Introduction
Neonatal hypoxic-ischemic encephalopathy (HIE) is
mainly triggered by perinatal asphyxia, leading to neural tissue
damage caused by deprivation of oxygen and glucose (1,2). The
pathophysiology of HIE is complex and may be related to
complications in the neonate, mother or placenta (3). Cerebral hypoxic ischemia causes a
strong neuroinflammatory response, which results in delayed cell
death. Neonatal HIE can cause mortality or major disability, as
well as cerebral palsy, neuromotor and cognitive delays, growth
restriction and epilepsy (4–8).
Hypothermic therapy is an effective treatment for
moderate and severe HIE by improving the neurological function of
survivors (8). Effective
hypothermia therapy can enhance neural repair, which is thought to
improve neurological outcomes (9).
However, current examinations of neonatal HIE have limitations and
different levels of effectiveness, as diagnosis largely depends on
clinical judgement and instrumental examination (10). Therefore, effective methods to
define the degree of insult of HIE are required urgently.
Circular RNAs (circRNAs) are a class of non-coding
RNAs that are produced from precursor mRNA back-splicing of exons
(11). The downstream 5′ splice
site and upstream 3′ splice site of circRNA are ligated by a 3′-5′
phosphodiester bond at the junction site (11,12).
Previous studies have shown that circRNAs play essential roles in
neuronal function, cell proliferation and immune responses
(13–15). Furthermore, circRNAs are usually
stable both in extracellular and intracellular environments
(16,17). However, the expression profiles and
functions of circRNAs in neonatal HIE have not been previously
reported.
In the present study, the expression profile of
circRNAs in three neonatal HIE samples and control samples were
detected by microarray analysis. Additionally, Gene Ontology (GO),
and Kyoto Encyclopedia of Genes and Genomes (KEGG) pathway analyses
results suggested that several processes were enriched in the HIE
group compared with the control group, and are related to the
cellular processes, immune system, signal transduction and cellular
community. Thus, the present results may provide a novel insight
into the progress of neonatal HIE.
Materials and methods
Sample collection
Samples were collected from Jingjiang People's
Hospital from Aug, 2017 to May, 2018. Peripheral blood samples (6
ml) from ten neonates with HIE and ten control infants without HIE
were collected. The clinical parameters of neonatal patients and
controls are listed in Table I. A
diagnosis of HIE was confirmed by routine examination; An Apgar
score of 2–6 for 1 min and <8 after first 5 min after birth
indicates HIE (18) (Table I). PAXgene Blood RNA tubes (Qiagen
GmbH) with 4 ml RNA protect reagent were used for collection of
peripheral blood samples. All HIE samples were collected prior to
hypothermia therapy. The present study was approved by the Medical
Ethics Committee of Jingjiang People's Hospital in China [approval
no. (2017)25].
 | Table I.Population demographics. |
Table I.
Population demographics.
| Characteristic | HIE (n=10) | Control (n=10) |
|---|
| Age, day | 1 (1–2) | 5 (2–10) |
| Sex, M/F | 6/4 | 4/6 |
| 1 min Apgar | 3 (2–6) | 10 (9–10) |
| 5 min Apgar | 7 (6–8) | 10 (10–10) |
| Cord pH | 7.34
(7.245–3.442) | – |
Total RNA extraction and
purification
Total RNA was extracted and purified from the whole
blood sample using the PAXgene Blood RNA kit (Qiagen, Inc.)
following the manufacturer's instructions. Total RNA was then
checked for RNA integration using an Agilent Bioanalyzer 2100
(Agilent Technologies, Inc.).
Microarray analysis
Microarray was performed using an Agilent microarray
platform (Shanghai Biotechnology Co., Ltd.), according to the
manufacturer's protocols. Total RNA was amplified and labeled using
a Low Input Quick Amp Labeling kit, One-Color (Agilent
Technologies, Inc.), following the manufacturer's instructions.
Then, the labeled cRNA were purified using a RNeasy mini kit
(Qiagen, Inc.). Each slide was hybridized with 1.65 µg Cy3-labeled
cRNA using a Gene Expression Hybridization kit (Agilent
Technologies, Inc.) in a hybridization oven at 65°C for 17 h
(Agilent Technologies, Inc.), according to the manufacturer's
protocol. After 17 h of hybridization, the slides were washed in
staining dishes (Thermo Shandon; Thermo Fisher Scientific, Inc.)
with Gene Expression Wash Buffer kit (Agilent Technologies, Inc.),
following the manufacturer's instructions. Then, slides were
scanned by an Agilent Microarray scanner (Agilent Technologies,
Inc.) with the following default settings: Dye channel, green; Scan
resolution=3 µm; photomultiplier tube 100%, 20 bit. Data were
extracted using Feature Extraction software 10.7 (Agilent
Technologies, Inc.). Raw data were normalized using a Quantile
algorithm, according to the manufacturer's instructions provided by
the microarray.
RNA-sequence analysis of HIF-1α
The microarray data discussed in the present study,
with regards to the expression of HIF-1α, have been
deposited in the National Center for Biotechnology Information Gene
Expression Omnibus (GEO) database. The GEO accession number is
GSE121178.
GO and KEGG pathway analysis
The GO and KEGG pathway was predicted using Amigo 2
(http://amigo.geneontology.org/; V2.5.12)
and GenomeNet website (http://www.genome.ad.jp/kegg/).
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted as described above. RT-qPCR
was performed using HiScript II Q RT SuperMix for qPCR kit (Vazyme
Biotech Co., Ltd.) and SYBR Green (Roche Diagnostics) method
following the manufacturer's protocol. To investigate the results
of the microarray analysis, the primers of circRNA were designed by
CircPrimer software (http://www.bioinf.com.cn; V1.2), and the sequences of
primers used are listed in Table
II. RT-qPCR was performed using an Applied Biosystems ViiA 7 Dx
system (Thermo Fisher Scientific, Inc.) with SYBR Green (Roche
Diagnostics). The qPCR conditions were as follows: Initial
denaturation at 50°C for 2 min, 95°C for 10 min, followed by 40
cycle at 95°C for 15 sec and 60°C for 30 sec. The expression of
circRNA was normalized to the 18S ribosomal RNA, using the
2−ΔΔCq method (19).
 | Table II.Primer sequences used for reverse
transcription-quantitative PCR of circular RNA expression. |
Table II.
Primer sequences used for reverse
transcription-quantitative PCR of circular RNA expression.
| CircID | Forward primer
(5′→3′) | Reverse primer
(5′→3′) |
|---|
|
has_circ_0005537 |
GGAGAAGACCAGGCAGAAGA |
TGGTCATGATTCATCCCAGCT |
|
hsa_circ_0050345 |
CCTGAGACCAAACTTACAGCC |
ACGTGGCAAGGTAGACAGAT |
|
hsa_circ_0050705 |
GCCACCACTTTGAGACACTG |
TGTAGTCCATCCGAACCCTG |
|
hsa_circ_0069578 |
TGGCTACTTTGGTTTCTGTCTG |
CATCATGGGCTGCCTGTATG |
|
hsa_circ_0070733 |
TGTGATGATGGCTGGACTGA |
CCACTGTGCCTTCAAACTCA |
circRNA and microRNA (miRNA)
interaction prediction
The circRNA and microRNA interaction was predicted
using miRanda (http://www.microrna.org/; V21.0). The network map of
circRNA and microRNA was drawn using Cytoscape (https://cytoscape.org; V3.7.2).
Statistical analysis
Data were analyzed using the SPSS 20.0 software
package (SPSS, Inc.) with an independent-sample t-test for
comparisons between the HIE and control group. All experiments were
repeated three times. Data are presented as the mean ± SD.
P<0.05 was considered to indicate a statistically significant
difference.
Results
circRNA microarray profiling
Peripheral blood samples were collected from ten
neonates with HIE and ten infants without HIE. The 10 HIE samples
included samples from six infants with moderate HIE and four with
severe HIE. Demographic data of the HIE group and non-HIE groups
are shown in Table I. To detect
the differentially expressed circRNAs in HIE, microarray profiling
was performed on three randomly selected HIE samples and controls.
A total of 88,750 circRNAs were detected, with 456 circRNAs found
to be differentially expressed between the HIE samples and control
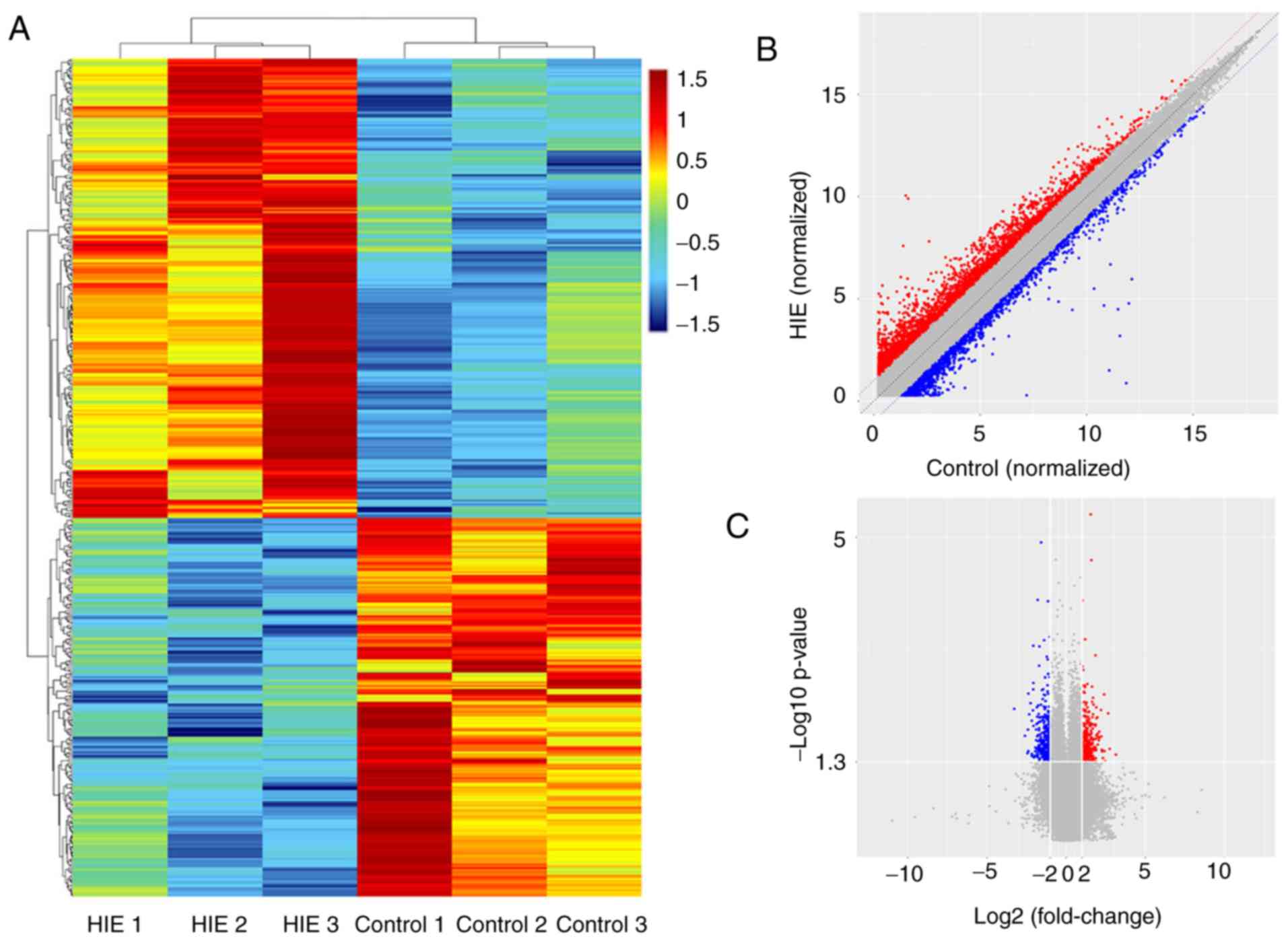
samples. Hierarchical clustering showed the total number of
differentially expressed circRNAs among the samples (Fig. 1A). A scatter plot was used to
visualize the circRNAs with a fold change of >2.0. The red and
green lines are the fold change lines for the differentially
expressed circRNAs (Fig. 1B).
Furthermore, a volcano plot was used to visualize the
differentially expressed circRNAs with fold changes, >2.0 and
with statistical significance (P<0.05; Fig. 1C). The present results suggested
that 456 circRNAs, including 250 upregulated and 206 downregulated
circRNAs, were differentially expressed in the HIE samples compared
with the non-HIE samples. Tables
III and IV demonstrated the
characteristics of the top 20 upregulated circRNAs and the top 20
downregulated circRNAs, respectively (fold change, ≥2;
P<0.05).
 | Table III.Top 20 upregulated circRNAs in the
hypoxic ischemic encephalopathy peripheral blood samples compared
with normal blood samples. |
Table III.
Top 20 upregulated circRNAs in the
hypoxic ischemic encephalopathy peripheral blood samples compared
with normal blood samples.
| circRNA | P-value | FC | circRNA_length
(bp) | Gene symbol |
|---|
|
hsa_circ_0015493 | 0.007985563 | 6.32061115 | 341 | QSOX1 |
|
hsa_circ_0084605 | 0.003891836 | 5.277928402 | 1661 | ASPH |
|
hsa_circ_0080208 | 0.032305489 | 4.829873143 | 6577 | GRB10 |
|
hsa_circ_0017745 | 0.031672451 | 4.771273975 | 313 | DHTKD1 |
|
hsa_circ_0085851 | 0.019112436 | 4.676079992 | 1269 | TSTA3 |
|
hsa_circ_0050345 | 0.007019217 | 4.596592582 | 617 | DPY19L3 |
|
hsa_circ_0005537 | 0.019407829 | 4.111283582 | 326 | CDC25C |
|
hsa_circ_0034212 | 0.009278198 | 3.938135751 | 2362 | HERC2 |
|
hsa_circ_0014223 | 0.009610085 | 3.769487918 | 523 | S100A8 |
|
hsa_circ_0088696 | 0.006352905 | 3.743285501 | 718 | STXBP1 |
|
hsa_circ_0050705 | 0.049517311 | 3.618324961 | 1452 | WDR62 |
|
hsa_circ_0043217 | 0.032711809 | 3.572406618 | 4544 | ACACA |
|
hsa_circ_0080599 | 0.02837951 | 3.530978904 | 522 |
GTF2IRD1 |
|
hsa_circ_0083857 | 0.012124746 | 3.307009337 | 708 | GSR |
|
hsa_circ_0033633 | 0.040119053 | 3.253147202 | 255 |
C14orf80 |
|
hsa_circ_0064377 | 0.014170302 | 3.212198165 | 3786 | CAND2 |
|
hsa_circ_0045827 | 0.043441186 | 3.178805121 | 1976 | MXRA7 |
|
hsa_circ_0085853 | 0.024352592 | 3.178538435 | 65 | TSTA3 |
|
hsa_circ_0010211 | 0.025240307 | 3.165611241 | 1788 |
ARHGEF10L |
|
hsa_circ_0067605 | 0.009250664 | 3.16140903 | 378 | GK5 |
 | Table IV.Top 20 downregulated circRNAs in the
hypoxic ischemic encephalopathy peripheral blood samples compared
with normal blood samples. |
Table IV.
Top 20 downregulated circRNAs in the
hypoxic ischemic encephalopathy peripheral blood samples compared
with normal blood samples.
| circRNA | P-value | FC | circRNA_length
(bp) | Gene symbol |
|---|
|
hsa_circ_0051858 | 0.006718587 | 9.691469839 | 1856 | TRPM4 |
|
hsa_circ_0077755 | 0.018710732 | 6.059898228 | 2896 | GJA1 |
|
hsa_circ_0012164 | 0.036270185 | 5.413530234 | 662 | KIF2C |
|
hsa_circ_0061590 | 0.014796495 | 5.306206578 | 4364 | SETD4 |
|
hsa_circ_0076770 | 0.008277684 | 5.021701979 | 2765 |
LOC730101 |
|
hsa_circ_0075624 | 0.010552438 | 4.49619749 | 271 | SYCP2L |
|
hsa_circ_0069578 | 0.040281981 | 4.492267813 | 617 | UCHL1 |
|
hsa_circ_0035951 | 0.003210402 | 4.217652612 | 729 | DENND4A |
|
hsa_circ_0062272 | 0.002515151 | 4.197587878 | 2975 |
SEPT5-GP1BB |
|
hsa_circ_0070733 | 0.042574192 | 4.072228574 | 790 | PRSS12 |
|
hsa_circ_0080184 | 0.039725995 | 3.946056497 | 11042 | TNS3 |
|
hsa_circ_0056518 | 0.033688712 | 3.852150499 | 126 | NCKAP5 |
|
hsa_circ_0073280 | 0.040374199 | 3.717489627 | 2346 | GPR98 |
|
hsa_circ_0009506 | 0.022064444 | 3.708353404 | 117 | ACOT7 |
|
hsa_circ_0008091 | 0.009072897 | 3.543094344 | 405 | NDUFS1 |
|
hsa_circ_0025111 | 0.019091997 | 3.526641269 | 241 | VWF |
|
hsa_circ_0034711 | 0.042187121 | 3.456252673 | 4006 | RPAP1 |
|
hsa_circ_0071519 | 0.038634183 | 3.451687321 | 2419 | STOX2 |
|
hsa_circ_0077888 | 0.031798526 | 3.429293915 | 1469 | EYA4 |
|
hsa_circ_0062751 | 0.005900349 | 3.389322219 | 2051 | GAS2L1 |
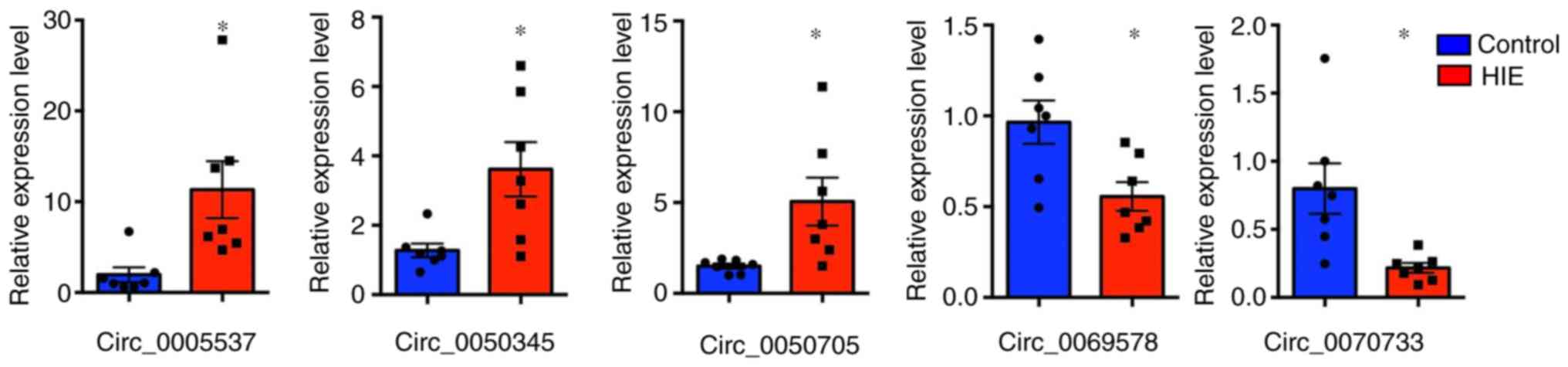
RT-qPCR validation
To further investigate the microarray analysis
results, the present study randomly selected five differentially
expressed circRNAs in the Tables
III and IV, including three
upregulated circRNAs (circRNA_0005537; circRNA_0050345;
circRNA_0050705) and two downregulated circRNAs (circRNA_0069578;
circRNA_0070733). RT-qPCR analysis was performed on additional HIE
samples and non-HIE samples. The present results suggested that the
mRNA expression levels of the selected circRNAs were significantly
different in the HIE samples compared with the control samples
(Fig. 2). Compared with the
control samples, the expression levels of circRNA_0005537,
circRNA_0050705 and circRNA_0050345 in the HIE samples was 5.21-,
2.32- and 2.59-fold higher, respectively. By contrast, the
expression levels of circRNA_0070733 and circRNA_0069578 in the HIE
samples was 3.58- and 1.84-fold lower, respectively, compared with
the control samples.
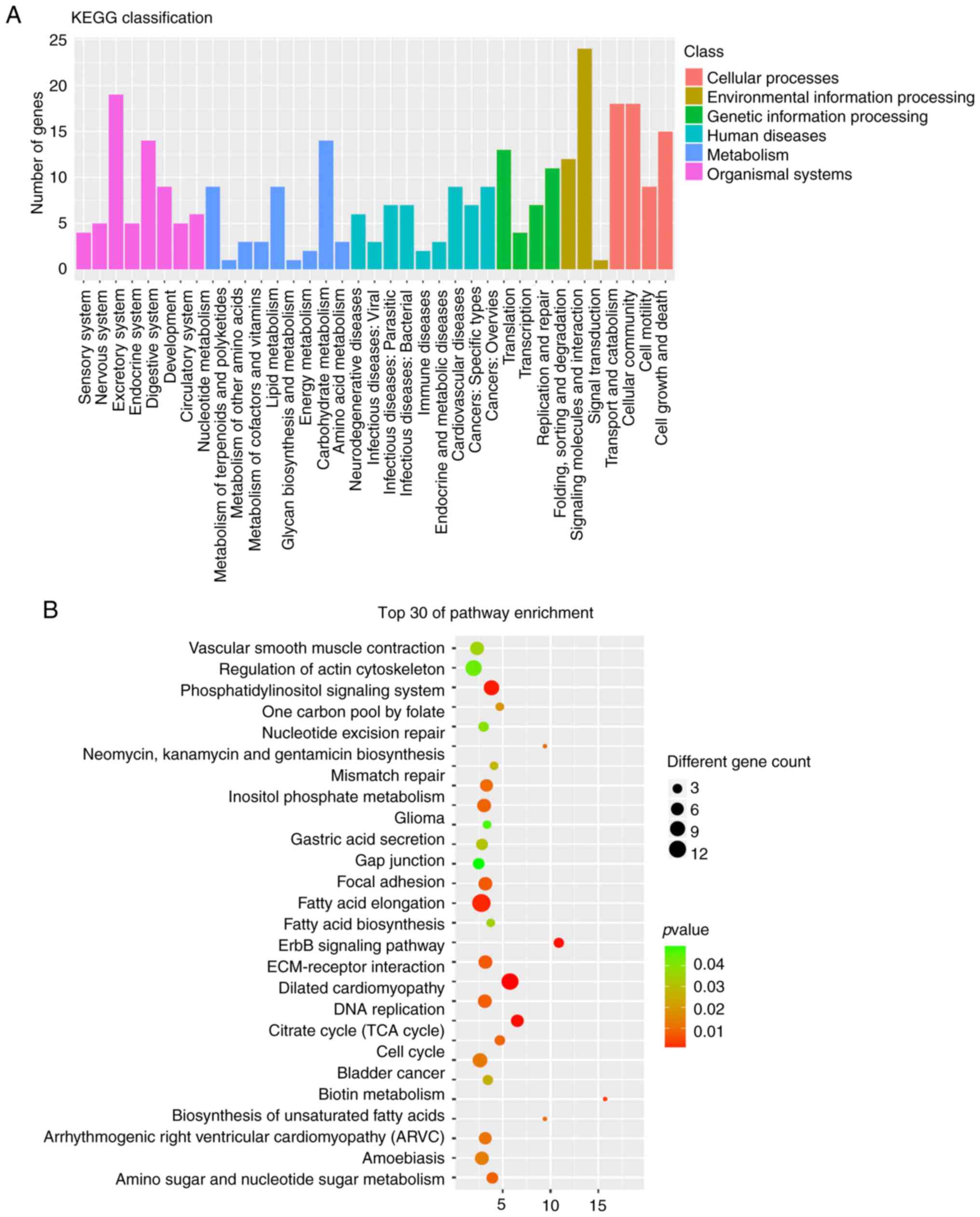
GO and KEGG pathway analysis of host
genes of the circRNAs in HIE
Previous studies have shown that circRNAs function
by regulating the expression levels of their parent genes at the
transcriptional level (20,21).
To further identify the function of host genes of the
differentially expressed circRNAs in HIE, GO and KEGG pathway
analysis of the host genes was performed (Figs. 3 and 4). The GO results demonstrated that the
host genes of the differentially expressed circRNAs are primarily
involved in cellular processes (22), cell, cell part and binding
(Fig. 3A), specifically in
regulation of phospholipase C activity, hindbrain morphogenesis and
fatty-acyl-CoA biosynthetic process (Fig. 3B). KEGG analysis results indicated
that the immune system, signal transduction and cellular community
(Fig. 4A), and in particular in
the regulation of actin cytoskeleton, focal adhesion and
ECM-receptor interaction (Fig.
4B), were related to the host genes of the differentially
expressed circRNAs.
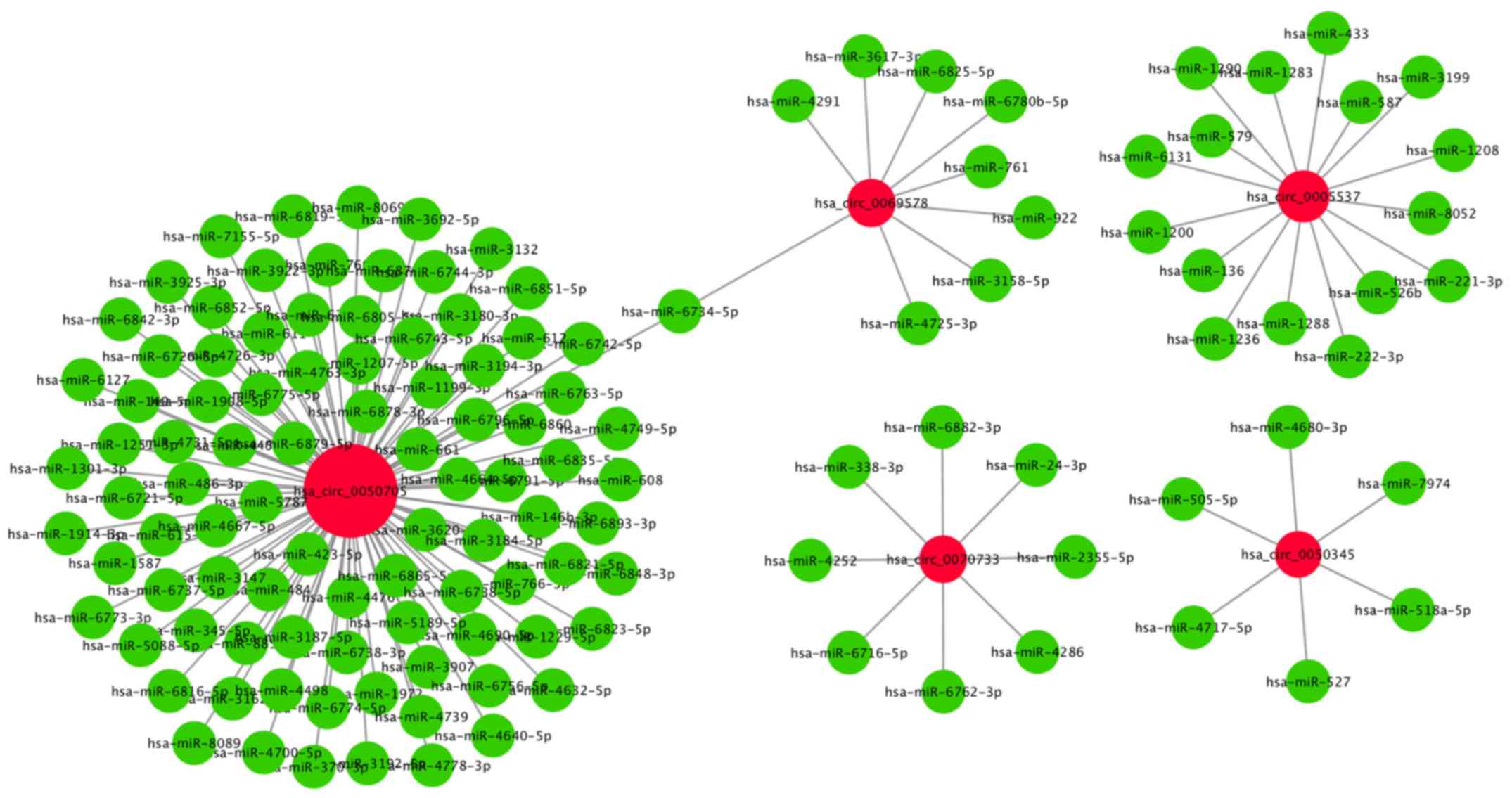
Interactions between circRNAs and
miRNAs
Previous studies have shown that circRNAs can bind
to miRNAs via miRNA response elements (MREs) and function as miRNA
sponges (13,23). To investigate the potential miRNAs
that may bind to circRNAs, miRanda was used to screen the MREs in
the five validated circRNAs. The present results indicated that
several miRNAs were associated with specific circRNAs (Table V). A total of 92 miRNAs were
identified to potentially bind to circRNA_0050705. It was also
found that 16 miRNAs could potentially bind to circRNA_0005537,
nine miRNAs could potentially bind to circRNA_0069578, eight miRNAs
could potentially bind to circRNA_0070733 and six miRNAs could
potentially bind to circRNA_0050345 (Fig. 5).
 | Table V.Interaction of circRNA and
miRNAs. |
Table V.
Interaction of circRNA and
miRNAs.
| circRNA | miRNA | miRNA number |
|---|
| circ_0005537 | miR-1200, miR-1208,
miR-1236, miR-1283, miR-1288, miR-1290, miR-136, miR-433, miR-526b,
miR-579, miR-587, miR-221-3p, miR-222-3p, miR-6131,miR-3199,
miR-8052 | 16 |
| circ_0050345 | miR-4717-5p,
miR-518a-5p, miR-527, miR-505-5p, miR-4680-3p, miR-7974 | 6 |
| circ_0050705 | miR-8089,
miR-4700-5p, miR-6738-3p, miR-637, miR-608, miR-6743-5p,
miR-3187-5p, miR-612, miR-6721-5p, miR-486-3p, miR-4731-5p,
miR-370-3p, miR-1207-5p, miR-6819-5p, miR-4739, miR-4690-5p,
miR-6878-3p, miR-6734-5p, miR-3692-5p, miR-6774-5p, miR-4632-5p,
miR-1199-3p, miR-1908-5p, miR-3162-5p, miR-6744-3p, miR-5088-5p,
miR-149-5p, miR-6835-5p, miR-6738-5p, miR-6876-5p,miR-8069,
miR-615-5p, miR-6773-3p, miR-6796-5p, miR-4640-5p,
miR-3132,miR-6737-5p, miR-3907, miR-6742-5p, miR-6763-5p, miR-6860,
miR-4726-3p,miR-6823-5p, miR-4778-3p, miR-6842-3p, miR-3192-5p,
miR-661, miR-4667-5p,miR-4459, miR-6726-5p, miR-4749-5p, miR-6127,
miR-3194-3p, miR-146b-3p,miR-6805-5p, miR-5189-5p, miR-4476,
miR-760, miR-4498, miR-611, miR-3180-3p, miR-345-5p, miR-6821-5p,
miR-7155-5p, miR-1229-5p, miR-1301-3p, miR-6791-5p,miR-5787,
miR-6893-3p, miR-6848-3p, miR-4664-5p, miR-1587,
miR-6816-5p,miR-4763-3p, miR-6865-5p, miR-1972, miR-6851-5p,
miR-423-5p, miR-3922-3p, miR-6879-5p, miR-6775-5p, miR-1251-3p,
miR-1914-3p, miR-484, miR-6756-5p,miR-3184-5p, miR-3620-5p,
miR-3147, miR-885-3p, miR-3925-3p, miR-766-3p, miR-6852-5p | 92 |
| circ_0069578 | miR-3158-5p,
miR-3617-3p, miR-922, miR-4291, miR-761, miR-4725-3p, miR-6734-5p,
miR-6780b-5p, miR-6825-5p | 9 |
| circ_0070733 | miR-2355-5p,
miR-6882-3p, miR-338-3p, miR-4286, miR-6762-3p, miR-24-3p,
miR-6716-5p, miR-4252 | 8 |
Discussion
Neonatal HIE is one of the common causes of death or
permanent disability, particularly in developing countries
(24,25). The crucial role of miRNAs (miRs)
and long non-coding RNAs in HIE pathologic processes have been
previously demonstrated (19,26).
The expression level of miR-374a is significantly downregulated in
the umbilical cord blood of neonatal HIE (26). Furthermore, miR-204 is reported to
participate in the pathogenesis of HIE via the regulation of
KLLN (27).
circRNAs are a novel type of non-coding RNA that
stably exist in peripheral blood (28) and play a crucial role in the
regulation of numerous pathological or biological processes
(29,30). The expression profiles of circular
RNAs in neonatal rats following hypoxic-ischemic brain damage
(HIBD) has been previously reported (31). Jiang et al (31) identified that 66 circRNAs are
differentially expressed in the early stages of HIBD. However, to
the best of our knowledge, circRNA profiles determined by
microarray analysis in the neonatal HIE have not been previously
reported. Therefore, the present study investigated differentially
expressed circRNAs in the peripheral blood of neonates with HIE and
healthy controls using microarray analysis. The microarray
expression profiles suggested that 250 upregulated circRNAs and 206
downregulated circRNAs were significantly differentially expressed
in patients with HIE. Therefore, the distinct expression profile of
circRNAs may participate in the pathogenesis of HIE and provide new
biomarkers for HIE diagnosis. To further investigate the microarray
data and identify potential clinically applicable biomarkers, the
present study assessed five significant differentially expressed
circRNAs in neonatal HIE. It was found that circRNA_0005537,
circRNA_0050345 and circRNA_0050705 were significantly upregulated
in patients with HIE compared with healthy controls, while
circRNA_0069578 and circRNA_0070733 were downregulated.
Hypoxia-inducible factor 1α (HIF-1α) is a sensitive
molecule regulated by oxygen tension and mediates the adaptive
response of the cell to a hypoxic environment (32). Zhu et al (33) reported that HIF-1α can regulate
BCL2 Interacting Protein 3 (BNIP3) by binding to the
BNIP3 promoter in hypoxia-induced neurons, and participates
in the process of HIE. The present results suggested that the
expression level of HIF-1α was upregulated in HIE group by
RNA-seq (fold change, >2; data submitted to GEO; accession no.
GSE121178). Previous studies have demonstrated that circRNAs may
bind to miRNAs and regulate mRNAs expression (34). The present results indicated that
miR-433 may potentially bind to circ_0005537, and that miR-338-3p
could potentially bind to circ_0070733. However, microRNA.org was used to predict the miRNAs that could
potentially bind to HIF-1α, and found that HIF-1α has
miR-433 and miR-338-3p specific binding sites (data not shown).
Therefore, the present results suggested that dysregulated
circ_0005537 and circ_0070733 in patients with HIE may regulate
HIF-1α expression, and participate in the pathological
process of HIE.
circRNA lack crucial elements for cap-dependent
translation, however, few endogenous circRNAs have been shown to
act as protein templated (35,36).
According to the open reading frame prediction and functional
internal ribosome entry site element prediction of the
circRNA_0005537, circRNA_0050345, circRNA_0050705, circRNA_0069578
and circRNA_0070733; it was suggested that these five circRNAs have
the possibility to translate into short proteins or peptides (data
not shown). However, the functional relevance of these five circRNA
requires further study.
Brain ischemia leads to a lack of ATP and initiates
a cascade of biochemical events, including cell death and the
induction of secondary brain injury in neonates (37). In addition, glucose and oxygen play
important roles in the pathogenesis of brain injury (1). Moreover, acetyl-CoA is essential in
generating ATP from ADP in the mitochondrion (1,38).
In the present study, GO and KEGG pathway analyses results
demonstrated that the host genes of dysregulated circRNAs
participate in hindbrain morphogenesis and fatty-acyl-CoA
biosynthetic processes. Therefore, the present results suggested
that the dysregulated circRNAs may be involved in the pathogenesis
of neonatal HIE.
In conclusion, microarray analysis was used to
detect differentially expressed circRNAs between neonates with HIE
and controls. To the best of our knowledge, the present study is
the first to examine the potential relationship between circRNAs
and HIE in neonates. In addition, the potential functions of the
host genes of the dysregulated circRNAs were predicted by GO and
KEGG pathway analyses. The GO and KEGG pathway analyses results
indicated that the abnormally expressed circRNAs may be involved in
the pathogenesis of HIE. However, whether the severity of HIE and
the effect of hypothermia therapy are related to differentially
expressed circRNAs in the progression of HIE requires further
investigation.
Acknowledgements
Not applicable.
Funding
The current study was supported by grants from the
National Natural Science Foundation of China (grant no. 81801503)
and the Natural Science Foundation of Jiangsu Province (grant no.
BK20180286).
Availability of data and materials
The microarray data that included the expression
profile of circular RNAs and the expression profile of long
non-coding RNAs (19) have been
deposited in the National Center for Biotechnology Information GEO
database (accession no. GSE121178). The datasets used and/or
analyzed during the current study are available from the
corresponding author on reasonable request.
Authors' contributions
YZ designed the project and edited the manuscript.
LY analyzed the data and wrote the manuscript. XD designed the
experiment and performed the bioinformatics analysis. SZ performed
the bioinformatics analysis. YH and XY performed sample collection
and statistical analysis. YF performed the data analysis. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
This work was approved by the Medical Ethics
Committee of Jingjiang People's Hospital in China [approval no.
(2017)25]. Informed consent was obtained from the guardian of
subjects for participation in this study.
Patient consent for publication
Informed consent was obtained from the guardian of
subjects for participation in this study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Verklan MT: The chilling details:
Hypoxic-ischemic encephalopathy. J Perinat Neonatal Nurs. 23:59–68;
quiz 69–70. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Qin X, Cheng J, Zhong Y, Mahgoub OK, Akter
F, Fan Y, Aldughaim M, Xie Q, Qin L, Gu L, et al: Mechanism and
treatment related to oxidative stress in neonatal hypoxic-ischemic
encephalopathy. Front Mol Neurosci. 12:882019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Rainaldi MA and Perlman JM:
Pathophysiology of birth asphyxia. Clin Perinatol. 43:409–422.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Shah PS, Ohlsson A and Perlman M:
Hypothermia to treat neonatal hypoxic ischemic encephalopathy:
Systematic review. Arch Pediatr Adolesc Med. 161:951–958. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Lai MC and Yang SN: Perinatal
hypoxic-ischemic encephalopathy. J Biomed Biotechnol.
2011:6098132011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gluckman PD, Wyatt JS, Azzopardi D,
Ballard R, Edwards AD, Ferriero DM, Polin RA, Robertson CM,
Thoresen M, Whitelaw A and Gunn AJ: Selective head cooling with
mild systemic hypothermia after neonatal encephalopathy:
Multicentre randomised trial. Lancet. 365:663–670. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Shankaran S, Laptook AR, Ehrenkranz RA,
Tyson JE, McDonald SA, Donovan EF, Fanaroff AA, Poole WK, Wright
LL, Higgins RD, et al: Whole-body hypothermia for neonates with
hypoxic-ischemic encephalopathy. N Engl J Med. 353:1574–1584. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Azzopardi DV, Strohm B, Edwards AD, Dyet
L, Halliday HL, Juszczak E, Kapellou O, Levene M, Marlow N, Porter
E, et al: Moderate hypothermia to treat perinatal asphyxial
encephalopathy. N Engl J Med. 361:1349–1358. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Azzopardi D, Strohm B, Marlow N,
Brocklehurst P, Deierl A, Eddama O, Goodwin J, Halliday HL,
Juszczak E, Kapellou O, et al: Effects of hypothermia for perinatal
asphyxia on childhood outcomes. N Engl J Med. 371:140–149. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Martinello K, Hart AR, Yap S, Mitra S and
Robertson NJ: Management and investigation of neonatal
encephalopathy: 2017 update. Arch Dis Child Fetal Neonatal Ed.
102:F346–F358. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen LL: The biogenesis and emerging roles
of circular RNAs. Nat Rev Mol Cell Biol. 17:205–211. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wilusz JE: A 360° view of circular RNAs:
From biogenesis to functions. Wiley Interdiscip Rev RNA.
9:e14782018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Li X, Yang L and Chen LL: The biogenesis,
functions, and challenges of circular RNAs. Mol Cell. 71:428–442.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang L, Fu J and Zhou Y: Circular RNAs and
their emerging roles in immune regulation. Front Immunol.
9:29772018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sekar S and Liang WS: Circular RNA
expression and function in the brain. Noncoding RNA Res. 4:23–29.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li Y, Zheng Q, Bao C, Li S, Guo W, Zhao J,
Chen D, Gu J, He X and Huang S: Circular RNA is enriched and stable
in exosomes: A promising biomarker for cancer diagnosis. Cell Res.
25:981–984. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Memczak S, Papavasileiou P, Peters O and
Rajewsky N: Identification and characterization of circular RNAs as
a new class of putative biomarkers in human blood. PLoS One.
10:e01412142015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Thornberg E, Thiringer K, Odeback A and
Milsom I: Birth asphyxia: Incidence, clinical course and outcome in
a Swedish population. Acta Paediatr. 84:927–932. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Dong X, Zhao Y, Huang Y, Yu L, Yang X and
Gao F: Analysis of long noncoding RNA expression profiles in the
whole blood of neonates with hypoxic-ischemic encephalopathy. J
Cell Biochem. Nov 26–2018.(Epub ahead of print).
|
|
20
|
Li Z, Huang C, Bao C, Chen L, Lin M, Wang
X, Zhong G, Yu B, Hu W, Dai L, et al: Exon-intron circular RNAs
regulate transcription in the nucleus. Nat Struct Mol Biol.
22:256–264. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang Y, Zhang XO, Chen T, Xiang JF, Yin
QF, Xing YH, Zhu S, Yang L and Chen LL: Circular intronic long
noncoding RNAs. Mol Cell. 51:792–806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The rosetta stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Johnston MV, Fatemi A, Wilson MA and
Northington F: Treatment advances in neonatal neuroprotection and
neurointensive care. Lancet Neurol. 10:372–382. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Azra Haider B and Bhutta ZA: Birth
asphyxia in developing countries: Current status and public health
implications. Curr Probl Pediatr Adolesc Health Care. 36:178–188.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Looney AM, Walsh BH, Moloney G, Grenham S,
Fagan A, O'Keeffe GW, Clarke G, Cryan JF, Dinan TG, Boylan GB and
Murray DM: Downregulation of umbilical cord blood levels of
miR-374a in neonatal hypoxic ischemic encephalopathy. J Pediatr.
167:269–273.e2. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Chen R, Wang M, Fu S, Cao F, Duan P and Lu
J: MicroRNA-204 may participate in the pathogenesis of
hypoxic-ischemic encephalopathy through targeting KLLN. Exp Ther
Med. 18:3299–3306. 2019.PubMed/NCBI
|
|
28
|
Jakobi T and Dieterich C: Deep
computational circular RNA analytics from RNA-seq data. Methods Mol
Biol. 1724:9–25. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Jiang L, Li H, Fan Z, Zhao R and Xia Z:
Circular RNA expression profiles in neonatal rats following
hypoxic-ischemic brain damage. Int J Mol Med. 43:1699–1708.
2019.PubMed/NCBI
|
|
32
|
Wang GL, Jiang BH, Rue EA and Semenza GL:
Hypoxia-inducible factor 1 is a basic-helix-loop-helix-PAS
heterodimer regulated by cellular O2 tension. Proc Natl Acad Sci
USA. 92:5510–5514. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhu L, Qi B and Hou D: Roles of HIF1α- and
HIF2α-regulated BNIP3 in hypoxia-induced injury of neurons. Pathol
Res Pract. 215:822–827. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Li Y, Huo C, Lin X and Xu J: Computational
identification of cross-talking ceRNAs. Adv Exp Med Biol.
1094:97–108. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Legnini I, Di Timoteo G, Rossi F, Morlando
M, Briganti F, Sthandier O, Fatica A, Santini T, Andronache A, Wade
M, et al: Circ-ZNF609 is a circular RNA that can be translated and
functions in myogenesis. Mol Cell. 66:22–37.e9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pamudurti NR, Bartok O, Jens M,
Ashwal-Fluss R, Stottmeister C, Ruhe L, Hanan M, Wyler E,
Perez-Hernandez D, Ramberger E, et al: Translation of circRNAs. Mol
Cell. 66:9–21.e7. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Thornton C, Leaw B, Mallard C, Nair S,
Jinnai M and Hagberg H: Cell death in the developing brain after
hypoxia-ischemia. Front Cell Neurosci. 11:2482017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vannucci RC, Brucklacher RM and Vannucci
SJ: Glycolysis and perinatal hypoxic-ischemic brain damage. Dev
Neurosci. 27:185–190. 2005. View Article : Google Scholar : PubMed/NCBI
|