Introduction
Skeletal muscle stem cells (SkMSCs) are capable of
self-renewal and muscle regeneration (1). Lineage progression directs quiescent
stem cells toward activation, proliferation and differentiation due
to muscle injury or pathological conditions through the activation
of multiple mitogenic factors (2–4).
Previous studies have suggested that key transcription factors
serve a significant role in the proliferation and differentiation
of SkMSCs (4,5). Progenitors expressing paired box
(Pax) proteins 3/7 are prerequisite factors for skeletal muscle
growth and are regarded as the source of adult SkMSCs (5–7). A
number of studies have suggested that the expression of Pax7 is
important for the maintenance of adult SkMSCs, and that the
activation of the myogenic differentiation markers (MyoD) gene
primes myogenesis (6,8,9).
Accumulating evidence suggests that a number of
microRNAs (miRNAs), such as miR-99a-5p (10), miR-9-5p (11), miR-208b (12), serve important roles in skeletal
muscle myogenesis by regulating gene expression, and that their
abnormal expression is associated with a number of muscle diseases
including, muscle atrophy and ischemic injury (6,13,14).
Regarding the molecular mechanism of miRNAs in myogenesis, studies
have reported a role for these molecules in the differentiation of
SkMSCs (13,15). Indeed, substantial evidence
supports the hypothesis that miRNAs are involved in regulating
muscle regeneration. A recent study has reported that miR-483-3p is
involved in the osteogenic differentiation of bone marrow-derived
mesenchymal stem cells (BMSCs) by targeting STAT1 and may serve as
a potential therapeutic target for bone loss due to aging (16). In addition, studies have revealed
that miR-7 regulates the neural differentiation of trabecular
meshwork mesenchymal stem cells (TMMSCs), and that the poly
(L-lactate) (PLLA)/poly (e-caprolactone) (PCL) scaffold, termed a
three-dimensional (3D) culture system, can promote their
differentiation towards glial and neural progenitor cells (14). Previous studies have reported that
miRNA-217-5p regulates pluripotent stem cell proliferation and
differentiation (17) and is
involved in metabolic processes in various cells, such as
endothelial (18) and colorectal
cancer (19) cells. These findings
provide insights into the application of miRNAs in regenerative and
cell therapy for muscle diseases (14). However, a limited number of studies
have explored the potential roles of miR-217-5p in SkMSCs.
Previous studies have reported that fibroblast
growth factor receptor 2 (FGFR2) exerts an important role in
embryogenesis and tissue regeneration, especially in bone and
vascular development (20,21). FGFR2 overexpression serves a
crucial role in the myogenesis of SkMSCs (22). Consistent with these findings,
owing to its association with the myogenesis of SkMSCs, FGFR2 is
considered a therapeutic target for muscle injury (23,24).
However, its role in the proliferation of SkMSCs remains unclear.
This present study aimed to investigate whether miR-217-5p may
mediate the expression of FGFR2 in SkMSCs.
Materials and methods
Animals and cell culture
Isolated single myofiber-associated cells were
prepared using limb muscles obtained from 2-week-old female Sprague
Dawley rats (n=5; 30–500 g) maintained in a 12:12 h light/dark
cycle at 23°C and 50–70% humidity, which were anesthetized with 50
mg/kg 1% sodium pentobarbital and euthanized by cervical
dislocation prior to the removal of the limb muscles. Animal
experiments were approved by The Institutional Animal Care and Use
Committee at The First Affiliated Hospital of Sun Yat-sen
University (Guangzhou, China). All animals were purchased from The
Guangdong Medical Laboratory Animal Center (Guangzhou, China). Rat
tibialis muscles were subjected to enzymatic dissociation (0.2%
collagenase, Sigma-Aldrich; Merck KGaA) at 37°C for 90 min. The
cell suspension was filtered through a 40-µm filter (Biosharp Life
Sciences). Following isolation, myofiber-associated cells were
stained for the isolation of particular cell populations by flow
cytometry and fluorescence-activated cell sorting (FACS). After
sorting, cells were cultured in DMEM (HyClone; GE Healthcare life
Sciences) supplemented with 20% FBS (Gibco; Thermo Fisher
Scientific, Inc.) and 1% chick embryo extract (Gemini Bio
Products). Myogenic differentiation was induced using DMEM with 2%
heat-inactivated horse serum (Gibco; Thermo Fisher Scientific,
Inc.). The NC group cells were cultured in DMEM supplemented with
10% FBS and the PG group cells were incubated in DMEM supplemented
with 20% FBS, 1% chick embryo extract and transforming growth
factor (TGF)β1. The cells were maintained in an incubator with a
humidified atmosphere of 95% air and 5% CO2 at 37°C.
RNA isolation and reverse
transcription-quantitative (RT-q) PCR amplification
RT-qPCR was performed as described previously
(25). Briefly, total RNA was
extracted using TRIzol® reagent (Thermo Fisher
Scientific, Inc.) and was reverse transcribed by
SuperScript® cDNA Synthesis Kit (Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
Briefly, an RT mixture containing 1 µl total RNA, 4 µl
deoxynucleoside triphosphates, 2 µl Primer Max, 4 µl RT bufer, 1 µl
SuperRT and diethyl pyrocarbonate (DEPC)-treated water to a final
volume of 20 µl were subjected to 37°C for 1 h, 95°C for 5 min and
4°C for cooling for 30 min. The primers used were as follows:
miR-217-5p forward, 5′-CGCGGATCCTATTGTATTACGGTAGGATG-3′ and
reverse, 5′-CCGCTCGAGCAGATAGCACGAACTTTT-3′; FGFR2 forward,
5′-GCGTCTCCAACGCCAAAGAGTCTTTCGTATATTATCAAAAT-3′ and reverse,
5′-CAGTGAATTTTGATAATATACGAAAGACTCTTTGGCGTTG-3′; Pax 7 forward,
5′-AGCCGAGTGCTCAGAATCAA-3′ and reverse, 5′-TCCTCTCGAAAGCCTTCTCC-3′;
MyoD forward, 5′-CGACTGCCTGTCCAGCATAG-3′ and reverse,
5′-GGACACTGAGGGGTGGAGTC-3′; myosin heavy chain (MyHC) forward,
5′-TGCCAAGACCGTGAGGAATG-3′ and reverse, 5′-AATGCATCACAGCTCCCGTG-3′;
and GAPDH forward, 5′-GGGTGATGCTGGTGCTGAGTATGT-3′ and reverse,
5′-AAGAATGGGAGTTGCTGTTGAAGTC-3′. Thermal cycling conditions were 2
min at 95°C followed by 40 cycles of 95°C for 15 sec and 60°C for
15 sec on a Real-Time PCR Detection System (Bio-Rad Laboratories,
Inc.), GAPDH was used as an internal control to normalize target
gene transcripts. Each sample was measured at least three times and
the 2−ΔΔCq method (26)
was used to assess relative levels of the mRNAs of the target
genes.
Target gene prediction
TargetScan (version 7.2l www.targetscan.org,) and miRNA.org (version
22.1; http://www.microrna.org) were used for
scanning the candidate targets of miR-217-5p. Basic information of
miR-217-5p was submitted online and the potential targets of
miR-217-5p were presented.
Luciferase reporter assay
The miR-217-5p mimics
(5′-UACUGCAUCAGGAACUGAUUGGC-3′), miR-217-5p antagomir
(5′-GCCAAUCAGUUCCUGAUGCAGUA-3′) NC-mimics
(5′-UUCUCCGAACGUGUCACGUTT-3′; 5′-ACGUGACACGUUCGGAGAATT-3′) and
NC-antagomir (5′-CAGUACUUUUGUGUAGUACAA-3′) were purchased from
Thermo Fisher Scientific, Inc. The amplified miR-192-5p mimic
sequence and miR-NC were transfected into the SkMSCs using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's instructions.
To identify the binding sequences and uniform
resource locator, luciferase reporter assay was used. The
miR-217-5p mimics, miR-NC, or miR-217-5p antagomir and the pRL-TK
vector (Promega Corporation) carrying the mutant (mut) or wild-type
(wt) FGFR2 3′ untranslated region (3′-UTR) were co-transfected into
SkMSCs using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Three days later, cells were lysed with
the Dual-Glo® Reagent (Promega Corporation), and
luciferase activity was measured using a Dual-Luciferase Reporter
Assay System (Promega Corporation). The firefly luciferase activity
was normalized to Renilla luciferase activity.
Western blot analysis
Protein was extracted using RIPA buffer with
protease inhibitor (Sigma-Aldrich; Merck KGaA), protein
concentration was determined by bicinchoninic assay (Thermo Fisher
Scientific, Inc.), and denatured for 5 min at 100°C prior to
electrophoresis using an 8–10% polyacrylamide gel and a mid-range
protein ladder (Beijing CoWin Biotech Co., Ltd.). Then, proteins
were transferred to a PVDF membrane (EMD Millipore), blocked with
2% goat serum (Gibco; Thermo Fisher Scientific, Inc.) for 1 h at
room temperature, and incubated with rabbit anti-FGFR2 (1:1,000;
cat. no. ab10648; Abcam), GAPDH (1:1,000; EPR1689; cat. no.
ab181602; Abcam), Pax7 (1:100; cat. no. ab199010; Abcam), MyoD
(1:250; cat. no. ab203383; Abcam) and MHC (1:200; cat. no. ab11083;
Abcam) primary antibodies overnight at 4°C. Membranes were then
washed using TBST + 0.5% Tween-20 (EMD Millipore) and incubated
with Alexa Fluor® 790-conjugated polyclonal goat
anti-rabbit IgG H&L (1:10,000; cat. no. ab175781; Abcam)
secondary antibody at room temperature for 1 h. Images were
acquired by scanning with LI-COR's Odyssey Infrared Imaging System
(LI-COR Biotechnology).
MTT assay
MTT assay was performed to evaluate the rate of
proliferation of SkMSCs. Following transfection with miR-217-5p
mimics, miR-217-5p antagomir and miR-NC, cells were seeded into
96-well plates (2×104 cells/well) and incubated at 37°C
for ~24 h. Then, 20 µl 5 mg/ml MTT solution was added into each
well, and the plates were incubated at 37°C for an additional 4 h.
After removing the medium, DMSO (160 µl/well) was added to each
well. The concentration of MTT formazan solubilized with PBS was
measured by a microplate reader (Tecan Group, Ltd.) at 490 nm
according to the manufacturer's instructions.
FACS
Flow cytometry analysis and cell sorting were
performed at the central laboratory of The First Affiliated
Hospital of Sun Yat-sen University (Guangzhou, China). Markers for
CD45 [BV510 mouse anti-rat CD45 Clone OX-1 (RUO); BD Pharmingen],
CD11b (rat CD11B APC WT.5; BD Pharmingen), anti-integrinβ1
[anti-integrin β1 (HMb1-1; PE/Cy7); cat. no. ab95622; Abcam] and
CD34 [anti-CD34 antibody (EP373Y); cat. no. ab81289; Abcam] were
used. The trypsinized cells were filtered using 200-mesh sieves and
incubated with the above antibodies at 4°C for 1 h. Then, the cells
were washed twice with PBS and resuspended in 200 ml PBS prior to
analysis using a BD Accuri C6 flow cytometer (BD Biosciences) and
flow cytometric data was analyzed using FlowJo software (version
7.6; Tree Star, Inc) according to the manufacturer's
instructions.
Cell proliferation assay
To verify the proliferation of SkMSCs, a
5-Ethynyl-2′deoxyuridine (EdU) Kit (Guangzhou RiboBio Co., Ltd.)
was used. SkMSCs were seeded at a density of 5×105
cells/well in 6-well plates coated with decellularized skeletal
muscle extracellular matrix hydrogels (Shanghai Linbo Scientific
Instruments Co., Ltd.) and cultured in the appropriate growth
medium for 24 h at 37°C for the following four groups: Negative
control (NC), miR217-5p, 2 µM AZD4547 and 2 µM miR217-5p + AZD4547.
The cells were treated with 10 µM EdU working solution growth
medium for 2 h in the dark. Then, the cells were treated with PBS
containing 4% paraformaldehyde for 20 min at room temperature,
followed by 2 mg/ml glycine and 0.5% Triton X-100 for 15 min at
room temperature. After Hoechst 33342 was added to each well, cells
were incubated for 30–40 min in the dark. Images were acquired at
×40 magnification using an Axiovison 4.8 camera attached to an Axio
Observer Z1 inverted microscope (Carl Zeiss, Inc.). Five image
fields were randomly captured for each sample.
Immunofluorescence staining and microscopy.
Immunofluorescence was performed as previously described (27). Samples were fixed with 4%
paraformaldehyde for 1 h at room temperature and then blocked with
2% goat serum (Gibco; Thermo Fisher Scientific, Inc.) for 1 h at
room temperature. DAPI (cat. no. ab228549, Abcam) was used to
identify the nuclei. The cells were incubated with primary
antibodies overnight at 4°C and were as follows: Pax7 (1:100, cat.
no. ab199010, Abcam), MyoD (1:250, cat. no. ab203383, Abcam) and
MHC (1:200, cat. no. ab11083, Abcam). Then the cells were incubated
with secondary antibodies for 1 h at room temperature and were as
follows: Alexa Fluor® 594-conjugated goat anti-mouse
(1:1,000, cat. no. ab150116, Abcam) and Alexa Fluor®
488-conjugated goat anti-rabbit IgG H&L (1:1,000, cat. no.
ab150077, Abcam). Cell images were acquired using an Axiovison 4.8
camera (magnification, ×40) attached to an Axio Observer Z1
inverted microscope (Carl Zeiss, Inc.) and the images were then
assembled using Adobe® Photoshop CS 6 software (Adobe
Systems, Inc.).
Confocal microscopy
Cells (1×104 cells/well) were seeded on
confocal dishes and maintained in an incubator for 24 h at 37°C.
The procedures were performed according to the manufacturer's
instructions (Thermo Fisher Scientific, Inc.). Cells were incubated
overnight at 4°C with anti- Pax7 (1:200; cat. no. ab199010; Abcam)
and MyoD (1:200; cat. no. ab203383; Abcam) primary antibodies, and
then incubated with Alexa Fluor® 594-conjugated goat
anti-mouse (1:1,000; cat. no. ab150116; Abcam) and Alexa
Fluor® 488-conjugated goat anti-rabbit IgG H&L
(1:1,000; cat. no. ab150077; Abcam) secondary antibodies for 1 h in
the dark at room temperature. Then, DAPI was added for 15 min at
room temperature. Finally, a confocal microscope (Nikon
Corporation) was used to acquire images at ×40 magnification.
Statistical analysis
Each experiment was repeated at least three times
and the data are presented as the mean ± standard deviation.
Statistical significance was determined by performing Student's
t-test for comparisons between two groups and one-way analysis of
variance followed by Tukey's post-hoc test for comparisons between
more than two groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Isolation and identification of
SkMSCs
FACS analysis results demonstrated SkMSCs to be
positive for CD34 and integrin β1 and negative for CD11b and CD45
(Fig. 1A). Cell colonies were
observed 3–5 days after the initial plating, and light microscopy
demonstrated the SkMSCs exhibited a fibroblast-like morphology.
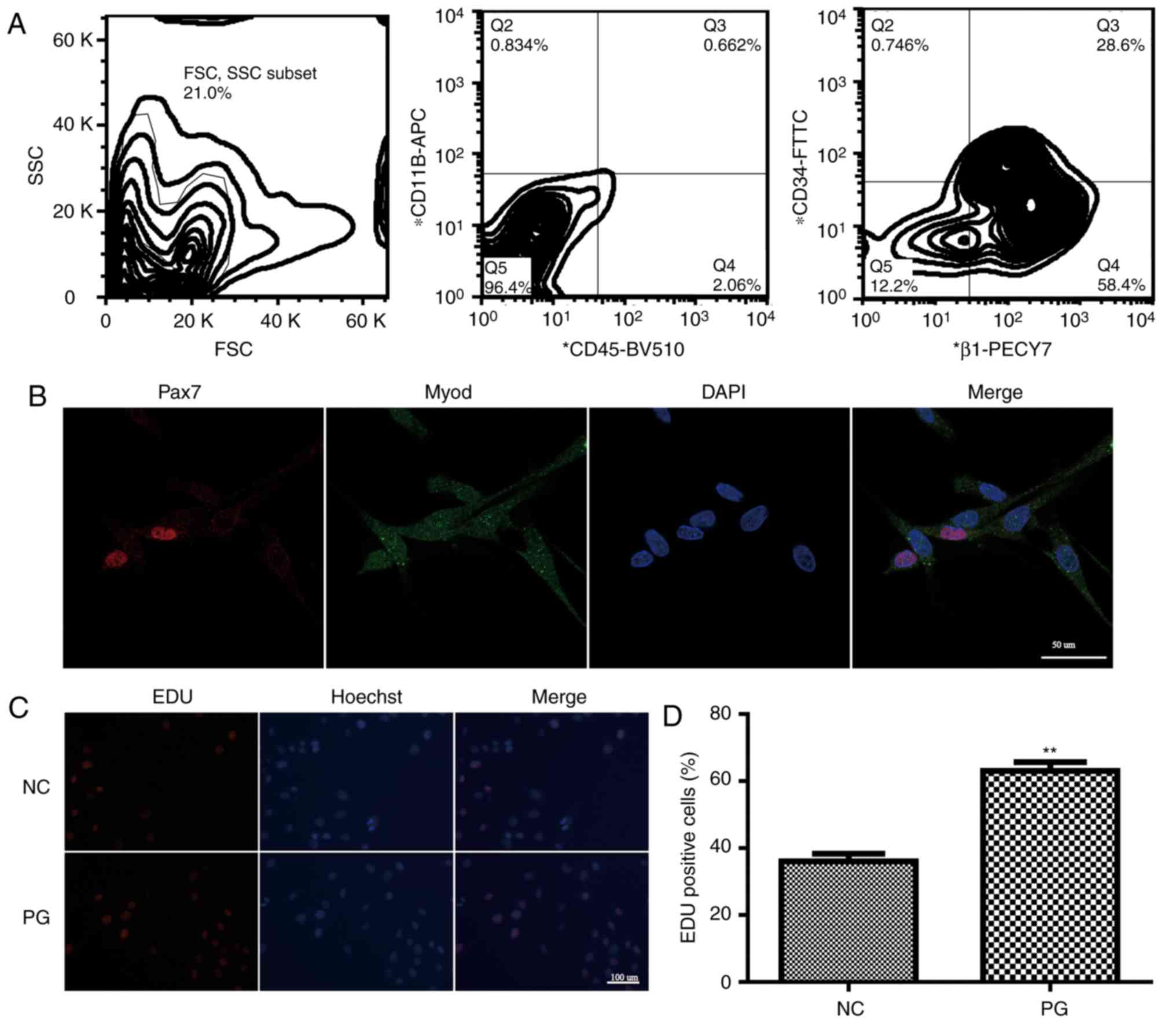 | Figure 1.Isolation and identification of
SkMSCs. (A) Double-sorted SkMSCs were positively stained with
antibodies against CD34 and integrin β1, but negative for CD11b and
CD45. (B) Immunofluorescence staining images for Pax7 (red) and
MyoD (green). Cell nuclei were stained by DAPI (blue). Scale bar,
10 mm. (C) EdU assays demonstrated the proliferation of SkMSCs in
the NC and PG groups. Scale bar, 50 mm (D) Quantification of
EdU-positive cells in the NC and PG groups. NC, control group
(SkMSCs cultured in DMEM supplemented with 10% FBS); PG,
high-proliferation group (SkMSCs cultured in DMEM supplemented with
20% FBS and 1% chick embryo extract with transforming growth factor
β1). **P<0.01. SkMSCs, skeletal muscle stem cells; Pax7, paired
box protein7 Pax7; MyoD, myogenic differentiation markers; EdU,
5-ethynyl-2′-deoxyuridine; NC, control group; PG,
high-proliferation group. |
To identify the characteristics of SkMSCs, the
expression of Pax7 and MyoD was examined. The majority of adhered
SkMSCs exhibited expression of Pax7 and MyoD (Fig. 1B). In addition, the PG group
exhibited a greater number of EdU-positive stained cells compared
with that in the NC group (Fig. 1C and
D), suggesting a high capacity for proliferation.
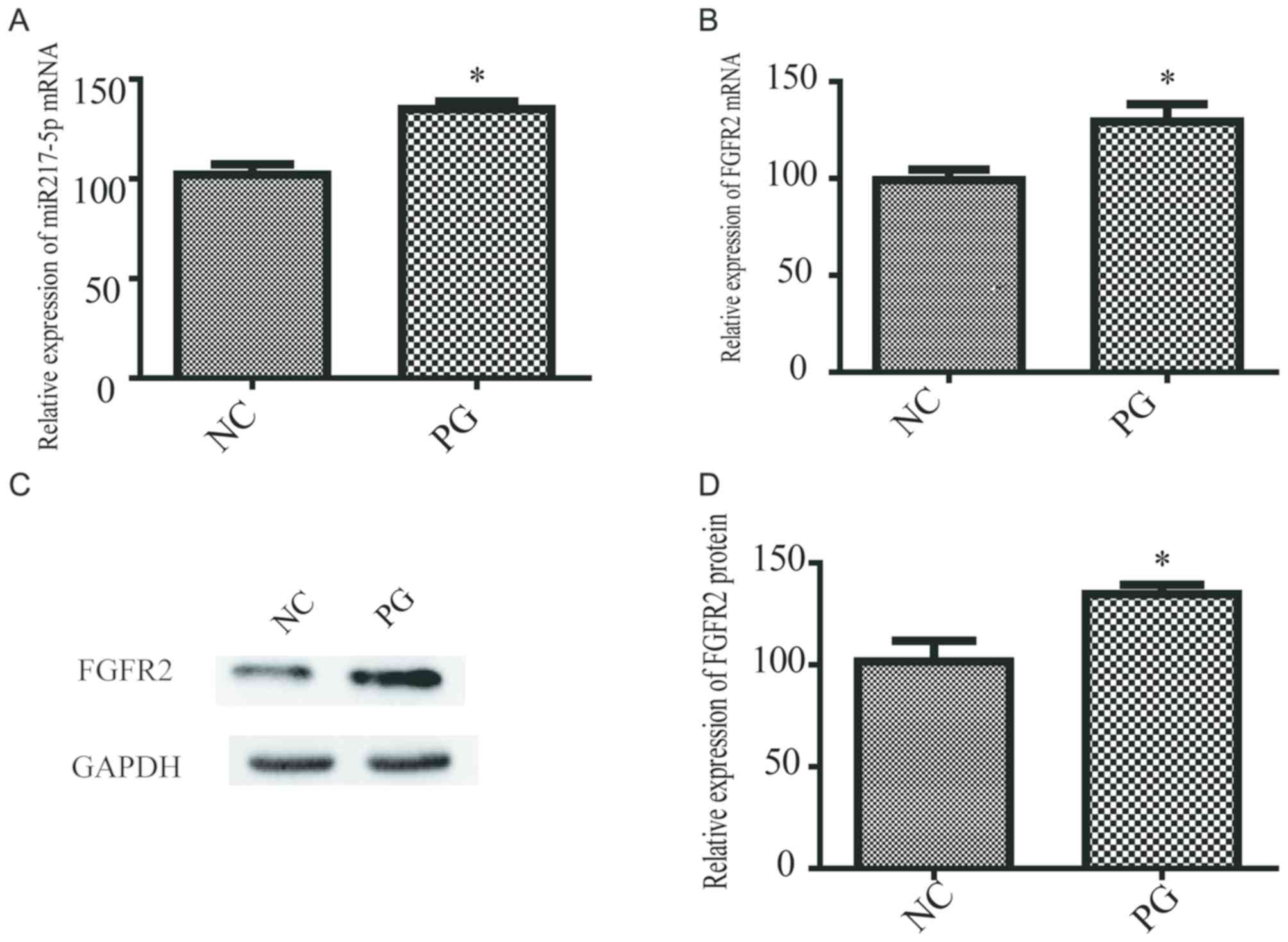
SkMSCs exhibiting an accelerated rate
of proliferation express high levels of miR-217-5p
miR-217-5p levels were measured in the medium
samples from PG and NC SkMSCs. RT-qPCR analysis demonstrated that
the expression of miR-217-5p was significantly higher in PG SkMSCs
compared with that in NC SkMSCs (Fig.
2A). In addition, the FGFR2 mRNA and protein expression was
significantly upregulated in PG SkMSCs compared with that in NC
SkMSCs (Fig. 2B-D). These results
suggested that miR-217-5p and FGFR2 expression was greater in
highly proliferating SkMSCs compared with normal SkMSCs.
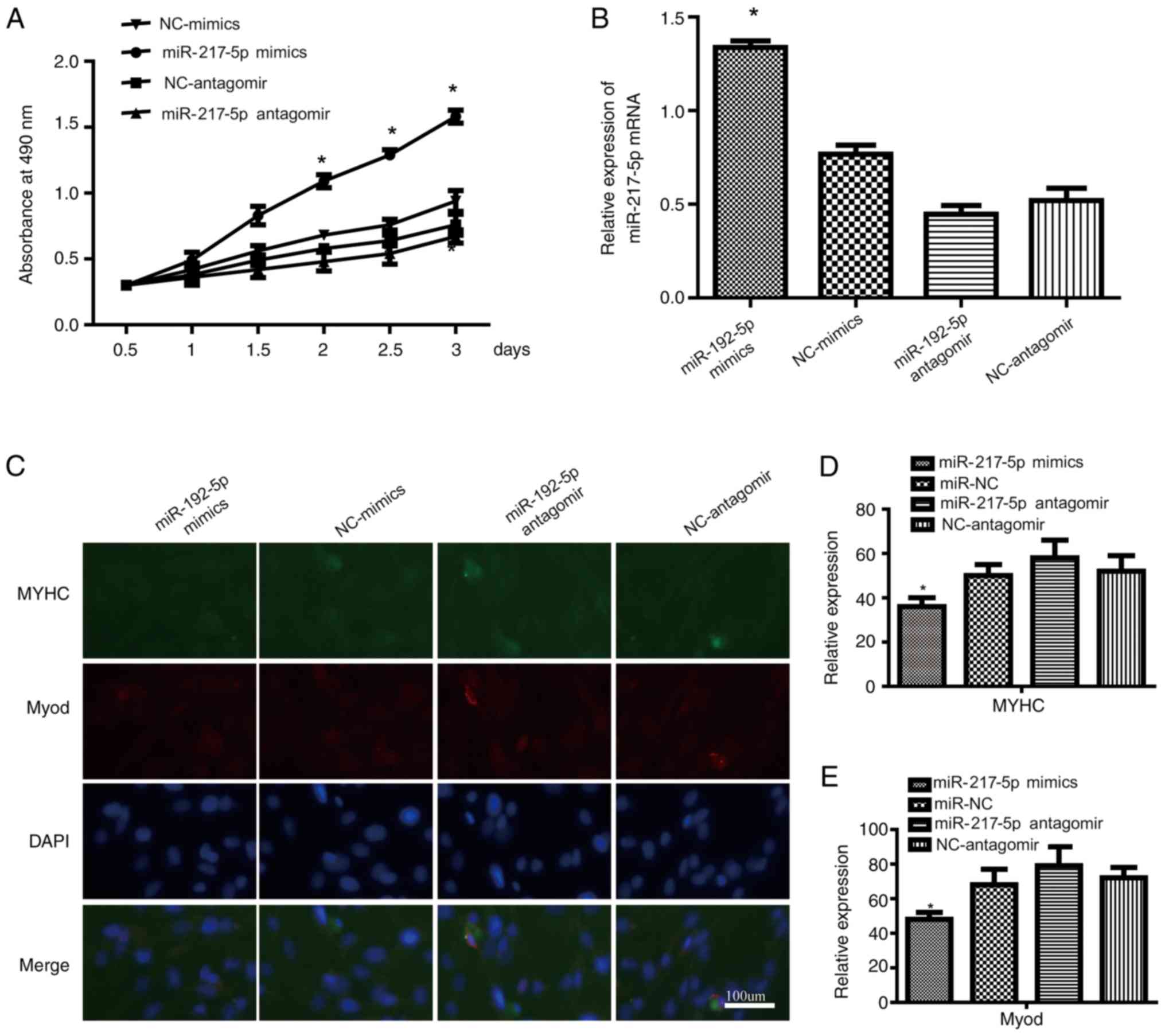
Ectopic expression of miR-217-5p
affects SkMSC proliferation and differentiation
miR-217-5p expression was modulated by transfecting
a miR-217-5p antagomir or miR-217-5p mimics into SkMSCs. MTT assay
demonstrated that the proliferation of SkMSCs was promoted by
miR-217-5p mimics and impeded by miR-217-5p antagomir compared with
that of the miR-NC group (Fig.
3A). RT-qPCR analysis was also performed to verify miR-217-5p
expression; miR-217-5p mimics significantly increased, whereas
miR-217-5p antagomir significantly decreased the expression levels
of miR-217-5p in SkMSCs compared with that of SkMSCs transfected
with miR-NC (Fig. 3B). In
addition, immunofluorescence staining demonstrated that the
expression levels of MyoD and MYHC were significantly lower in the
miR-217-5p mimic group and significantly higher in the miR-217-5p
antagomir group compared with those of the miR-NC group (Fig. 3C-E). These data suggested that
miR-217-5p enhanced the proliferation and inhibited the
differentiation of SkMSCs.
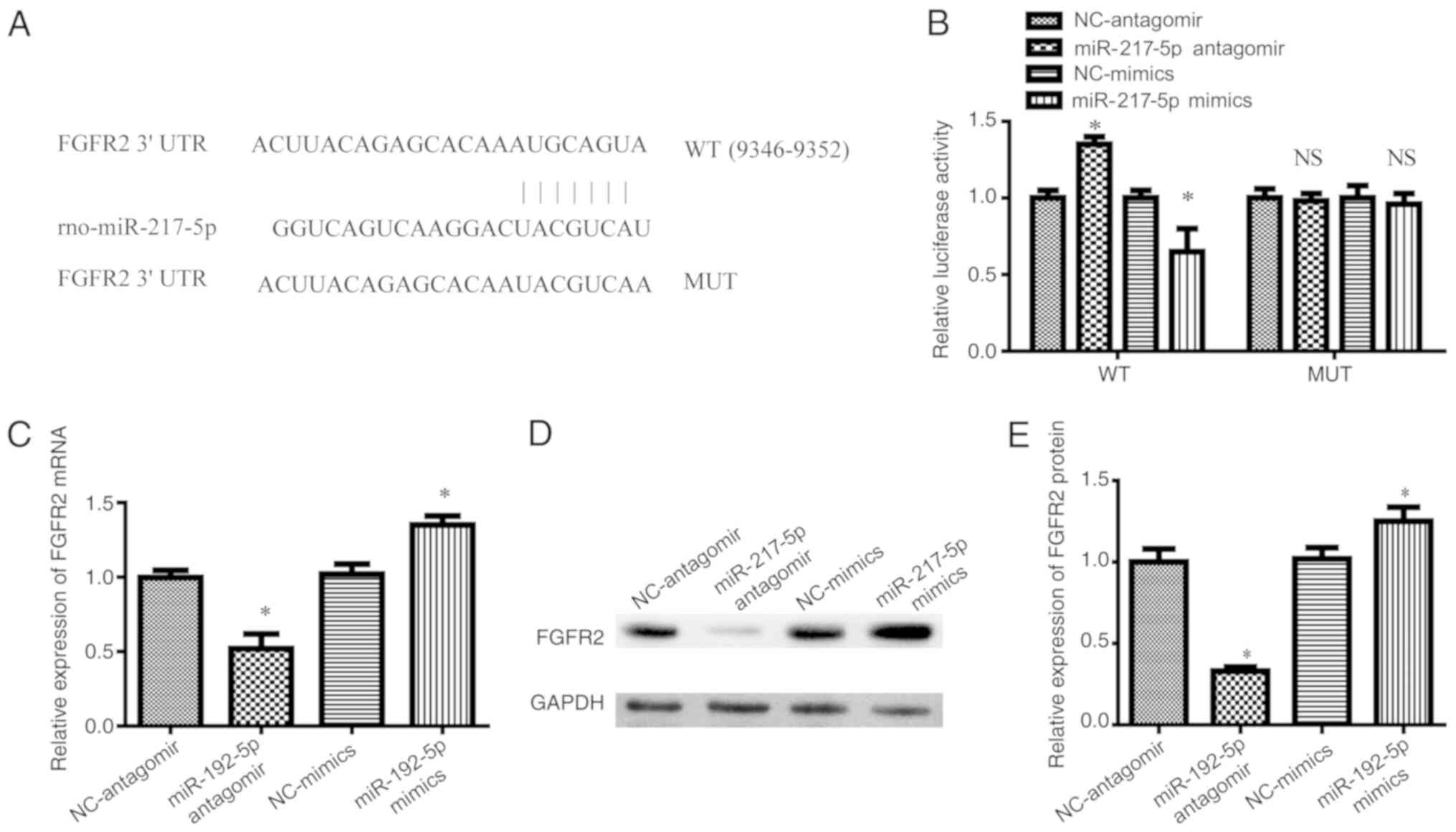
FGFR2 is a direct target of miR-217-5p
in SkMSCs
TargetScan and miRNA databases were used to predict
the downstream targets of miR-217-5p and further explore the
underlying molecular mechanism involved in the proliferation of
SkMSCs. As binding between the 3′-UTR of FGFR2 and miR-217-5p was
predicted (Fig. 4A), luciferase
reporter assay was performed to investigate whether FGFR2 was
directly targeted by miR-217-5p. Transfection of SkMSCs with
miR-217-5p mimics significantly decreased the luciferase activity
of the wild-type, but not the mutant, 3′-UTR of FGFR2 compared to
that of SkMSCs transfected with miR-NC (Fig. 4B). By contrast, the miR-217-5p
antagomir significantly increased the luciferase activity of
wild-type FGFR2 in SkMSCs compared with that of SkMSCs transfected
with miR-NC and miR-217-5p mimics (Fig. 4B). In addition, the results also
demonstrated that miR-217-5p mimics significantly increased and
miR-217-5p antagomir significantly decreased the mRNA and protein
expression levels of FGFR2 in SkMSCs compared with those of miR-NC
transfected SkMSCs (Fig. 4C-E).
These findings suggested that FGFR2 was a direct target of
miR-217-5p in SkMSCs.
miR-217-5p regulates the proliferation
and differentiation of SkMSCs by targeting FGFR2
To verify the mechanism of miR-217-5p in myogenesis,
SkMSCs were transfected with miR-217-5p mimics or miR-NC. RT-qPCR
results demonstrated that FGFR2 mRNA expression was increased in
SkMSCs transfected with miR-217-5p mimics compared with that in
SkMSCs transfected with miR-NC (Fig.
5A). In addition, RT-qPCR and western blotting results
demonstrated that a selective FGFR inhibitor (AZD4547) suppressed
the mRNA and protein expression of FGFR2 compared with that in the
miR-NC group, whereas miR-217-5p overexpression (miR-217-5p mimics
+ AZD4547) reduced this suppression (Fig. 5A-C). Additionally, MTT assay
demonstrated that the proliferation of SkMSCs was suppressed by
AZD4547 compared with miR-NC, and that miR-217-5p mimics + AZD4547
reduced the reversal (Fig. 5D).
Furthermore, to study the role of miR-192-5p in differentiation of
SkMSCs, the expression of MYHC and MyoD was examined. Western
blotting analysis demonstrated that the protein expression levels
of MYHC and MyoD was suppressed by miR-217-5p mimics but enhanced
by AZD4547 compared with those in the miR-NC group (Fig. 5E and F). These results suggested
that miR-217-5p may regulate the myogenesis of SkMSCs by targeting
FGFR2.
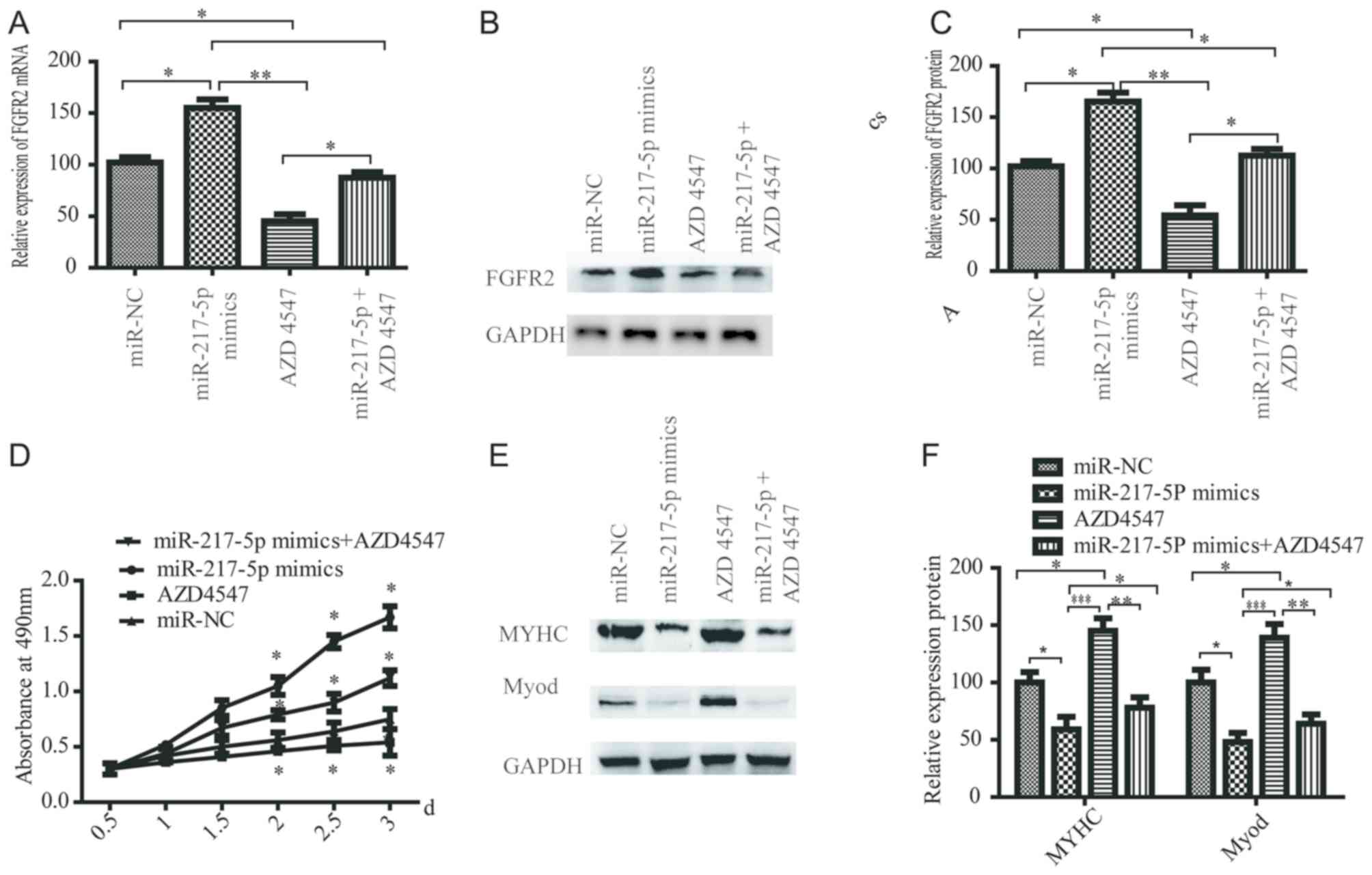 | Figure 5.miR-217-5p regulates the
proliferation and differentiation of SkMSCs by targeting FGFR2.
(A-C) SkMSCs were transfected with miR-192-5p antagomir, miR-NC,
miR-217-5p mimics or miR-217-5P mimics + AZD4547 and the (A) mRNA
and (B and C) protein expression levels of FGFR2 were examined. The
level of FGFR2 was normalized to that of GAPDH. (D) MTT assay
results suggested that compared with that of the miR-NC group the
proliferation of SkMSCs was suppressed by AZD4547, and miR-217-5p
reduced this effect. (E and F) The protein expression levels of
MYHC and MyoD was suppressed by miR-217-5p mimics but enhanced by
AZD4547 compared with the miR-NC group. The level of MYHC and MyoD
was normalized to that of GAPDH. *P<0.05, **P<0.01,
***P<0.001. miR, microRNA; NC, control group; SkMSCs, skeletal
muscle stem cells; FGFR2, fibroblast growth factor receptor 2;
MyoD, myogenic differentiation markers; MyHC, myosin heavy
chain. |
Discussion
The regenerative capacity of adult skeletal muscle
is attributed to SkMSCs. Which have the ability to proliferate,
differentiate and self-renew (1,7).
SkMSCs are involved in muscle formation and regeneration in
response to acute or chronic injury (7,28).
The results of the present study demonstrated that the expression
levels of miR-217-5p were increased in SkMSC culture medium and
that miR-217-5p mimics promoted the proliferation and suppressed
the differentiation of SkMSCs. In addition, miR-217-5p may have the
potential to facilitate the proliferation of SkMSCs possibly by
targeting FGFR2.
A miRNA is a type of small noncoding RNA 20–30
nucleotides in length that regulates gene expression through the
inhibition of translation or promotion of the degradation of target
mRNA by binding to its 3′-UTR (17,19,29–32).
Recent studies have reported that the prerequisite for the myogenic
differentiation of quiescent SkMSCs is the activation of myogenic
markers and MyoD expression (7,33–35).
Previous studies have reported that miRNAs are involved in
regeneration and differentiation of SkMSCs (36–39).
Increasing evidence indicates that several miRNAs promote or
inhibit stem cell progression (19). miR-217-5p has been implicated in
the apoptosis of colorectal cancer cells by directly targeting
protein kinase c iota type I (PRKCI), BAG family molecular
chaperone regulator 3 (BAG3), integrin subunit alpha v (ITGAV) and
mitogen-activated protein kinase 1 (MAPK1) (19). Further studies have revealed that
miR-217-5p regulates pluripotent stem cell proliferation and
differentiation by LPS (17–19).
However, the role of miR-217-5p in SkMSCs is still unclear. The
present study explored the effects of miR217-5p on the
proliferation of SkMSCs. The results of the present study
demonstrated that the expression levels of miR-217-5p were
increased in SkMSC culture medium and promoted SkMSCs proliferation
compared with that of the miR-NC group.
FGFR2 is associated with breast, lung and clear cell
renal cell carcinomas (40–42).
In addition, FGFR2 has recently been identified as a therapeutic
target for carcinoma owing to its association with tumorigenesis
(43–45). Studies have reported that FGFR2
promotes the proliferation of stem cells (46,47)
and that a novel circular RNA of FGFR2 may serve a role in
enhancing skeletal muscle proliferation and differentiation by
targeting miR-133a-5p and miR-29b-1-5p (6). A recent study has revealed that
miR-142-3p suppresses the induction of the FGFR2-driven oncogenic
process by directly binding transient receptor potential ankyrin-1
(TRPA1) (20).
The results of the present study suggested that
miR-217-5p may directly target FGFR2 and enhance the expression of
FGFR2, indicating a positive regulatory effect of miR-217-5p on
this target gene. Although miRNAs have a predominantly negative
effect on the expression of the protein encoded by the target gene,
several reports have demonstrated a positive effect of miRNAs
(48–51). Similarly, the results of the
present study suggested that miR-217-5p promoted the target gene
expression.
In the present study, TargetScan and miRNA databases
were used to predict the downstream targets of miR-217-5p and
further explore the molecular mechanism underlying the
proliferation and differentiation of SkMSCs. SkMSCs have
self-renewal properties and can regenerate muscle (28). However, a previous study has
suggested that the SkMSC population contains undifferentiated cells
that can differentiate into several other types of mesenchymal
cells, such as adipocytes, chondrocytes and osteocytes (52). SkMSCs can be activated during
muscle repair; however, they also have the potential to
differentiate into other phenotypes. The results of the present
study demonstrated that miR-217-5p mimics induced the upregulation
of FGFR2, promoting myogenesis of SkMSCs.
There were several limitations to the present study.
First, only one cell type was used in the present study, and
additional cell types may be required. Second, although the present
study indicated that miR-217-5p levels were increased in SkMSC
culture medium, which promoted SkMSCs proliferation by targeting
FGFR2, the underlying mechanism remained unclear. Third, the
results of this study were not validated in vivo,
necessitating further exploration of the role of miR-217-5p in
SkMSCs. Further study to analyze the mechanism of skeletal muscle
regeneration is necessary.
In summary, the results of the present study
suggested that miR-217-5p maintains an appropriate proliferation
rate and suppresses differentiation into a non-muscle cell
phenotype, thus regulating the myogenesis of SkMSCs by targeting
FGFR2, which may reflect a promising therapeutic strategy for the
treatment of muscle injuries.
Acknowledgements
Not applicable.
Funding
This study was supported by The National Natural
Science Foundation Item (grant nos. 81871787 and 81601057) and The
Natural Science Foundation of Guangdong Province (grant nos.
2018A030310254 and 2015A030310350).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
MZ and YY carried out experiments, data analysis and
wrote the manuscript. BQ and GC performed experiments and helped
with data quantifications. LG and JY designed the project and
supervised the experiment. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Animal care was approved by the Institutional Animal
Care and Use Committee at The First Affiliated Hospital of Sun
Yat-sen University (approval no. SYSU-IACUC-2020-000052).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
MyHC
|
myosin heavy chain
|
|
MyoD
|
myogenic differentiation markers
|
|
Pax7
|
paired box protein7
|
|
SkMSCs
|
skeletal muscle stem cells
|
|
TGFβ1
|
transforming growth factor-β1
|
|
EdU
|
5-Ethynyl-2′-deoxyuridine
|
|
FACS
|
fluorescence-activated cell
sorting
|
|
UTR
|
untranslated region
|
References
|
1
|
De Micheli AJ, Laurilliard EJ, Heinke CL,
Ravichandran H, Fraczek P, Soueid-Baumgarten S, De Vlaminck I,
Elemento O and Cosgrove BD: Single-cell analysis of the muscle stem
cell hierarchy identifies heterotypic communication signals
involved in skeletal muscle regeneration. Cell reports.
30:3583–3595.e5. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sacco A, Doyonnas R, Kraft P, Vitorovic S
and Blau HM: Self-renewal and expansion of single transplanted
muscle stem cells. Nature. 456:502–506. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sheehan SM and Allen RE: Skeletal muscle
satellite cell proliferation in response to members of the
fibroblast growth factor family and hepatocyte growth factor. J
Cell Physiol. 181:499–506. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cerletti M, Jurga S, Witczak CA, Hirshman
MF, Shadrach JL, Goodyear LJ and Wagers AJ: Highly efficient,
functional engraftment of skeletal muscle stem cells in dystrophic
muscles. Cell. 134:37–47. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Madaro L, Torcinaro A, De Bardi M, Contino
FF, Pelizzola M, Diaferia GR, Imeneo G, Bouchè M, Puri PL and De
Santa F: Macrophages fine tune satellite cell fate in dystrophic
skeletal muscle of mdx mice. PLoS Genet. 15:e10084082019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Chen X, Ouyang H, Wang Z, Chen B and Nie
Q: A novel circular RNA generated by FGFR2 gene promotes myoblast
proliferation and differentiation by sponging miR-133a-5p and
miR-29b-1-5p. Cells. 7:E1992018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Fukuda S, Kaneshige A, Kaji T, Noguchi YT,
Takemoto Y, Zhang L, Tsujikawa K, Kokubo H, Uezumi A, Maehara K, et
al: Sustained expression of HeyL is critical for the proliferation
of muscle stem cells in overloaded muscle. Elife. 8:e482842019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Tanaka KK, Hall JK, Troy AA, Cornelison
DD, Majka SM and Olwin BB: Syndecan-4-expressing muscle progenitor
cells in the SP engraft as satellite cells during muscle
regeneration. Cell Stem Cell. 4:217–225. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kumar P, Ciftci S, Barthes J,
Knopf-Marques H, Muller CB, Debry C, Vrana NE and Ghaemmaghami AM:
A composite Gelatin/hyaluronic acid hydrogel as an ECM mimic for
developing mesenchymal stem cell derived epithelial tissue patches.
J Tissue Eng Regen Med. 14:45–57. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Cao X, Tang S, Du F, Li H, Shen X, Li D,
Wang Y, Zhang Z, Xia L, Zhu Q and Yin H: miR-99a-5p regulates the
proliferation and differentiation of skeletal muscle satellite
cells by targeting MTMR3 in chicken. Genes (Basel). 11:E3692020.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yin H, He H, Shen X, Zhao J, Cao X, Han S,
Cui C, Chen Y, Wei Y, Xia L, et al: miR-9-5p inhibits skeletal
muscle satellite cell proliferation and differentiation by
targeting IGF2BP3 through the IGF2-PI3K/Akt signaling pathway. Int
J Mol Sci. 21:E16552020. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu L, Wang H, Liao Y, Zhou P, Xu Y, Zhao
Y, Xie S, Zhao S and Li X: miR-208b modulating skeletal muscle
development and energy homoeostasis through targeting distinct
targets. RNA Biol. 17:743–754. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Guan X, Gao Y, Zhou J, Wang J, Zheng F,
Guo F, Chang A, Li X and Wang B: miR-223 regulates adipogenic and
osteogenic differentiation of mesenchymal stem cells through a
C/EBPs/miR-223/FGFR2 regulatory feedback loop. Stem Cells.
33:1589–1600. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jedari B, Rahmani A, Naderi M and Nadri S:
MicroRNA-7 promotes neural differentiation of trabecular meshwork
mesenchymal stem cell on nanofibrous scaffold. J Cell Biochem.
121:2818–2827. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Phelps M, Stuelsatz P and Yablonka-Reuveni
Z: Expression profile and overexpression outcome indicate a role
for βKlotho in skeletal muscle fibro/adipogenesis. FEBS J.
283:1653–1668. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Xiao Y, Guo Q, Jiang TJ, Yuan Y, Yang L,
Wang GW and Xiao WF: miR4833p regulates osteogenic differentiation
of bone marrow mesenchymal stem cells by targeting STAT1. Mol Med
Rep. 20:4558–4566. 2019.PubMed/NCBI
|
|
17
|
Zeng ZL, Lin XL, Tan LL, Liu YM, Qu K and
Wang Z: MicroRNAs: Important regulators of induced pluripotent stem
cell generation and differentiation. Stem Cell Rev Rep. 14:71–81.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang X, Wang Z, Li W, Huang R, Zheng D
and Bi G: MicroRNA-217-5p ameliorates endothelial cell apoptosis
induced by ox-LDL by targeting CLIC4. Nutr Metab Cardiovasc Dis.
30:523–533. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Flum M, Kleemann M, Schneider H, Weis B,
Fischer S, Handrick R and Otte K: miR-217-5p induces apoptosis by
directly targeting PRKCI, BAG3, ITGAV and MAPK1 in colorectal
cancer cells. J Cell Commun Signal. 12:451–466. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Berrout J, Kyriakopoulou E, Moparthi L,
Hogea AS, Berrout L, Ivan C, Lorger M, Boyle J, Peers C, Muench S,
et al: TRPA1-FGFR2 binding event is a regulatory oncogenic driver
modulated by miRNA-142-3p. Nat Commun. 8:9472017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Li X, Nie C, Tian B, Tan X, Han W, Wang J,
Jin Y, Li Y, Guan X, Hong A and Chen X: miR-671-5p blocks the
progression of human esophageal squamous cell carcinoma by
suppressing FGFR2. Int J Biol Sci. 15:1892–1904. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Nakano S, Nakamura K, Teramoto N,
Yamanouchi K and Nishihara M: Basic fibroblast growth factor is
pro-adipogenic in rat skeletal muscle progenitor clone, 2G11 cells.
Anim Sci J. 87:99–108. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Du YE, Tu G, Yang G, Li G, Yang D, Lang L,
Xi L, Sun K, Chen Y, Shu K, et al: MiR-205/YAP1 in activated
fibroblasts of breast tumor promotes VEGF-independent angiogenesis
through STAT3 signaling. Theranostics. 7:3972–3988. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kwiatkowski BA, Kirillova I, Richard RE,
Israeli D and Yablonka-Reuveni Z: FGFR4 and its novel splice form
in myogenic cells: Interplay of glycosylation and tyrosine
phosphorylation. J Cell Physiol. 215:803–817. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhu M, Liu C, Li S, Zhang S, Yao Q and
Song Q: Sclerostin induced tumor growth, bone metastasis and
osteolysis in breast cancer. Sci Rep. 7:113992017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zou P, Zhu M, Lian C, Wang J, Chen Z,
Zhang X, Yang Y, Chen X, Cui X, Liu J, et al: miR-192-5p suppresses
the progression of lung cancer bone metastasis by targeting TRIM44.
Sci Rep. 9:196192019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Schultz SS and Lucas PA: Human stem cells
isolated from adult skeletal muscle differentiate into neural
phenotypes. J Neurosci Methods. 152:144–155. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chu Q, Sun Y, Bi D, Cui J and Xu T:
Up-regulated of miR-8159-5p and miR-217-5p by LPS stimulation
negatively co-regulate TLR1 in miiuy croaker. Dev Comp Immunol.
67:117–125. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Erdos Z, Barnum JE, Wang E, DeMaula C, Dey
PM, Forest T, Bailey WJ and Glaab WE: Evaluation of the relative
performance of pancreas specific microRNAs in rat plasma as
biomarkers of pancreas injury. Toxicol Sci. 173:5–18. 2019.
View Article : Google Scholar
|
|
31
|
Du W, Tang H, Lei Z, Zhu J, Zeng Y, Liu Z
and Huang JA: miR-335-5p inhibits TGF-β1-induced
epithelial-mesenchymal transition in non-small cell lung cancer via
ROCK1. Respir Res. 20:2252019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Choi YJ, Kim H, Kim JW, Song CW, Kim DS,
Yoon S and Park HJ: Phthalazinone pyrazole enhances the hepatic
functions of human embryonic stem cell-derived hepatocyte-like
cells via suppression of the epithelial-mesenchymal transition.
Stem Cell Rev Rep. 14:438–450. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Schmidt M, Schüler SC, Hüttner SS, von
Eyss B and von Maltzahn J: Adult stem cells at work: Regenerating
skeletal muscle. Cell Mol Life Sci. 76:2559–2570. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Dumont NA, Bentzinger CF, Sincennes MC and
Rudnicki MA: Satellite cells and skeletal muscle regeneration.
Compr Physiol. 5:1027–1059. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Feige P, Brun CE, Ritso M and Rudnicki MA:
Orienting muscle stem cells for regeneration in homeostasis, aging,
and disease. Cell Stem Cell. 23:653–664. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Sato T, Yamamoto T and Sehara-Fujisawa A:
miR-195/497 induce postnatal quiescence of skeletal muscle stem
cells. Nat Commun. 5:45972014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tirone M, Giovenzana A, Vallone A, Zordan
P, Sormani M, Nicolosi PA, Meneveri R, Gigliotti CR, Spinelli AE,
Bocciardi R, et al: Severe heterotopic ossification in the skeletal
muscle and endothelial cells recruitment to chondrogenesis are
enhanced by monocyte/macrophage depletion. Front Immunol.
10:16402019. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao Y, Chen M, Lian D, Li Y, Li Y, Wang
J, Deng S, Yu K and Lian Z: Non-coding RNA regulates the myogenesis
of skeletal muscle satellite cells, injury repair and diseases.
Cells. 8:E9882019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Kletukhina S, Neustroeva O, James V,
Rizvanov A and Gomzikova M: Role of mesenchymal stem cell-derived
extracellular vesicles in epithelial-mesenchymal transition. Int J
Mol Sci. 20:E48132019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sadej R, Lu X, Turczyk L, Novitskaya V,
Lopez-Clavijo AF, Kordek R, Potemski P, Wakelam MJO,
Romanska-Knight H and Berditchevski F: CD151 regulates expression
of FGFR2 in breast cancer cells via PKC-dependent pathways. J Cell
Sci. 131:jcs2206402018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Li L, Zhang S, Wei L, Wang Z, Ma W, Liu F
and Qian Y: FGF2 and FGFR2 in patients with idiopathic pulmonary
fibrosis and lung cancer. Oncol Lett. 16:2490–2494. 2018.PubMed/NCBI
|
|
42
|
Vanmechelen M, Lambrechts D, Van Brussel
T, Verbiest A, Couchy G, Schöffski P, Dumez H, Debruyne PR, Lerut
E, Machiels JP, et al: Fibroblast growth factor receptor-2
polymorphism rs2981582 is correlated with progression-free survival
and overall survival in patients with metastatic clear-cell renal
cell carcinoma treated with sunitinib. Clin Genitourin Cancer.
17:e235–e246. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Maehara O, Suda G, Natsuizaka M, Ohnishi
S, Komatsu Y, Sato F, Nakai M, Sho T, Morikawa K, Ogawa K, et al:
Fibroblast growth factor-2-mediated FGFR/Erk signaling supports
maintenance of cancer stem-like cells in esophageal squamous cell
carcinoma. Carcinogenesis. 38:1073–1083. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wang D, Yang L, Yu W and Zhang Y:
Investigational fibroblast growth factor receptor 2 antagonists in
early phase clinical trials to treat solid tumors. Expert Opin
Investig Drugs. 28:903–916. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Wang Y and Qin H: miR-338-3p targets RAB23
and suppresses tumorigenicity of prostate cancer cells. Am J Cancer
Res. 8:2564–2574. 2018.PubMed/NCBI
|
|
46
|
Shanmuganathan S and Angayarkanni N:
Chebulagic acid and Chebulinic acid inhibit TGF-β1 induced fibrotic
changes in the chorio-retinal endothelial cells by inhibiting ERK
phosphorylation. Microvasc Res. 121:14–23. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xu Y, Xiao H, Luo H, Chen Y, Zhang Y, Tao
L, Jiang Y, Chen Y and Shen X: Inhibitory effects of oxymatrine on
TGF-β1-induced proliferation and abnormal differentiation in rat
cardiac fibroblasts via the p38MAPK and ERK1/2 signaling pathways.
Mol Med Rep. 16:5354–5362. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Huang V, Place RF, Portnoy V, Wang J, Qi
Z, Jia Z, Yu A, Shuman M, Yu J and Li LC: Upregulation of Cyclin B1
by miRNA and its implications in cancer. Nucleic Acids Res.
40:1695–1707. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Vasudevan S: Posttranscriptional
upregulation by microRNAs. Wiley Interdiscip Rev RNA. 3:311–330.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Lu H, Buchan RJ and Cook SA: MicroRNA-223
regulates Glut4 expression and cardiomyocyte glucose metabolism.
Cardiovasc Res. 86:410–420. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Saifi M, Yogindran S, Nasrullah N, Nissar
U, Gul I and Abdin MZ: Co-expression of anti-miR319g and miRStv_11
lead to enhanced steviol glycosides content in Stevia rebaudiana.
BMC Plant Biol. 19:2742019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Alessandri G, Pagano S, Bez A, Benetti A,
Pozzi S, Iannolo G, Baronio M, Invernici G, Caruso A, Muneretto C,
et al: Isolation and culture of human muscle-derived stem cells
able to differentiate into myogenic and neurogenic cell lineages.
Lancet. 364:1872–1883. 2004. View Article : Google Scholar : PubMed/NCBI
|