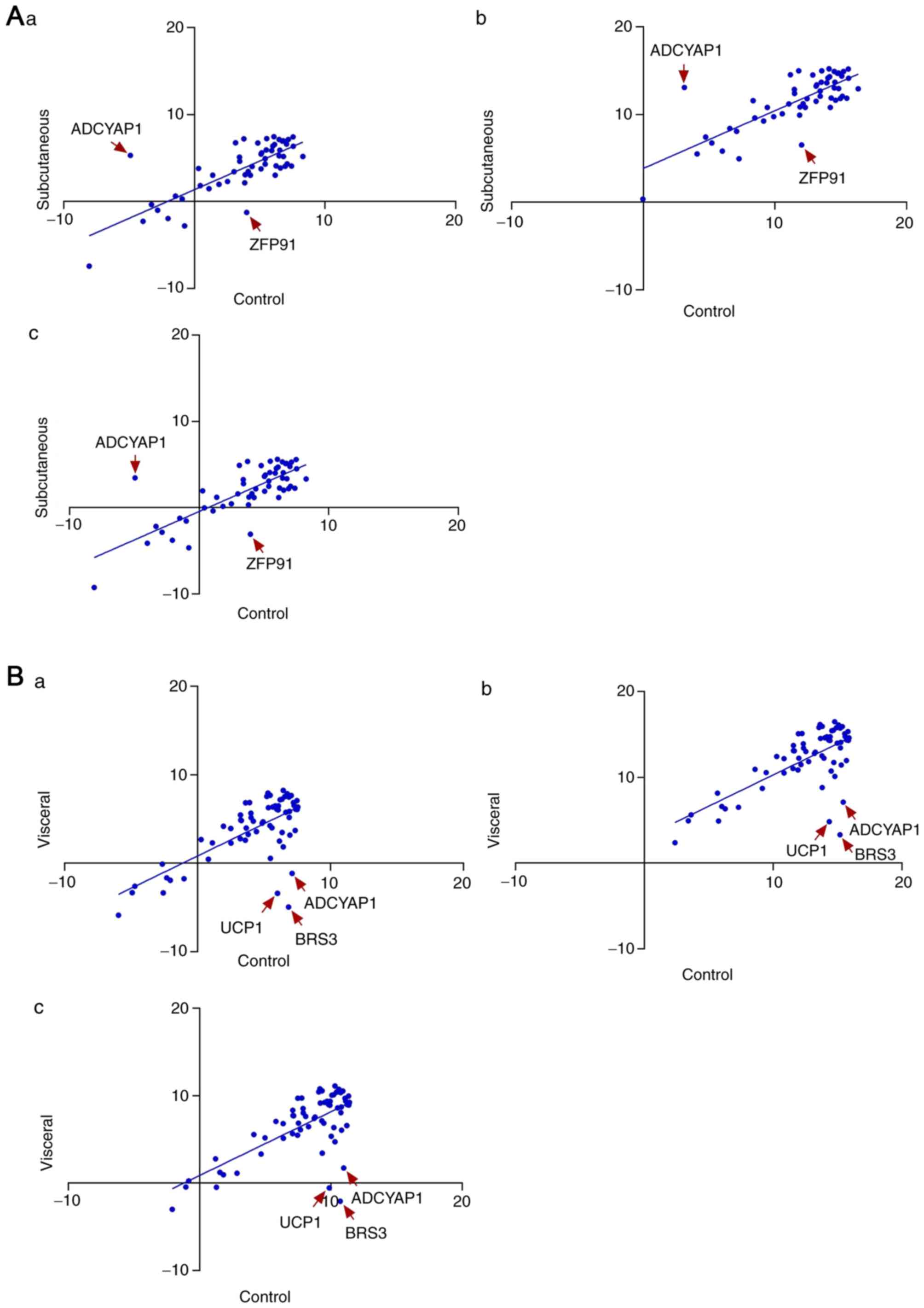
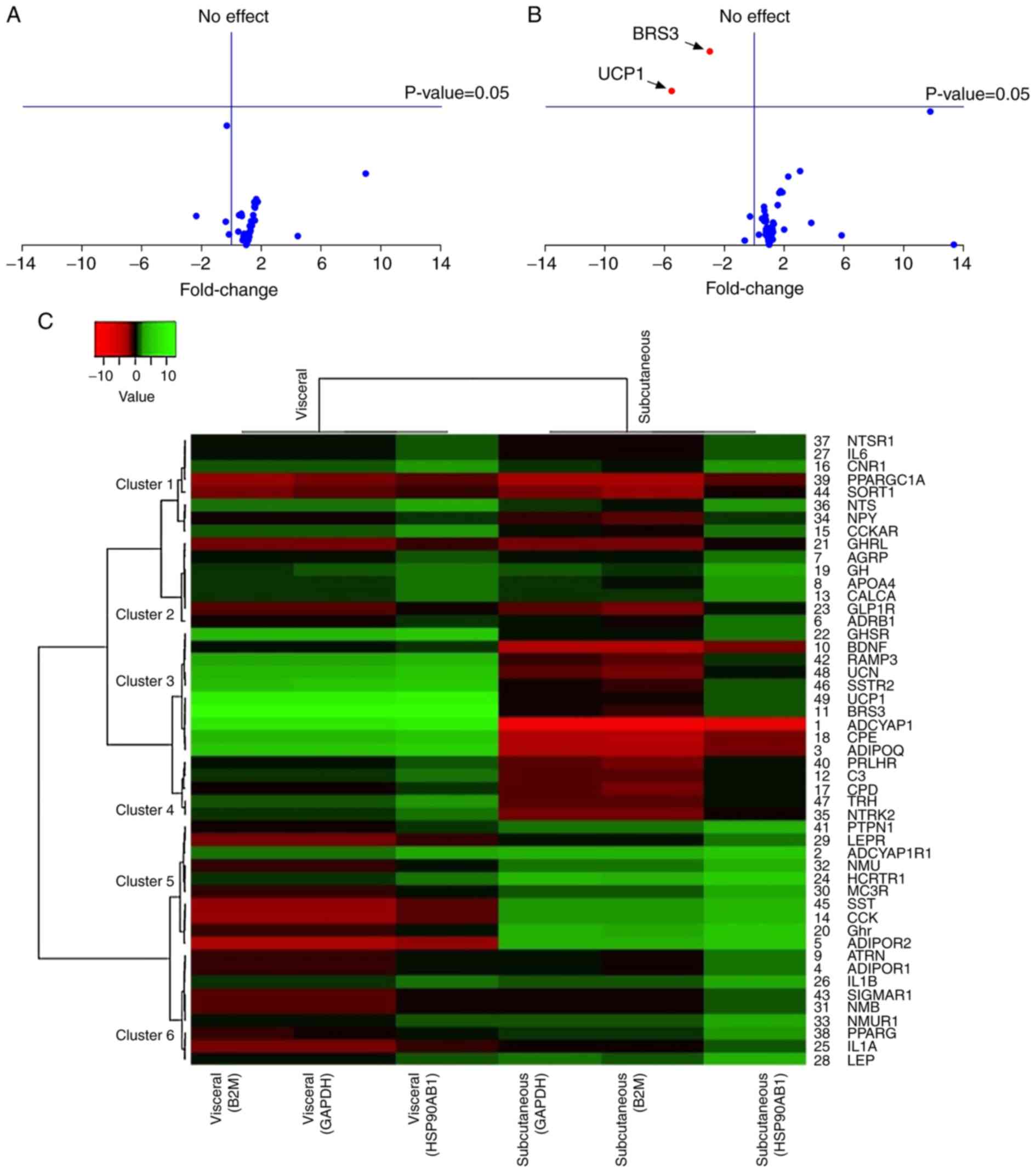
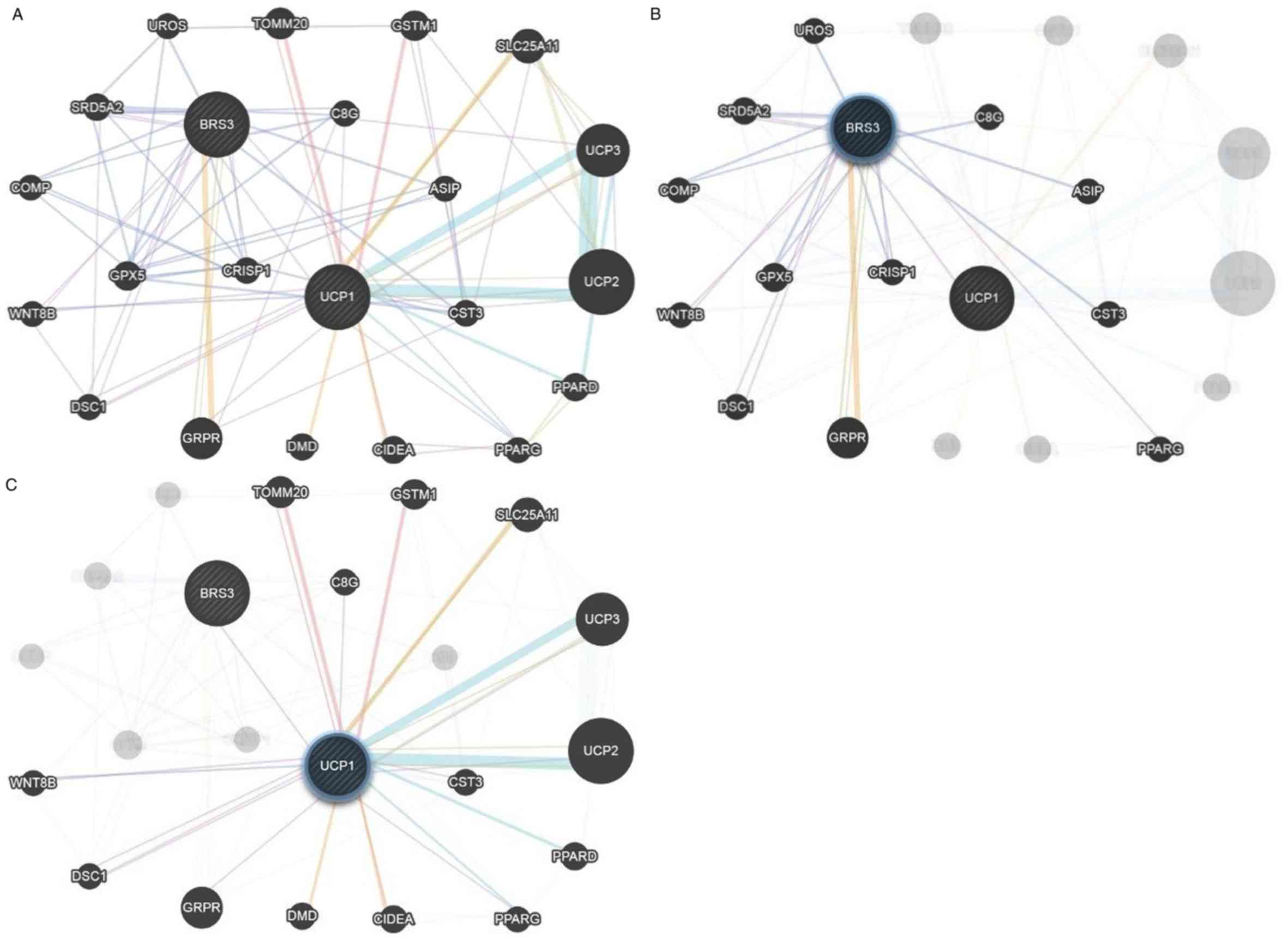
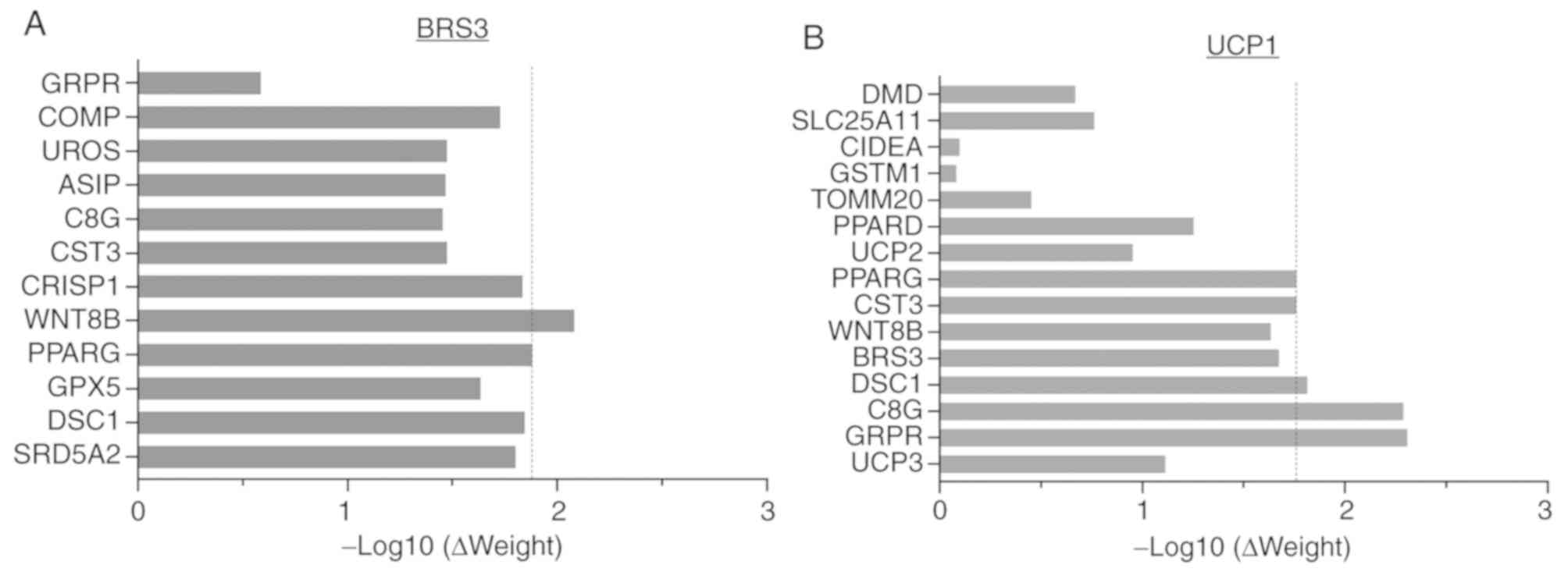
|
1
|
Sharma AM and Staels B: Review: Peroxisome
proliferator-activated receptor gamma and adipose
tissue-understanding obesity-related changes in regulation of lipid
and glucose metabolism. J Clin Endocrinol Metab. 92:386–395. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wajchenberg BL: Subcutaneous and visceral
adipose tissue: Their relation to the metabolic syndrome. Endocr
Rev. 21:697–738. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Bjørndal B, Burri L, Staalesen V, Skorve J
and Berge RK: Different adipose depots: Their role in the
development of metabolic syndrome and mitochondrial response to
hypolipidemic agents. J Obes. 2011:4906502011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shuster A, Patlas M, Pinthus JH and
Mourtzakis M: The clinical importance of visceral adiposity: A
critical review of methods for visceral adipose tissue analysis. Br
J Radiol. 85:1–10. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Pascot A, Lemieux S, Lemieux I, Prud'homme
D, Tremblay A, Bouchard C, Nadeau A, Couillard C, Tchernof A,
Bergeron J and Després JP: Age-related increase in visceral adipose
tissue and body fat and the metabolic risk profile of premenopausal
women. Diabetes Care. 22:1471–1478. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Romieu I, Dossus L, Barquera S, Blottière
HM, Franks PW, Gunter M, Hwalla N, Hursting SD, Leitzmann M,
Margetts B, et al: Energy balance and obesity: What are the main
drivers? Cancer Causes Control. 28:247–258. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Swinburn BA, Caterson I, Seidell JC and
James WP: Diet, nutrition and the prevention of excess weight gain
and obesity. Public Health Nutr. 7:123–146. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Fonseca DC, Sala P, Ferreira BAM, Reis J,
Torrinhas RS, Bendavid I and Waitzberg DL: Body weight control and
energy expenditure. Clin Nutr Exp. 20:55–59. 2018. View Article : Google Scholar
|
|
9
|
ParÏõÂzkova J: Dietary habits and
nutritional status in adolescents in central and eastern europe.
Eur J Clin Nutr. 54 (Suppl 1):S36–S40. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Racette SB, Deusinger SS and Deusinger RH:
Obesity: Overview of prevalence, etiology, and treatment. Phys
Ther. 83:276–288. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
De Lorenzo A, Soldati L, Sarlo F, Calvani
M, Di Lorenzo N and Di Renzo L: New obesity classification criteria
as a tool for bariatric surgery indication. World J Gastroenterol.
22:681–703. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Cozzo AJ, Fuller AM and Makowski L:
Contribution of adipose tissue to development of cancer. Compr
Physiol. 8:237–282. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li L, Wang G, Li N, Yu H, Si J and Wang J:
Identification of key genes and pathways associated with obesity in
children. Exp Ther Med. 14:1065–1073. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Choe SS, Huh JY, Hwang IJ, Kim JI and Kim
JB: Adipose tissue remodeling: Its role in energy metabolism and
metabolic disorders. Front Endocrinol (Lausanne). 7:302016.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luo L and Liu M: Adipose tissue in control
of metabolism. J Endocrinol. 231:R77–R99. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Khoo BY, Najimudin N and Muhammad TS: The
PPARgamma coding region and its role in visceral obesity. Biochem
Biophys Res Commun. 371:177–179. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Longo M, Zatterale F, Naderi J, Parrillo
L, Formisano P, Raciti GA, Beguinot F and Miele C: Adipose tissue
dysfunction as determinant of obesity-associated metabolic
complications. Int J Mol Sci. 20:E23582019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yogarajah T, Bee YT, Noordin R and Yin KB:
Increased peroxisome proliferator-activated receptor γ expression
levels in visceral adipose tissue, and serum CCL2 and interleukin-6
levels during visceral adipose tissue accumulation. Mol Med Rep.
11:515–520. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Division of Research and Innovation, .
Guidelines for the care and use of animals for scientific purposes.
Universiti Sains Malaysia; 2016, http://www.research.usm.my/default.asp?tag=10&f=1&k=1
|
|
20
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The Gene
Ontology Consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
The Gene Ontology Consortium, . The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mi H, Muruganujan A, Ebert D, Huang X and
Thomas PD: PANTHER version 14: More genomes, a new PANTHER GO-slim
and improvements in enrichment analysis tools. Nucleic Acids Res.
47:D419–D426. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nadarajan K, Balaram P and Khoo BY: MK886
inhibits the pioglitazone-induced anti-invasion of MDA-MB-231 cells
is associated with PPARα/γ, FGF4 and 5LOX. Cytotechnology.
68:1771–1787. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Desvergne B and Wahli W: Peroxisome
proliferator-activated receptors: Nuclear control of metabolism.
Endocr Rev. 20:649–688. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang WL and Frucht H: Activation of the
PPAR pathway induces apoptosis and COX-2 inhibition in HT-29 human
colon cancer cells. Carcinogenesis. 22:1379–1383. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Lee YH, Mottillo EP and Granneman JG:
Adipose tissue plasticity from WAT to BAT and in between. Biochim
Biophys Acta Mol Basis Dis. 1842:358–369. 2014. View Article : Google Scholar
|
|
28
|
Giralt M and Villarroya F: White, brown,
beige/brite: Different adipose cells for different functions?
Endocrinology. 154:2992–3000. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Park HS, Ju UI, Park JW, Song JY, Shin DH,
Lee KH, Jeong LS, Yu J, Lee HW, Cho JY, et al: PPARγ neddylation
essential for adipogenesis is a potential target for treating
obesity. Cell Death Differ. 23:1296–1311. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ghaben AL and Scherer PE: Adipogenesis and
metabolic health. Nature Reviews Molecular Cell Biology volume.
20:242–258. 2019. View Article : Google Scholar
|
|
31
|
Lateef DM, Xiao C, Brychta RJ, Diedrich A,
Schnermann J and Reitman ML: Bombesin-like receptor 3 regulates
blood pressure and heart rate via a central sympathetic mechanism.
Am J Physiol Heart Circ Physiol. 310:H891–H898. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zheng Q, Lin J, Huang J, Zhang H, Zhang R,
Zhang X, Cao C, Hambly C, Qin G, Yao J, et al: Reconstitution of
UCP1 using CRISPR/Cas9 in the white adipose tissue of pigs
decreases fat deposition and improves thermogenic capacity. Proc
Natl Acad Sci USA. 114:E9474–E9482. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Villarroya F, Iglesias R and Giralt M:
PPARs in the control of uncoupling proteins gene expression. PPAR
Res. 2007:743642007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yamada K, Ohki-Hamazaki H and Wada K:
Differential effects of social isolation upon body weight, food
consumption, and responsiveness to novel and social environment in
bombesin receptor subtype-3 (BRS-3) deficient mice. Physiol Behav.
68:555–561. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Blücher C and Stadler SC: Obesity and
breast cancer: Current insights on the role of fatty acids and
lipid metabolism in promoting breast cancer growth and progression.
Front Endocrinol (Lausanne). 8:2932017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Faber J and Fonseca LM: How sample size
influences research outcomes. Dental Press J Orthod. 19:27–29.
2014. View Article : Google Scholar : PubMed/NCBI
|