Introduction
At present, lung cancer is the leading cause of
malignant tumor-associated mortality worldwide. Despite
improvements in preventative and therapeutic strategies in recent
decades, the 5-year survival rate is still low of only 15–20%
(1). Non-small cell lung cancer
(NSCLC) accounts for 85% of lung cancer cases (2). The survival of patients with NSCLC
has significantly improved following the development of
chemotherapy and molecular targeted therapy. However, due to the
high recurrence and metastasis rate, the long-term survival rate
remains poor (3,4). Therefore, it has been hypothesized
that further investigation into the anticancer functions of small
molecule anticancer compounds may identify novel diagnostic and
prognostic markers, thereby improving the survival rate of patients
(5–7).
Advanced glycosylation end-product specific receptor
(AGER) is a member of the immunoglobulin superfamily of cell
surface receptors. AGER protein is a multi-ligand receptor that
interacts with a wide range of ligands, including advanced
glycosylation end products (AGEs), β-sheet fibrils, S100 proteins
(S100B, S100P, S100A4, S100A6, S100A8/9 and S100A11-13), high
mobility family protein-1 and prion (7,8).
AGER expression is associated with diabetic angiopathies and thymic
hyperplasia, and functions via the Toll-like receptor 4 and
AGE/AGER signaling pathways (9).
AGER proteins mediate macrophages under normal conditions (10), whereas the cross-linking reaction
between AGER protein and the extracellular matrix is enhanced under
pathological conditions, resulting in an increased thickness and
permeability of the endangium (11). Substantial evidence has suggested
that abnormal AGER expression is closely associated with the immune
inflammatory response and tumorigenesis (12). A number of studies have also
reported that the expression and mutation rate of AGER are highly
increased in esophageal cancer (13), as well as in other types of cancer,
including breast, gastric and endometrial cancer (14–16).
However, a number of studies have reported that AGER expression is
significantly downregulated in lung cancer (13,17–21).
In addition, AGER is a highly polymorphic gene with single
nucleotide polymorphisms, which may be associated with lung
diseases, including chronic obstructive pulmonary disease and acute
respiratory distress syndrome (22). Furthermore, high expression of AGER
protein is associated with pulmonary inflammation and the
deterioration of other lung diseases (23). For example, Caraher et al
(24) reported the absence of RAGE
mitigated acute deleterious effects of particulate matter and may
be a biologically plausible mediator of PM-related lung disease.
The study of Machahua et al (25) has demonstrated that serum AGE/RAGEs
are potential biomarkers of idiopathic pulmonary fibrosis
pneumonia. It has also been reported that AGER is closely
associated with the low survival rate of patients with lung cancer,
based on the analysis of an oncogene microarray and The Cancer
Genome Atlas (TCGA) database (26). Therefore, the abnormal expression
of AGER in lung cancer tissues and cells indicates that AGER serves
an important role in lung cancer, which suggests that AGER may
represent as a potential therapeutic target during the development
of lung cancer.
The present study aimed to explore the effects of
AGER on the biological behavior of the NSCLC.
Materials and methods
Bioinformatics analysis
The NSCLC microarray dataset GSE27262 (27)was obtained from the Gene Expression
Omnibus (GEO) database (www.ncbi.nlm.nih.gov/geo). R language 3.5.3 software
(https://cran.r-project.org/bin/windows/base/old/3.5.3/)
was used to conduct differential analysis. R package ‘pheatmap’ was
used to create the heatmap of differentially expressed genes
(DEGs). The expression level of AGER was validated using TCGA
database (ualcan.path.uab.edu/cgi-bin/ualcan-res.pl).
Cell culture and transfection
The human normal lung BEAS-2B cell line, the NSCLC
H1299 cell line and the human embryonic kidney 293T cell line were
purchased from the Cell Bank of Type Culture Collection of the
Chinese Academy of Sciences. BEAS-2B and H1299 cell lines were
cultured in RPMI-1640 medium (Sigma-Aldrich; Merck KGaA)
supplemented with 5% fetal bovine serum (FBS, Gibco; Thermo Fisher
Scientific, Inc.), 293T cell line were cultured in Dulbecco's
Modified Eagle's Medium (DMEM,Gibco, Rockville, MD, USA),
supplemented with 10% fetal bovine serum, 100 U/ml penicillin, and
100 µg/ml streptomycin. All cells were cultured at 37°C with 5%
CO2. At 70–80% confluence, cells were digested with
0.25% trypsin for 3 min at room temperature for passage. Cells in
the logarithmic growth phase were selected for subsequent
experiments and were divided into four groups: lentivirus
(LV)-negative control (NC), LV-AGER, small interfering RNA
(siRNA/si)-NC, and si-AGER. AGER cDNA was cloned into the
pLenti-C-mGFP vector (OriGene Technologies, Inc.). The
pLenti-C-mGFP-AGER plasmid (LV-AGER; Invitrogen; Thermo Fisher
Scientific, Inc.) and the corresponding pLenti-C-mGFP-NC (LV-NC;
Invitrogen; Thermo Fisher Scientific, Inc.) were used with two
packaging vectors pspax2 (Invitrogen; Thermo Fisher Scientific,
Inc.) and pMD2.G (Invitrogen; Thermo Fisher Scientific, Inc.)
co-transfected into 293T cells (cell density: 1.5×104)
at a final concentration of 50 nM at room temperature for at least
5 min using Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.). Lentiviral particles were harvested and
filtered to infect H1299 cells (1×105 cells/well), and
transfected for 48 h at room temperature for subsequent
experiments.
The AGER siRNA and its negative control sequences
were designed using BLOCK-iT™ RNAi Designer (www.invitrogen.com/rnai): siRNA-NC
(5′-TGCCCTACCCTAGTGTGAT-3′), AGER-siRNA1
(5′-TGCTGATCCTCCCTGAGAT-3′) AGER-siRNA2 (5′-GCTGATCCTCCCTGAGATA-3′)
and AGER-siRNA3 (5′-GCCTTATCCCTAACAGCCA-3′). H1299 cells
(1×105 cells/well) were seeded into a 6-well culture
plate and cultured to 60–70% confluency at room temperature.
Subsequently, 8 µl siRNA (20 µmol/l) was diluted in 250 µl
serum-free DMEM and incubated for 5 min at room temperature.
Lipofectamine® 2000 reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) was diluted in serum-free DMEM and added to the
diluted siRNAs for 20 min at room temperature. siRNA-NC, and
AGER-siRNA complexes were added to cells for 48 h at room
temperature. Transfection efficiency was measured using reverse
transcription-quantitative PCR (RT-qPCR). Interference efficiency
was detected using RT-qPCR.
RT-qPCR
Total RNA was extracted from H1299 cells according
to TRIzol® reagent instructions (Invitrogen; Thermo
Fisher Scientific, Inc.). RNA concentration was determined via UV
spectrophotometry. Total RNA was reversely transcribed into cDNA
using the PrimeScript RT kit (Takara Biomedical Technology Co.,
Ltd.) according to the manufacturer's instructions. Subsequently,
qPCR was performed according to the instructions of SYBR Green PCR
Kit (Qiagen, Hilden, Germany). The following primer pairs were used
for qPCR: AGER forward, 5′-GTGTCCTTCCCAACGGCTC-3′ and reverse,
5′-ATTGCCTGGCACCGGAAAA-3′; and β-actin forward,
5′-GTGGGGCGCCCCAGGCACCA-3′ and reverse,
5′-CTCCTTAATGTCACGCACGATTTC-3′. The following thermocycling
conditions were used for qPCR: initial denaturation at 95°C for 5
min; followed by 30 cycles of 95°C for 40 sec, 57°C for 40 sec and
72°C for 40 sec, with a final extension at 72°C for 10 min. AGER
mRNA levels were quantified using the 2−ΔΔCt method
(28) and normalized to the
internal reference gene β-actin. RT-qPCR was performed in
triplicate.
Western blotting
Total protein was extracted from the H1299 cells
using cold NP40 lysis buffer or RIPA buffer (Beyotime Institute of
Biotechnology). The protein was quantified using a bicinchoninic
acid assay, and 30 µg of total proteins were separated via 12%
SDS-PAGE at 30 mA for 120 min and transferred to nitrocellulose
membranes. Subsequently, the membranes were blocked with 5% skim
milk powder (dissolved in TBS+0.1% Tween-20) for 60 min at room
temperature. The membranes were incubated overnight at 4°C with
primary antibodies targeted against: AGER (cat. no. ab3611;
1:1,000; Abcam), Bax (cat. no. ab32503; 1:1,000; Abcam), Bcl-2
(cat. no. ab32124; 1:500; Abcam) and GAPDH (cat. no. ab181602;
1:2,500; Abcam). Following primary incubation, the membranes were
incubated with horseradish peroxidase-conjugated anti-rabbit IgG
H&L secondary antibodies (cat. no. ab6721; 1:2,000; Abcam) at
room temperature for 120 min. Proteins were visualized using an ECL
luminescent kit (Beijing Solarbio Science & Technology Co.,
Ltd.). Western blotting was performed in triplicate and protein
expression was quantified using Quantity One 4.6.6 software
(Bio-Rad Laboratories, Inc.) with GAPDH as the internal
reference.
Cell proliferation assay
Cell proliferation was measured using MTT assay
(Sigma-Aldrich; Merck KGaA), according to the manufacturer's
instruction. Briefly, NSCLC H1299 cells were seeded in 96-well
plates at a density of 2×103 cells/well and incubated at
37°C for 24, 48 and 72 h. Subsequently, 20 µl MTT (5 mg/ml) was
added to each well and incubated for 4 h at 37°C. Following the MTT
incubation, 150 µl DMSO was added to dissolve the purple formazan
crystals for 15 min at room temperature. The absorbance of each
well at a wavelength of 570 nm was determined using a microplate
reader. The assay was performed in triplicate.
Colony formation assay
H1299 cells were digested with 0.25% trypsin to
individual cells and suspended in culture medium. Cells were seeded
at 200 cells per dish and cultured for 3 weeks at 37°C. When
macroscopic clones appeared in the culture dish, cells were fixed
with 4% paraformaldehyde for 15 min and stained with 0.1% crystal
violet stain for 10 min at room temperature. The number of colonies
(≥50 cells/colony) was calculated as follows: Colony formation
rate=(number of colonies/200) × 100%. The assay was performed in
triplicate.
Flow cytometry
Early and late apoptotic cells were detected using
flow cytometry with the Annexin V-FITC Apoptosis Detection kit (BD
Biosciences), according to the manufacturer's protocol. Briefly,
H1299 cells were washed twice with cold PBS and resuspended in 200
µl PBS. Subsequently, Annexin V-FITC and propidium iodide solution
was added to the cells, and the cells were incubated at room
temperature for 15 min in the dark. Finally, flow cytometry (Becton
and Dickinson Company) was utilized to detect the apoptotic cells.
CELLQuest 3.0 software (Becton and Dickinson Company) was utilized
to analyze the data. Flow cytometry was performed in
triplicate.
Wound healing assay
A 200 µl medium pipette tip was used to scrape a
single wound into the H1299 cell monolayer. The monolayer was
washed three times with PBS to remove scratched cells. Cells were
cultured in DMEM (Thermo Fisher Scientific, Inc.) medium at 37°C
with 5% CO2. At 0 and 24 h time points, five randomly
selected fields were observed using an optical inverted microscope
(magnification, ×100). The migratory rate of the cells was
calculated using the following formula: (0 h trace width–24 h trace
width)/0 h trace width. The assay was performed in triplicate.
Transwell invasion assay
Each group of H1299 cells was suspended in FBS-free
DMEM medium and seeded at a density of 2×105 cells/well
in the upper chamber of a 24-well Transwell chamber pretreated with
Matrigel for 30 min at 37°C. The cells were incubated at 37°C for 4
h. DMEM containing 10% FBS (500 µl) was plated in the lower chamber
of the Transwell plates. After 24-h incubation at 37°C, cells on
the upper surface of the Transwell membrane were removed using a
cotton swab. Invaded cells were fixed with 4% paraformaldehyde for
20 min and crystal violet staining for 15 min at room temperature.
Stained cells in six fields of view were observed and counted using
an optical inverted microscope at ×100 magnification. The assay was
performed in triplicate.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 21.0; IBM Corp.). Data were presented as the mean
± standard deviation. Comparisons between two groups were analyzed
using paired Student's t-test and multigroup comparisons were made
using analysis of variance (ANOVA) with Tukey's post hoc test.
P<0.05 was considered to indicate a statistically
significant difference.
Results
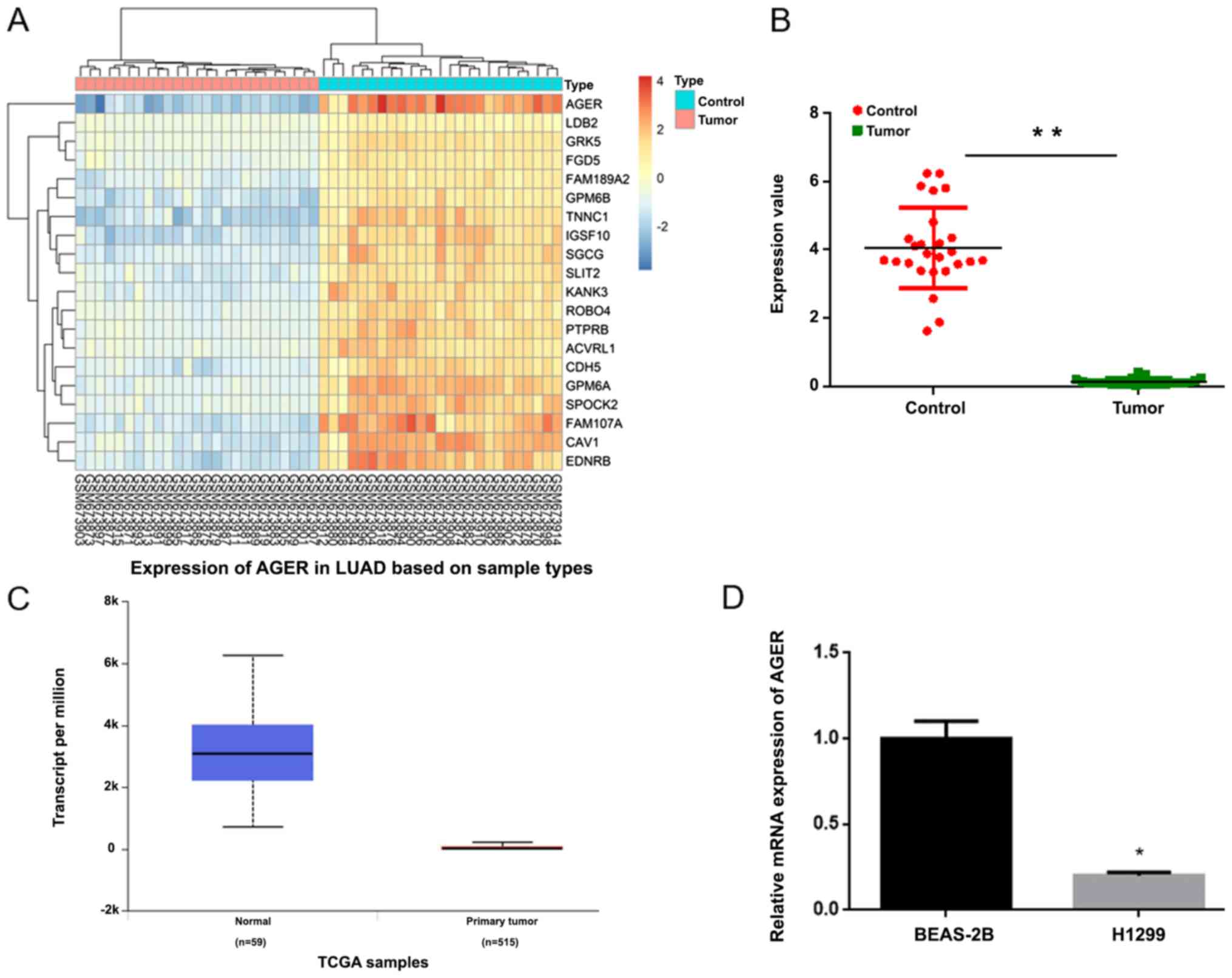
AGER expression is downregulated in
NSCLC tissues and cells
The microarray GSE27262 dataset was used to analyze
DEGs. The results suggested that AGER was the most significant DEG
and AGER expression was significantly downregulated in NSCLC
tissues compared with control tissues (Fig. 1A and B). AGER was further validated
as a DEG using TCGA database (Fig.
1C). Furthermore, RT-qPCR was performed to measure the
expression of AGER in the NSCLC cell line. Compared with the normal
lung BEAS-2B cell line, AGER expression was significantly decreased
in the H1299 cell line (Fig. 1D;
P<0.05).
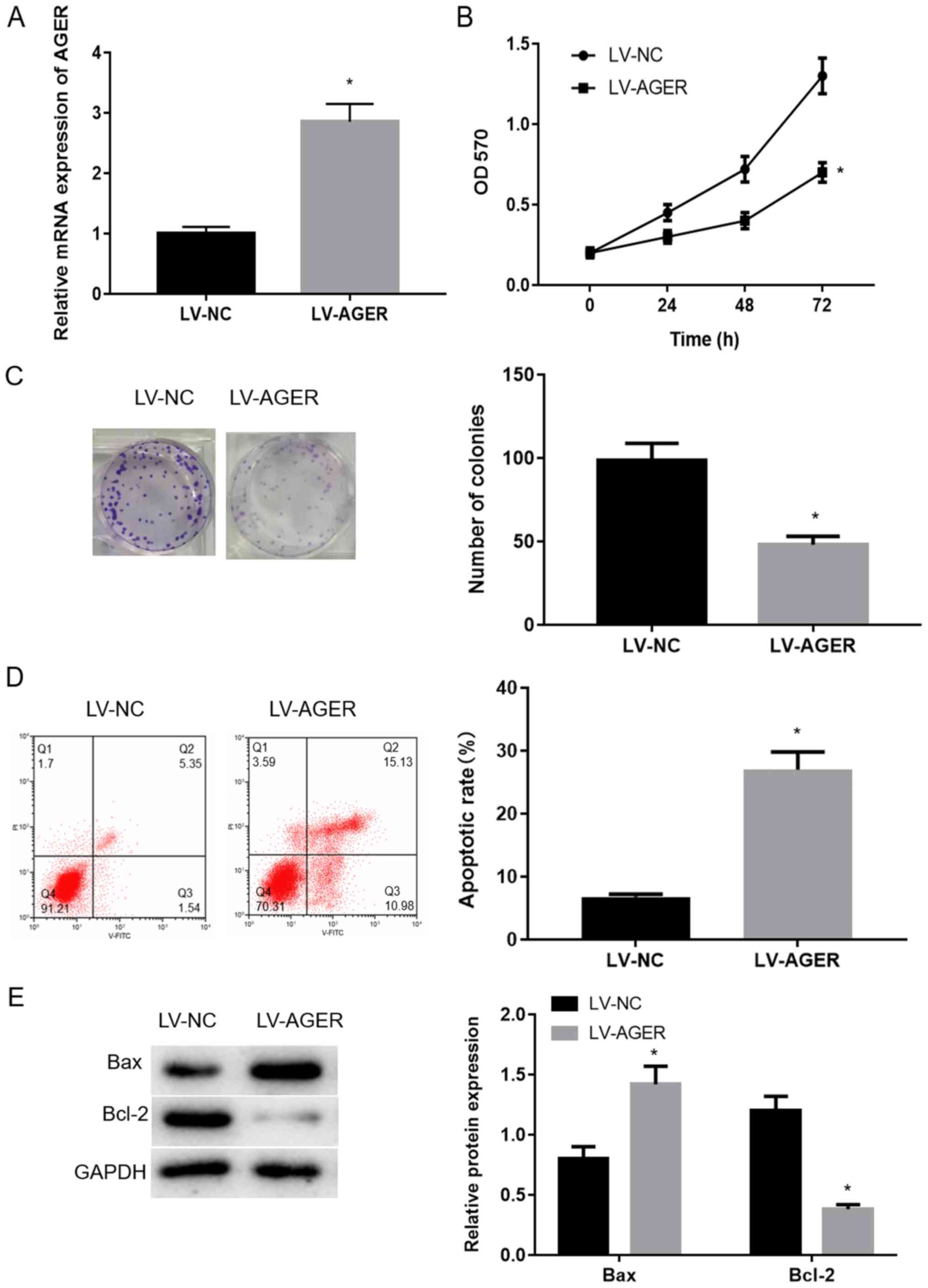
AGER overexpression decreases
proliferation and promotes apoptosis of H1299 cells
AGER overexpression efficiency in the LV-NC and
LV-AGER groups was measured via RT-qPCR. AGER expression in the
LV-AGER group was significantly increased compared with the LV-NC
group (Fig. 2A; P<0.05).
To further investigate the effect of AGER on the biological
function of H1299 cells, MTT assay was performed to measure H1299
cell viability. The results suggested that proliferation in the
LV-AGER group was significantly decreased compared with the LV-NC
group (Fig. 2B; P<0.05).
Colony formation assays were used to assess alterations in cell
clonality, and the results demonstrated that the colony formation
ability of the LV-AGER group was decreased compared with the LV-NC
group (Fig. 2C;
P<0.05).
Apoptotic cells were detected by Annexin V-FITC flow
cytometry. The apoptotic rate was significantly increased in the
LV-AGER group compared with the LV-NC group (Fig. 2D; P<0.05). The protein
expression of bcl-2 and bax has been confirmed to be closely
related to the apoptosis of cancer cells. When the ratio of bcl-2
and bax is down-regulated, it can significantly induce the
apoptosis of cancer cells (29,30).
The western blotting analysis results indicated that the
antiapoptotic protein Bcl-2 was downregulated and the proapoptotic
protein Bax was upregulated in the LV-AGER group compared with the
LV-NC group (Fig. 2E). The results
indicated that AGER overexpression decreased H1299 cell
proliferation and promoted apoptosis.
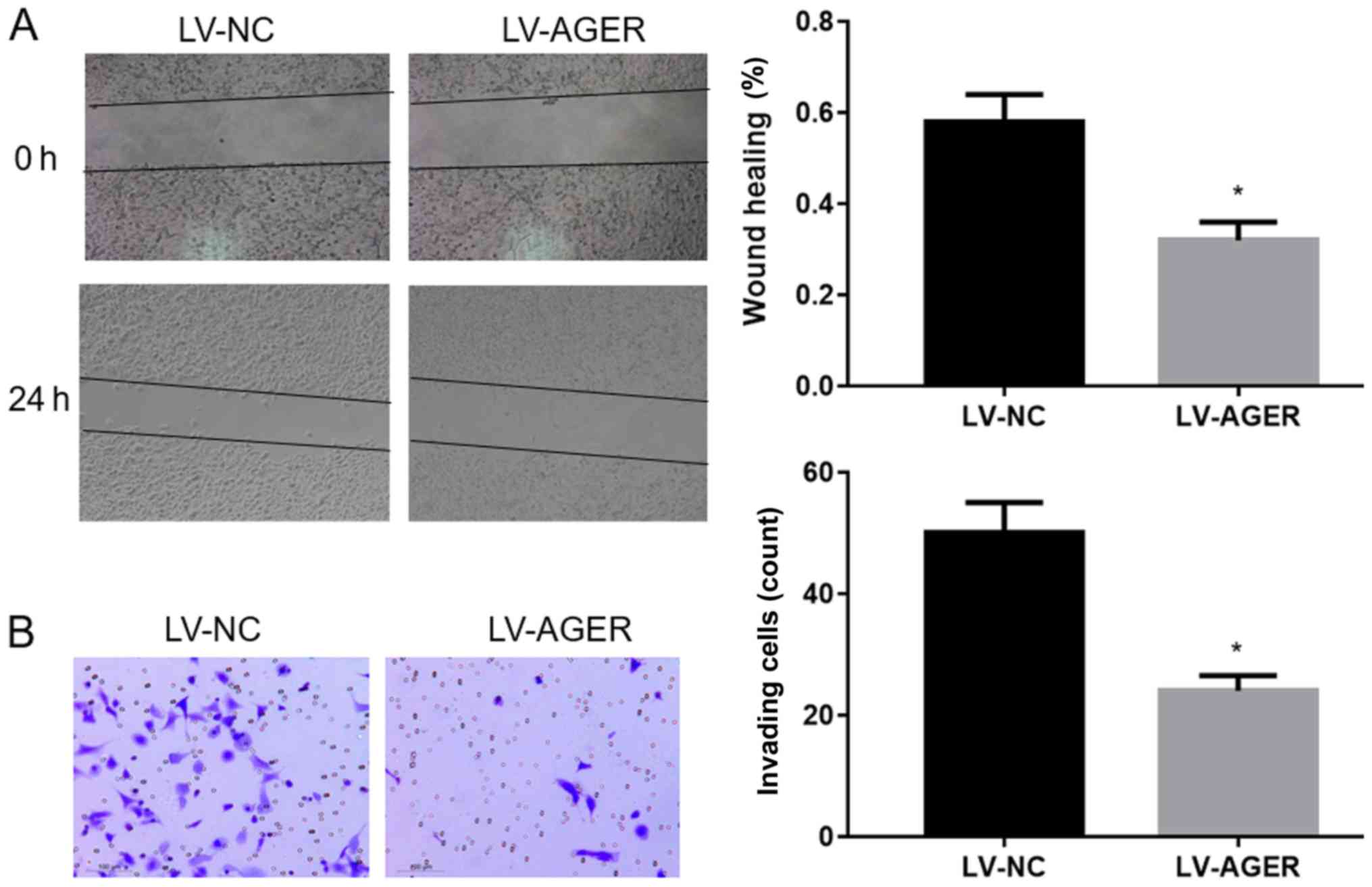
AGER overexpression decreases the
migration and invasion of H1299 cells
The migratory ability of H1299 cells was assessed
using a wound healing assay, and the results demonstrated that cell
migration was significantly decreased in the LV-AGER group compared
with that in the LV-NC group (Fig.
3A; P<0.05). Furthermore, Transwell invasion assays
were conducted to investigate the invasive ability of H1299 cells.
Compared with the LV-NC group, the invasive ability of the LV-AGER
group was significantly decreased (Fig. 3B; P<0.05). The results
indicated that AGER overexpression decreased the invasion and
migration abilities of NSCLC cells.
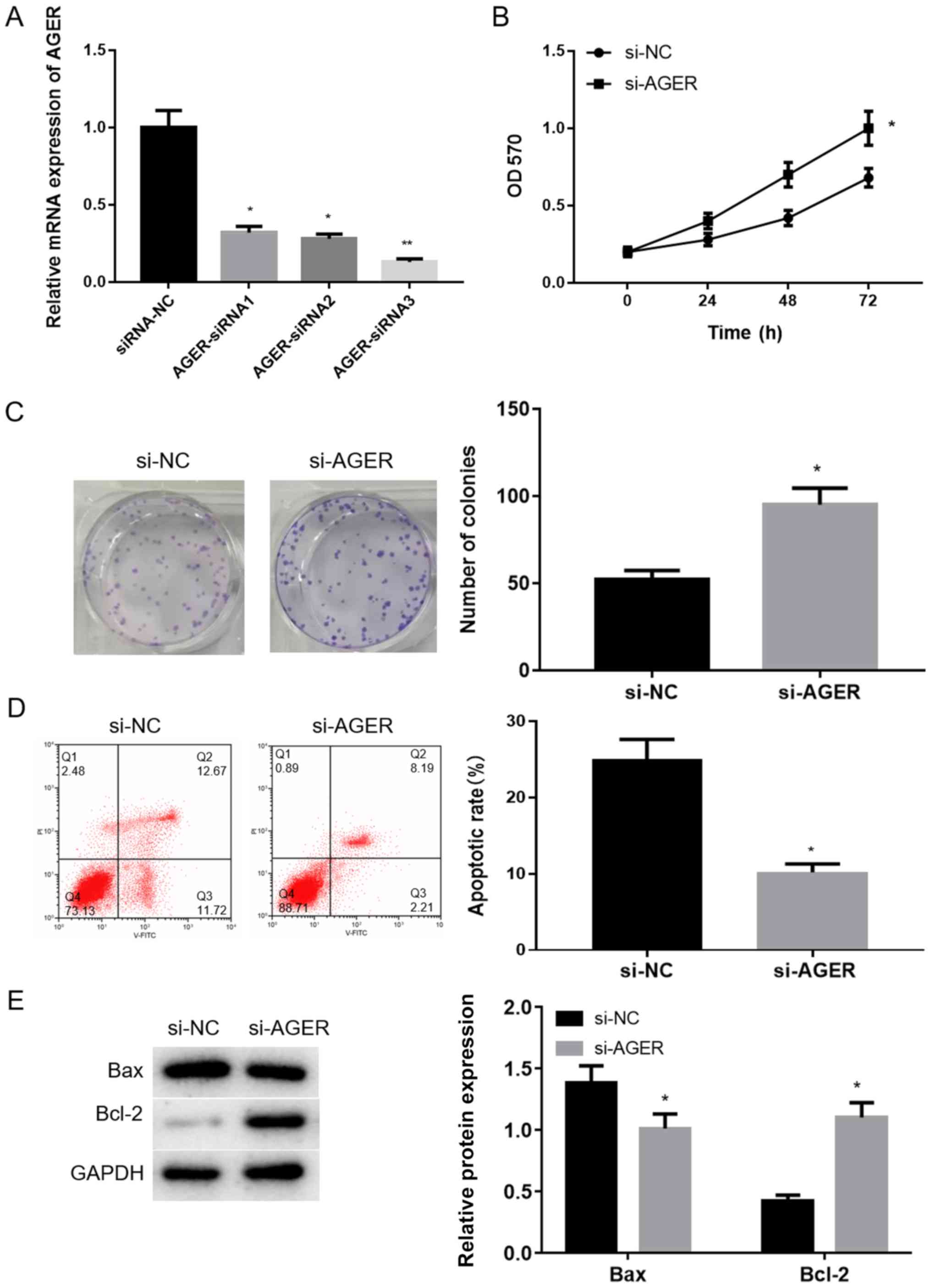
AGER knockdown increases proliferation
and decreases apoptosis of H1299 cells
The effects of AGER on NSCLC cells were further
investigated using AGER knockdown cells. RT-qPCR was performed to
measure the transfection efficiency. AGER-siRNA1 and AGER-siRNA2
significantly decreased AGER mRNA expression compared with the
siRNA-NC group (Fig. 4A;
P<0.05). The most significant reduction in AGER mRNA was
observed in the AGER-siRNA3 group compared with the siRNA-NC group
(Fig. 4A; P<0.01).
Therefore, AGER-siRNA3 transfected cells were selected for
subsequent experiments.
The results of the MTT assay indicated that cells
transfected with AGER-siRNA3 exhibited significantly increased cell
viability compared with cells transfected with siRNA-NC (Fig. 4B; P<0.05). The colony
formation assay results (Fig. 4C)
suggested that the colony formation ability of the AGER-siRNA3
group was significantly increased compared with that of the si-NC
group (P<0.05). Flow cytometry results indicated that the
rate of apoptosis was significantly decreased in the si-AGER group
compared with the si-NC group (Fig.
4D; P<0.05). Western blotting analysis also suggested
that Bax protein expression was significantly downregulated, and
Bcl-2 protein expression was significantly upregulated in the
si-AGER group compared with the si-NC group (Fig. 4E; P<0.05).
The aforementioned results further indicated that
AGER knockdown promoted the proliferation and colony formation
ability of H1299 cells but decreased apoptosis.
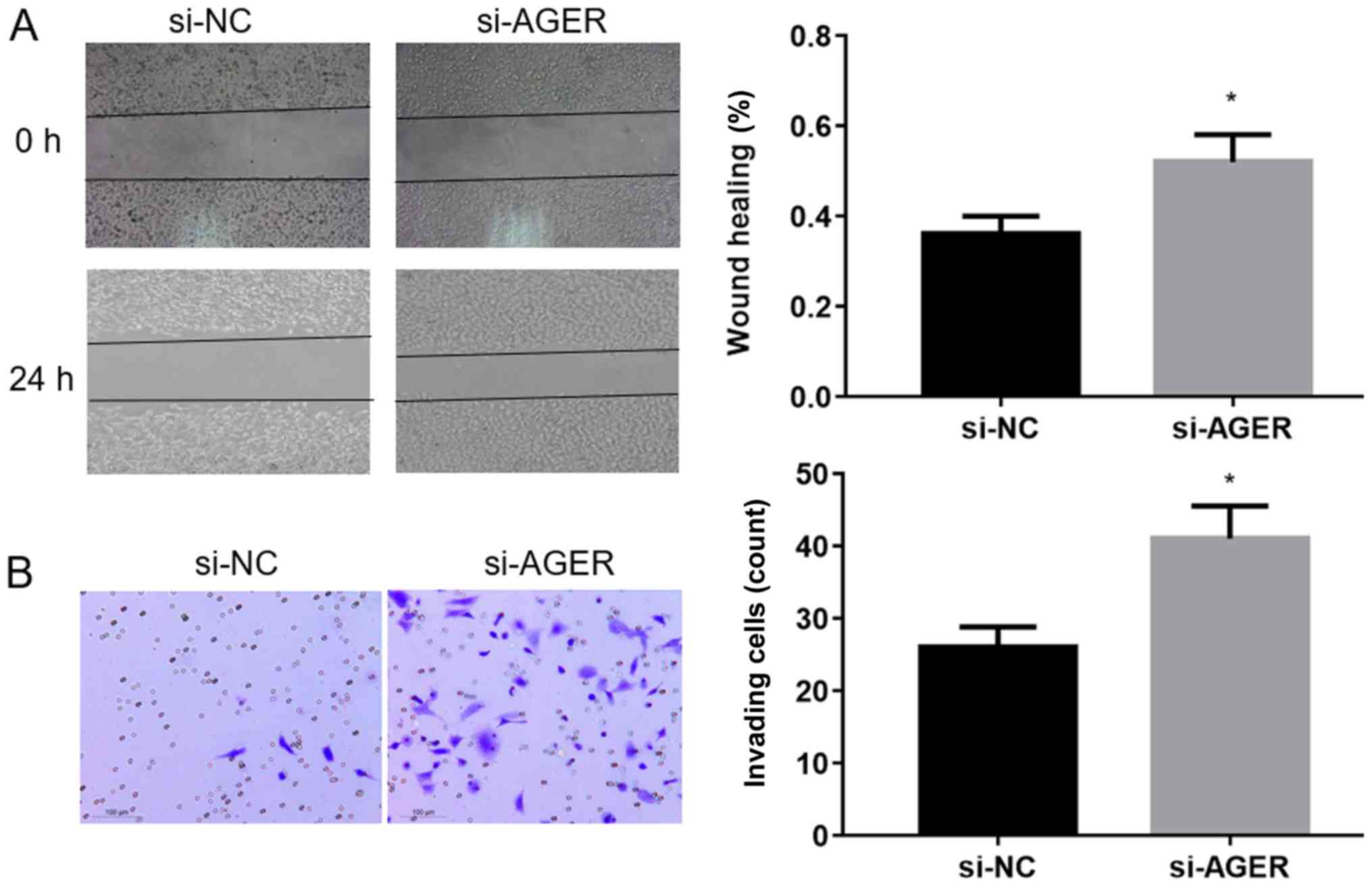
AGER knockdown increases the migration
and invasion abilities of H1299 cells
The wound healing assay was performed to assess the
migration ability of H1299 cells transfected with AGER-siRNA3. The
results suggested that the migration ability of H1299 cells
transfected with AGER-siRNA3 was significantly increased compared
with the si-NC group (Fig. 5A;
P<0.05). The Transwell invasion assay was conducted to
investigate the invasive ability of H1299 cells transfected
AGER-siRNA3. The AGER-siRNA3 group displayed significantly
increased invasive abilities compared with the si-NC group
(Fig. 5B; P<0.05). The
results indicated that AGER knockdown increased the migration and
invasion of H1299 cells.
Discussion
Analysis of the GEO and TCGA databases indicated
that AGER was differentially expressed in NSCLC tissues and
downregulated in patients with NSCLC compared with normal controls.
Previous studies have also reported that AGER may be used as a
potential prognostic biomarker for lung adenocarcinoma (17,22,31),
which supports the results of the present study.
To the best of our knowledge, the association
between AGER and NSCLC has only been analyzed using bioinformatics
analysis in the relevant literature (17,26),
and the effects of AGER on the biological behavior of NSCLC have
not been reported. Therefore, the effects of AGER on the
proliferation, apoptosis and migration of the NSCLC H1299 cell line
were investigated in the present study. AGER overexpression reduced
the proliferation and colony formation ability of H1299 cells, and
increased the rate of apoptosis. AGER overexpression also
significantly upregulated Bax protein expression and downregulated
Bcl-2 protein expression in H1299 cells. It has been documented
that upregulating Bax or downregulating Bcl-2 promotes apoptosis
(30). These results indicated
that AGER overexpression decreased the proliferation and increased
the apoptosis of H1299 cells.
In addition, the results of the wound healing and
Transwell invasion assays indicated that AGER overexpression
decreased the migration and invasion of H1299 cells. Transfection
experiments were performed to knockdown AGER expression. AGER
knockdown increased the proliferation, migration and invasion of
H1299 cells and decreased apoptosis. The results further indicated
the effects of AGER on the biological behavior of the H1299 cell
line. However, the molecular mechanism underlying the effects of
AGER on lung cancer requires further investigation. A previous
study has reported that AGER may be downregulated in A549 cells due
to oxidative-dependent activation of p38 MAPK and NF-kB (26). Another study has also reported that
LINC00173 regulates NSCLC via the AGER/NF-κB signaling pathway
(18). Therefore, it was
hypothesized that AGER may exert its effects on lung cancer via the
NF-κB signaling pathway, which requires further investigation.
In present study, the effect of AGER on the
proliferation, apoptosis and migration of H1299 cells was
investigated. The results further suggested that AGER may serve as
a potential therapeutic target for NSCLC. However, there were
certain limitations to the present study. For example, the
regulatory effect of AGER on NSCLC has not been observed in
vivo, and the specific mechanism of AGER action has not been
studied in depth. However, the present provided further research
evidence for AGER as a potential therapeutic target for NSCLC.
In conclusion, the results of the present study
demonstrated that AGER was significantly downregulated in the H1299
cell line compared with the normal lung cell line. Functional tests
revealed the antioncogenic characteristics of AGER in NSCLC cells,
and AGER overexpression decreased the proliferation and migration,
and increased apoptosis of NSCLC cells.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The dataset (GSE27262) analyzed during the current
study is available in the Gene Expression Omnibus (GEO) database
(www.ncbi.nlm.nih.gov/geo). The
expression level of AGER was validated using TCGA database
(ualcan.path.uab.edu/cgi-bin/ualcan-res.pl).
Authors' contributions
HL and QW contributed to the study design. WZ
conducted the literature search. GX and MD acquired the data from
GEO database and performed data analysis. QW wrote the manuscript.
JC revised the manuscript. HL gave the final approval of the
version to be submitted. All authors contributed to data analysis,
drafting and revising the manuscript and agreed to be accountable
for all aspects of the work. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhang F, Yang R, Zhang G, Cheng R, Bai Y,
Zhao H, Lu X, Li H, Chen S, Li J, et al: Anticancer function of
alpha-solanine in lung adenocarcinoma cells by inducing
microRNA-138 expression. Tumour Biol. 37:6437–6446. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Sun B, Liu HF, Ding Y and Li Z: Evaluating
the diagnostic and prognostic value of serum miR-770 in non-small
cell lung cancer. Eur Rev Med Pharmacol Sci. 22:3061–3066.
2018.PubMed/NCBI
|
|
3
|
Shi H, Bi H, Sun X, Dong H, Jiang Y, Mu H,
Li W, Liu G, Gao R and Su J: Tubeimoside-1 inhibits the
proliferation and metastasis by promoting miR-126-5p expression in
non-small cell lung cancer cells. Oncol Lett. 16:3126–3134.
2018.PubMed/NCBI
|
|
4
|
Torre LA, Siegel RL and Jemal A: Lung
Cancer Statistics. Adv Exp Med Biol. 893:1–19. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chu K, Gao G, Yang X, Ren S, Li Y, Wu H,
Huang Y and Zhou C: MiR-512-5p induces apoptosis and inhibits
glycolysis by targeting p21 in non-small cell lung cancer cells.
Int J Oncol. 48:577–586. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wei J, Ma Z, Li Y, Zhao B, Wang D and Jin
Y and Jin Y: miR-143 inhibits cell proliferation by targeting
autophagy-related 2B in non-small cell lung cancer H1299 cells. Mol
Med Rep. 11:571–576. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhou YL, Xu YJ and Qiao CW: MiR-34c-3p
suppresses the proliferation and invasion of non-small cell lung
cancer (NSCLC) by inhibiting PAC1/MAPK pathway. Int J Clin Exp
Pathol. 8:6312–6322. 2015.PubMed/NCBI
|
|
8
|
Li X, Li M, Chen D, Shi G and Zhao H:
PAQR3 inhibits proliferation via suppressing PI3K/AKT signaling
pathway in non-small cell lung cancer. Arch Med Sci. 14:1289–1297.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li Q, Xia S, Yin Y, Guo Y, Chen F and Jin
P: miR-5591-5p regulates the effect of ADSCs in repairing diabetic
wound via targeting AGEs/AGER/JNK signaling axis. Cell Death Dis.
9:5662018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xu Y, Toure F, Qu W, Lin L, Song F, Shen
X, Rosario R, Garcia J, Schmidt AM and Yan SF: Advanced glycation
end product (AGE)-receptor for AGE (RAGE) signaling and
up-regulation of Egr-1 in hypoxic macrophages. J Biol Chem.
285:23233–23240. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chong SF, Lee JH, Zelikin AN and Caruso F:
Tuning the permeability of polymer hydrogel capsules: an
investigation of cross-linking density, membrane thickness, and
cross-linkers. Langmuir. 27:1724–1730. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bongarzone S, Savickas V, Luzi F and Gee
AD: Targeting the Receptor for Advanced Glycation Endproducts
(RAGE): A Medicinal Chemistry Perspective. J Med Chem.
60:7213–7232. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Jing RR, Cui M, Sun BL, Yu J and Wang HM:
Tissue-specific expression profiling of receptor for advanced
glycation end products and its soluble forms in esophageal and lung
cancer. Genet Test Mol Biomarkers. 14:355–361. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Nankali M, Karimi J, Goodarzi MT, Saidijam
M, Khodadadi I, Razavi AN and Rahimi F: Increased Expression of the
Receptor for Advanced Glycation End-Products (RAGE) Is Associated
with Advanced Breast Cancer Stage. Oncol Res Treat. 39:622–628.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang D, Li T, Ye G, Shen Z, Hu Y, Mou T,
Yu J, Li S, Liu H and Li G: Overexpression of the receptor for
advanced glycation endproducts (RAGE) is associated with poor
prognosis in gastric cancer. PLoS One. 10:e01226972015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zheng L, Li D, Zhou YM, Yang H, Cheng D
and Ma XX: Effects of receptor for advanced glycation endproducts
on microvessel formation in endometrial cancer. BMC Cancer.
16:932016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhang W, Fan J, Chen Q, Lei C, Qiao B and
Liu Q: SPP1 and AGER as potential prognostic biomarkers for lung
adenocarcinoma. Oncol Lett. 15:7028–7036. 2018.PubMed/NCBI
|
|
18
|
Yang Q, Tang Y, Tang C, Cong H, Wang X,
Shen X and Ju S: Diminished LINC00173 expression induced miR-182-5p
accumulation promotes cell proliferation, migration and apoptosis
inhibition via AGER/NF-κB pathway in non-small-cell lung cancer. Am
J Transl Res. 11:4248–4262. 2019.PubMed/NCBI
|
|
19
|
Fu J, Khaybullin R, Liang X, Morin M, Xia
A, Yeh A and Qi X: Discovery of gene regulation pattern in lung
cancer by gene expression profiling using human tissues. Genom
Data. 3:112–115. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Pan Z, Liu L, Nie W, Miggin S, Qiu F, Cao
Y, Chen J, Yang B, Zhou Y, Lu J and Yang L: Long non-coding RNA
AGER-1 functionally upregulates the innate immunity gene AGER and
approximates its anti-tumor effect in lung cancer. Mol Carcinog.
57:305–318. 2018. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Serveaux-Dancer M, Jabaudon M, Creveaux I,
Belville C, Blondonnet R, Gross C, Constantin JM, Blanchon L and
Sapin V: Pathological implications of receptor for advanced
glycation end-product (AGER) gene polymorphism. Dis Markers.
2019:20673532019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liu W, Ouyang S, Zhou Z, Wang M, Wang T,
Qi Y, Zhao C, Chen K and Dai L: Identification of genes associated
with cancer progression and prognosis in lung adenocarcinoma:
Analyses based on microarray from Oncomine and The Cancer Genome
Atlas databases. Mol Genet Genomic Med. 7:e005282019.PubMed/NCBI
|
|
23
|
Beucher J, Boelle PY, Busson PF,
Muselet-Charlier C, Clement A and Corvol H; French C F Modifier
Gene Study Investigators, : AGER-429T/C is associated with an
increased lung disease severity in cystic fibrosis. PLoS One.
7:e419132012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Caraher EJ, Kwon S, Haider SH, Crowley G,
Lee A, Ebrahim M, Zhang L, Chen LC, Gordon T, Liu M, et al:
Receptor for advanced glycation end-products and World Trade Center
particulate induced lung function loss: A case-cohort study and
murine model of acute particulate exposure. PLoS One.
12:e01843312017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Machahua C, Montes-Worboys A,
Planas-Cerezales L, Buendia-Flores R, Molina-Molina M and
Vicens-Zygmunt V: Serum AGE/RAGEs as potential biomarker in
idiopathic pulmonary fibrosis. Respir Res. 19:2152018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
de Bittencourt Pasquali MA, Gelain DP,
Zeidan-Chulia F, Pires AS, Gasparotto J, Terra SR and Moreira JC:
Vitamin A (retinol) downregulates the receptor for advanced
glycation endproducts (RAGE) by oxidant-dependent activation of p38
MAPK and NF-kB in human lung cancer A549 cells. Cell Signal.
25:939–954. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wei TY, Juan CC, Hisa JY, Su LJ, Lee YC,
Chou HY, Chen JM, Wu YC, Chiu SC, Hsu CP, et al: Protein arginine
methyltransferase 5 is a potential oncoprotein that upregulates G1
cyclins/cyclin-dependent kinases and the phosphoinositide
3-kinase/AKT signaling cascade. Cancer Sci. 103:1640–1650. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Campbell KJ and Tait SWG: Targeting BCL-2
regulated apoptosis in cancer. Open Biol. 8(pii): 1800022018.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Hassan M, Watari H, AbuAlmaaty A, Ohba Y
and Sakuragi N: Apoptosis and molecular targeting therapy in
cancer. Biomed Res Int. 2014:150845. 2014. View Article : Google Scholar
|
|
31
|
Yu DH, Huang JY, Liu XP, Ruan XL, Chen C,
Hu WD and Li S: Effects of hub genes on the clinicopathological and
prognostic features of lung adenocarcinoma. Oncol Lett.
19:1203–1214. 2020.PubMed/NCBI
|