Introduction
Breast cancer (BC) is the most frequently diagnosed
malignant tumor and the second leading cause of cancer-related
mortality in women (1). Despite
advances in early detection and treatment, ~30% of BC patients
develop tumor metastasis during treatment (2,3). Due
to the diversity of breast tumors, the discovery of effective
treatments is challenging. Although currently available
chemotherapies have led to an increase in overall survival,
numerous breast tumors may acquire resistance following the initial
response (4,5). It is therefore important to identify
and investigate new molecular targets that inhibit BC progression
and metastasis by affecting gene pathways and restoring drug
susceptibility.
A previous study indicated that non-coding RNA plays
an important regulatory role in BC development and progression
(6). MicroRNAs (miRNAs/miRs)
belong to a class of evolutionarily conserved RNAs that influence
gene expression at the post-transcriptional level (7). Experimental studies have shown that
abnormal expression of miRNA is associated with tumorigenesis and
cancer metastasis (8,9).
miR-1 is abundantly expressed in muscle, where it
inhibits the proliferation of progenitor cells and promotes
myogenesis. It is expressed in the cytoplasm of carcinoma cells and
had been shown to suppress tumor growth in numerous cancer types
(10). For example, miR-1 is the
lowest expressed miRNA in prostate tumors and is considered a
candidate tumor suppressor and prognostic marker for prostate
cancer (11).
Previous reports suggest that miR-1 may suppress
tumor proliferation in BC (12,13).
The aim of the present study was to determine a novel molecular
network involved in BC development by investigating the functional
role of miR-1 using in vitro and in vivo
experiments.
Materials and methods
Patient samples
Breast cancer tissues and adjacent normal breast
tissues were obtained from the Breast Surgery Department of the
Hospital of Shanghai, Jiaotong University. A total number of 47
women aged from 30–75 years old with histologically confirmed
invasive ER-positive subtype BC were included. Patients were
recruited from 2015 September to 2017 August. Immunohistochemical
staining of ER/progesterone receptor (PR) and fluorescent in
situ hybridization of human epidermal growth factor receptor
(HER) 2 after surgery was performed as a regular procedure by
pathologists. There was no radiotherapy or chemotherapy prior to
surgery. All BC patients provided written consent for the use of
their specimens in the present study, which was approved by the
independent ethics committee of Renji Hospital, School of Medicine,
Shanghai Jiaotong University (China). Breast tissue samples were
collected immediately following resection and then stored at −80°C
before RNA extraction.
RNA isolation and reverse
transcription-quantitative (RT-q) PCR
Total RNA from 1×106 cultured cells and
tissue samples was isolated using TRIzol reagent (Invitrogen;
Thermo Fisher Scientific, Inc.). The quality and concentration of
RNA was detected with NanoDrop 2000 (Thermo Fisher Scientific,
Inc.). A miRcute miRNA first-strand cDNA synthesis kit and a
miRcute miRNA qPCR kit were used to detect the miRNA expression
level according to the manufacturer's protocols (Tiangen Biotech
Co., Ltd.). The primers used for U6 snRNA (CD201-0145) and miR-1
(CD201-0003) were also obtained from Tiangen Biotech Co., Ltd. The
mRNA expression level was detected by RT-PCR using a Takara Reverse
Transcriptase kit and a SYBR Green PCR kit according to the
manufacturer's protocols (Takara Biotechnology Co., Ltd.). The
following thermocycling conditions were used for the qPCR: Initial
denaturation at 95°C for 30 sec; followed by 40 cycles of
denaturation at 95°C for 5 sec, annealing at 60°C for 10 sec and
extension at 72°C for 30 sec. Each sample was tested in triplicate.
Expression levels were quantified using the 2−ΔΔCq
method (14) and normalized to the
internal reference gene, U6 for miR-1 expression or GAPDH for other
genes. The primer sequences were as follows: GAPDH forward,
5′-GAAGGTGAAGGTCGGAGTC-3′ and reverse 5′-GAAGATGGTGATGGGATTTC-3′;
and Bcl-2 forward, 5′-AGTCTGGGAATCGATCTGGA-3′ and reverse
5′-GCAACGATCCCATCAATCTT-3′. The primers used for miR-1 and U6 were
described in a previous report (15).
Cell culture
Human breast cancer cell lines (MCF-7 and ZR-7530)
were cultured in DMEM (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% fetal bovine serum (Gibco; Thermo Fisher
Scientific, Inc.) and 1% (v/v) penicillin/streptomycin. All cells
were cultured in a humidified atmosphere of 5% CO2 and
95% air at 37°C. The cells were purchased from The Type Culture
Collection of the Chinese Academy of Sciences.
Overexpression of miR-1 in cells
The pLV-hsa-miR-1 plasmid and the negative control
pLV-miRNA-vector were purchased from Shanghai GenePharma Co., Ltd.
The viruses were packaged in 293T (Type Culture Collection of the
Chinese Academy of Sciences) cells according to standard protocols
and the virus particles were harvested 72 h later. The packaged
lentiviruses were termed LV-miR-1, while the empty lentiviral
vector LV-ctrl was used as a control. Cells were infected with
viral particles (MOI = 10 for each) for 24 h and 5 µg/ml Polybrene,
which was followed by selection with puromycin (2 µg/ml) for ≤7
days. Polybrene and puromycin were purchased from Shanghai
GenePharma Co., Ltd.
Cell proliferation assay
The Cell Counting Kit-8 method was used according to
the manufacturer's protocol to determine cell proliferation
(Dojindo Molecular Technologies, Inc.). MCF-7 and ZR-7530 cells
transfected with either LV-miR-1 or LV-ctrl were seeded into
96-well plates at a density of 5×103 cells/well, and
then 10 µl CCK-8 solution was added to each well so that the final
volume of culture medium was 200 µl. To perform the clonogenic
assay, 5×103 cells were seeded in a 10 cm dish. Visible
colonies were fixed in 4% formaldehyde at room temperature for 30
min and after 10 days, the colonies were stained with 0.1% crystal
violet at room temperature for 30 min. EdU incorporation assay was
performed using EdU incorporation assay kit (Guangzhou RiboBio Co.,
Ltd.) according to the manufacturer's instructions.
Cell migration and invasion
assays
Cell migration and invasion capacity were determined
using a Transwell assay using Transwell permeable supports
(Corning, Inc.). A total of 1×105 breast cancer
cells/well were plated in the upper chambers of Transwell plates
with serum-free DMEM medium, of which membranes were or were not
precoated with Matrigel (BD Biosciences) for the invasion and
migration assay, respectively. Modified Boyden chambers containing
Matrigel were placed in a 24-well culture plate. Matrigel was
precoated for 12 h at 37°C. A total of 800 µl DMEM medium
supplemented with 10% FBS was plated in the lower chambers. After
24 h incubation in wells without Matrigel or 48 h with Matrigel,
migration and invasion were assessed, respectively. Then, cells in
the lower chambers were fixed in 4% paraformaldehyde at room
temperature for 30 min and stained with a 0.5% crystal violet
fixative solution for 30 min at room temperature, following which
the cells were counted using an inverted fluorescent microscope
(magnification, ×200; Leica Microsystems, Inc.).
Apoptosis assay
Cells transfected with the LV-miR-1 or the negative
control (NC) were collected and stained with Annexin-V-FITC and
7-AAD using an FITC Annexin V apoptosis detection kit (PE Annexin V
Apoptosis Detection kit; BD Pharmingen; BD Biosciences) according
to the manufacturer's protocol. Apoptosis was subsequently detected
using a FACScan flow cytometer (BD Biosciences). Flow cytometry
data was analyzed by BD FACSDiva software V6.1.3 (BD Biosciences).
The cells were divided into dead cells, early apoptotic cells,
apoptotic cells and viable cells and the percentages of apoptotic
cells calculated. Cells considered viable were PE Annexin V- and
7-AAD-negative, early apoptotic cells were PE Annexin V-positive
and 7-AAD-negative, and late apoptotic cells and dead cells were
positive for both PE Annexin V and 7-AAD.
Western blotting
Breast cancer cells were harvested 3 days after
transfection and were lysed in RIPA buffer (Beyotime Institute of
Biotechnology). Total protein was quantified using a bicinchoninic
acid assay kit. A total of 30 µg protein/lane was separated by 8 or
12% SDS-PAGE followed by transfer of the proteins to a
nitrocellulose filter membrane. After blocking in 5% BSA (Shanghai
Shenggong Biology Engineering Technology Service, Ltd.) at 25°C for
2 h, the membranes were subjected to incubation with primary
antibodies (1:1,000) overnight at 4°C. Following the primary
antibody incubation, membranes were incubated with horseradish
peroxidase-conjugated goat anti-rabbit secondary antibodies
(1:5,000; cat. no. 7074; Cell Signaling Technology, Inc.). β-actin
served as the loading control. The protein immunoreactive bands
were visualized by the ECL detection instrument (Thermo Fisher
Scientific, Inc.) plus chemiluminescent substrate. Densitometry
analysis was performed using ImageJ software (version 1.8.0;
National Institutes of Health) with β-actin as the internal
control. Detailed information on the antibodies used is given in
Table SI.
Dual luciferase reporter assay
The Bcl-2 3′UTR sequences to which miR-1 potentially
bind were searched by TargetScan (http://www.targetscan.org). Then, the DNA fragments of
the 3′UTR of Bcl-2 were cloned into the dual-luciferase expression
vector, pmirGLO (Promega Corporation), which was termed wt-Bcl2.
The mutant vector was generated by mutating the miR-1 seed region
binding site and this vector was termed mut-Bcl2. The miR-1 mimics,
the miR-1 control, the firefly luciferase reporter plasmid and the
Renilla luciferase vector (pRL-SV40, Promega Corporation)
were co-transfected into BC cells using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.) and the
luciferase activity was evaluated using a dual-luciferase reporter
assay kit (Promega Corporation).
Analysis of the epigenetic silencing
of miR-1
5-Azacytidine (5-AzaC) and trichostatin A (TSA) were
purchased from Sigma-Aldrich (Merck KGaA). Briefly, ZR7530 and
MCF-7 cells were plated at a density of 1×106 cells per
10 cm and were then treated with 5-AzaC (5 µM) and/or TSA (0.3 µM).
Cells were cultured in this medium at 37°C for 2 days before they
were harvested for RNA extraction.
Effect of miR-1 on paclitaxel and
cis-platinum sensitivities
The MCF-7 cells transfected with either LV-miR-1 or
LV-ctrl were seeded into 96-well plates(1×104
cells/well). After 24 h, the appropriate cytotoxic drug was added:
10 nM paclitaxel or 0.2 µM cisplatin (DDP). Drug sensitivities were
measured using a CCK-8 assay, as described above. DDP-resistant
MDA-MB-231 cells were obtained from Shanghai Longhua Hospital. The
IC50 of MDA-MB-231 and MDA-MB-231/DDP to DDP was 17.72
and 173.70 µg/ml, respectively, and the fold-resistance of
MDA-MB-231/DDP cells to DDP was 9.80 (16).
Animal studies
A total of 16 female BALB/C athymic nude mice (age,
6 weeks; weight, 23 g; SLAC, Shanghai, China) were housed and
manipulated according to the protocols approved by the Renji
Hospital Medical Experimental Animal Care Commission. The nude mice
were housed in pathogen-free environment with 12-h light/dark
cycle, controlled humidity (50%) and temperature (28°C) and had
free access to food and water. For the in vivo studies,
5×106 LV-miR-1 or LV-miR-ctrl cells were injected
subcutaneously in the backs of nude mice. The length and width of
the tumors was measured every week and tumor volumes were
calculated using the following formula: Volume
(mm3)=length (L; mm) × width (W; mm)2/2.
Then, 4 weeks following injection, the mice were euthanized by
cervical dislocation and the tumors removed and weighed. The mice
were maintained under specific pathogen-free conditions and fed
sterile chow. Mice were housed and manipulated in accordance with
the principles and guidelines approved by the Renji Hospital
Medical Experimental Animal Care Commission.
Analysis of The Cancer Genome Atlas
(TCGA) dataset
A normalized mRNA and miRNA expression dataset for
BC was downloaded from the TCGA (http://www.tcga.org) (17) and Spearman's correlation
coefficient was calculated for the BC samples. The expression of
miR-1 in tumor and normal tissues was analyzed using data from TCGA
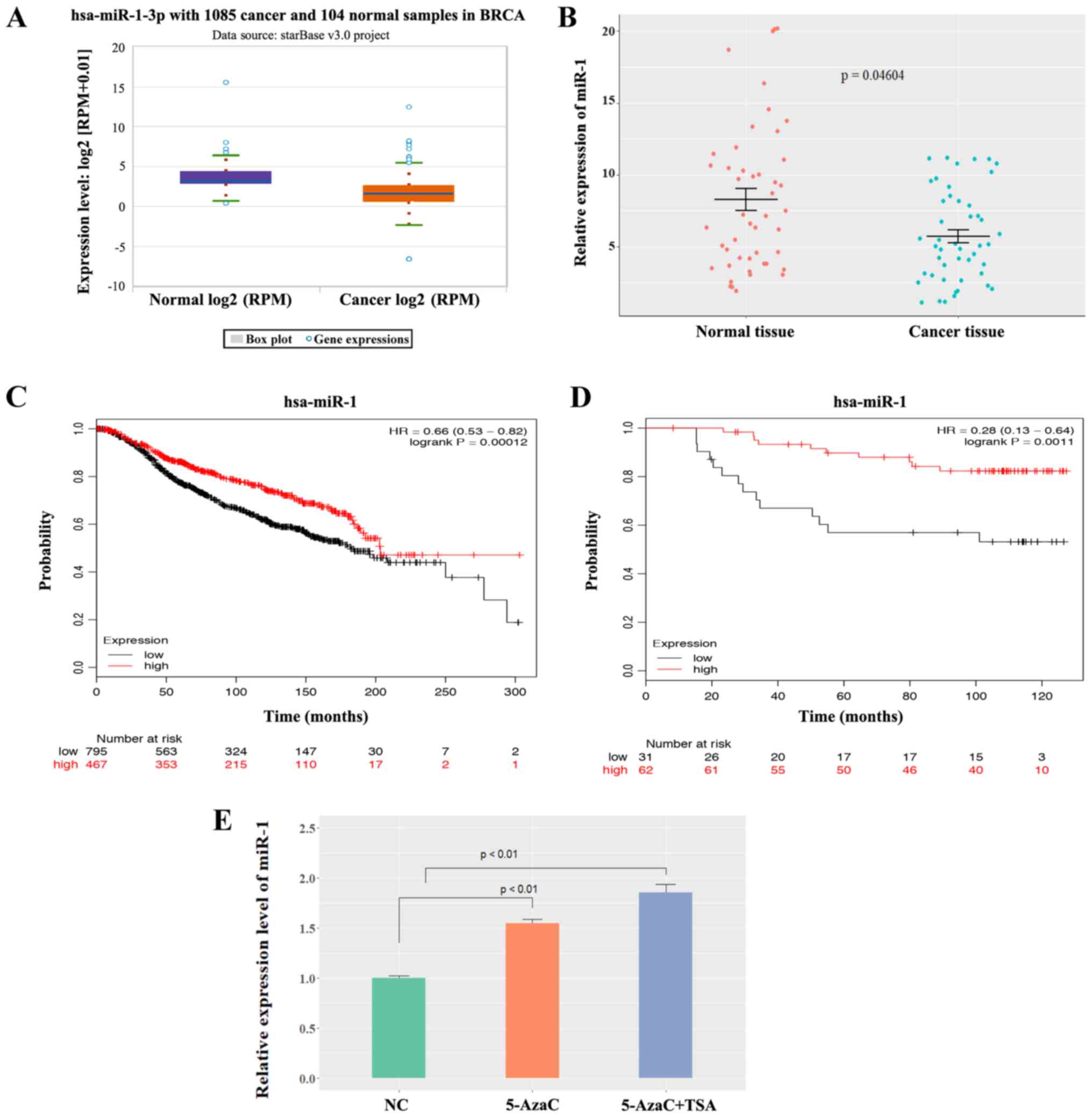
project. The box plot in Fig. 1A
presenting the median and range was download from the ENCORI
Pan-Cancer Analysis Platform (http://starbase.sysu.edu.cn/panCancer.php).
Statistical analysis
Data are presented as the mean ± SEM from three
independent experiments. Statistical calculations were performed
with R program (version 3.4; Rstudio, Inc.). The differences
between the means were tested by Student's t-test or one-way ANOVA
followed by Tukey post hoc test. Spearman's correlation analyses
were used to identify the correlation between miR-1 and Bcl-2.
P<0.05 was considered to indicate a statistically significant
difference. StarBase3 database was used to analyze the expression
of miR-1 in breast cancer tissues and healthy tissues (http://starbase.sysu.edu.cn/). The Kaplan-Meier
analysis of patient survival data in METABRIC and the Dataset
GSE19783 were performed on Kaplan Meier-plotter (https://kmplot.com/analysis/).
Results
miR-1 expression is downregulated in
BC and the low expression of miR-1 is correlated with poor
survival
To determine whether miR-1 was involved in the
tumorigenesis of BC, the expression of miR-1 was analyzed using
StarBase3 database and the data showed that miR-1 was expressed at
significantly low levels in BC samples compared with normal samples
(P<0.0001; Fig. 1A). Patient
survival was analyzed according to the different patient groups and
it was found that low miR-1 expression was correlated with poor
overall survival only in luminal-type (ER-positive) patients, but
not in human epidermal growth factor receptor 2-positive or
triple-negative patients (Fig.
S1). miR-1 expression was compared in 47 pairs of ER-positive
BC tissues and normal breast tissues using the patient samples
described above. The data demonstrated that miR-1 was expressed at
low levels in BC tissues compared with normal breast tissues
(P<0.05; Fig. 1B), which
indicated that miR-1 was a potential tumor suppressor in BC. A
Kaplan-Meier analysis of patient survival data in METABRIC
(Fig. 1C) and the Dataset GSE19783
(Fig. 1D) revealed that low miR-1
expression was correlated with poor overall survival. In
hepatocellular carcinoma and prostate cancer, it has been reported
that low expression of miR-1 is mediated by methylation (18,19).
To investigate whether miR-1 is epigenetically silenced in BC,
MCF-7 cells were treated with either 5-AzaC or TSA. 5-AzaC is a
well-known DNA hypomethylating agent and TSA is a histone
deacetylase inhibitor. It was observed that miR-1 expression was
activated following treatment with 5-AzaC and TSA (P<0.01;
Fig. 1E), which indicates that
miR-1 may be epigenetically silenced in BC cells and may be why
miR-1 is downregulated in BC. This experiment demonstrated that
miR-1 is expressed at low levels in breast cancer tissues and that
a low level of miR-1 is correlated with poor survival.
Upregulation of miR-1 inhibits BC cell
proliferation
To investigate the role of miR-1 in cancer cell
growth, BC cells (ZR-7530, MCF-7) were transfected with the
LV-miR-1 and the expression of miR-1 was significantly increased
(Fig. 2A). A CCK-8 assay
demonstrated that overexpression of miR-1 significantly inhibited
the growth capacity of BC cells (Fig.
2B). Similarly, a clonogenic survival assay of BC cells
demonstrated that upregulation of miR-1 significantly inhibited BC
cell colony formation efficiency (Fig.
2C). A EdU proliferation assay also revealed that the capacity
of BC cell proliferation was decreased in the LV-miR-1 group
(Fig. 2D). Taken together, the
data showed that enhanced miR-1 expression inhibits the
proliferation of BC cells.
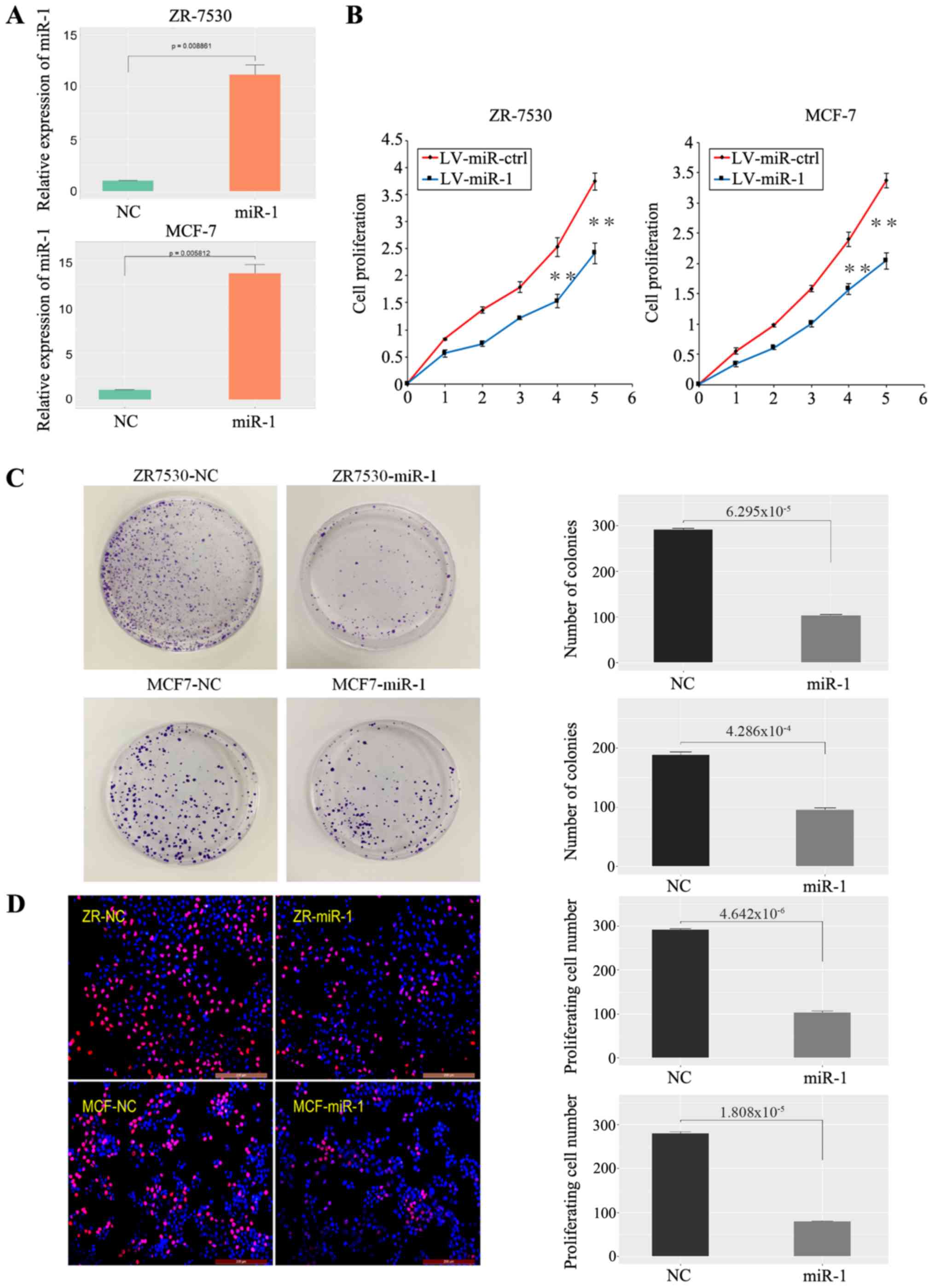 | Figure 2.miR-1 inhibits BC cell growth. (A)
Validation of miR-1 overexpression in BC cells (n=3, P<0.01).
(B) A high level of miR-1 inhibited BC cell proliferation, as shown
by CCK-8 assay (1-5: 12, 24, 48, 72 and 96 h), n=6, **P<0.01.
(C) A high level of miR-1 inhibited BC cell clone formation, n=3,
P<0.01. (D) A high level of miR-1 inhibited BC cell
proliferation, as demonstrated by EdU essay, n=3, P<0.01. miR,
microRNA; BC, breast cancer; NC, normal control; ctrl, control; LV,
lentiviral. |
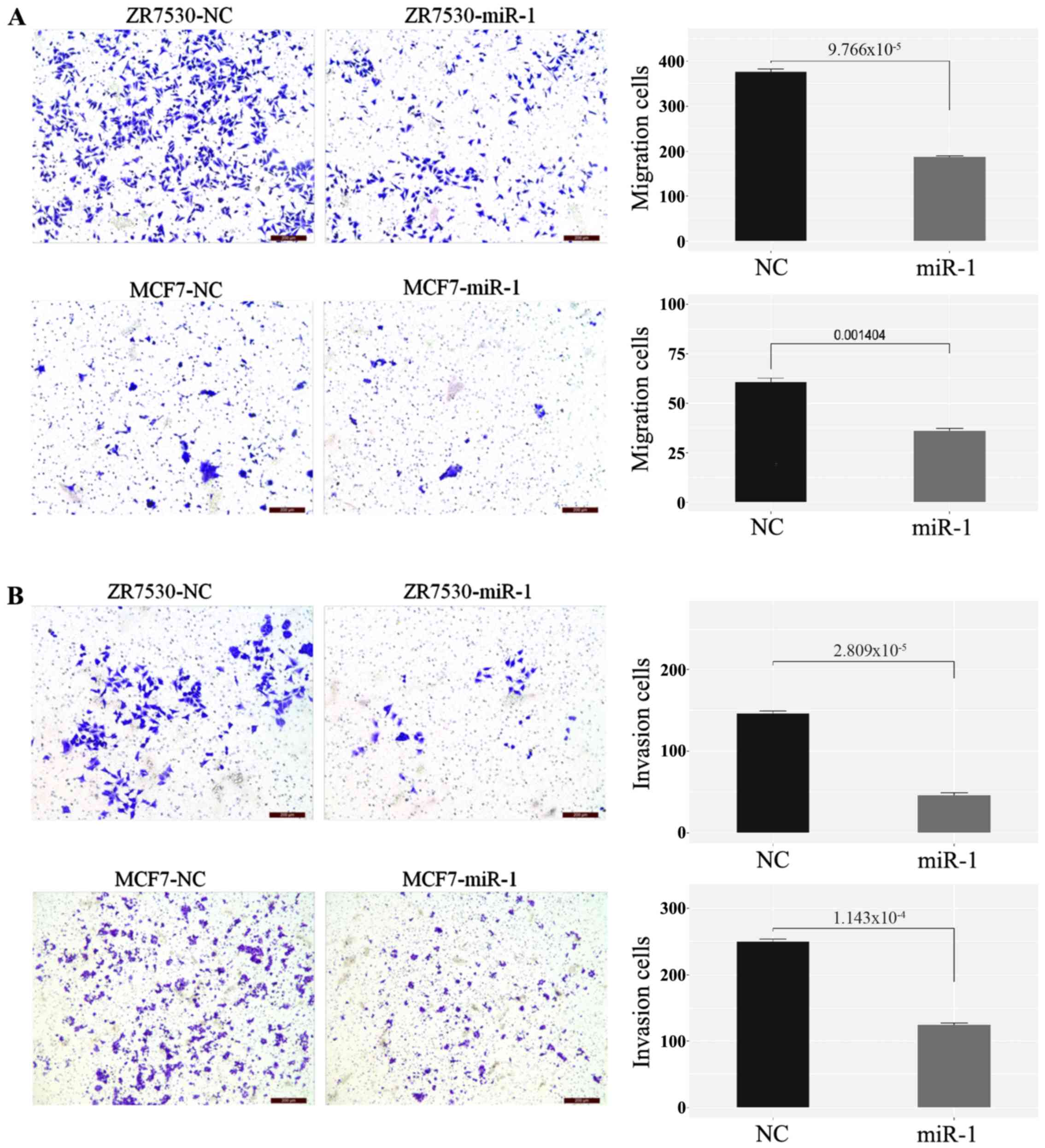
Upregulation of miR-1 inhibits cell
migration and invasiveness of BC
Transwell migration and invasion assays were used to
evaluate the effect of miR-1 on BC cell metastasis. As shown in the
figures, migration (Fig. 3A) and
invasion (Fig. 3B) were both
significantly decreased in miR-1-transfected cells compared with
the negative control. These results indicated that overexpression
of miR-1 inhibits BC cell migration and invasion potential.
Upregulation of miR-1 can promote BC
cell apoptosis
The flow cytometric analysis demonstrated that
upregulation of miR-1 increased the rate of apoptosis in MCF-7
cells compared with the control (Fig.
4A). In addition, the expression levels of Bax, Bad, cleaved
poly ADP ribose polymerase (PARP), and cleaved caspase-3, which are
well-defined protein markers of apoptosis, were increased in
miR-1-overexpressing cells (Fig.
4B). The expression levels of proliferation markers, such as
p-AKT, p-ERK1/2, myeloid cell leukemia 1 (Mcl-1) and anti-binding
immunoglobulin protein (BIP), were decreased in
miR-1-overexpressing cells (Fig.
4C). Genes that regulate the cell cycle and DNA damage were
also examined (Fig. S2A).
However, for cyclin dependent kinase (CDK)4, CDK6 and BRCA2, no
significant difference was observed between the BC cells that
stably expressed miR-1 and the negative control cells. However,
increased expression of p21 was observed in miR-1-overexpressing BC
cells. Furthermore, the cell adhesion proteins E-cadherin and
claudin-1 were upregulated, while the epithelial-mesenchymal
transition (EMT)-related protein zinc finger E-box binding homeobox
(ZEB)1 was downregulated in miR-1-overexpressing cells (Fig. 4D). The above results demonstrated
that miR-1 may inhibit BC progression and metastasis by promoting
the apoptosis of BC cells and by inhibiting cell proliferation and
EMT.
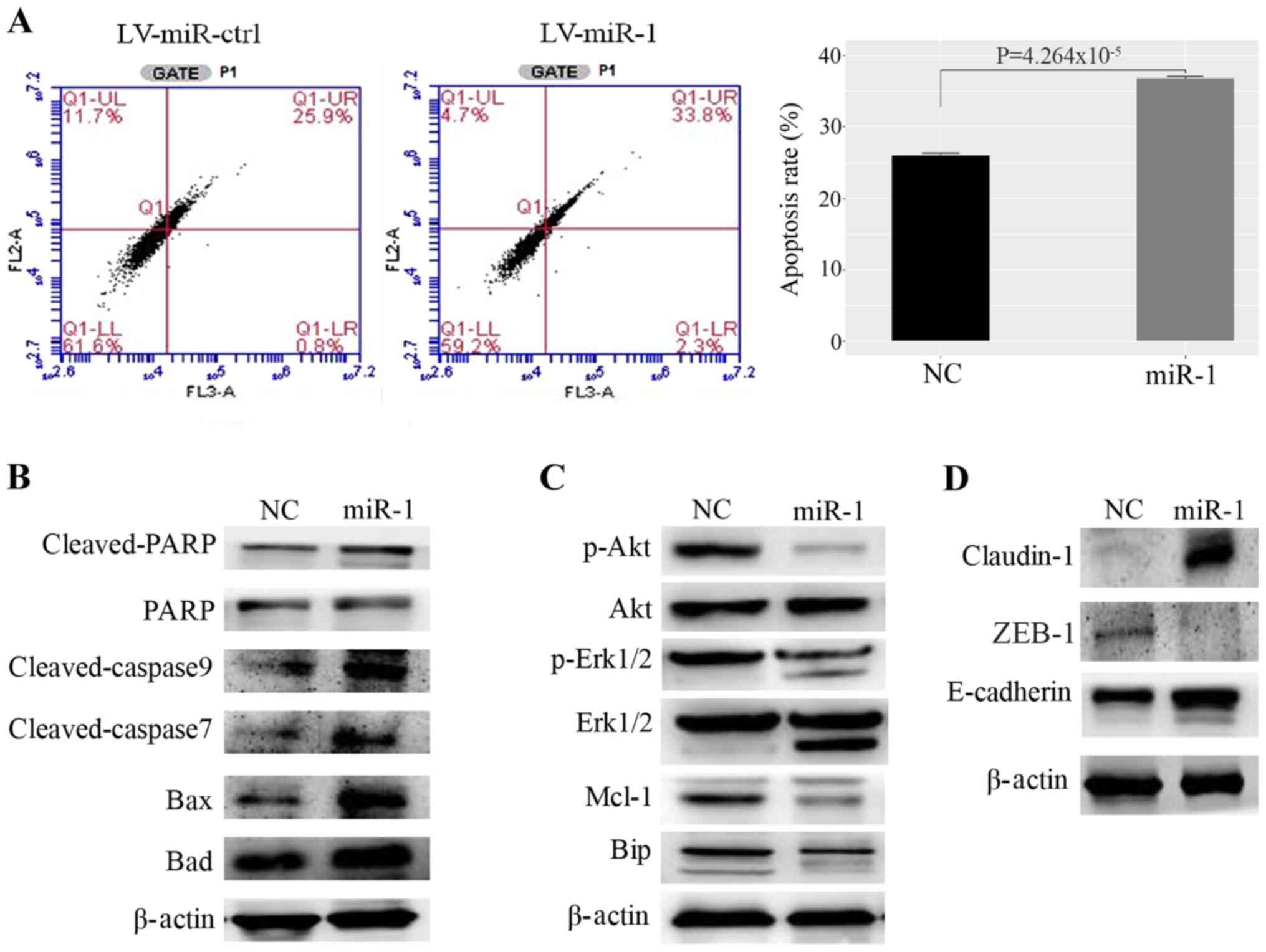 | Figure 4.Upregulation of miR-1 promotes MCF-7
cell apoptosis. (A) miR-1 promotes apoptosis of MCF7 cells, as
demonstrated by fluorescence activated cell sorting, n=3,
P<0.01. (B) Proteins that promote apoptosis were upregulated in
miR-1-overexpressing cells. (C) Proteins that inhibit apoptosis and
induce proliferation were decreased in miR-1- overexpressing cells.
(D) The cell adhesion protein claudin-1 was upregulated and the
EMT-related proteins ZEB1 and E-cadherin were upregulated in
miR-1-overexpressing cells. miR, microRNA; ctrl, control; NC,
normal control; PARP, poly ADP ribose polymerase; Mcl-1, myeloid
cell leukemia 1; Bip, binding immunoglobulin protein; EMT,
epithelial-mesenchymal transition; ZEB1, zinc finger E-box binding
homeobox 1.; LV, lentiviral. |
Bcl-2 is a target of miR-1
The TargetScan search showed that the 3′UTR of the
Bcl-2 gene contains miR-1 binding sites (Fig. 5A). Bcl-2 expression was analyzed in
patient samples and Bcl-2 was highly expressed in BC tissues in
contrast to normal breast tissues (Fig. S2B). Then, whether miR-1 can
regulate the expression level of Bcl-2 was investigated and it was
found that miR-1 downregulated Bcl-2 expression at the mRNA and
protein levels in MCF-7 (Fig. 5B and
C) and ZR-7530 cells (Fig. S2C
and D). The wild-type and mutant Bcl-2 3′UTR were subcloned
downstream of the luciferase gene and named wt-Bcl2 and mut-Bcl2,
respectively and were then transfected together with miR-1-3p
mimics. It was observed that the luciferase activities of the
wt-Bcl2 vector was reduced following treatment with miR-1 mimics.
In addition, luciferase activity in cells transfected with the
mut-Bcl2 and miR-1 mimic was almost comparable with that of the
control cells (Fig. 5D). In
addition, it was found that the expression level of miR-1 and Bcl-2
in BC tissues were negatively correlated according to data
downloaded from the TCGA database (Fig. 5E), which also demonstrated that
Bcl-2 may be a target of miR-1. Bcl-2 in MCF-7 cells were also
modulated in the presence or absence of miR-1 mimics and then
migration and invasion evaluated, as shown in Fig. S3A and B. The number of migrating
and invading cells returned to levels similar to that of the NC
group following ectopic Bcl-2 expression in miR-1-overexpressing
cells. This finding demonstrated that overexpression of miR-1
inhibited cell migration and invasion, which is reversed by Bcl-2
overexpression. These data also showed that miR-1 can bind directly
to Bcl-2 through their respective miRNA recognition sites. Thus, it
was demonstrated that Bcl-2 is a target gene of miR-1.
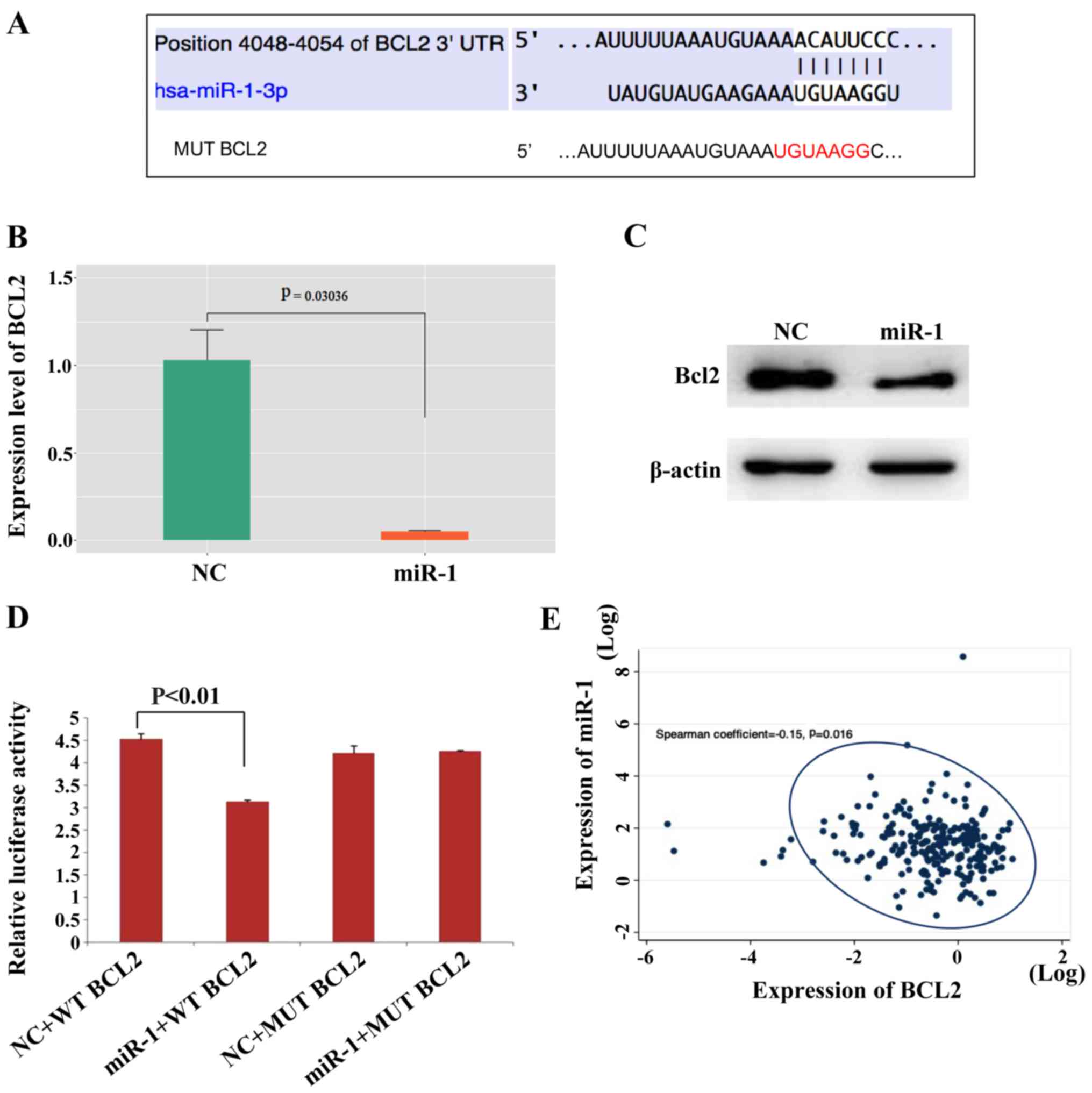 | Figure 5.Bcl-2 is a direct target of miR-1.
(A) The Bcl-2 5′UTR contains binding sites for miR-1 and the
mutated sequence of the Bcl-2 3′UTR. (B) Bcl2 mRNA was decreased in
miR-1- overexpressing MCF-7 cells, n=3, P<0.01. (C) Bcl2 protein
was also decreased in miR-1-overexpressing MCF-7 cells. (D)
Luciferase reporter assays in MCF7 cells showed that miR-1 can
directly bind to the 3′UTR of WT Bcl2 and decreased the luciferase
activity, n=3, P<0.01. (E) The expression levels of miR-1 and
Bcl-2 in BC tissues were negatively correlated according to data
downloaded from the TCGA, P=0.016. miR, microRNA; BC, breast
cancer; NC, normal control; MUT, mutant; WT, wild type. |
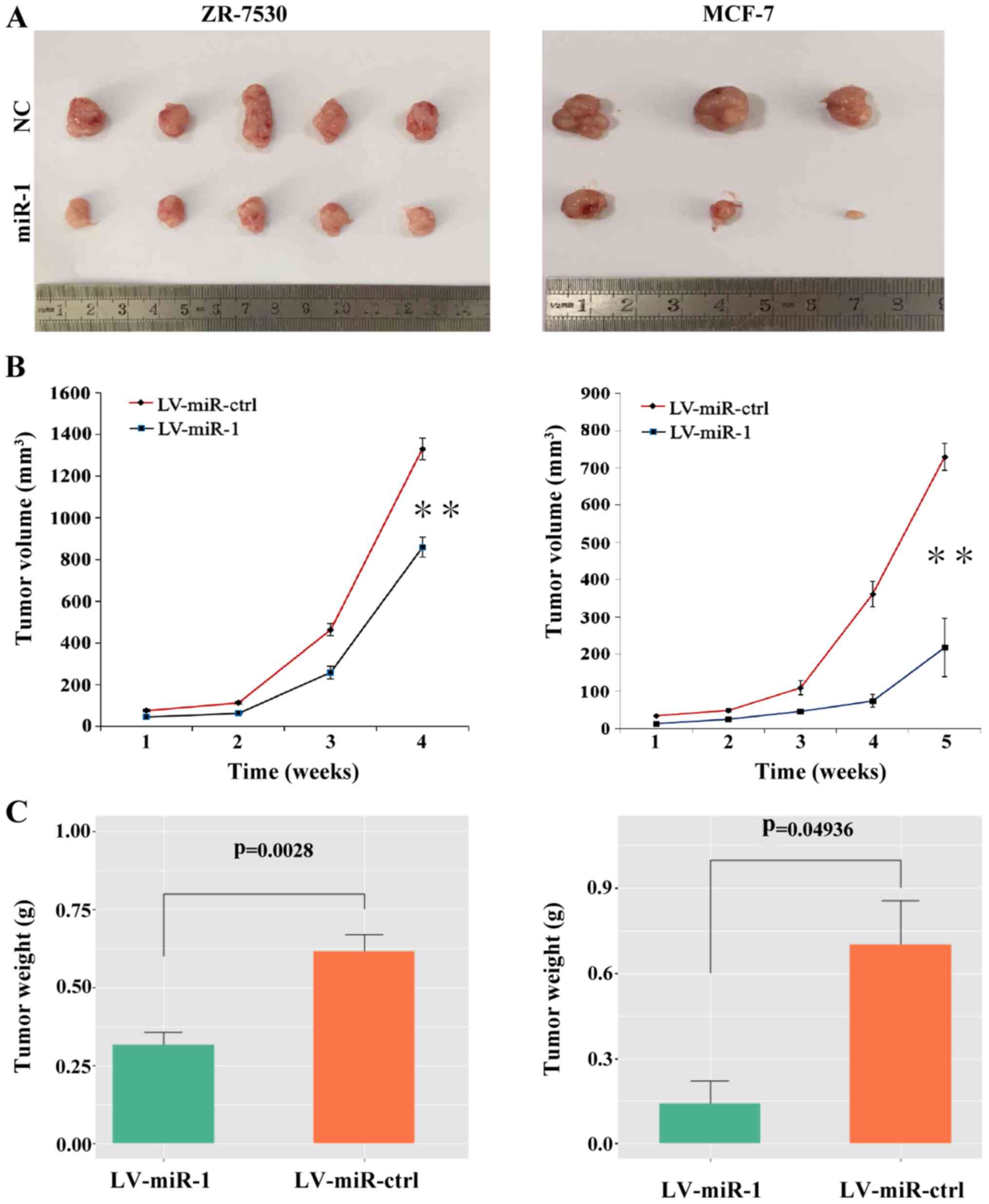
In vivo tumor growth is suppressed by
upregulation of miR-1
To explore the role of miR-1 in tumor growth in
vivo, BC cells with a high level of miR-1 or the negative
control cells were injected subcutaneously into nude mice. After 4
to 5 weeks, the mice were euthanized and intact tumors were removed
(Fig. 6A). The tumor volumes and
weight were measured and the results showed that the average tumor
volumes were decreased in the miR-1 group compared with the
negative control group (Fig. 6B).
The tumor weight was also significantly decreased in the
miR-1-overexpressing group (Fig.
6C). These results indicated that upregulation of miR-1 can
suppress the growth of BC cells in vivo.
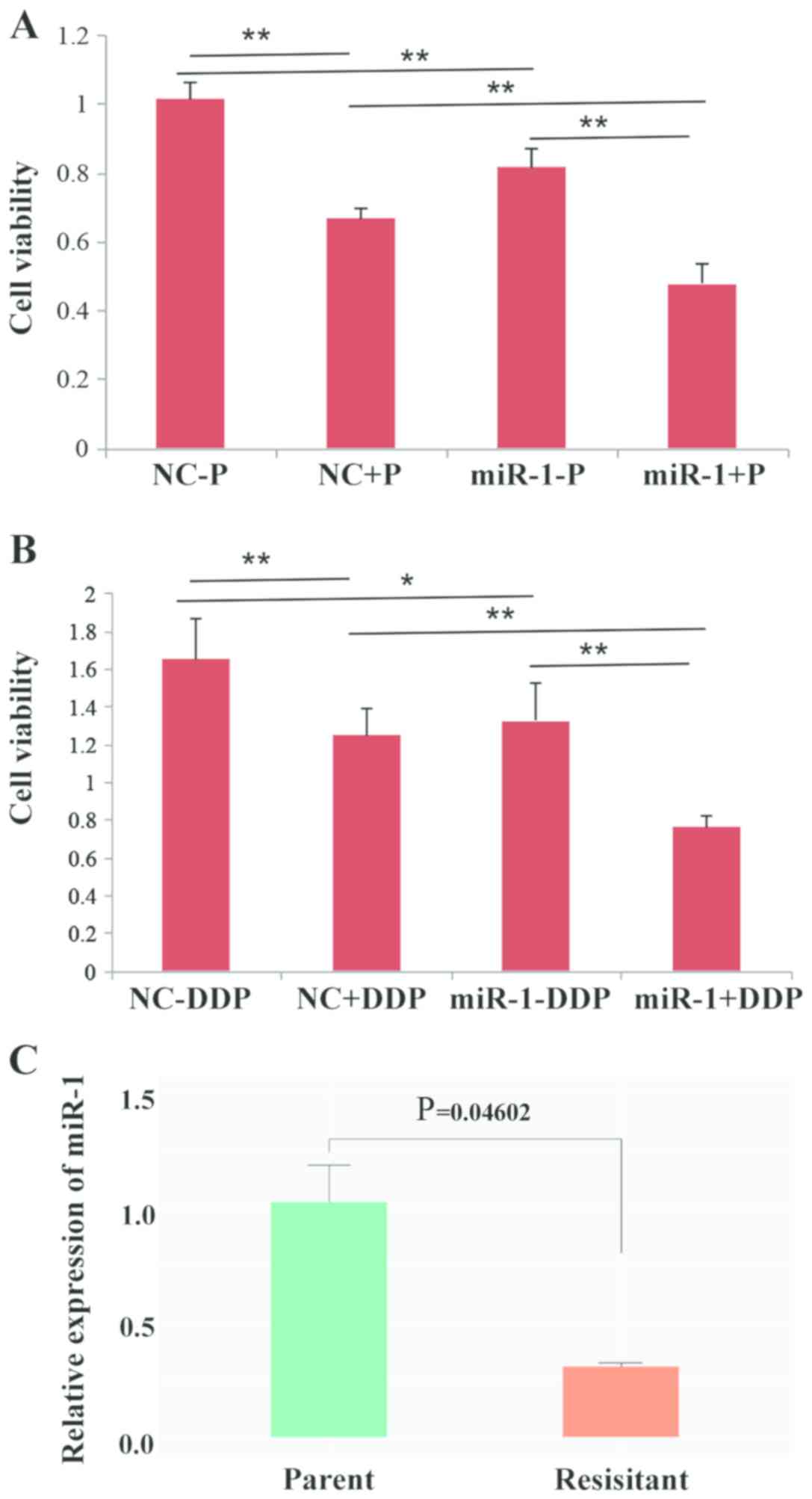
Upregulation of miR-1 enhances drug
sensitivity in BC cells
It has been reported that overexpression of Bcl-2
and its close relatives is a major component of chemo-resistance
(20), and since it was observed
that miR-1 could downregulate Bcl-2, it was hypothesized that miR-1
may influence the sensitivity of BC cells to drugs. Thus, cell
viability was observed following treatment with paclitaxel or DDP.
The results showed that overexpression of miR-1 can enhance the
sensitivity of BC cells to paclitaxel and DDP because cell
proliferation was decreased in the miR-1-overexpressing group
compared with the control group (Fig.
7A and B). The expression level of miR-1 in DDP-resistant BC
cells was also tested and the results demonstrated that the
expression of miR-1 is significantly decreased in resistant BC
cells compared with parental BC cells (P=0.046; Fig. 7C). Taken together, the data from
the present study indicated that overexpression of miR-1 could
enhance drug sensitivity in BC cells.
Discussion
miR-1 has been shown to be downregulated in some
cancer types, where it inhibits tumor growth (21,22).
However, the specific role and mechanism of miR-1 in BC requires
further elucidation. It was revealed that miR-1 was expressed at a
low level in BC tissues and that overexpression of miR-1 inhibited
BC cell proliferation and migration. For the first time, to the
best of the authors' knowledge the present study illustrated the
interaction between miR-1 and Bcl-2 in BC cells and showed that
miR-1 promoted cell apoptosis through Bcl-2 downregulation. It also
demonstrated for the first time that miR-1-overexpressing BC cells
are more sensitive to paclitaxel and DDP.
The present study found that miR-1 was expressed at
low levels in BC tissues. Therefore, it was hypothesized that miR-1
may be a tumor suppressor gene in BC. In vitro experiments
indicated that transfection of MCF-7 and ZR-7530 cells with miR-1
can inhibit cell growth. Flow cytometry analysis revealed that
overexpression of miR-1 can promote cell apoptosis. These results
were consistent with those of a previous study, which reported that
a high level of miR-1 can inhibit the growth of hepatocellular
carcinoma cells by promoting apoptosis (18). Western blotting data also showed
that proteins that promote apoptosis such as Bax, cleaved PARP and
cleaved caspase-3, were increased in miR-1-overexpressing cells. In
contrast, p-AKT, Bip and Mcl-1, which are important for
proliferation, were decreased in the miR-1 overexpression group. To
evaluate the role of miR-1 in the regulation of the cell cycle or
DNA damage in BC cells, the expression of related genes was
analyzed, but no significant difference was observed between BC
cells in which miR-1 was stably expressed and negative control
cells. Only p21, which mediates the p53 tumor suppressor gene, was
upregulated in LV-miR-1 cells. Notably, in agreement with the
findings of the present study, p21 has also been shown to be
regulated by miR-1 in rhabdomyosarcoma (23). Further investigation revealed that
overexpression of miR-1 can inhibit cell migration and invasion. In
addition, the EMT-related protein ZEB1 was downregulated in
miR-1-overexpressing cells. These data implied that miR-1 may act
as a tumor suppressor gene in BC by promoting apoptosis and
inhibiting EMT.
The potential target genes of miR-1 were
investigated next. Bcl-2 belongs to a group of related proteins
that play key roles in apoptosis, or programmed cell death
(20,24). The over-expression of Bcl-2 protein
has been identified in numerous types of tumors, including BC.
Western blot analysis revealed that miR-1 can bind to Bcl-2 and
decrease Bcl-2 protein levels in BC cells. A luciferase assay also
provided evidence that miR-1 can directly regulate Bcl-2 mRNA
expression by targeting the Bcl-2 3′-UTR. Consistent with the
results of the present study, Tang et al (25) reported that miR-1 is closely
related to ischemia/reperfusion injury in a rat model and that the
level of miR-1 is inversely correlated with Bcl-2 expression in
cardiomyocytes in a I/R rat model.
Finally, the function of miR-1 in drug resistance
was investigated. The over-expression of Bcl-2 has been shown to
regulate the development of drug resistance in BC (26,27).
Paclitaxel and DDP are among the well-known chemotherapy drugs used
to treat numerous types of tumors, including BC (28). The authors' previous study
demonstrated that neoadjuvant chemotherapy consisting of a
paclitaxel and DDP combination was highly effective for BC patients
(29). However, drug resistance is
a key obstacle to the success of chemotherapy (30). It was therefore hypothesized that
miR-1 may sensitize BC cells to anticancer drugs by targeting
Bcl-2, which protects BC cells from apoptosis. Indeed, the data
from the present study showed that paclitaxel- and DDP-induced
apoptosis was increased in BC cells that overexpressed miR-1.
Similar the current study, Hua et al (31) reported that miR-1 overexpression
improved DDP sensitivity by inhibiting ATG3-mediated autophagy in
non-small cell lung cancer cells. Thus, miR-1 may have a role in
the treatment of BC patients who are administered paclitaxel and
DDP, but the specific mechanism requires further investigation in
the future.
The present study illustrated that miR-1 is
expressed at low levels in BC samples and can regulate cell
proliferation and migration by downregulating Bcl-2. The data also
showed that miR-1 can increase the sensitivity of BC cells to
paclitaxel and DDP. In conclusion, miR-1 is a candidate tumor
suppressor in human BC and increasing the expression of miR-1 may
be a potential treatment strategy for BC.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
The present study was supported by the
Multidisciplinary Cross Research Foundation of Shanghai Jiaotong
University (grant nos. ZH2018QNA42 and YG2017QN49) and the Shanghai
Natural Science Foundation (grant no. 19ZR1431100), the Shanghai
Municipal Commission of Health and Family Planning (grant no.
201640006), the Clinical Research Plan of SHDC (grant no. 12016231
and 16CR3065B), and the Incubating Program for Clinical Research
and Innovation of Renji Hospital (grant no. PYMDT-002).
Availability of data and materials
All datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
LZ and JL provided the concept and designed the
designed the experiments of the study. JP and CY performed the
experiments. ZW and WY performed the statistical and bioinformatics
analysis. YW and YL collected the clinical samples and contributed
to the interpretation of the data. JP wrote and YW helped revised
the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The Ethics Committee of Renji Hospital affiliated
with Shanghai Jiaotong University of Medicine approved the protocol
and written informed consent was provided by the included patients
and healthy controls.
Patient consent for publication
All BC patients provided written informed consent
for the use of their specimens in this study.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Minemura H, Takagi K, Miki Y, Shibahara Y,
Nakagawa S, Ebata A, Watanabe M, Ishida T, Sasano H and Suzuki T:
Abnormal expression of miR-1 in breast carcinoma as a potent
prognostic factor. Cancer Sci. 106:1642–1650. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Gerratana L, Fanotto V, Bonotto M,
Bolzonello S, Minisini AM, Fasola G and Puglisi F: Pattern of
metastasis and outcome in patients with breast cancer. Clin Exp
Metastasis. 32:125–133. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Higgins MJ and Baselga J: Targeted
therapies for breast cancer. J Clin Invest. 121:3797–3803. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ali S and Coombes RC: Endocrine-responsive
breast cancer and strategies for combating resistance. Nat Rev
Cancer. 2:101–112. 2002. View
Article : Google Scholar : PubMed/NCBI
|
|
6
|
Piao HL and Ma L: Non-coding RNAs as
regulators of mammary development and breast cancer. J Mammary
Gland Biol Neoplasia. 17:33–42. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Tavazoie SF, Alarcon C, Oskarsson T, Padua
D, Wang Q, Bos PD, Gerald WL and Massague J: Endogenous human
microRNAs that suppress breast cancer metastasis. Nature.
451:147–152. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nohata N, Hanazawa T, Enokida H and Seki
N: microRNA-1/133a and microRNA-206/133b clusters: Dysregulation
and functional roles in human cancers. Oncotarget. 3:9–21. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Hudson RS, Yi M, Esposito D, Watkins SK,
Hurwitz AA, Yfantis HG, Lee DH, Borin JF, Naslund MJ, Alexander RB,
et al: MicroRNA-1 is a candidate tumor suppressor and prognostic
marker in human prostate cancer. Nucleic Acids Res. 40:3689–3703.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Liu C, Zhang S, Wang Q and Zhang X: Tumor
suppressor miR-1 inhibits tumor growth and metastasis by
simultaneously targeting multiple genes. Oncotarget. 8:42043–42060.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu T, Hu K, Zhao Z, Chen G, Ou X, Zhang
H, Zhang X, Wei X, Wang D, Cui M and Liu C: MicroRNA down-regulates
proliferation and migration of breast cancer stem cells by
inhibiting the Wnt/β-catenin pathway. Oncotarget. 6:41638–41649.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu J, Zhao FP, Peng ZL, Zhang MM, Lin SX,
Liang BJ, Zhang B, Liu X, Wang L, Li G, et al: EZH2 promotes
angiogenesis through inhibition of miR-1/Endothelin-1 axis in
nasopharyngeal carcinoma. Oncotarget. 5:11319–11332. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sheng JY, Shi BL and Chen HF:
Establishment and appraisal of DDP resistant variant of triple
negative breast cancer cell line MDA-MB-231. Cancer Res Prev Treat.
43:175–180. 2016.
|
|
17
|
Cancer Genome Atlas Network, .
Comprehensive molecular portraits of human breast tumours. Nature.
490:61–70. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Datta J, Kutay H, Nasser MW, Nuovo GJ,
Wang B, Majumder S, Liu CG, Volinia S, Croce CM, Schmittgen TD, et
al: Methylation mediated silencing of MicroRNA-1 gene and its role
in hepatocellular carcinogenesis. Cancer Res. 68:5049–5058. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chang YS, Chen WY, Yin JJ,
Sheppard-Tillman H, Huang J and Liu YN: EGF receptor promotes
prostate cancer bone metastasis by downregulating miR-1 and
activating TWIST1. Cancer Res. 75:3077–3086. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Adams JM and Cory S: The Bcl-2 apoptotic
switch in cancer development and therapy. Oncogene. 26:1324–1337.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nasser MW, Datta J, Nuovo G, Kutay H,
Motiwala T, Majumder S, Wang B, Suster S, Jacob ST and Ghoshal K:
Down-regulation of micro-RNA-1 (miR-1) in lung cancer. Suppression
of tumorigenic property of lung cancer cells and their
sensitization to doxorubicin-induced apoptosis by miR-1. J Biol
Chem. 283:33394–33405. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yoshino H, Chiyomaru T, Enokida H,
Kawakami K, Tatarano S, Nishiyama K, Nohata N, Seki N and Nakagawa
M: The tumour-suppressive function of miR-1 and miR-133a targeting
TAGLN2 in bladder cancer. Br J Cancer. 104:808–818. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yan D, Dong Xda E, Chen X, Wang L, Lu C,
Wang J, Qu J and Tu L: MicroRNA-1/206 targets c-Met and inhibits
rhabdomyosarcoma development. J Biol Chem. 284:29596–29604. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Cory S, Huang DC and Adams JM: The Bcl-2
family: Roles in cell survival and oncogenesis. Oncogene.
22:8590–8607. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Tang Y, Zheng J, Sun Y, Wu Z, Liu Z and
Huang G: MicroRNA-1 regulates cardiomyocyte apoptosis by targeting
Bcl-2. Int Heart J. 50:377–387. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Davis JM, Navolanic PM,
Weinstein-Oppenheimer CR, Steelman LS, Hu W, Konopleva M,
Blagosklonny MV and McCubrey JA: Raf-1 and Bcl-2 induce distinct
and common pathways that contribute to breast cancer drug
resistance. Clin Cancer Res. 9:1161–1170. 2003.PubMed/NCBI
|
|
27
|
Zhou M, Liu Z, Zhao Y, Ding Y, Liu H, Xi
Y, Xiong W, Li G, Lu J, Fodstad O, et al: MicroRNA-125b confers the
resistance of breast cancer cells to paclitaxel through suppression
of pro-apoptotic Bcl-2 antagonist killer 1 (Bak1) expression. J
Biol Chem. 285:21496–21507. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Duan Z, Lamendola DE, Duan Y, Yusuf RZ and
Seiden MV: Description of paclitaxel resistance-associated genes in
ovarian and breast cancer cell lines. Cancer Chemother Pharmacol.
55:277–285. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhou L, Xu S, Yin W, Lin Y, Du Y, Jiang Y,
Wang Y, Zhang J, Wu Z and Lu J: Weekly paclitaxel and cisplatin as
neoadjuvant chemotherapy with locally advanced breast cancer: A
prospective, single arm, phase II study. Oncotarget. 8:79305–79314.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Siddik ZH: Cisplatin: Mode of cytotoxic
action and molecular basis of resistance. Oncogene. 22:7265–7279.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hua L, Zhu G and Wei J: MicroRNA-1
overexpression increases chemosensitivity of non-small cell lung
cancer cells by inhibiting autophagy related 3-mediated autophagy.
Cell Biol Int. 42:1240–1249. 2018. View Article : Google Scholar : PubMed/NCBI
|