomarker and therapeutic target that can be used in GBM.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HF provided the concept and designed the experiments
of the study. JZ and GD collected the clinical samples, acquired
and analyzed the data. JZ also designed and supervised the study.
HF prepared, edited and reviewed the manuscript.
Ethics approval and consent to
participate
All patients provided written informed consent prior
to the study and the Ethics Committee of North Sichuan Medical
College supervised and ethically approved the present study.
Patient consent for publication
All patients provided written informed consent to
participate the study and agreed to publish data presented in the
paper.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gladson CL, Prayson RA and Liu WM: The
pathobiology of glioma tumors. Annu Rev Pathol. 5:33–50. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Zeng A, Hu Q, Liu Y, Wang Z, Cui X, Li R,
Yan W and You Y: IDH1/2 mutation status combined with Ki-67
labeling index defines distinct prognostic groups in glioma.
Oncotarget. 6:30232–30238. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Van Gool S, Maes W, Ardon H, Verschuere T,
Van Cauter S and De Vleeschouwer S: Dendritic cell therapy of
high-grade gliomas. Brain Pathol. 19:694–712. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Stupp R, Mason WP, van den Bent MJ, Weller
M, Fisher B, Taphoorn MJ, Belanger K, Brandes AA, Marosi C, Bogdahn
U, et al: Radiotherapy plus concomitant and adjuvant temozolomide
for glioblastoma. N Engl J Med. 352:987–996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Cai T, Liu Y and Xiao J: Long noncoding
RNA MALAT1 knockdown reverses chemoresistance to temozolomide via
promoting microRNA-101 in glioblastoma. Cancer Med. 7:1404–1415.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang Z, Xu X, Liu N, Cheng Y, Jin W, Zhang
P, Wang X, Yang H, Liu H and Tu Y: SOX9-PDK1 axis is essential for
glioma stem cell self-renewal and temozolomide resistance.
Oncotarget. 9:192–204. 2017.PubMed/NCBI
|
|
7
|
Sethi A and Sholl LM: Emerging evidence
for MicroRNAs as regulators of cancer stem cells. Cancers (Basel).
3:3957–3971. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bartel DP: MicroRNAs: Genomics,
biogenesis, mechanism, and function. Cell. 116:281–297. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Mendell JT: MicroRNAs: Critical regulators
of development, cellular physiology and malignancy. Cell Cycle.
4:1179–1184. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Markopoulos GS, Roupakia E, Tokamani M,
Alabasi G, Sandaltzopoulos R, Marcu KB and Kolettas E: Roles of
NF-kB signaling in the regulation of miRNAs impacting on
inflammation in cancer. Biomedicines. 6:E402018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li S, Chowdhury R, Liu F, Chou AP, Li T,
Mody RR, Lou JJ, Chen W, Reiss J, Soto H, et al: Tumor-suppressive
miR148a is silenced by CpG island hypermethylation in IDH1-mutant
gliomas. Clin Cancer Res. 20:5808–5822. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang R, Luo H, Wang S, Chen W, Chen Z,
Wang HW, Chen Y, Yang J, Zhang X, Wu W, et al: MicroRNA-377
inhibited proliferation and invasion of human glioblastoma cells by
directly targeting specificity protein 1. Neuro Oncol.
16:1510–1522. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang Y, Dutta A and Abounader R: The role
of microRNAs in glioma initiation and progression. Front Biosci
(Landmark Ed). 17:700–712. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sumazin P, Yang X, Chiu HS, Chung WJ, Iyer
A, Llobet-Navas D, Rajbhandari P, Bansal M, Guarnieri P, Silva J
and Califano A: An extensive microRNA-mediated network of RNA-RNA
interactions regulates established oncogenic pathways in
glioblastoma. Cell. 147:370–381. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Würdinger T, Tannous BA, Saydam O, Skog J,
Grau S, Soutschek J, Weissleder R, Breakefield XO and Krichevsky
AM: miR-296 regulates growth factor receptor overexpression in
angiogenic endothelial cells. Cancer Cell. 14:382–393. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee H, Hwang SJ, Kim HR, Shin CH, Choi KH,
Joung JG and Kim HH: Neurofibromatosis 2 (NF2) controls the
invasiveness of glioblastoma through YAP-dependent expression of
CYR61/CCN1 and miR-296-3p. Biochim Biophys Acta. 1859:599–611.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bai Y, Liao H, Liu T, Zeng X, Xiao F, Luo
L, Guo H and Guo L: MiR-296-3p regulates cell growth and multi-drug
resistance of human glioblastoma by targeting ether-à-go-go (EAG1).
Eur J Cancer. 49:710–724. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Jiang Y, Ren W, Wang W, Xia J, Gou L, Liu
M, Wan Q, Zhou L, Weng Y, He T and Zhang Y: Inhibitor of β-catenin
and TCF (ICAT) promotes cervical cancer growth and metastasis by
disrupting E-cadherin/β-catenin complex. Oncol Rep. 38:2597–2606.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Koyama T, Tago K, Nakamura T, Ohwada S,
Morishita Y, Yokota J and Akiyama T: Mutation and expression of the
beta-catenin-interacting protein ICAT in human colorectal tumors.
Jpn J Clin Oncol. 32:358–362. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Imai M, Nakamura T, Akiyama T and Horii A:
Infrequent somatic mutations of the ICAT gene in various human
cancers with frequent 1p-LOH and/or abnormal nuclear accumulation
of beta-catenin. Oncol Rep. 12:1099–1103. 2004.PubMed/NCBI
|
|
21
|
Zhang Y, Li T, Guo P, Kang J, Wei Q, Jia
X, Zhao W, Huai W, Qiu Y, Sun L and Han L: MiR-424-5p reversed
epithelial-mesenchymal transition of anchorage-independent HCC
cells by directly targeting ICAT and suppressed HCC progression.
Sci Rep. 4:62482014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang K, Zhu S, Liu Y, Dong X, Shi Z,
Zhang A, Liu C, Chen L, Wei J, Pu P, et al: ICAT inhibits
glioblastoma cell proliferation by suppressing Wnt/β-catenin
activity. Cancer Lett. 357:404–411. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tan Z, Zheng H, Liu X, Zhang W, Zhu J, Wu
G, Cao L, Song J, Wu S, Song L and Li J: MicroRNA-1229
overexpression promotes cell proliferation and tumorigenicity and
activates Wnt/β-catenin signaling in breast cancer. Oncotarget.
7:24076–24087. 2016.PubMed/NCBI
|
|
24
|
Louis DN, Ohgaki H, Wiestler OD, Cavenee
WK, Burger PC, Jouvet A, Scheithauer BW and Kleihues P: The 2007
WHO classification of tumours of the central nervous system. Acta
Neuropathol. 114:97–109. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Betel D, Wilson M, Gabow A, Marks DS and
Sander C: The microRNA.org resource: Targets and expression.
Nucleic Acids Res. 36:D149–D153. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Huang DW, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
Bioinformatics Resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ramis G, Villalonga-Planells R,
Serra-Sitjar M, Brell M, Fernández de Mattos S and Villalonga P:
The tumor suppressor FOXO3a mediates the response to EGFR
inhibition in glioblastoma cells. Cell Oncol (Dordr). 42:521–536.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Peng Y, He X, Chen H, Duan H, Shao B, Yang
F, Li H, Yang P, Zeng Y, Zheng J, et al: Inhibition of
microRNA-299-5p sensitizes glioblastoma cells to temozolomide via
the MAPK/ERK signaling pathway. Biosci Rep. 38:BSR201810512018.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Zhang W, Shen C, Li C, Yang G, Liu H, Chen
X, Zhu D, Zou H, Zhen Y, Zhang D and Zhao S: miR-577 inhibits
glioblastoma tumor growth via the Wnt signaling pathway. Mol
Carcinog. 55:575–585. 2015. View
Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zeng A, Yin J, Li Y, Li R, Wang Z, Zhou X,
Jin X, Shen F, Yan W and You Y: miR-129-5p targets Wnt5a to block
PKC/ERK/NF-kappaB and JNK pathways in glioblastoma. Cell Death Dis.
9:3942018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhang G, Chen L, Khan AA, Li B, Gu B, Lin
F, Su X and Yan J: miRNA-124-3p/neuropilin-1(NRP-1) axis plays an
important role in mediating glioblastoma growth and angiogenesis.
Int J Cancer. 143:635–644. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Chen W, Kong KK, Xu XK, Chen C, Li H, Wang
FY, Peng XF, Zhang Z, Li P, Li JL and Li FC: Downregulation of
miR205 is associated with glioblastoma cell migration, invasion,
and the epithelial-mesenchymal transition, by targeting ZEB1 via
the Akt/mTOR signaling pathway. Int J Oncol. 52:485–495.
2018.PubMed/NCBI
|
|
35
|
Huang SW, Ali ND, Zhong L and Shi J:
MicroRNAs as biomarkers for human glioblastoma: Progress and
potential. Acta Pharmacol Sin. 39:1405–1513. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yuan GQ, Wei NL, Mu LY, Wang XQ, Zhang YN,
Zhou WN and Pan YW: A 4-miRNAs signature predicts survival in
glioblastoma multiforme patients. Cancer Biomark. 20:443–452. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lu D, Li Y, Liu QR, Wu Q, Zhang H, Xie P
and Wang Q: Wls promotes the proliferation of breast cancer cells
via Wnt signaling. Med Oncol. 32:1402015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jang GB, Hong IS, Kim RJ, Lee SY, Park SJ,
Lee ES, Park JH, Yun CH, Chung JU, Lee KJ, et al: Wnt/β-catenin
small-molecule inhibitor CWP232228 preferentially inhibits the
growth of breast cancer stem-like cells. Cancer Res. 75:1691–1702.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gong A and Huang S: FoxM1 and
Wnt/β-catenin signaling in glioma stem cells. Cancer Res.
72:5658–5662. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Shin J, Shin Y, Oh SM, Yang H, Yu WJ, Lee
JP, Huh SO, Lee SH, Suh YH, Chung S and Kim HS: MiR-29b controls
fetal mouse neurogenesis by regulating ICAT-mediated Wnt/β-catenin
signaling. Cell Death Dis. 5:e14732014. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Liu J, Han P, Gong J, Wang Y, Chen B, Liao
J and Tian D: Knockdown of KIAA1199 attenuates growth and
metastasis of hepatocellular carcinoma. Cell Death Discov.
4:1022018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Gan CP, Sam KK, Yee PS, Zainal NS, Lee
BKB, Abdul Rahman ZA, Patel V, Tan AC, Zain RB and Cheong SC:
IFITM3 knockdown reduces the expression of CCND1 and CDK4 and
suppresses the growth of oral squamous cell carcinoma cells. Cell
Oncol (Dordr). 42:477–490. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Deng X, Liu Z, Liu X, Fu Q, Deng T, Lu J,
Liu Y, Liang Z, Jiang Q, Cheng C and Fang W: miR-296-3p negatively
regulated by nicotine stimulates cytoplasmic translocation of c-Myc
via MK2 to suppress cell growth, metastasis and chemotherapy
resistance. Mol Ther. 26:1066–1081. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Fu Q, Song X, Liu Z, Deng X, Luo R, Ge C,
Li R, Li Z, Zhao M, Chen Y, et al: miRomics and proteomics reveal a
miR-296-3p/PRKCA/FAK/Ras/c-Myc feedback loop modulated by
HDGF/DDX5/β-catenin complex in lung adenocarcinoma. Clin Cancer
Res. 23:6336–6350. 2017. View Article : Google Scholar : PubMed/NCBI
|