Introduction
Dilated cardiomyopathy (DCM), a primary
cardiomyopathy with significant enlargement of the ventricle
(1), is one of the most
intractable diseases in the cardiovascular field (2). DCM can progressively develop into
severe congestive heart failure and seriously threatens the
survival rate of the patient (3).
However, due to its unknown etiology and underlying mechanisms, DCM
cannot be efficiently treated by existing therapeutic strategies,
except for heart transplantation (4). Thus, identifying the specific
molecular mechanism of DCM is important to facilitate its diagnosis
and treatment, and could improve the prognosis of patients with
DCM.
With the development of databases such as The Cancer
Genome Atlas and the Gene Expression Omnibus (GEO), genetic
research on DCM has become feasible and is currently ongoing
(5). Huang et al (6) analyzed 102 samples from the GEO
database (GSE5406), and identified module and hub genes of
differentially expressed genes (DEGs) that are related to the
progression of DCM. In addition, Xiao et al (7) used RNA-Seq data (GSE116250) and gene
annotation of the Ensembl database to find the key module involved
in DCM, and identified five co-expression modules that may have
important functions in DCM occurrence. Thus, these previous studies
provide possible research directions for elucidating the
pathogenesis of DCM.
However, due to limited sample quantities,
insufficient analysis methods and the lack of experimental
verification, a number of previous DEG results for DCM may not be
accurate (8). Therefore, to define
the related genes in DCM, the present study performed an integrated
bioinformatics analysis on four DCM microarrays from GEO, and a
protein-protein interaction (PPI) network was established to
investigate the interactions among DEGs. Then, the present study
examined these hub genes in vivo using reverse
transcription-quantitative PCR (RT-qPCR) in a mouse DCM model,
which was established by intraperitoneal injection of doxorubicin
(DOX).
Materials and methods
Raw transcriptional data acquisition
and preprocessing
‘Dilated cardiomyopathy’ and ‘human’ were used as
key words to search the GEO database (http://www.ncbi.nlm.nih.gov/geo/), and raw data (CEL
files) from five datasets (GSE17800, GSE21610, GSE42955, GSE79962
and GSE3585), consisting of myocardium transcriptional expression
data from 89 patients with DCM and 37 healthy controls were
downloaded (details in Table I)
for further analysis. Raw data were first normalized using the RMA
function in the R package affy (v1.64.0) (9). Probes in GSE17800, GSE21610 and
GSE79962 were then screened by the MAS5CALLS algorithm packaged in
affy, and probes detected as ‘absent’ across all samples were
filtered out. The R packages hgu133plus2.db (v3.2.3) (10), hugene10sttranscriptcluster.db
(v8.7.0) (11) and hgu133a.db
(v3.2.3) (12) were used to
annotate probes with gene symbols according to the microarray
platform.
 | Table I.Details of the microarrays used. |
Table I.
Details of the microarrays used.
| GEO series
number | Control | DCM | Tissue | Platform |
|---|
| GSE17800 | 8 | 40 | Myocardium |
GPL570,[HG-U133_Plus_2] Affymetrix Human
Genome U133 Plus 2.0 Array |
| GSE21610 | 8 | 21 | Myocardium |
GPL570,[HG-U133_Plus_2] Affymetrix Human
Genome U133 Plus 2.0 Array |
| GSE42955 | 5 | 12 | Myocardium |
GPL6244,[HuGene-1_0-st] Affymetrix Human
Gene 1.0 ST Array [transcript (gene) version] |
| GSE79962 | 11 | 9 | Myocardium |
GPL6244,[HuGene-1_0-st] Affymetrix Human
Gene 1.0 ST Array [transcript (gene) version] |
| GSE3585 | 5 | 7 | Myocardium | GPL96,[HG-U133A]
Affymetrix Human Genome U133A Array |
Meta-analysis of DEGs
The expression matrixes of GSE17800, GSE21610,
GSE42955 and GSE79962 were merged by common gene symbols. MetaMA (v
3.1.2) (13) is an R package for
microarray DEG meta-analyses that uses moderated effect size and
P-value combinations, while RankProd (v3.12.0) (14) uses a non-parametric rank product
method. The merged expression matrix was processed using metaMA and
RankProd according to the reference manuals, and overlapping genes
from the two methods were deemed as DEGs.
Functional annotation of the DEGs
Lists of upregulated DEGs and downregulated DEGs
were uploaded to the Database for Annotation, Visualization and
Integrated Discovery (DAVID; v6.8) (15,16)
website for Gene Ontology (GO, v2019-07) (17) and Kyoto Encyclopedia of Genes and
Genomes (KEGG, v91.0) (18)
pathway enrichment analysis. The results were downloaded from the
website and then visualized using the R package ggplot2 (v3.2.0)
(19).
PPI network construction
The DEG list was uploaded to the Search Tool for the
Retrieval of Interacting Genes/Proteins (STRING; https://string-db.org/cgi/input.pl; v11.0)
(20) database, and then a PPI
network was established with the minimum required interaction score
set as the highest confidence (>0.9). Cytoscape (https://cytoscape.org, v3.7.1) was used to visualize
the PPI network, and the MCODE (v1.6) (21) plug-in was used to screen important
modules of the network.
Identification of hub genes
CytoHubba (v0.1) (22), a plug-in of Cytoscape software, was
used to identify hub genes of the PPI network via four different
algorithms. The algorithms used for analysis included degree,
Maximum Neighborhood Component (MNC), Density of Maximum
Neighborhood Component (DMNC) and Maximal Clique Centrality (MCC).
A Venn diagram was constructed and consisted of genes ranked in the
top 20 of each method, and genes overlapping in the four groups
were deemed hub genes.
DCM animal model establishment
All animal studies were approved by the Animal Care
and Ethics Committee of Nanjing Medical University. The animal
experiments were performed according to the Guide for the Care and
Use of Laboratory Animals (23).
The DCM mouse model was established by intraperitoneal injection of
DOX as previously described (24,25).
A total of 12 male C57BL/6 mice (age, 8 weeks; weight, 22–24 g)
were purchased from Shanghai SLAC Laboratory Animal Co., Ltd. All
mice were housed with sterile rodent chow and water available ad
libitum in the animal holding room maintained at 26°C and 50%
relative humidity, under a 12-h light/dark cycle. Mice were
randomly divided into two groups: Control (n=6) and DOX (n=6)
groups. Mice in the DOX group were intraperitoneally injected with
DOX hydrochloride (cat. no. HY-15142; MedChemExpress; 4 mg/kg) once
a week for 5 consecutive weeks. The control group received an equal
volume (0.1 ml) of normal saline (0.9%). Mice that presented with
progressive cardiac functional decline and ventricular dilation
were successfully established as the DCM mouse model.
Transthoracic echocardiography
Then, 7 days after last injection, mice were sedated
with 5% isoflurane-O2 (cat. no. 792632; Sigma-Aldrich;
Merck KGaA) balanced mixture (maintained at 1.5%). Transthoracic
echocardiography (Vevo 1100; VisualSonics, Inc.) was performed in
M-mode using a 30-MHz transducer immediately after anesthetization.
The left ventricular (LV) echocardiogram was assessed in both
parasternal long-axis and short-axis views. End-systolic and
end-diastolic dimensions were defined as the phases corresponding
to the electrocardiogram T wave and to the R wave, respectively
(26). M-mode LV internal diameter
end systole/diastole (LVIDs/d), LV posterior wall end diastole
(LVPWd) and interventricular septal end diastole (IVSd) were
averaged from 3–5 beats. Ejection fraction and fractional
shortening were calculated as previously described (26).
RNA extraction and RT-qPCR
After echocardiography detection all mice were
anaesthetized by breathing a 5% isoflurane-O2 balanced
mixture (maintained at 1.5%). Mice were confirmed to be deeply
anesthetized after they were immobile for 1 min. Then, mice were
euthanatized by inhalation of 25% CO2, until respiratory
and cardiac arrest occurred. Hearts were isolated and ground into 1
mm3 pieces, and then dissolved in pure TRI
Reagent® (cat. no. 93289; Sigma-Aldrich; Merck KGaA).
RNA was extracted using the RNAprep Pure Tissue kit (cat. no.
DP431; Tiangen Biotech Co., Ltd.) according to the manufacturer's
instructions. RT was performed using the PrimeScript RT reagent kit
with gDNA Eraser (cat. no. RR047A; Takara Bio, Inc.) according to
the manufacturer's instructions. gDNA was removed at 42°C for 2
min, then cDNA was synthesized at 37°C for 15 min and 85°C for 5
sec. qPCR was conducted using the CellAmp Direct TB
Green® RT-qPCR kit (cat. no. 3735A; Takara Bio, Inc.)
and an ABI ViiA 7 system (cat. no. 4453536; Thermo Fisher
Scientific, Inc.). The following thermocycling conditions were
used: Melting at 95°C for 30 sec, annealing at 95°C for 3 sec, and
extension at 60°C for 30 sec for 40 cycles. The primers (Table II) for β-actin, proenkephalin
(PENK), chordin like 1 (CHRDL1), calumenin (CALU), insulin-like
growth factor binding protein 3 (IGFBP3) and ceruloplasmin (CP)
were synthesized by Generay Biotech Co., Ltd. All samples were
detected in triplicate, and gene expression values were normalized
to the values of β-actin using 2−ΔΔCq method (27).
 | Table II.Primer sequences. |
Table II.
Primer sequences.
| Gene | Forward
(5′→3′) | Reverse
(5′→3′) |
|---|
| PENK |
GGACTGCGCTAAATGCAGCTA |
GAAGCCTCCGTACCGTTTCAT |
| CHRDL1 |
AACCTCCAAGCCAAAACTTTGA |
CCAGTGCTACTTTTCTGGTTGTC |
| CALU |
AATGCTGATGGGTTCATTGATCT |
GGTTCTTATCTCGAAACTCCACG |
| IGFBP3 |
CACACCGAGTGACCGATTCC |
GTGTCTGTGCTTTGAGACTCAT |
| CP |
CTTAGCCTTGGCAAGAGATAAGC |
GATGACAGGGCCTAAAAACCC |
| Actb |
GGCTGTATTCCCCTCCATCG |
CCAGTTGGTAACAATGCCATGT |
Statistical analysis
SPSS software (v16.0, SPSS, Inc.) was used for
statistical analysis, and GraphPad Prism software (v5, GraphPad
Software, Inc.) was used for mapping. Data are presented as the
mean ± SD. For two-group comparisons, an independent sample t-test
was used. P<0.05 was considered to indicate a statistically
significant difference.
Results
Identification of DEGs between
patients with DCM and healthy controls
The raw data of GSE17800, GSE21610, GSE42955 and
GSE79962, consisting of myocardium transcriptional expression data
from 82 patients with DCM and 32 healthy controls, were downloaded
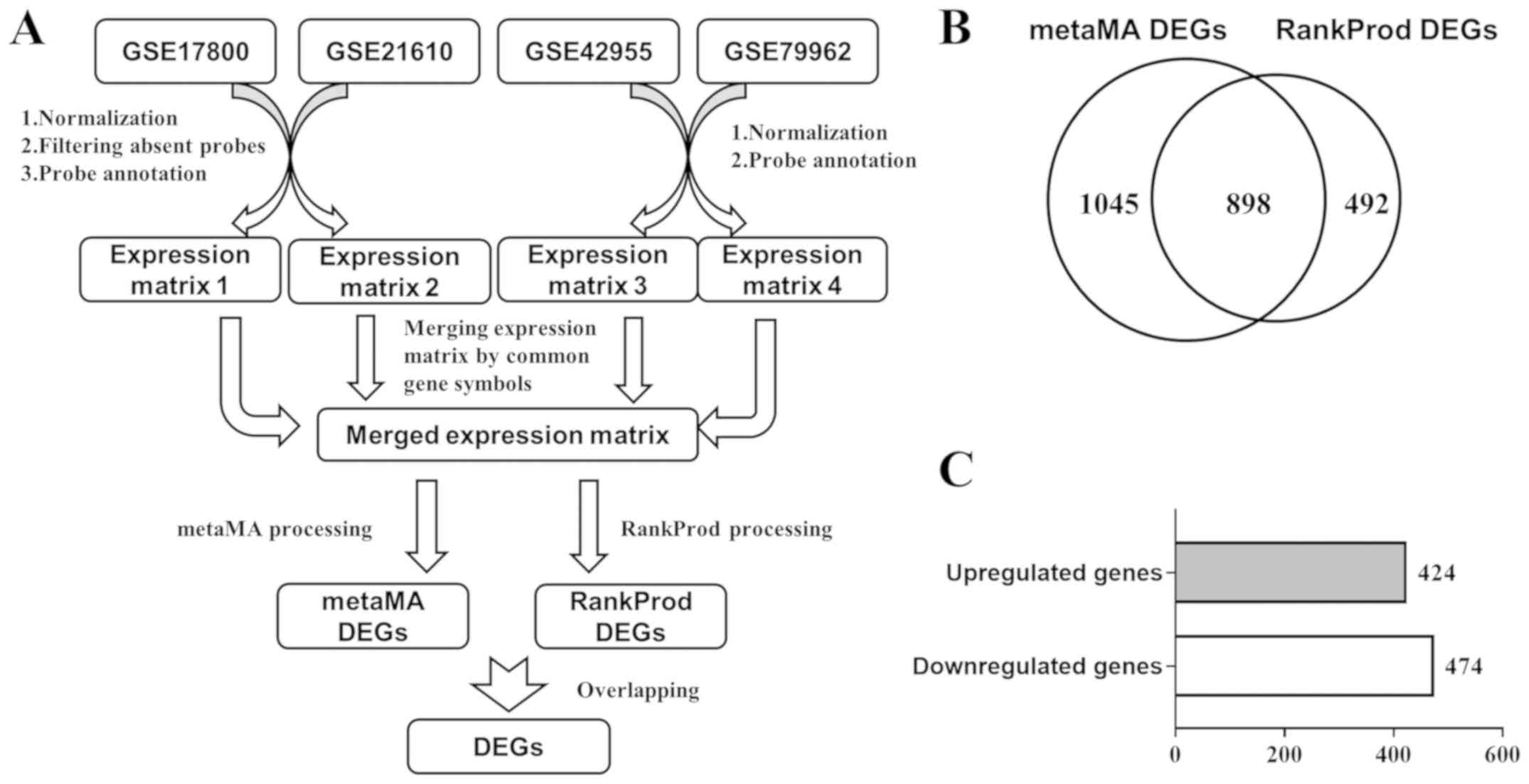
from the GEO database. After preprocessing (Fig. 1A), the merged expression matrix of
these four datasets was used for downstream analysis. The merged
matrix was inputted into metaMA and RankProd, which are two R
packages designed for meta-analysis of DEGs. A total of 1,943 genes
(metaMA DEGs) were found to be differentially expressed by the
metaMA package, while 1,390 genes (RankProd DEGs) were found to be
differentially expressed by the RankProd package. A total of 898
overlapping genes between the metaMA DEGs and RankProd DEGs were
considered DEGs (Fig. 1B), among
which 424 genes were upregulated and 474 genes were downregulated
(Fig. 1C). The top 30 upregulated
genes and downregulated genes are listed in Tables III and IV, respectively. The most significantly
upregulated genes were natriuretic peptide A (NPPA) and NPPB, while
the most significantly downregulated gene was myosin heavy chain 6
(MYH6). In addition, the upregulation of NPPA and NPPB, and
downregulation of MYH6 are well-established markers of heart
dysfunction (28).
 | Table III.Top 30 upregulated differentially
expressed genes. |
Table III.
Top 30 upregulated differentially
expressed genes.
| Upregulated
gene | Fold change | Percentage of false
prediction |
|---|
| NPPA | 5.57 |
6.41×10−49 |
| NPPB | 4.60 |
5.28×10−40 |
| SFRP4 | 2.46 |
2.61×10−25 |
| SMOC2 | 2.28 |
4.41×10−23 |
| HBB | 2.53 |
1.40×10−22 |
| EIF1AY | 2.45 |
1.82×10−22 |
| PHLDA1 | 1.99 |
4.61×10−22 |
| LTBP2 | 2.06 |
7.48×10−20 |
| THBS4 | 2.05 |
2.12×10−19 |
| FRZB | 2.07 |
5.42×10−19 |
| POSTN | 1.91 |
5.71×10−19 |
| MXRA5 | 1.82 |
1.01×10−18 |
| CCN2 | 1.96 |
6.47×10−18 |
| OMD | 2.00 |
6.13×10−18 |
| TNNT1 | 1.91 |
1.12×10−17 |
| PRELP | 2.06 |
1.34×10−17 |
| ASPN | 1.81 |
5.31×10−17 |
| FMOD | 1.94 |
7.19×10−17 |
| CCDC80 | 1.79 |
1.42×10−16 |
| RGS4 | 1.84 |
1.04×10−15 |
| SERPINE2 | 1.70 |
2.58×10−15 |
| USP9Y | 1.68 |
2.78×10−15 |
| STAT4 | 1.90 |
2.77×10−15 |
| PLCE1 | 1.75 |
2.67×10−15 |
| UCHL1 | 1.80 |
3.19×10−15 |
| OGN | 1.74 |
1.08×10−14 |
| HSPA2 | 1.72 |
2.10×10−14 |
| NAP1L3 | 1.77 |
3.88×10−14 |
| MFAP4 | 1.68 |
4.80×10−14 |
| JAK2 | 1.70 |
1.27×10−13 |
 | Table IV.Top 30 downregulated differentially
expressed genes. |
Table IV.
Top 30 downregulated differentially
expressed genes.
| Downregulated
gene | Fold change | Percentage of false
prediction |
| MYH6 | 0.32 |
2.33×10−38 |
| ETNPPL | 0.39 |
6.74×10−29 |
| SERPINA3 | 0.38 |
1.88×10−27 |
| DHRS7C | 0.45 |
1.51×10−25 |
| AQP4 | 0.52 |
1.29×10−20 |
| CCL2 | 0.47 |
1.89×10−17 |
| LYVE1 | 0.58 |
2.19×10−16 |
| LSAMP | 0.53 |
2.69×10−16 |
| DLK1 | 0.53 |
6.81×10−16 |
| CORIN | 0.53 |
1.04×10−15 |
| HOPX | 0.55 |
1.92×10−15 |
| CA14 | 0.55 |
4.94×10−15 |
| RARRES1 | 0.54 |
8.04×10−15 |
| CD163 | 0.62 |
9.97×10−15 |
| KCNIP2 | 0.60 |
1.01×10−14 |
| C1orf105 | 0.57 |
1.20×10−14 |
| CD14 | 0.56 |
3.45×10−14 |
| F13A1 | 0.61 |
1.53×10−13 |
| F5 | 0.82 |
2.01×10−13 |
| TOGARAM2 | 0.60 |
2.15×10−13 |
| CPNE4 | 0.59 |
2.05×10−12 |
| AQP3 | 0.62 |
4.89×10−12 |
| VSIG4 | 0.67 |
1.31×10−11 |
| HEY2 | 0.63 |
5.73×10−11 |
| PLIN2 | 0.62 |
6.50×10−11 |
| SELE | 0.57 |
1.46×10−10 |
| ADAMTS9 | 0.63 |
1.71×10−10 |
| NAMPT | 0.65 |
1.90×10−10 |
| SLCO5A1 | 0.60 |
2.63×10−10 |
| HINT3 | 0.60 |
9.40×10−10 |
Validation of DEGs
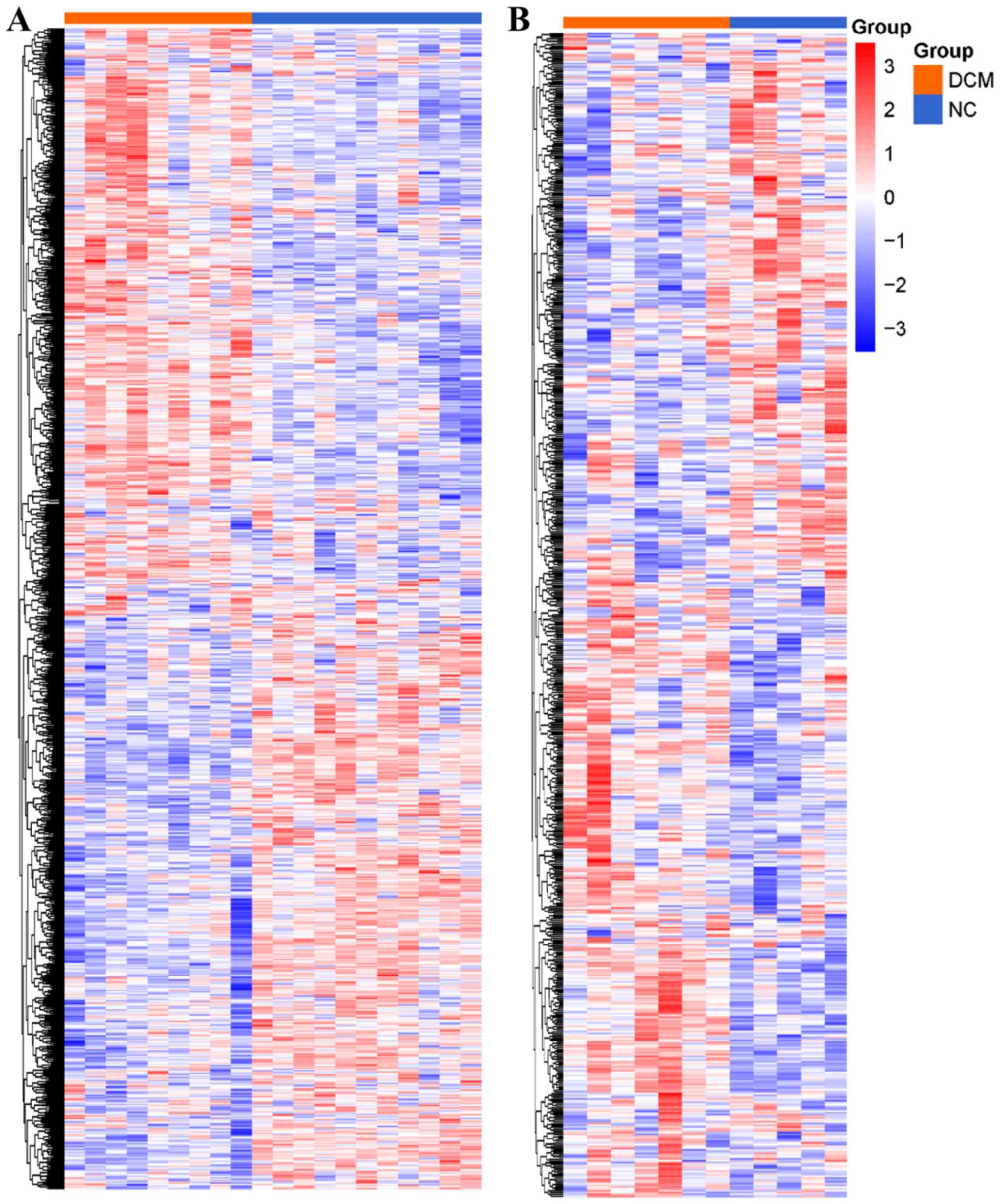
To assess the robustness of the DEGs, GSE79962 was
randomly selected for clustering heat map analysis with all 898
DEGs involved (Fig. 2A). For
further validation, a fifth microarray dataset (GSE3585) out of the
training data was downloaded. After normalization, filtering and
annotation, the expression matrix of GSE3585 (healthy controls=5;
patients with DCM=7) was extracted. In total, 659 genes of the 898
DEGs were found in the GSE3585 expression matrix, and these 659
genes were used to plot a clustering heat map (Fig. 2B). These two clustering heat maps
demonstrated that the DEGs were differentially expressed in the
myocardium from patients with DCM compared with the healthy
controls.
GO annotation of DEGs
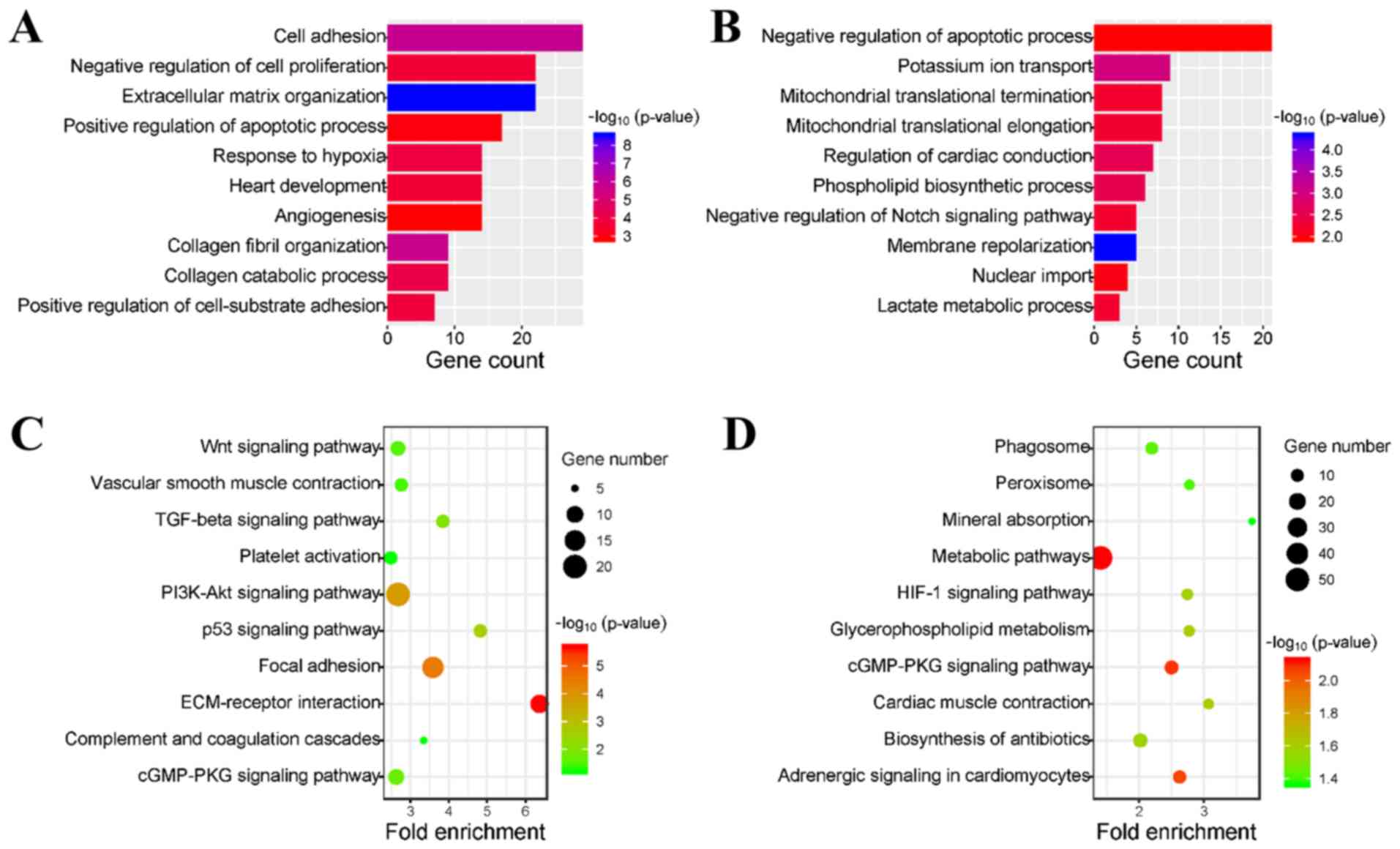
To identify an overview of the biological functions
of the DEGs, upregulated and downregulated DEGs were uploaded to
the DAVID database for GO enrichment analysis. GO analysis results
indicated that the upregulated genes were mainly involved in the
following biological processes: Cell adhesion, negative regulation
of cell proliferation, extracellular matrix (ECM) organization and
positive regulation of apoptotic process (Fig. 3A). In addition, downregulated genes
were primarily enriched in the negative regulation of the apoptotic
process, potassium ion transport, mitochondrial translational
termination and mitochondrial translational elongation (Fig. 3B). Furthermore, all these
biological processes are closely related to the occurrence and
development of DCM. In addition, it was revealed that both the
upregulated and downregulated genes were involved in the process of
apoptosis regulation, which indicates the importance of apoptosis
in DCM.
KEGG pathway enrichment of the
DEGs
The present study investigated the KEGG pathways
involved in the DEGs. It was demonstrated that upregulated DEGs
were mainly enriched in the ECM-receptor interaction, the p53
signaling pathway, focal adhesion and the transforming growth
factor-β signaling pathway (Fig.
3C). In addition, downregulated DEGs were primarily enriched in
cardiac muscle contraction, the hypoxia-inducible factor-1
signaling pathway, adrenergic signaling in cardiomyocytes and the
cGMP-protein kinase G signaling pathway (Fig. 3D). Therefore, the present results
indicated that these pathways may play vital roles in the
progression of DCM.
Construction of the PPI network, and
identification of important modules and hub genes
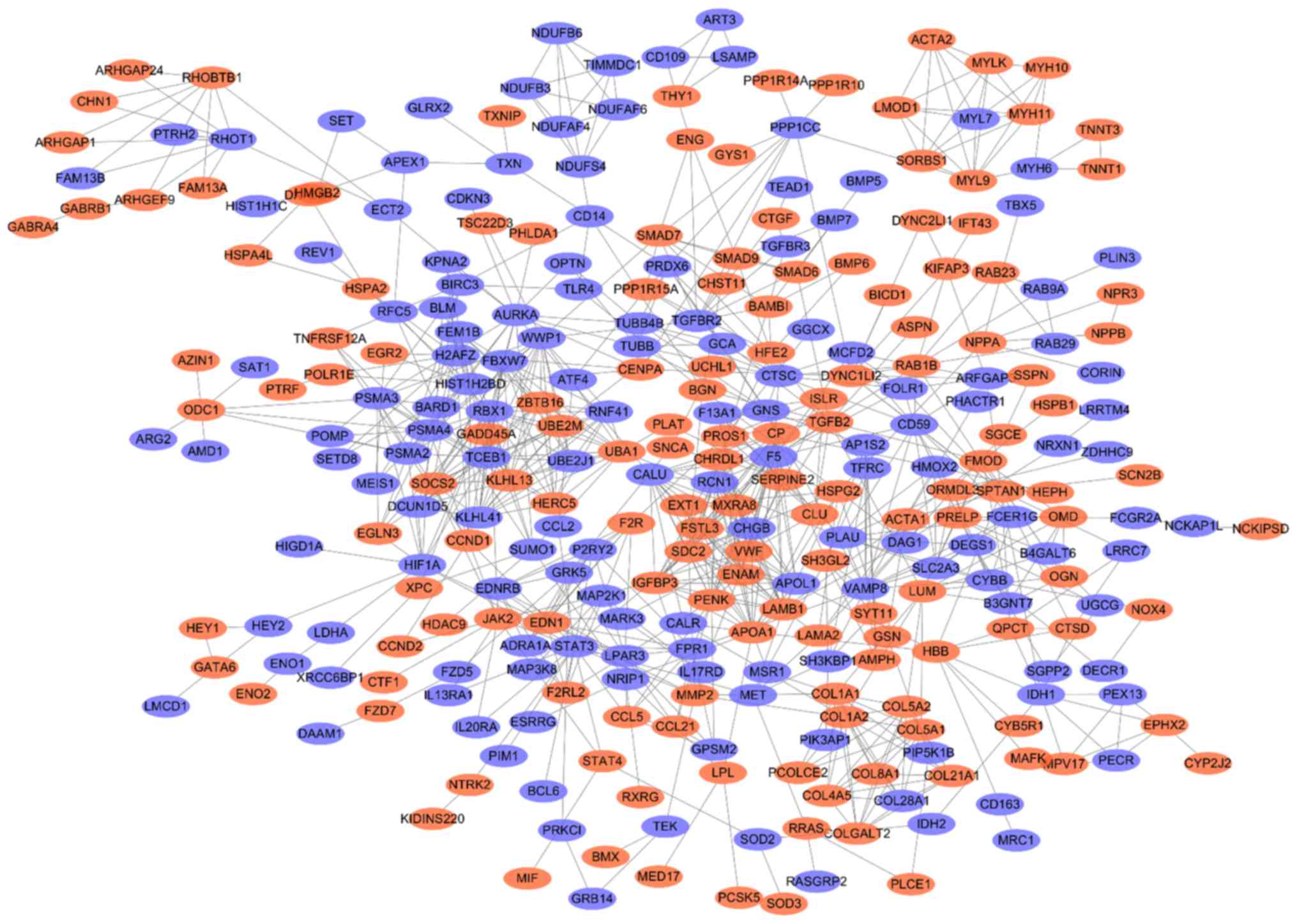
To assess the interactions among the proteins
encoded by the DEGs, DEGs were uploaded to the STRING database to
establish the PPI network (Fig.
4). It was demonstrated that there were 300 nodes and 852 edges
in the network. Furthermore, among the 300 nodes, 151 nodes were
upregulated, while 149 were downregulated. The MCODE plug-in was
used to identify key modules of the PPI network, and the top three
modules are presented in Fig.
5A-C. GO analysis results indicated that DEGs in module 2 were
mainly enriched in protein ubiquitination-related biological
processes, while the DEGs in module 3 were collagen- and
ECM-related (Fig. 5D). Using four
algorithms, the cytoHubba plug-in was used to detect hub genes of
the PPI network. In addition, the top 20 genes were identified by
degree, MCC, MNC and DMNC, which were then used for Venn diagram
analysis (Fig. 5E). It was
revealed that PENK, CHRDL1, CALU, apolipoprotein L1 (APOL1), IGFBP3
and CP overlapped among the four groups, and thus were deemed the
hub genes that may play important roles in DCM (Table V).
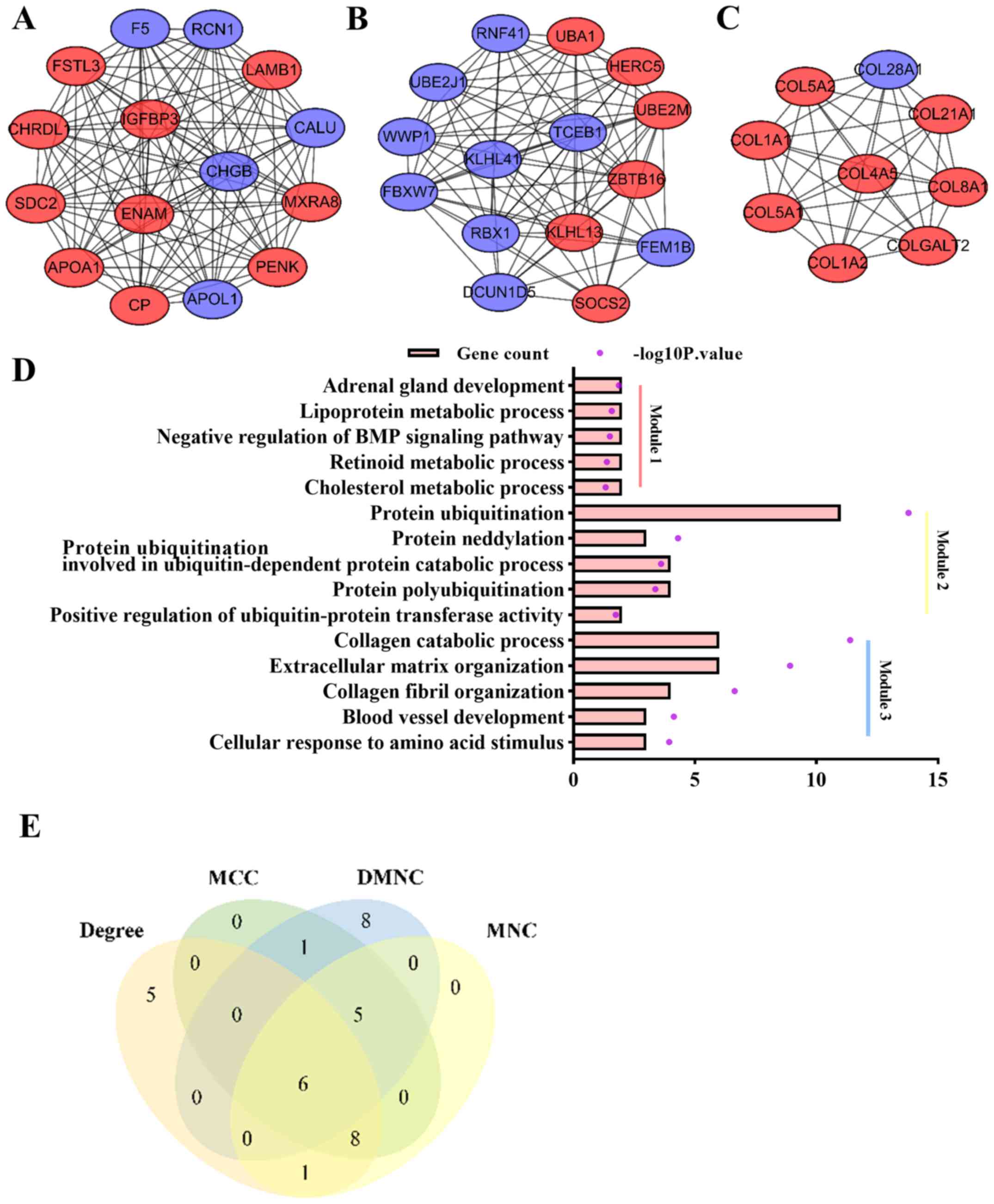 | Figure 5.Identification of important modules
and hub genes in the PPI network. (A) The first module, (B) the
second module and (C) the third module of the PPI network detected
by the MCODE plug-in and ranked from largest to smallest by score.
Red nodes are upregulated DEGs, while blue nodes are downregulated
DEGs. Edges between nodes represent interactions of DEGs. (D) GO
analysis of the DEGs in module 1, module 2 and module 3. The orange
bar represents the count of DEGs enriched in the GO term, and the
purple dot represents the -log10P-value. (E) Venn diagram of the
top 20 genes calculated by four algorithms. DEGs, differentially
expressed genes; PPI, protein-protein interaction; GO, Gene
Ontology; BMP, bone morphogenetic protein; MNC, Maximum
Neighborhood Component; DMNC, Density of Maximum Neighborhood
Component; MCC, Maximal Clique Centrality. |
 | Table V.Topological properties of hub
genes. |
Table V.
Topological properties of hub
genes.
| Hub gene | Degree | MCC | DMNC | MNC | Fold change | Percentage of false
prediction | Expression
change |
|---|
| PENK | 19 |
8.72×1010 | 1.02476 | 14 | 1.71 |
8.04×10−13 | Upregulated |
| CHRDL1 | 16 |
8.72×1010 | 1.02476 | 14 | 1.31 |
8.04×10−6 | Upregulated |
| CALU | 16 |
8.72×1010 | 0.92137 | 15 | 0.84 |
1.85×10−2 | Downregulated |
| APOL1 | 15 |
8.72×1010 | 0.92137 | 15 | 0.77 |
5.89×10−3 | Downregulated |
| IGFBP3 | 15 |
8.72×1010 | 0.92137 | 15 | 1.37 |
9.86×10−6 | Upregulated |
| CP | 15 |
8.72×1010 | 1.02476 | 14 | 1.32 |
2.98×10−6 | Upregulated |
Validation of the hub genes in a mouse
DCM model
Mice receiving repeated DOX injections were used as
a DCM in vivo model. As previously reported (29,30),
DOX mice that presented with progressive cardiac functional decline
and ventricular dilation were successfully established as the DCM
mouse model. Mice injected with cumulative 20 mg/kg DOX exhibited
impaired heart function (Fig.
6A-C), dilated LV (Fig. 6D and
E) and thin ventricular walls (Fig. 6F and G), which imitated the
pathological manifestation of human DCM (31). APOL1 does not have a homologous
gene in mice (32), but the mRNA
expression levels of PENK, CHRDL1, CALU, IGFBP3 and CP in mouse
hearts were detected with RT-qPCR. The present results indicated
that the mRNA expression levels of PENK, CHRDL1, IGFBP3 and CP were
significantly increased, and the expression of CALU was
significantly decreased, in the hearts of mice treated with DOX
compared with the saline controls (Fig. 6H). In addition, the changes in the
expression of these hub genes observed in vivo were
consistent with the microarray data, thus indicating an potential
connection between hub genes and DCM.
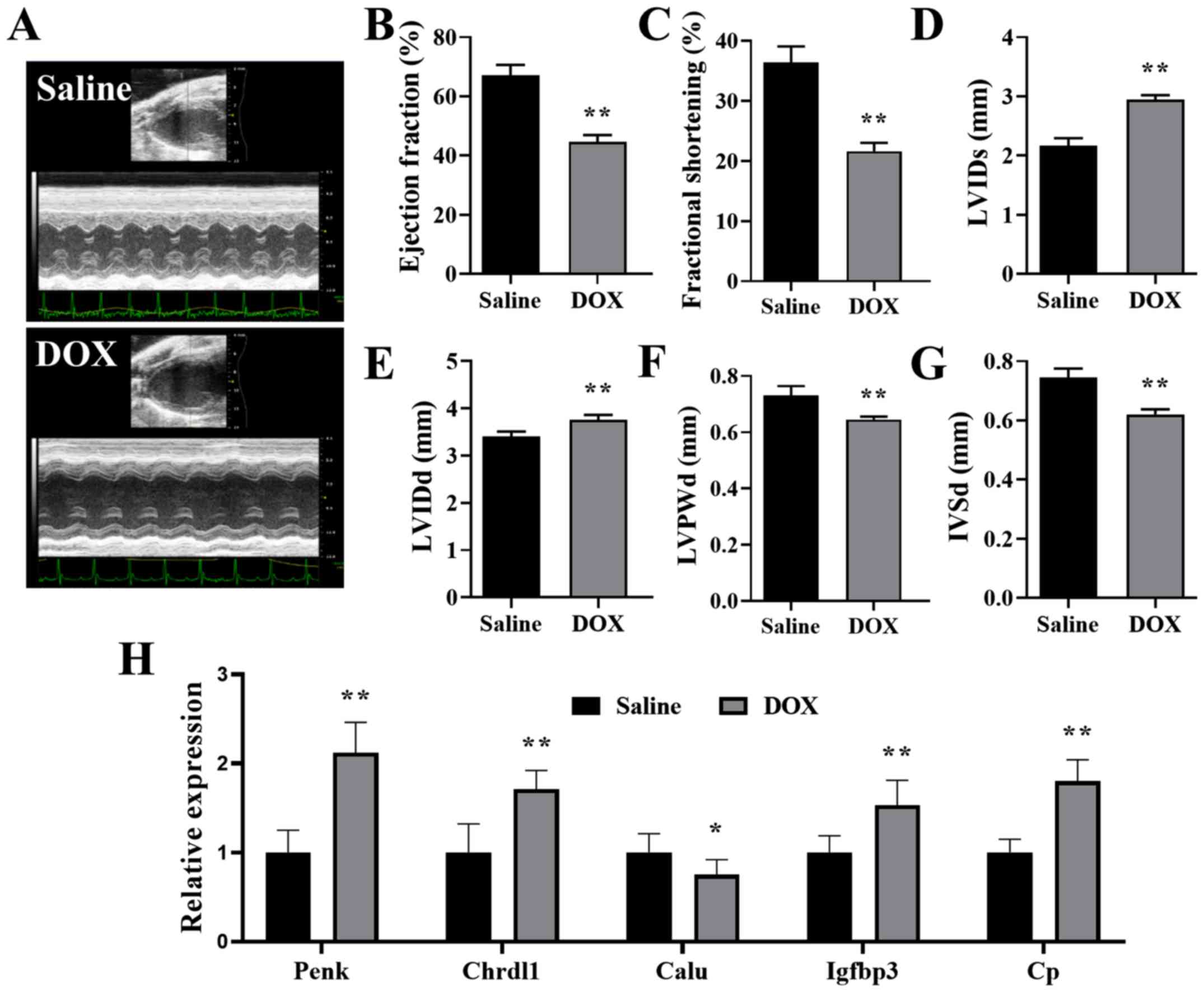 | Figure 6.Validation of the hub genes in the
mouse DCM model. (A) Representative echocardiography images of mice
treated with saline and DOX. (B) Ejection fraction, (C) fractional
shortening, (D) LVIDs, (E) LVIDd, (F) LVPWd and (G) IVSd of mice
treated with saline or DOX. (H) Relative expression of PENK,
CHRDL1, CALU, IGFBP3 and CP in the hearts of mice treated with DOX,
normalized to the expression of β-actin. n=6. *P<0.05,
**P<0.01 vs. saline. DOX, doxorubicin; LVIDs/d, left ventricular
internal diameter end systole/diastole; LVPWd, left ventricular
posterior wall end diastole; IVSd, interventricular septal end
diastole; PENK, proenkephalin; CHRDL1, chordin like 1; CALU,
calumenin; IGFBP3, insulin-like growth factor binding protein 3;
CP, ceruloplasmin. |
Discussion
The present study analyzed four DCM microarrays from
GEO, which to the best of the authors' knowledge is the largest
sample size that has been investigated, compared with the former
researches on gene expression of DCM (6,7,33,34),
using integrated bioinformatics analysis. With the establishment of
a PPI network, six hub genes associated with DCM were identified,
including upregulated PENK, CHRDL1, IGFBP3 and CP, as well as
downregulated CALU and APOL1. For further assessment of these hub
genes, RT-qPCR was performed in a DOX-induced DCM mouse model. In
line with the results of bioinformatics analysis, similar gene
changes were observed in DOX mouse hearts; except for APOL1, which
lacks homologous genes in mice (32). Furthermore, compared with mice in
the control group, the expression levels of PENK, CHRDL1, IGFBP3
and CP were significantly increased in DOX mice, while the
expression of CALU was significantly decreased.
It is speculated that some of the identified hub
genes may serve as biological markers for early DCM screening in
the clinic. CP, encoding the metalloprotein that binds the copper
in plasma, is involved in the peroxidation of Fe(II) transferrin to
Fe(III) transferrin (35). In
addition, CP is associated with cardiovascular disease by
decreasing nitric oxide bioavailability in blood (36). In a previous clinical study, CP was
revealed to be independently related to cardiac function in
patients with heart failure (37).
CALU produces a Ca2+-binding protein that is localized
in the endoplasmic reticulum (ER) (38). Furthermore, CALU is mainly
expressed in the heart, and facilitates protein folding and sorting
in the ER (38). During the
excitation-contraction coupling process, CALU regulates
Ca2+ uptake (39) and
plays an important role in maintaining normal heart function
(38). Thus, aberrant expression
levels of CP and CALU could provide a foundation for the
progressive decline of cardiac function in DCM, and have potential
value in the early screening of DCM (40).
In addition, the DEGs identified in the present
study may be valuable for the prognosis of DCM. PENK, encoding a
preproprotein of pentapeptide opioids, mainly shows biased
expression in adrenal tissue (41). Previous studies have also revealed
that PENK is an important biomarker for renal dysfunction (42–44).
In addition, APOL1, encoding a secreted high-density lipoprotein
that binds to apolipoprotein A-I to promote lipid exchange, is
reported to be associated with kidney disease when it is mutated
(45). With regards to the
interaction between the cardiovascular and renal systems, PENK and
APOL1 are also of potential value for cardiac disease research
(46). Kanagala et al
(47) revealed a relationship
between PENK and mortality rate in patients with heart failure.
Furthermore, the aberrant expression of APOL1 has been revealed to
play an important role in the development of hypertension (48). In line with these previous studies,
the present results identified upregulation of PENK and
downregulation of APOL1 in patients with DCM and DCM-induced mice,
which indicated that these may provide prognostic information for
the outcome in the progression of DCM.
Furthermore, the present study may provide a novel
therapeutic target for the mechanism and treatment of DCM. CHRDL1
is an important gene that expresses the antagonist of bone
morphogenetic protein 4 (BMP4) (49). In cardiac disease, BMP4 expression
varies with cardiac function (50,51).
In addition, Wu et al (51)
revealed that recombinant BMP4 plays a protective role in mouse
cardiomyocytes. The present study found increased expression of
CHRDL1 in DCM, which could potentially antagonize the protective
effect of BMP4 and accelerate DCM progression. Thus, suppressing
the expression of CHRDL1 may be a novel treatment target for
attenuating DCM development. IGFBP3 is a member of the IGFBP
family, and encodes a protein with an IGFBP domain and a
thyroglobulin type-I domain (52).
Previous studies have revealed that under hypoxic conditions,
IGFBP3 can promote mitochondria-dependent cardiomyocyte apoptosis
by inhibiting the IGF1R/PI3K/Akt survival pathway (53). In the present study, an increased
expression of IGFBP3 was identified in DCM, indicating that
aberrant IGFBP3 expression may be an important mechanism of DCM
occurrence.
To investigate the identified hub genes, a DCM mouse
model was established by intraperitoneal injection of DOX. DCM is
the final common response of myocardium to diverse genetic and
environmental insults (1). The
etiological causes of DCM are various, and it is generally
recognized that anthracycline cardiotoxicity is a common cause of
DCM (54,55). In addition, the clinical diagnosis
of DCM depends on the clinical feature of progressively aggravated
LV dilatation and systolic dysfunction (54). The DOX mouse model also exhibits
progressive cardiac functional decline and ventricular dilation
(29), which is consistent with
the diagnosis of DCM. In addition, while the specific pathogenesis
of DCM and the mechanism of its progressive progression remain
unknown, previous studies revealed that the development of DCM may
be associated with decreased mitochondrial function (56), abnormal oxidative stress (57,58),
as well as excessive myocardial apoptosis and necrosis (59,60).
In line with these possible causes, it has also been reported that
the DOX model is accompanied by similar mitochondrial dysfunction
(29,61), oxidative stress injury (62) and apoptotic changes (63). Thus, the present study used the DOX
mouse model to imitate the process of DCM for assessment of the hub
genes. Consistent with the bioinformatics analysis results, similar
genetic changes were observed in the DOX mouse model. It was
determined that the expression levels of PENK, CHRDL1, IGFBP3 and
CP were significantly increased in the hearts of DOX mice, while
the expression of CALU was significantly decreased, which is
consistent with the results of integrated microarray analysis.
However, while these hub genes in DCM have been
identified, the specific underlying mechanisms of the genes
involved in DCM development are not fully understood. Along with
the involvement in autophagy, apoptosis and oxidation, which have
been previously identified (64),
these hub genes may also interact with each other, which further
affects the survival and function of myocardial cells. Thus,
further studies are still required to establish the causative
correlations between these genes and the occurrence of DCM.
In conclusion, the present study identified six hub
genes related to DCM, including PENK, CHRDL1, IGFBP3, CP, CALU and
APOL1. In addition, these hub genes may provide a mechanism for
DCM, and could serve as biomarkers for screening and diagnosis in
the clinic.
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Nature Science Foundation of China (grant no. 81770333).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HZ, JH and QS designed the present study, which was
performed by JH, HZ and WJ. JH and WJ made substantial
contributions to acquisition and analysis of data. HZ and WJ also
made contributions to interpretation of data. JH wrote the initial
draft of the manuscript. HZ revised it critically for important
intellectual content. QS has given final approval of the version to
be published. All authors have participated sufficiently in the
work to take public responsibility for appropriate portions of the
content and approved the manuscript, as well as agreed to be
accountable for all aspects of the work. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
All animal studies were approved by the Animal Care
and Ethics Committee of Nanjing Medical University. The animal
experiments were performed according to the Guide for the Care and
Use of Laboratory Animals (National Institutes of Health
publication, 8th edition, 2011).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Jefferies JL and Towbin JA: Dilated
cardiomyopathy. Lancet. 375:752–762. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fatkin D, Huttner IG, Kovacic JC, Seidman
JG and Seidman CE: Precision medicine in the management of dilated
cardiomyopathy: JACC state-of-the-art review. J Am Coll Cardiol.
74:2921–2938. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Richardson P, McKenna W, Bristow M, Maisch
B, Mautner B, O'Connell J, Olsen E, Thiene G, Goodwin J, Gyarfas I,
et al: Report of the 1995 world health organization/international
society and federation of cardiology task force on the definition
and classification of cardiomyopathies. Circulation. 93:841–842.
1996. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hunt SA, Abraham WT, Chin MH, Feldman AM,
Francis GS, Ganiats TG, Jessup M, Konstam MA, Mancini DM, Michl K,
et al: 2009 Focused update incorporated into the ACC/AHA 2005
guidelines for the diagnosis and management of heart failure in
adults a report of the American college of cardiology
foundation/american heart association task force on practice
guidelines developed in collaboration with the international
society for heart and lung transplantation. J Am Coll Cardiol.
53:e1–e90. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rosenbaum AN, Agre KE and Pereira NL:
Genetics of dilated cardiomyopathy: Practical implications for
heart failure management. Nat Rev Cardiol. 17:286–297. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Huang H, Luo B, Wang B, Wu Q, Liang Y and
He Y: Identification of potential gene interactions in heart
failure caused by idiopathic dilated cardiomyopathy. Med Sci Monit.
24:7697–7709. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xiao J, Li F, Yang Q, Zeng XF and Ke ZP:
Co-expression analysis provides important module and pathways of
human dilated cardiomyopathy. J Cell Physiol. 235:494–503. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhuang Y, Gong YJ, Zhong BF, Zhou Y and
Gong L: Bioinformatics method identifies potential biomarkers of
dilated cardiomyopathy in a human induced pluripotent stem
cell-derived cardiomyocyte model. Exp Ther Med. 14:2771–2778. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of Affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Carlson M: hgu133plus2.db: Affymetrix
Human Genome U133 Plus 2.0 Array annotation data (chip
hgu133plus2), R package version 3.2.3. 2016.
|
|
11
|
MacDonald JW:
hugene10sttranscriptcluster.db: Affymetrix hugene10 annotation data
(chip hugene10sttranscriptcluster). R package version 8.7.0.
2017.
|
|
12
|
Carlson M: hgu133a.db: Affymetrix Human
Genome U133 Set annotation data (chip hgu133a). R package version
3.2.3. 2016.
|
|
13
|
Marot G, Foulley JL, Mayer CD and
Jaffrézic F: Moderated effect size and P-value combinations for
microarray meta-analyses. Bioinformatics. 25:2692–2699. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Del Carratore F, Jankevics A, Eisinga R,
Heskes T, Hong F and Breitling R: RankProd 2.0: A refactored
bioconductor package for detecting differentially expressed
features in molecular profiling datasets. Bioinformatics.
33:2774–2775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Huang da W, Sherman BT and Lempicki RA:
Systematic and integrative analysis of large gene lists using DAVID
bioinformatics resources. Nat Protoc. 4:44–57. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Huang da W, Sherman BT and Lempicki RA:
Bioinformatics enrichment tools: Paths toward the comprehensive
functional analysis of large gene lists. Nucleic Acids Res.
37:1–13. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
The Gene Ontology Consortium, . The gene
ontology resource: 20 years and still GOing strong. Nucleic Acids
Res. 47(D1): D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45(D1):D353–D361. 2017.
View Article : Google Scholar
|
|
19
|
Wickham H: ggplot2: Elegant graphics for
data analysis. Springer. 2016.
|
|
20
|
Szklarczyk D, Morris JH, Cook H, Kuhn M,
Wyder S, Simonovic M, Santos A, Doncheva NT, Roth A, Bork P, et al:
The STRING database in 2017: Quality-controlled protein-protein
association networks, made broadly accessible. Nucleic Acids Res.
45(D1): D362–D368. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Bader GD and Hogue CW: An automated method
for finding molecular complexes in large protein interaction
networks. BMC Bioinformatics. 4:22003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chin CH, Chen SH, Wu HH, Ho CW, Ko MT and
Lin CY: cytoHubba: Identifying hub objects and sub-networks from
complex interactome. BMC Syst Biol. 8 (Suppl 4):S112014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
National Research Council (US) Committee
for the Update of the Guide for the Care and Use of Laboratory
Animals, . Guide for the Care and Use of Laboratory Animals. (8th).
National Academies Press (US) National Academy of Sciences.
(Washington (DC)). 2011.
|
|
24
|
Sun A, Cheng Y, Zhang Y, Zhang Q, Wang S,
Tian S, Zou Y, Hu K, Ren J and Ge J: Aldehyde dehydrogenase 2
ameliorates doxorubicin-induced myocardial dysfunction through
detoxification of 4-HNE and suppression of autophagy. J Mol Cell
Cardiol. 71:92–104. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xia Y, Chen Z, Chen A, Fu M, Dong Z, Hu K,
Yang X, Zou Y, Sun A, Qian J and Ge J: LCZ696 improves cardiac
function via alleviating Drp1-mediated mitochondrial dysfunction in
mice with doxorubicin-induced dilated cardiomyopathy. J Mol Cell
Cardiol. 108:138–148. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Stypmann J, Engelen MA, Troatz C,
Rothenburger M, Eckardt L and Tiemann K: Echocardiographic
assessment of global left ventricular function in mice. Lab Anim.
43:127–137. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Gonzalez-Valdes I, Hidalgo I, Bujarrabal
A, Lara-Pezzi E, Padron-Barthe L, Garcia-Pavia P, Gómez-del Arco P,
Redondo JM, Ruiz-Cabello JM, Jimenez-Borreguero LJ, et al: Bmi1
limits dilated cardiomyopathy and heart failure by inhibiting
cardiac senescence. Nat Commun. 6:64732015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li S, Wang W, Niu T, Wang H, Li B, Shao L,
Lai Y, Li H, Janicki JS, Wang XL, et al: Nrf2 deficiency
exaggerates doxorubicin-induced cardiotoxicity and cardiac
dysfunction. Oxid Med Cell Longev. 2014:7485242014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Gomes AC, Falcão-Pires I, Pires AL,
Brás-Silva C and Leite-Moreira AF: Rodent models of heart failure:
An updated review. Heart Fail Rev. 18:219–249. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bozkurt B, Colvin M, Cook J, Cooper LT,
Deswal A, Fonarow GC, Francis GS, Lenihan D, Lewis EF, McNamara DM,
et al: Current diagnostic and treatment strategies for specific
dilated cardiomyopathies: A scientific statement from the American
heart association. Circulation. 134:e579–e646. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bruggeman LA, Wu Z, Luo L, Madhavan SM,
Konieczkowski M, Drawz PE, Thomas DB, Barisoni L, Sedor JR and
O'Toole JF: APOL1-G0 or APOL1-G2 transgenic models develop
preeclampsia but not kidney disease. J Am Soc Nephrol.
27:3600–3610. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Zhao J, Lv T, Quan J, Zhao W, Song J, Li
Z, Lei H, Huang W and Ran L: Identification of target genes in
cardiomyopathy with fibrosis and cardiac remodeling. J Biomed Sci.
25:632018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang H, Yu Z, He J, Hua B and Zhang G:
Identification of the molecular mechanisms underlying dilated
cardiomyopathy via bioinformatic analysis of gene expression
profiles. Exp Ther Med. 13:273–279. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Vasilyev VB: Looking for a partner:
Ceruloplasmin in protein- protein interactions. Biometals.
32:195–210. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Cabassi A, Binno SM, Tedeschi S, Ruzicka
V, Dancelli S, Rocco R, Vicini V, Coghi P, Regolisti G, Montanari
A, et al: Low serum ferroxidase I activity is associated with
mortality in heart failure and related to both
peroxynitrite-induced cysteine oxidation and tyrosine nitration of
ceruloplasmin. Circ Res. 114:1723–1732. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hammadah M, Fan Y, Wu Y, Hazen SL and Tang
WH: Prognostic value of elevated serum ceruloplasmin levels in
patients with heart failure. J Card Fail. 20:946–952. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Sahoo SK and Kim do H: Characterization of
calumenin in mouse heart. BMB Rep. 43:158–163. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Sahoo SK and Kim DH: Calumenin interacts
with SERCA2 in rat cardiac sarcoplasmic reticulum. Mol Cells.
26:265–269. 2008.PubMed/NCBI
|
|
40
|
Mazzarotto F, Tayal U, Buchan RJ,
Midwinter W, Wilk A, Whiffin N, Govind R, Mazaika E, de Marvao A,
Dawes TJW, et al: Reevaluating the genetic contribution of
monogenic dilated cardiomyopathy. Circulation. 141:387–398. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ng LL, Squire IB, Jones DJ, Cao TH, Chan
DCS, Sandhu JK, Quinn PA, Davies JE, Struck J, Hartmann O, et al:
Proenkephalin, renal dysfunction, and prognosis in patients with
acute heart failure: A GREAT network study. J Am Coll Cardiol.
69:56–69. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Hollinger A, Wittebole X, François B,
Pickkers P, Antonelli M, Gayat E, Chousterman BG, Lascarrou JB,
Dugernier T, Di Somma S, et al: Proenkephalin A 119–159 (Penkid) is
an early biomarker of septic acute kidney injury: The kidney in
sepsis and septic shock (Kid-SSS) study. Kidney Int Rep.
3:1424–1433. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Emmens JE, Ter Maaten JM, Damman K, van
Veldhuisen DJ, de Boer RA, Struck J, Bergmann A, Sama IE, Streng
KW, Anker SD, et al: Proenkephalin, an opioid system surrogate, as
a novel comprehensive renal marker in heart failure. Circ Heart
Fail. 12:e0055442019. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Marino R, Struck J, Hartmann O, Maisel AS,
Rehfeldt M, Magrini L, Melander O, Bergmann A and Di Somma S:
Diagnostic and short-term prognostic utility of plasma
pro-enkephalin (pro-ENK) for acute kidney injury in patients
admitted with sepsis in the emergency department. J Nephrol.
28:717–724. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Shah S, Shapiro R, Murphy B and Menon MC:
APOL1 high-risk genotypes and renal transplantation. Clin
Transplant. 33:e135822019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yogasundaram H, Chappell MC, Braam B and
Oudit GY: Cardiorenal syndrome and heart failure-challenges and
opportunities. Can J Cardiol. 35:1208–1219. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kanagala P, Squire IB, Jones DJL, Cao TH,
Chan DCS, McCann G, Sandhu JK, Quinn PA, McAdam J, Marsh AM, et al:
Proenkephalin and prognosis in heart failure with preserved
ejection fraction: A GREAT network study. Clin Res Cardiol.
108:940–949. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Robinson TW and Freedman BI: The Impact of
APOL1 on chronic kidney disease and hypertension. Adv Chronic
Kidney Dis. 26:131–136. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Webb TR, Matarin M, Gardner JC, Kelberman
D, Hassan H, Ang W, Michaelides M, Ruddle JB, Pennell CE, Yazar S,
et al: X-linked megalocornea caused by mutations in CHRDL1
identifies an essential role for ventroptin in anterior segment
development. Am J Hum Genet. 90:247–259. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Sun B, Huo R, Sheng Y, Li Y, Xie X, Chen
C, Liu HB, Li N, Li CB, Guo WT, et al: Bone morphogenetic protein-4
mediates cardiac hypertrophy, apoptosis, and fibrosis in
experimentally pathological cardiac hypertrophy. Hypertension.
61:352–360. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Wu X, Sagave J, Rutkovskiy A, Haugen F,
Baysa A, Nygård S, Czibik G, Dahl CP, Gullestad L, Vaage J and
Valen G: Expression of bone morphogenetic protein 4 and its
receptors in the remodeling heart. Life Sci. 97:145–154. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ranke MB: Insulin-like growth factor
binding-protein-3 (IGFBP-3). Best Pract. Res Clin Endocrinol Metab.
29:701–711. 2015.
|
|
53
|
Feng CC, Lin CC, Lai YP, Chen TS,
Marthandam Asokan S, Lin JY, Lin KH, Viswanadha VP, Kuo WW and
Huang CY: Hypoxia suppresses myocardial survival pathway through
HIF-1α-IGFBP-3-dependent signaling and enhances cardiomyocyte
autophagic and apoptotic effects mainly via FoxO3a-induced BNIP3
expression. Growth Factors. 34:73–86. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Japp AG, Gulati A, Cook SA, Cowie MR and
Prasad SK: The diagnosis and evaluation of dilated cardiomyopathy.
J Am Coll Cardiol. 67:2996–3010. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Ky B, Putt M, Sawaya H, French B, Januzzi
JL Jr, Sebag IA, Plana JC, Cohen V, Banchs J, Carver JR, et al:
Early increases in multiple biomarkers predict subsequent
cardiotoxicity in patients with breast cancer treated with
doxorubicin, taxanes, and trastuzumab. J Am Coll Cardiol.
63:809–816. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Chen L and Knowlton AA: Mitochondrial
dynamics in heart failure. Congest Heart Fail. 17:257–261. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Pang XF, Lin X, Du JJ and Zeng DY: LTBP2
knockdown by siRNA reverses myocardial oxidative stress injury,
fibrosis and remodelling during dilated cardiomyopathy. Acta
Physiol (Oxf). 228:e133772020. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Lubrano V and Balzan S: Role of oxidative
stress-related biomarkers in heart failure: Galectin 3,
α1-antitrypsin and LOX-1: New therapeutic perspective? Mol Cell
Biochem. 464:143–152. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Liu L, Sun K, Zhang X, Tang Y and Xu D:
Advances in the role and mechanism of BAG3 in dilated
cardiomyopathy. Heart Fail Rev. Dec 6–2019.(Epub ahead of print).
View Article : Google Scholar
|
|
60
|
Mazelin L, Panthu B, Nicot AS, Belotti E,
Tintignac L, Teixeira G, Zhang Q, Risson V, Baas D, Delaune E, et
al: mTOR inactivation in myocardium from infant mice rapidly leads
to dilated cardiomyopathy due to translation defects and
p53/JNK-mediated apoptosis. J Mol Cell Cardiol. 97:213–225. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Ni C, Ma P, Wang R, Lou X, Liu X, Qin Y,
Xue R, Blasig I, Erben U and Qin Z: Doxorubicin-induced
cardiotoxicity involves IFNγ-mediated metabolic reprogramming in
cardiomyocytes. J Pathol. 247:320–332. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Goyal V, Bews H, Cheung D, Premecz S,
Mandal S, Shaikh B, Best R, Bhindi R, Chaudhary R, Ravandi A, et
al: The cardioprotective role of N-Acetyl cysteine amide in the
prevention of doxorubicin and trastuzumab-mediated cardiac
dysfunction. Can J Cardiol. 32:1513–1519. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Liu D, Ma Z, Di S, Yang Y, Yang J, Xu L,
Reiter RJ, Qiao S and Yuan J: AMPK/PGC1α activation by melatonin
attenuates acute doxorubicin cardiotoxicity via alleviating
mitochondrial oxidative damage and apoptosis. Free Radic Biol Med.
129:59–72. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Schultheiss HP, Fairweather D, Caforio
ALP, Escher F, Hershberger RE, Lipshultz SE, Liu PP, Matsumori A,
Mazzanti A, McMurray J and Priori SG: Dilated cardiomyopathy. Nat
Rev Dis Primers. 5:322019. View Article : Google Scholar : PubMed/NCBI
|