Introduction
Immunoglobulin A (IgA) nephropathy (IgAN), one of
the most common causes of primary glomerulonephritis in pediatrics,
was conventionally considered a benign disease (1). However, IgAN is now known to be a
progressive renal disease, with 20–30% of patients developing
end-stage renal failure over 20–30 years (2,3).
Although the pathogenesis of IgAN remains unknown,
it has been reported that aberrant glycosylation and glomerular
mesangial deposition of IgA1 serve critical roles in its
pathogenesis (4–6). Deficiency of core 1
β3-galactosyltransferase (C1β3Gal-T) is associated with aberrant
glycosylation of IgA1 (7).
Moreover, Ju and Cummings (8)
demonstrated that the unique molecular chaperone of C1β3Gal-T,
C1β3Gal-T-specific molecular chaperone (C1GALT1C1/COSMC) is vital
for the activity of C1β3Gal-T (8–10).
Previous studies have reported that COSMC expression levels in B
lymphocytes were lower in patients with IgAN compared with healthy
controls and patients with other renal diseases (9,11).
However, the mechanisms underlying COSMC expression are not fully
understood (10). Serino et
al (12,13) indicated that microRNA (miRNA)-148b
targeted and regulated C1β3Gal-T expression. Moreover, as a
molecular chaperone, the regulation of COSMC is crucial for its
biological functions, but the mechanisms remain largely unknown
(10). Therefore, the present
study aimed to investigate the effect of miRNAs on COSMC
expression.
Materials and methods
Screening for miRNA expression levels
in pediatric patients with IgAN and healthy controls
The study cohort included 9 pediatric patients with
IgAN (IgAN group), who were independently diagnosed by two
experienced pathologists, and 6 healthy pediatrics (control group)
that were recruited by Beijing Children's Hospital between January
2015 and December 2018. There were no significant differences
between the two groups in terms of age and sex (P>0.05; Table I). Peripheral blood samples (5 ml)
were from the veins of the patients and healthy controls. Plasma
was obtained by centrifugation at 1,710 × g at 4°C for 5 min.
Samples were subsequently stored at −80°C until required for
subsequent analysis. B lymphocytes were isolated, and miRNAs were
extracted for single-end second-generation sequencing. Briefly,
total RNA was extracted from the plasma using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and a miRNeasy
Mini kit (Qiagen, Inc.). Hybridization and ligation were performed
for small RNA fractions of <50 nucleotide using a
NEBNext® Multiplex Oligos for Illumina®
(Methylated Adaptor, Index Primers Set 1) kit (cat. no. E7535L; New
England Biolabs, Inc.). The miRNAs were subsequently sequenced
using the Illumina HiSeq 2000 Sequencing system. All procedures
performed involving human participants were approved by the Medical
Ethics Committee of Beijing Children's Hospital, Capital Medical
University (IRB approval no. 2014-12). Informed consent was
obtained from the parents of all participants prior to the
study.
 | Table I.Clinical data of the IgAN and healthy
control groups. |
Table I.
Clinical data of the IgAN and healthy
control groups.
| Variables | IgAN group | Healthy control
group | P-value |
|---|
| Cases | 9 | 6 |
|
| Age (years) | 9.03±2.99 | 9.68±2.57 | 0.607 |
| Sex |
|
| 0.659 |
| Male | 7 | 5 |
|
|
Female | 2 | 1 |
|
Lymphocyte isolation
EDTA-anticoagulated blood samples (5 ml) from the
IgAN group were collected the day after kidney biopsy and
immediately sent for cell isolation and processing. Fresh blood
samples (2 ml) were mixed with Hanks' balanced salt solution (2 ml)
at room temperature for 30 min. Subsequently, AstroMACS Separation
Buffer (Miltenyi Biotec, Inc.) was added to the liquid surface and
centrifuged at room temperature at 3,000 rcf for 20 min. The second
layer of cells was then collected and mixed with 5 ml Hanks'
balanced salt solution, and the mixture was centrifuged twice at
room temperature and 2,000 rcf for 15 min to separate the
lymphocytes. Then, 1 ml Dynabeads (Thermo Fisher Scientific, Inc.)
was added to 1 ml PBS and mixed by shaking. The tube containing the
suspension was placed in a magnetic system at room temperature for
1 min. The supernatant was collected, and the process was repeated
2–3 times. The separated lymphocytes were resuspended and cell
counts were determined to adjust the cell concentration to
1×105/ml. The suspension was shaken for 25 min at 4°C
before placing the tube in the magnetic system for 2 min. The
supernatant was removed, 1 ml PBS placed in the magnetic system for
1 min was added and the supernatant was collected; this procedure
was repeated four times. To purify CD19+ cells, the
magnetic bead suspension was resuspended and 10 µl DETACHaBEAD
reagent (Thermo Fisher Scientific, Inc.) was added per 25 µl
magnetic bead suspension and then mixed. Subsequently, after
washing with 1 ml 10% PBS, the supernatant was collected by
centrifugation at room temperature and 600 rcf for 6 min to remove
the residual DETACHaBEAD reagent. The cells were then resuspended
for subsequent experiments.
miRNA target prediction
The bioinformatics software PicTar (http://www.pictar.org/) and TargetScan version 7.1
(http://www.targetscan.org) were used to
analyze the corresponding miRNAs of COSMC.
Validation tests of the IgAN and
control groups
To expand the sample size, a further 15 pediatric
patients with IgAN and 15 controls (four healthy controls and 11
pediatric patients with other renal diseases) were recruited
between January 2013 to December 2018 from Beijing Children's
Hospital. No significant differences were observed among these
three groups in terms of age, sex and 24 h urine total protein
results, except the urine proteins in the healthy control group
(Table II). B lymphocytes were
isolated from 5 ml peripheral blood samples with 5 ml EDTA as an
anticoagulant. Total RNA was extracted using an miRNeasy kit (cat.
no. 217184; Qiagen GmbH) according to the manufacturer's
instructions. A spectrophotometer was used to detect the absorbance
at 450 nm of the extracted RNA for analysis. RNA was reverse
transcribed into cDNA using the PrimeScript RT Reagent kit (Takara
Biotechnology Co., Ltd.) at 37°C for 1 h. Expression levels were
detected by qPCR using the miScript SYBR® Green PCR kit
(Qiagen, Inc.), according to the manufacturer's protocol. The
following primers were used for the qPCR: MiRNA-196b-5p forward,
5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCAACAA-3′ and reverse,
5′-ACACTCCAGCTGGGTAGGTAGTTTCCTGTT-3′; and RNU6 forward,
5′-CTCATGACCACAGTCCATGCC-3′ and reverse,
5′-GGCATGGACTGTGGTCATGAG-3′. The thermocycling conditions for the
qPCR were as follows: Initial denaturation at 95°C for 30 sec; and
40 cycles of 95°C for 5 sec and 60°C for 30 sec. The relative
miRNA-196b expression levels were calculated using the
2−ΔΔCq method (14).
 | Table II.Clinical data of the IgAN and control
groups. |
Table II.
Clinical data of the IgAN and control
groups.
| Variables | IgAN group | Healthy control
group | Other renal disease
group | P-value |
|---|
| Cases | 15 | 4 | 11 |
|
| Average age
(years) |
9.60±2.55a | 8.68±2.80 | 8.80±2.92 | 0.470 |
| Sex |
|
|
| 0.907 |
|
Male | 12 | 3 | 8 |
|
|
Female | 3 | 1 | 3 |
|
| 24 h urine total
protein (mg) |
819.4±233.0b |
| 751±511 | 0.328 |
Western blotting analysis of effect of
miRNA-196b on the expression of COSMC
Isolated B lymphocytes of pediatric patients with
IgAN from the present study were divided into three groups: i)
Control group (untreated); ii) miRNA-196b group which contained the
miRNA-196b-transfected B lymphocytes; and iii) the negative control
(NC) group, which contained the NC-transfected B lymphocytes. Total
cellular proteins were extracted from 1×106 B
lymphocytes/ml using RIPA lysis buffer [50 mM Tris (pH 7.4), 150 mM
NaCl, 1% Triton X-100, 1% sodium deoxycholate and 0.1% SDS;
Sigma-Aldrich; Merck KGaA] and quantified using the bicinchoninic
acid protein assay. The total proteins (20 µg/lane) obtained from
the cells were separated using 12% SDS-PAGE and then transferred to
PVDF membranes (0.45 µm; Roche Diagnostics). After blocking with 5%
non-fat dry milk at 4°C overnight, the membranes were incubated
with antibodies against anti-COSMC (1:2,000; cat. no. 68310; Abcam)
or β-actin (1:3,000; cat. no. 4970; Cell Signaling Technology,
Inc.) at 4°C overnight. Then, the membranes were incubated at room
temperature for 1 h with secondary antibodies labeled with
horseradish peroxidase (HRP)-conjugated goat anti-rabbit IgG-HRP
(cat. no. sc-2005; 1:5,000; Santa Cruz Biotechnology, Inc.) or goat
anti-mouse IgG-HRP (cat. no. sc-2004; 1:5,000; Santa Cruz
Biotechnology, Inc.). An enhanced chemiluminescence kit (Amersham;
GE Healthcare Life Sciences) was used to visualize the protein
bands.
Comparative analysis after miRNA-196b
transfection
In our previous study (11), three groups of patients were
included: i) 26 biopsy-proven pediatric patients with IgAN; ii) 13
healthy pediatrics (control group); and iii) 11 pediatric patients
with other renal diseases, including hereditary nephritis and
membranous nephropathy (second control group). In the present
study, B lymphocytes of pediatric patients with IgAN from the above
cohort (26 biopsy-proven pediatric patients with IgAN) were
transfected with miRNA-196b mimics (Thermo Fisher Scientific, Inc.;
forward, 5′-CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCCCAACAA-3′ and
reverse, 5′-ACACTCCAGCTGGGTAGGTAGTTTCCTGTT-3′) or its negative
control (NC; Thermo Fisher Scientific, Inc.; forward,
5′-CTCATGACCACAGTCCATGCC-3′ and reverse,
5′-GGCATGGACTGTGGTCATGAG-3′). A total of 20 µg/ml miRNA-196b or the
NC was transfected into 1×106 B lymphocytes/ml in the
IgAN group using Lipofectamine® 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol, for 48 h prior to subsequent
experimentation. Then, Gd-IgA1 expression levels were determined
using a Gd-IgA1 ELISA kit (cat. no. F10079; Shanghai Fanke Industry
Co., Ltd.), according to the manufacturer's protocol. Informed
consent was provided by the patients for the use of their samples
in this study.
Correlations between miRNA-196b
expression levels and clinical data of patients
In addition to the detection of miRNA-196b
expression levels, the clinical data of the IgAN group (n=14; 1
patient was excluded due to in insufficient clinical data) was
recorded and correlation analysis was performed using Pearson's
correlation coefficient. Renal function was determined by detecting
the levels of serum blood urea nitrogen (BUN) using UREA reagent
(cat. no. OSR6134; Beckman Coulter, Inc.), according to the
manufacturer's protocol, the serum creatinine (sCr) levels using a
Creatinine Test kit [cat. no. MC1212B; Merit Choice Bioengineering
(Beijing) Co., Ltd.], according to the manufacturer's protocol, and
the glomerular filtration rate (GFR) using the Schwartz formula, as
previously described (15). The 24
h urine total protein concentration was determined using Total
protein UC FS reagent (Diasys Diagnostic Systems GmbH), according
to the manufacturer's protocol, and urine red blood cells were
collected from urine by centrifugation (500 × g; 5 min; 20–25°C)
and counted using a phase contrast microscope (magnification,
×400).
CD3, CD4 and CD8 expression levels in peripheral
blood T cells were assessed using flow cytometric analysis.
Briefly, 2 ml EDTA anti-coagulation peripheral blood was collected
from the patients with IgAN group and 100 µl whole blood from the
sample was added to each tube. Samples were blocked with 3% FBS
(cat. no. 04-001-1; Biological industries) at room temperature for
10 min and then, 5 µl C-S® tetraCHROME™
CD45-FITC/CD4-RD1/CD8-ECD/CD3-PC5 antibody (cat. no. 6607013;
Beckman Coulter, Inc.) was added and incubated in the dark for 10
min at room temperature. Subsequently, 500 µl hemolysin was
added/tube, mixed by shaking and incubated at room temperature for
10 min in the dark, followed by incubation at room temperature for
10 min in the dark of 500 µl PBS. T cells were visualized using a
Beckman Coulter Cytomics FC-500 Flow Cytometer (Beckman Coulter,
Inc.) and data was analyzed using CXP analysis software (version
2.0; Beckman Coulter, Inc.).
Histochemistry
Renal histological manifestations were observed by
histochemical staining and graded according to Lee's glomerular
grading system (16). Briefly,
renal tissues from the right kidney of the 14 patients mentioned
above were collected following renal biopsy and fixed in 10%
formalin for 4 h at room temperature. Tissues were embedded in
paraffin and cut into 3-µm thick sections. Sections were
deparaffinized with xylene, rehydrated in a descending series of
ethanal and routinely stained hematoxylin and eosin, periodic
acid-Schiff (PAS), PAS + methenamine, and Masson's trichrome.
Stained sections were observed using a light microscope
(magnification, ×40, ×100, ×200 or ×400) to determine patient renal
pathology. Based on the results, the patients were divided into two
groups: IgAN with grade I–II renal damage as group A and IgAN with
grade III–V renal damage as group B.
Statistical analysis
Data are presented as the mean ± SD of ≥2 averaged
replicates of independent experiments. Quantitative data were
compared using one-way ANOVA followed by Least Significant
Difference post hoc test (Table
II) or Tukey's test (Fig. 1),
or Student's t-test (Table III).
Crosstabs and c2 tests were used to compare frequencies.
Pearson's correlation coefficient was used to identify correlations
among factors. All data analyses were performed using the SPSS
software (version 18.0; SPSS, Inc.). P<0.05 was considered to
indicate a statistically significant difference.
 | Table III.Correlation between miRNA-196b
expression levels and clinical data. |
Table III.
Correlation between miRNA-196b
expression levels and clinical data.
| Variables in IgAN
group | Mean | SD or
SEMc | r | n | P-value |
|---|
| 24 h urine total
protein, mg | 821.47 | 235.6c | −0.590 |
|
0.021a |
| CD3 | 70.83 | 7.47 | 0.361 |
| 0.186 |
| CD4 | 29.51 | 6.35 | −0.044 |
| 0.875 |
| CD8 | 33.55 | 5.06 | 0.066 |
| 0.814 |
| BUN, mmol/l | 4.79 | 2.24 | −0.340 |
| 0.215 |
| sCr, µmol/l | 52.65 | 16.97 | −0.422 |
| 0.117 |
| GFR, ml/min/1.73
m2 | 103.14 | 20.97 | 0.317 |
| 0.250 |
| RBC, /HP | 80.8 | 22.61 | 0.478 |
| 0.071 |
|
Pathologyb |
|
|
|
| 0.609 |
| Grade
I–II |
|
|
| 4 |
|
| Grade
III–V |
|
|
| 10 |
|
Results
miRNA expression levels in pediatric
patients with IgAN and healthy controls
B lymphocytes were isolated from the peripheral
blood samples of 9 pediatric patients with IgAN and 6 healthy
controls. miRNAs extracted from these cells were subjected to
second-generation sequencing. Bioinformatics analysis detected 205
miRNAs with significant differences in expression levels between
the IgAN and control groups (P<0.05;
P=0.013–1.41×10−22; data not shown).
miRNA target prediction
The target miRNAs of COSMC were investigated and
predicted using PicTar and TargetScan. Among the differentially
expressed miRNAs, two miRNAs were selected, miRNA-196b-5p and
miRNA-33a-3p, and the target gene of these miRNAs was predicted to
be COSMC (data not shown). The present results suggested that there
were significant differences miRNA-196b and miRNA-33a expression
levels between the IgAN and control groups (P=4.73×10−7
and P=0.001, respectively; data not shown). In addition, as there
was a greater difference in the miRNA-196b expression levels
compared with the miRNA-33a expression levels between the two
groups, miRNA-196b was selected for further analysis in this
study.
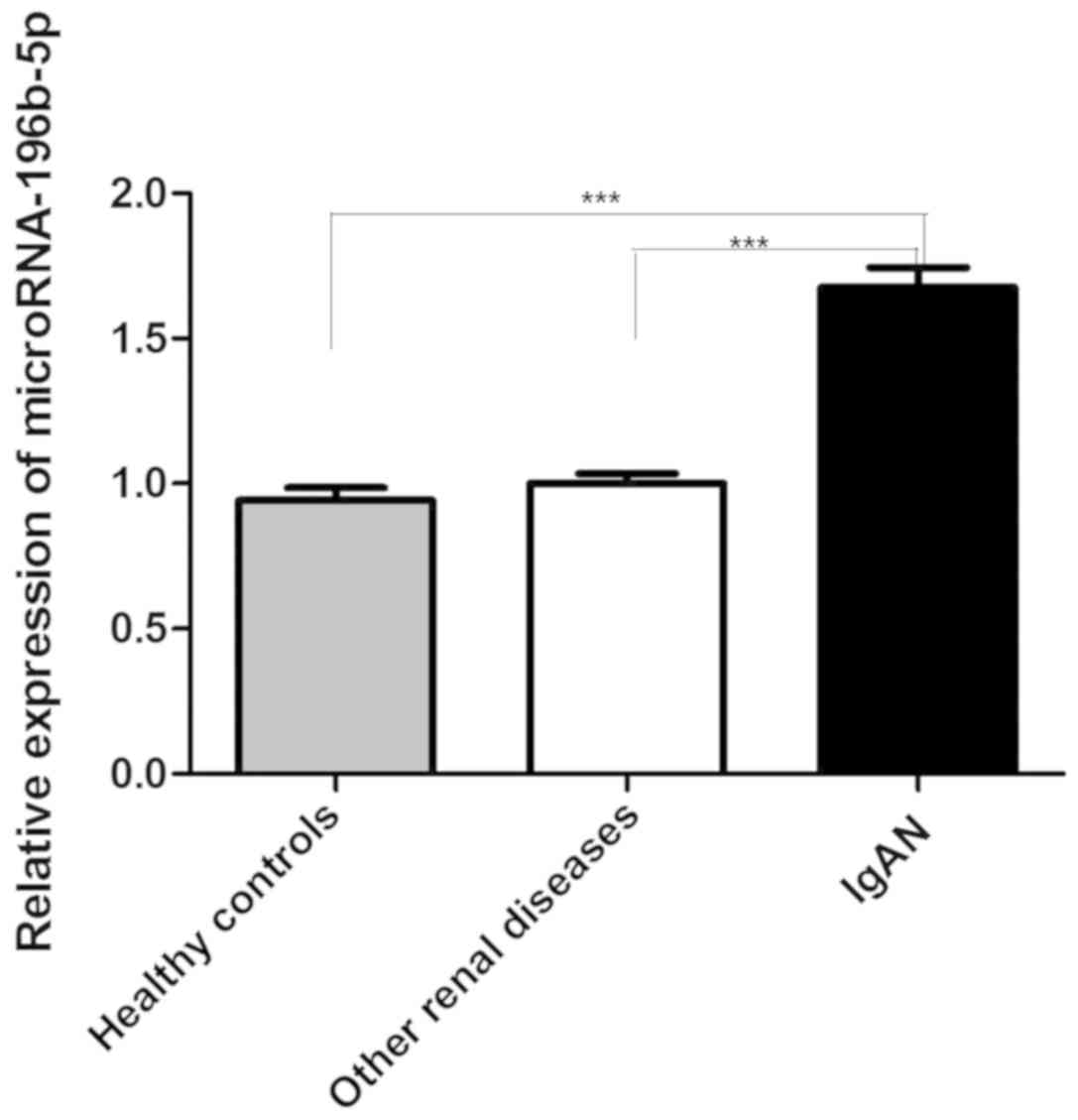
miRNA-196b expression levels between
the IgAN and control groups
B lymphocytes were isolated from the peripheral
blood samples of 15 pediatric patients with IgAN and 15 controls
(healthy and other renal diseases). The expression levels of
miRNA-196b-5p in the IgAN group were significantly higher compared
with the healthy control group and the other renal diseases group
(P<0.0001; Fig. 1).
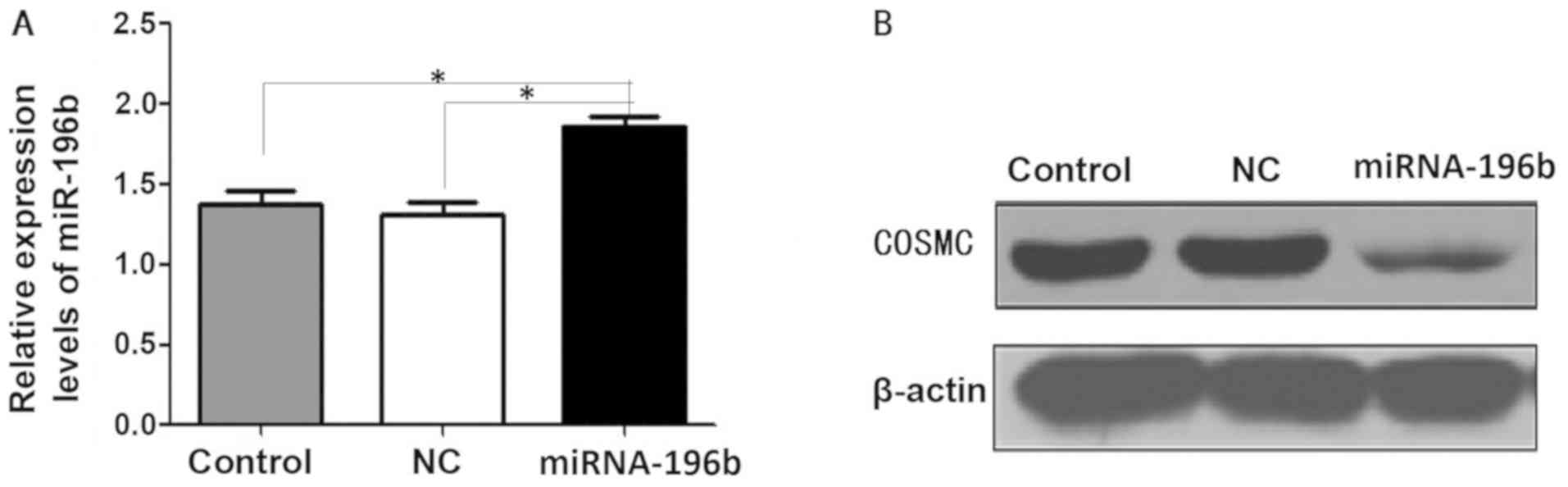
Cellular studies
The transfection of miRNA-196b was demonstrated to
be successful as significantly increased expression levels of
miRNA-196b were observed in the miRNA-196b group (P<0.05;
Fig. 2A). Compared with the
control and NC group, COSMC expression levels were decreased in the
miRNA-196b group after the addition of the miRNA-196b mimic
(Fig. 2B).
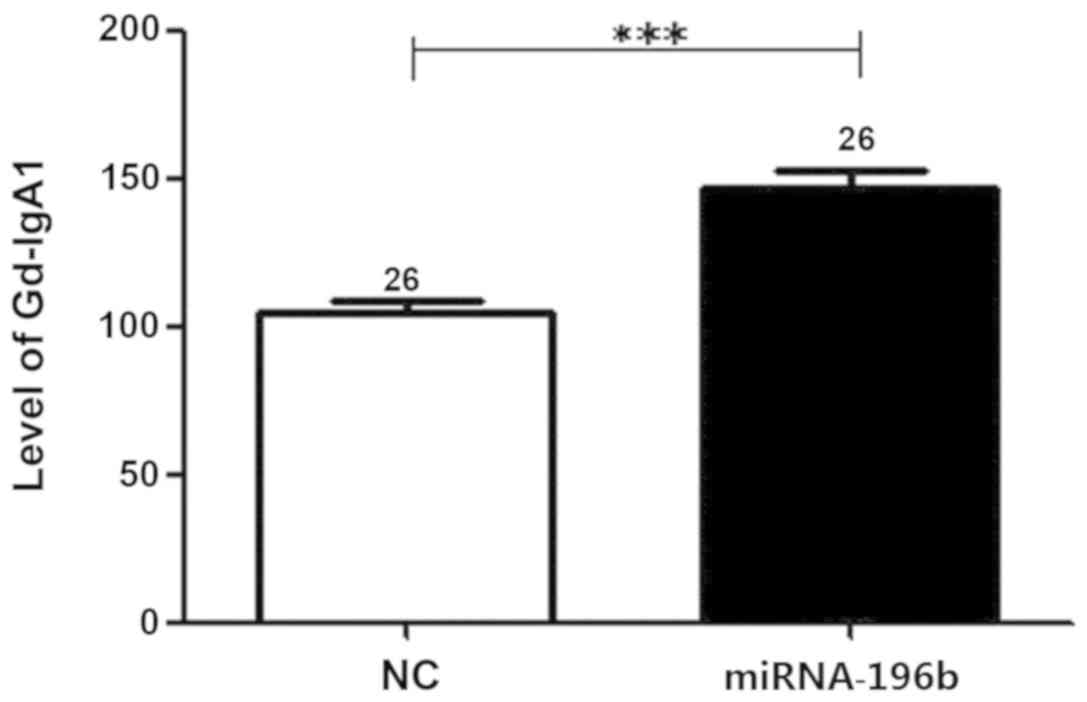
Gd-IgA1 expression levels
In our previous study, compared with the healthy
control group (n=13) and other renal disease group (n=11), Gd-IgA1
expression levels in the supernatants of B lymphocytes isolated
from peripheral blood samples, were significantly increased in the
IgAN group (11). In the present
study, B lymphocytes from the IgAN group transfected with
miRNA-196b mimic (n=26) exhibited significantly increased Gd-IgA1
expression levels compared with the group with no intervention
(P=4.56×10−6; Fig.
3).
Correlations between miRNA-196b
expression levels and clinical data of patients
The results of the present study suggested that
there were no significant differences between the IgAN with grade
I–II renal damage and the IgAN with grade III–V renal damage groups
in terms of miRNA-196b expression levels (P=0.609; Table III). Correlation analysis results
suggested that there were no correlations between miRNA-196b
expression levels and CD3, CD4 and CD8 expression levels, serum
BUN, sCR and GFR levels (P=0.186, 0.875, 0.814, 0.215, 0.117 and
0.250, respectively). In addition, there was no significant
correlation between miRNA-196b expression and urine red blood cell
levels. However, there was a significant correlation between
miRNA-196b expression levels and the 24 h urine total protein level
(P=0.021; Table III).
Discussion
miRNAs are highly conserved non-coding small
single-stranded RNAs, ~22 nucleotides in length, that play crucial
roles in gene expression and regulation (17). Different types of miRNAs regulate
different target genes (18). In
the human genome, 60–70% of protein-coding genes are regulated by
miRNAs and miRNAs have been shown to be associated with various
important physiological and pathological processes, such as
development, organogenesis, apoptosis, cell proliferation and
tumorigenesis (19–22). Previous studies have reported that
miRNAs may be involved in the pathophysiology of numerous renal
diseases (23–26). Serino et al (13) reported that in patients with IgAN,
miRNA-148b expression was downregulated in peripheral blood
mononuclear cells and C1β3Gal-T-1 expression level was decreased
compared with normal healthy controls. Moreover, miRNA-148b
expression was negatively correlated with C1β3Gal-T-1 expression in
the same patient cohort (13).
These studies suggested that miRNA-148b overexpression in patients
with IgAN may inhibit C1β3Gal-T-1 expression, thereby leading to
aberrant glycosylation of IgA1.
Gd-IgA1 can easily aggregate and form immune
complexes that are not cleared as efficiently, which leads to
mesangial deposition, cell proliferation and expansion of the
extracellular matrix (2,13); this subsequently results in
inflammation and glomerular injury (27). However, it remains unknown whether
the function of COSMC, the key chaperone of C1β3Gal-T, is regulated
by miRNAs (13). Therefore, in the
present study, the global expression profile of peripheral blood
mononuclear cells obtained from pediatric patients with IgAN was
analyzed. Bioinformatics analysis identified 205 miRNAs that were
differentially regulated in this patient cohort and suggested that
miRNAs may be involved in the pathogenesis of IgAN. In addition,
COSMC was identified as one of the target genes of miRNA-196b and
miRNA-33a, which were among the 205 detected miRNAs. The present
results suggested that there were significant differences
miRNA-196b and miRNA-33a expression levels between the IgAN and
control groups. As there was a greater difference in terms of
miRNA-196b expression levels compared with miRNA-33a expression
levels between the two groups, miRNA-196b was selected for further
analysis. Future studies will investigate miRNA-33a in depth.
The present study demonstrated that miRNA-196b
expression increased in human peripheral blood B lymphocytes from
the IgAN group compared with the healthy control and other renal
disease groups, thus indicating that the abnormal expression was
specific to the IgAN group. The other renal disease control group
included patients with hereditary nephritis and membranous
nephropathy. As the pathogenesis of purpura nephritis is similar to
that of IgAN (28), pediatric
patients with purpura nephritis were not included in the control
groups. Moreover, future studies will investigate miRNA-196b
expression levels between pediatric patients with IgAN and patients
with purpura nephritis.
In addition, the present results suggested that
there were no correlations between miRNA-196b expression levels,
and CD3, CD4, CD8, BUN, Cr or urine red blood cell levels.
Moreover, there were no correlations between miRNA-196b expression
levels, and GFR or renal pathology, while there was a significant
correlation between miRNA-196b expression levels and 24 h urine
total protein. As 24 h urine total protein is one of the
independent factors of disease severity and prognosis (1), miRNA-196b expression levels may be
used as a non-invasive diagnostic marker of IgAN progression.
However, due to the limited sample size of the present study, no
significant association was identified between miRNA-196b
expression level and renal pathological grading. Therefore, future
studies with larger sample numbers are underway to investigate the
possible correlations between miRNA-196b expression levels and
renal pathology with new grading standards.
To investigate whether miRNA-196b can modulate the
target gene COSMC expression levels, B lymphocytes of pediatric
patients with IgAN were transfected with miRNA-196b mimics. The
present results suggested that COSMC expression levels
significantly decreased and Gd-IgA1 expression levels increased
after miRNA-196b transfection. Therefore, it is hypothesized that
increased miRNA-196b expression levels may downregulate COSMC
expression levels. Moreover, as the chaperone protein of a key
enzyme, reduced COSMC expression levels may affect the normal
function of C1β3Gal-T, which could result in the formation,
accumulation and deposition of deglycosylated IgA1 in serum and the
kidneys, subsequently leading to the onset of IgAN.
However, the present study is only a preliminary
study, thus the specific mechanism underlying the regulation of
COSMC expression by miRNA-196b is not fully understood. Therefore,
further cytological experiments are required. In addition, due to
the limited number of samples in the present study, further
clinical tests with greater numbers of patient samples are needed.
In the present study, due to the limited blood samples, it is
difficult to perform multiple experimental tests. Future studies
will involve animal experiments to investigate the effects in
vivo.
Collectively, the present results suggested that
miRNA-196b expression levels in B lymphocytes were significantly
higher in pediatric patients with IgAN compared with patients with
other renal diseases and healthy controls. Therefore, miRNA-196b
levels may be related to the clinical characteristics of pediatric
patients with IgAN. The transfection of isolated B lymphocytes with
miRNA-196b mimics significantly downregulated COSMC expression and
increased Gd-IgA1 expression levels. The present results suggested
that miRNA-196b may play crucial roles in the formation of Gd-IgA1
and pathogenesis of IgAN by targeting and regulating COSMC
expression. However, the specific underlying mechanisms require
further investigation.
Acknowledgements
Not applicable.
Funding
The present study was supported by National Natural
Science Fund (grant no. 81600551), the Beijing Municipal Science
and Technology Commission Major Research Project (grant no.
D181100000118006), the Capital Health Research and Development of
Special Grant (grant no. 2016-2-2094) and the Application of
Capital Clinical Characteristics Program of Beijing Municipal
Science and Technology Commission (grant no. Z161100000516106).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QS and XL analyzed and interpreted the data and were
major contributors in the writing of the manuscript. JL, HZ, NZ and
YL performed the clinical and laboratory experiments. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
All procedures performed in studies involving human
participants were approved by Medical Ethics Committee of Beijing
Children's Hospital, Capital Medical University (IRB approval no.
2014-12). Informed consent was obtained from the parents of all
participants prior to the study.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interest.
Glossary
Abbreviations
Abbreviations:
|
miRNA
|
microRNA
|
|
IgAN
|
immunoglobulin A nephropathy
|
|
C1β3Gal-T
|
core1β3-galactosyltransferase
|
|
COSMC
|
C1β3Gal-T-specific molecular
chaperone
|
References
|
1
|
Seikrit C, Rauen T and Floege J:
Immunoglobulin A nephropathy. Internist (Berl). 60:432–439. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ebefors K, Liu P, Lassén E, Elvin J,
Candemark E, Levan K, Haraldsson B and Nyström J: Mesangial cells
from patients with IgA nephropathy have increased susceptibility to
galactose-deficient IgA1. BMC Nephrol. 17:402016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sanders JT, Hastings MC, Moldoveanu Z,
Novak J, Julian BA, Bursac Z and Wyatt RJ: Serial
galactose-deficient IgA1 levels in children with IgA nephropathy
and healthy controls. Int J Nephrol. 2017:82106412017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Coppo R: Biomarkers and targeted new
therapies for IgA nephropathy. Pediatr Nephrol. 32:725–731. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Kim MJ, Schaub S, Molyneux K, Koller MT,
Stampf S and Barratt J: Effect of immunosuppressive drugs on the
changes of serum galactose-deficient IgA1 in patients with IgA
nephropathy. PLoS One. 11:e01668302016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lai KN, Tang SC, Schena FP, Novak J,
Tomino Y, Fogo AB and Glassock RJ: IgA nephropathy. Nat Rev Dis
Primers. 2:160012016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shin DH, Lim BJ, Han IM, Han SG, Kwon YE,
Park KS, Lee MJ, Oh HJ, Park JT, Han SH, et al: Glomerular IgG
deposition predicts renal outcome in patients with IgA nephropathy.
Mod Pathol. 29:743–752. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ju T and Cummings RD: A unique molecular
chaperone Cosmc required for activity of the mammalian core 1 beta
3-galactosyltransferase. Proc Natl Acad Sci USA. 99:16613–16618.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Suzuki H, Yasutake J, Makita Y, Tanbo Y,
Yamasaki K, Sofue T, Kano T and Suzuki Y: IgA nephropathy and IgA
vasculitis with nephritis have a shared feature involving
galactose-deficient IgA1-oriented pathogenesis. Kidney Int.
93:700–705. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Placzek WJ, Yanagawa H, Makita Y, Renfrow
MB, Julian BA, Rizk DV, Suzuki Y, Novak J and Suzuki H: Serum
galactose-deficient-IgA1 and IgG autoantibodies correlate in
patients with IgA nephropathy. PLoS One. 13:e01909672018.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun Q, Zhang J, Zhou N, Liu X and Shen Y:
DNA methylation in Cosmc promoter region and aberrantly
glycosylated IgA1 associated with pediatric IgA nephropathy. PLoS
One. 10:e01123052015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Serino G, Pesce F, Sallustio F, De Palma
G, Cox SN, Curci C, Zaza G, Lai KN, Leung JC, Tang SC, et al: In a
retrospective international study, circulating miR-148b and let-7b
were found to be serum markers for detecting primary IgA
nephropathy. Kidney Int. 89:683–692. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Serino G, Sallustio F, Cox SN, Pesce F and
Schena FP: Abnormal miR-148b expression promotes aberrant
glycosylation of IgA1 in IgA nephropathy. J Am Soc Nephrol.
23:814–824. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Schwartz GJ, Muñoz A, Schneider MF, Mak
RH, Kaskel F, Warady BA and Furth SL: New equations to estimate GFR
in children with CKD. J Am Soc Nephrol. 20:629–637. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lee HS, Lee MS, Lee SM, Lee SY, Lee ES,
Lee EY, Park SY, Han JS, Kim S and Lee JS: Histological grading of
IgA nephropathy predicting renal outcome: Revisiting H. S. Lee's
glomerular grading system. Nephrol Dial Transplant. 20:342–348.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Runtsch MC, Hu R, Alexander M, Wallace J,
Kagele D, Petersen C, Valentine JF, Welker NC, Bronner MP, Chen X,
et al: MicroRNA-146a constrains multiple parameters of intestinal
immunity and increases susceptibility to DSS colitis. Oncotarget.
6:28556–28572. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chandrasekaran K, Karolina DS, Sepramaniam
S, Armugam A, Wintour EM, Bertram JF and Jeyaseelan K: Role of
microRNAs in kidney homeostasis and disease. Kidney Int.
81:617–627. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang G, Kwan BC, Lai FM, Chow KM, Li PK
and Szeto CC: Elevated levels of miR-146a and miR-155 in kidney
biopsy and urine from patients with IgA nephropathy. Dis Markers.
30:171–179. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pauley KM, Cha S and Chan EK: MicroRNA in
autoimmunity and autoimmune diseases. J Autoimmun. 32:189–194.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bi Y, Liu G and Yang R: MicroRNAs: Novel
regulators during the immune response. J Cell Physiol. 218:467–472.
2009. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Pauley KM and Chan EK: MicroRNAs and their
emerging roles in immunology. Ann N Y Acad Sci. 1143:226–239. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Denby L and Baker AH: Targeting non-coding
RNA for the therapy of renal disease. Curr Opin Pharmacol.
27:70–77. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lai JY, Luo J, O'Connor C, Jing X, Nair V,
Ju W, Randolph A, Ben-Dov IZ, Matar RN, Briskin D, et al:
MicroRNA-21 in glomerular injury. J Am Soc Nephrol. 26:805–816.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Serino G, Sallustio F, Curci C, Cox SN,
Pesce F, De Palma G and Schena FP: Role of let-7b in the regulation
of N-acetylgalactosaminyltransferase 2 in IgA nephropathy. Nephrol
Dial Transplant. 30:1132–1139. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Denby L, Ramdas V, Lu R, Conway BR, Grant
JS, Dickinson B, Aurora AB, McClure JD, Kipgen D, Delles C, et al:
MicroRNA-214 antagonism protects against renal fibrosis. J Am Soc
Nephrol. 25:65–80. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yeo SC, Cheung CK and Barratt J: New
insights into the pathogenesis of IgA nephropathy. Pediatr Nephrol.
33:763–777. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Mao S, Xuan X, Sha Y, Zhao S, Zhu C, Zhang
A and Huang S: Clinico-pathological association of
Henoch-Schoenlein purpura nephritis and IgA nephropathy in
children. Int J Clin Exp Pathol. 8:2334–2342. 2015.PubMed/NCBI
|