Introduction
Sevoflurane, a volatile anesthetic agent, is widely
used for pediatric anesthesia in the clinic, and is characterized
by rapid onset and offset, and low airway irritation and blood/gas
partition coefficient (1).
Previous evidence has suggested that long-term exposure to volatile
anesthetics have side effects on brain development. It is reported
that sevoflurane administration can impair memory processes,
spatial memory, and the ability of the hippocampus to learn tasks
in human and animal brains (2–4).
Thus, concerns regarding the side effects of sevoflurane in
children undergoing surgery have been highlighted.
An increasing number of studies have explored the
molecular mechanisms that underlie the effects of sevoflurane
exposure on the brain (1,4–6). The
neurotoxicity of sevoflurane was shown to decrease guanylate kinase
concentration in glutamatergic synapses in the development of rat
brains (1). Sevoflurane-induced
memory impairment is closely related with the suppression of
glycogen synthase in the hippocampus (4). In addition, memory impairment
following sevoflurane exposure is also reported to occur due to the
decreased cytosolic calcium concentration and µ-calpain activity
(5). Previous studies suggest that
sevoflurane exposure alters the expression of genes involved in
cognitive function-related metabolic pathways (6). Another previous study suggested that
sevoflurane exposure leads to changes in the expression of
receptors and enzymes involved in amyloid β clearance, which
contributes to Alzheimer's disease development (7,8). The
cognitive dysfunction induced by sevoflurane has been shown to be
relieved by mediating the Toll-like receptor 4/myeloid
differentiation primary response 88/NF-κB signaling pathway
(8).
MicroRNAs (miRNAs/miRs) have been reported to be
involved in neuropsychiatric disorders and impairments in cognitive
function (9,10). However, the role of miRNAs in
sevoflurane-induced neurotoxicity has not been fully clarified. The
binding of Wnt ligands to receptors/co-receptors promotes Wnt
signaling activation (11).
Previous studies have revealed that the WNT signaling pathway is
one of the main signaling pathways involved in numerous types of
disease, including osteoporosis, cancer and diabetes (12,13).
Moreover, it has been identified that miRNAs serve important
regulatory roles in the Wnt signaling pathway (14). However, the association between
miRNAs and Wnt signaling in sevoflurane-induced neurotoxicity
remains largely unknown. In this paper, miRNA expression in
hippocampus samples from newborn mice was characterized by
microarray technology. The miRNAs with differential expression were
determined, followed by function and pathway enrichment analysis.
The aim of this study was to explore the mechanism underlying the
effect of sevoflurane on the developing brain and facilitates the
discovery of safe anesthetic strategies.
Materials and methods
Animals
A total of four 15-day-old pregnant Institute of
Cancer Research (ICR) mice were purchased from Shanghai Sipubikai
Laboratory Animal Co., Ltd. The pregnant mice were maintained in an
animal house at a temperature of 20±2°C and humidity of 55±5%, with
a 12-h light/dark cycle, and free access to food and water. After
parturition, the healthy 7-day-old ICR mice were used for further
analysis (15).
Approval was obtained from Animal Care and Ethics
Committee of Zhejiang Chinese Medical University and all the animal
procedures were performed according to the ethical standards.
Experimental groups. A total of six 7-day-old male
ICR mice (weight, 250±10 g) were randomly assigned into two groups
(n=3/group): The Sevoflurane group and Control group. Mice in the
Sevoflurane group were administered with 2.4% sevoflurane for 6 h
consecutively between 9:00 am and 3:00 pm according to previously
described methods (16). Animals
in the Control group were treated with the same dose of normal
saline solution via a venous catheter at a rate of 1.0 ml/h. After
treatment, all the animals were breast-fed for 14 days followed by
a Morris water maze test (17).
Morris water maze
The water maze was comprised of a cylindrical pool
(height, 50 cm; diameter, 80 cm) and a platform (diameter, 10 cm).
The water surface was 2 cm higher than the platform and water was
maintained at 22±0.5°C. The animals were trained to find the
platform and stayed for 2 min in the water maze twice/day for two
consecutive days. The pool was divided into four quadrants. Rats
were randomly delegated into the four quadrants, and then were
placed in the water, facing the wall of the pool. If the mice
failed to find the platform within 2 min, mice were placed on the
platform for 20 sec. The second trial was conducted after a delay
of 5–10 sec. The Morris water maze experiments were monitored by
videos recorded on a computer. After training, the time for mice to
reach to platform (escape latency) was recorded within 120 sec.
Then, a probe trial was conducted after removing platform from the
pool, and the mice were placed in a given quadrant and allowed to
explore the maze for 120 sec. The cross-platform path length,
percentage of the total trial path length that passed through the
platform location, duration that the mouse stayed on the platform
and the number of times across the platform were recorded.
RNA isolation
After the multiple behavioral tests on each mouse,
mice were sacrificed, and hippocampal tissues were isolated. Total
RNA was extracted by TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.) according to the manufacturer's
instructions. The purity of RNA was detected by a NanoDrop ND-2000
(NanoDrop Technologies; Thermo Fisher Scientific, Inc.) and the
quality was assessed by an Agilent Bioanalyzer 2100 (Agilent
Technologies, Inc.).
miRNA profiling with microarray
Total RNA samples with an RNA integrity number >9
were used for microarray analysis. RNA (250 ng) from each sample
was dephosphorylated, degenerated and labeled with Cyanine-3-CTP
(Cy3) according to the manufacturer's instructions for the Human
miRNA Microarray kit (Agilent Technologies, Inc.). After purifying,
RNA was hybridized to gene arrays at 55°C for 20 h and scanned on
an Agilent Scanner G2505C system (Agilent Technologies, Inc). The
raw data were exported to .txt format by Agilent Feature Extraction
(FE) software (version 9.5.3; Agilent Technologies, Inc.) for
further analysis.
Data preprocessing and differential
expression analysis
The text format data were transformed by Affy
package v1.50.0 (18) in R
(http://www.bioconductor.org/packages/release/bioc/html/affy.html)
and preprocessed by the robust multi-array average method (19,20),
including background correction, normalization and expression
calculation.
Compared with controls, the miRNAs with differential
expression in the Sevoflurane group were assessed by limma v3.26.9
(http://bioconductor.org/packages/release/bioc/html/limma.html)
(21). The P-values and fold
change (FC) in expression of miRNAs were calculated. P<0.01 and
|log2FC|>0.263 was considered to indicate a
statistically significant difference (22,23).
Hierarchical clustering of differentially expressed miRNAs was
performed by pheatmap package version 1.0.8 (https://cran.r-project.org/web/packages/pheatmap).
Prediction of miRNA target genes
The differentially expressed miRNAs were uploaded to
the miRNA-Gene Targets module of miRWalk 2.0 (http://zmf.umm.uni-heidelberg.de/apps/zmf/mirwalk2/miRretsys-self.html)
(24,25). Subsequently, the miRNA targets were
also predicted by miRanda version 3.0 (http://cbio.mskcc.org/microrna), miRDB version 4.0
(http://mirdb.org/miRDB), PITA (http://genie.weizmann.ac.il/pubs/mir07/mir07_data.html),
RNA22 version 2.0 (https://cm.jefferson.edu/rna22), RNAhybrid version
2.12 (https://bibiserv.cebitec.uni-bielefeld.de/rnahybrid)
and TargetScan version 7.0 (http://www.targetscan.org/) databases.
In order to explore the biological functions of
differentially expressed miRNAs, the predicted target genes were
subjected to Gene Ontology (GO) (26) and Kyoto Encyclopedia of Genes and
Genomes (KEGG) pathway enrichment analysis (27) using clusterProfiler version 2.4.3
(https://bioconductor.org/packages/release/bioc/html/clusterProfiler.html)
(28). P<0.05 was set as the
threshold value.
miRNA-mRNA regulatory network
construction
The predicted miRNA-target pairs were identified,
and the miRNA-target regulatory network was constructed by
Cytoscape version 2.8 software (29). The topology of the miRNA regulatory
network was analyzed and the hub nodes with significant degrees
were screened out.
Statistical analysis
The data are expressed as mean ± SD. Differences
between groups were compared using t-tests. P<0.05 was
considered to indicate a statistically significant difference.
Results
Sevoflurane exposure affects learning
and memory function in newborn mice
During learning, following exposure to sevoflurane,
mice in the Sevoflurane group (119.33±4.04 sec) showed longer mean
latency time to reach the platform than those in the Control group
(13.67±2.08 sec; P<0.001; Table
I). During the probe trial, the cross-platform path length was
significantly shorter in sevoflurane-treated mice (71.19±2.45 mm),
compared with Controls (92.06±2.09 mm; P<0.001). Similarly, the
percentage of the total path length that went through the platform
location was significantly lower in sevoflurane-exposed mice
(0.62±0.02) compared with the Control group (0.83±0.03;
P<0.001). The platform duration in mice exposed to sevoflurane
was significantly declined (0.65±0.04 sec) compared with the
Control group (0.83±0.06 sec; P=0.0028). In addition, the mean
number of times across platform for mice treated with sevoflurane
(1.33±0.58) was not significantly lower than the Control group
(2.67±0.58; P=0.47).
 | Table I.Sevoflurane exposure affects the
learning and memory ability in mice. |
Table I.
Sevoflurane exposure affects the
learning and memory ability in mice.
| Phases of
trial | Index | Sevoflurane
group | Control group | P-value |
|---|
| Learning trial | Mean escape
latency, sec | 119.33±4.04 | 13.67±2.08 | <0.001 |
| Probe trial | Cross-platform path
length, mm | 71.19±2.45 | 92.06±2.09 | <0.001 |
|
| Percentage of total
path length in platform | 0.62±0.02 | 0.83±0.03 | <0.001 |
|
| Platform duration,
sec | 0.65±0.04 | 0.83±0.06 | 0.0028 |
|
| Platform entries,
number of events | 1.33±0.58 | 2.67±0.58 | 0.47 |
Data preprocessing
Based on the raw data, expression information for
52,044 miRNAs was available. The expression profiles of 49,880
miRNAs were obtained, followed by data preprocessing. After miRNA
overlaps were removed, the mean expression values of 1,247 mature
miRNAs were calculated following previously published descriptions
(30).
Differentially expressed miRNAs
With P<0.01 and |log2FC|>0.263
(18,19), the expression of 18 miRNAs,
including 11 upregulated miRNAs (miR-1897-5p, miR-188-5p,
miR-3098-5p, miR-3095-3p, miR-5107-5p, miR-3470a, miR-705,
miR-5126, miR-149-3p, miR-1187 and miR-1982-5p) and seven
downregulated miRNAs (miR-425-5p, miR-101b-3p, miR-92a-3p,
miR-338-3p, miR-467a-3p, miR-219-5p and miR-219-2-3p) were found to
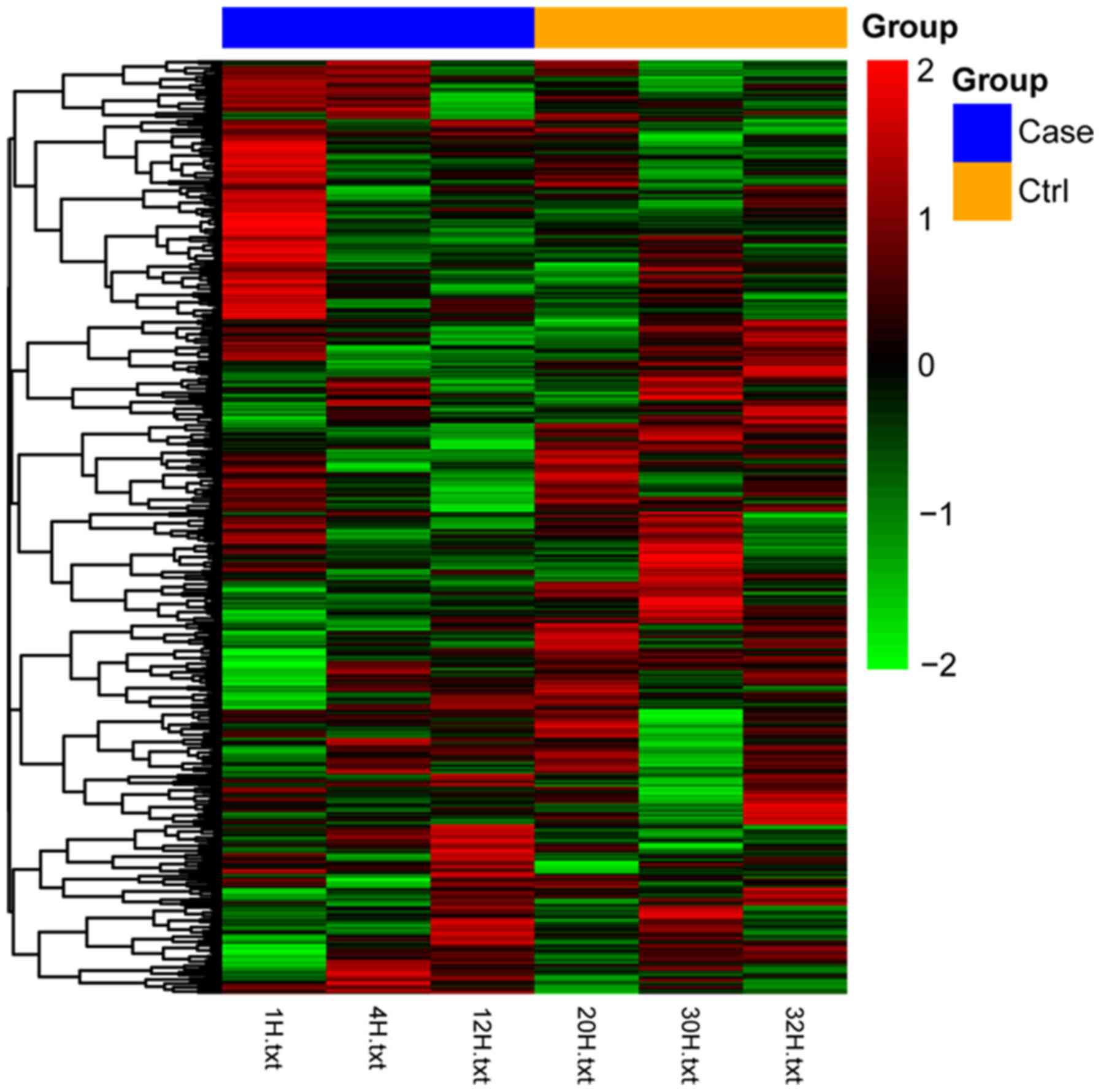
be significantly affected by sevoflurane administration. The
heatmap of differentially expressed miRNAs, which illustrate that
the samples in different groups can be distinguished by the
expression profiles of differentially expressed miRNAs, is
presented in Fig. 1. Upregulated
miRNAs in the sevoflurane group are shown in red, whereas
downregulated miRNAs are presented as green.
miRNA-target genes and function
enrichment analysis
Among 18 differentially expressed miRNAs, 3
upregulated miRNAs (miR-1187, miR-188-5p and miR-705) and 6
downregulated miRNAs (miR-101b-3p, miR-219a-5p, miR-338-3p,
miR-425-5p, miR-467a-3p and miR-92a-3p) were identified to have
1,252 target genes based on the information of seven miRNA related
databases. After the overlaps were removed, 1,095 target genes were
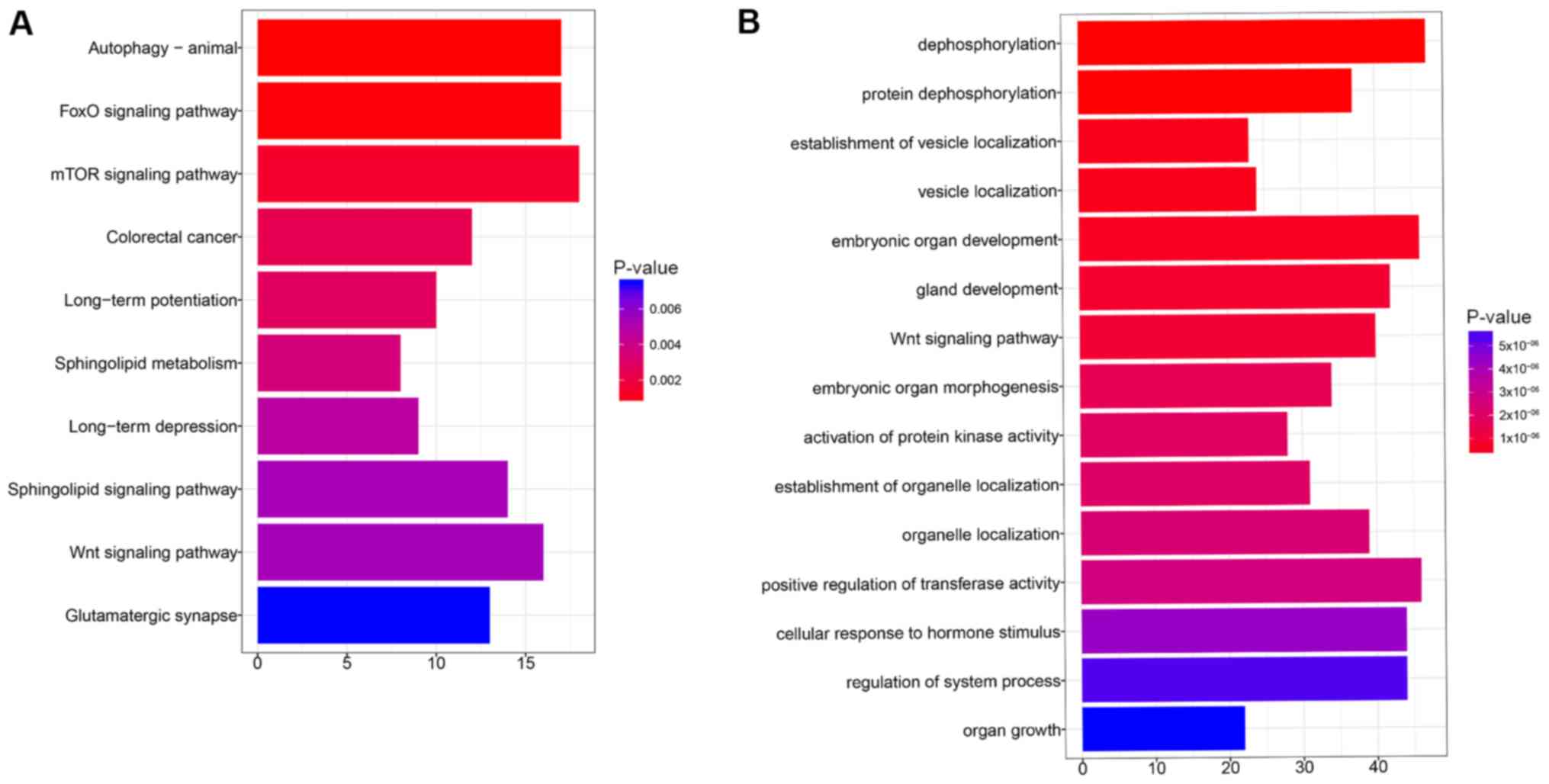
obtained. Pathway enrichment analysis showed that the target genes
were closely involved in the ‘FoxO signaling pathway’, ‘mTOR
signaling pathway’ and ‘Wnt signaling pathway’ (Fig. 2A). Target genes clustered into
different function groups such as ‘dephosphorylation’, ‘vesicle
localization’ and the ‘Wnt signaling pathway’ (Fig. 2B).
miRNA-related functions and
pathways
Based on the miRNA-target gene information, the
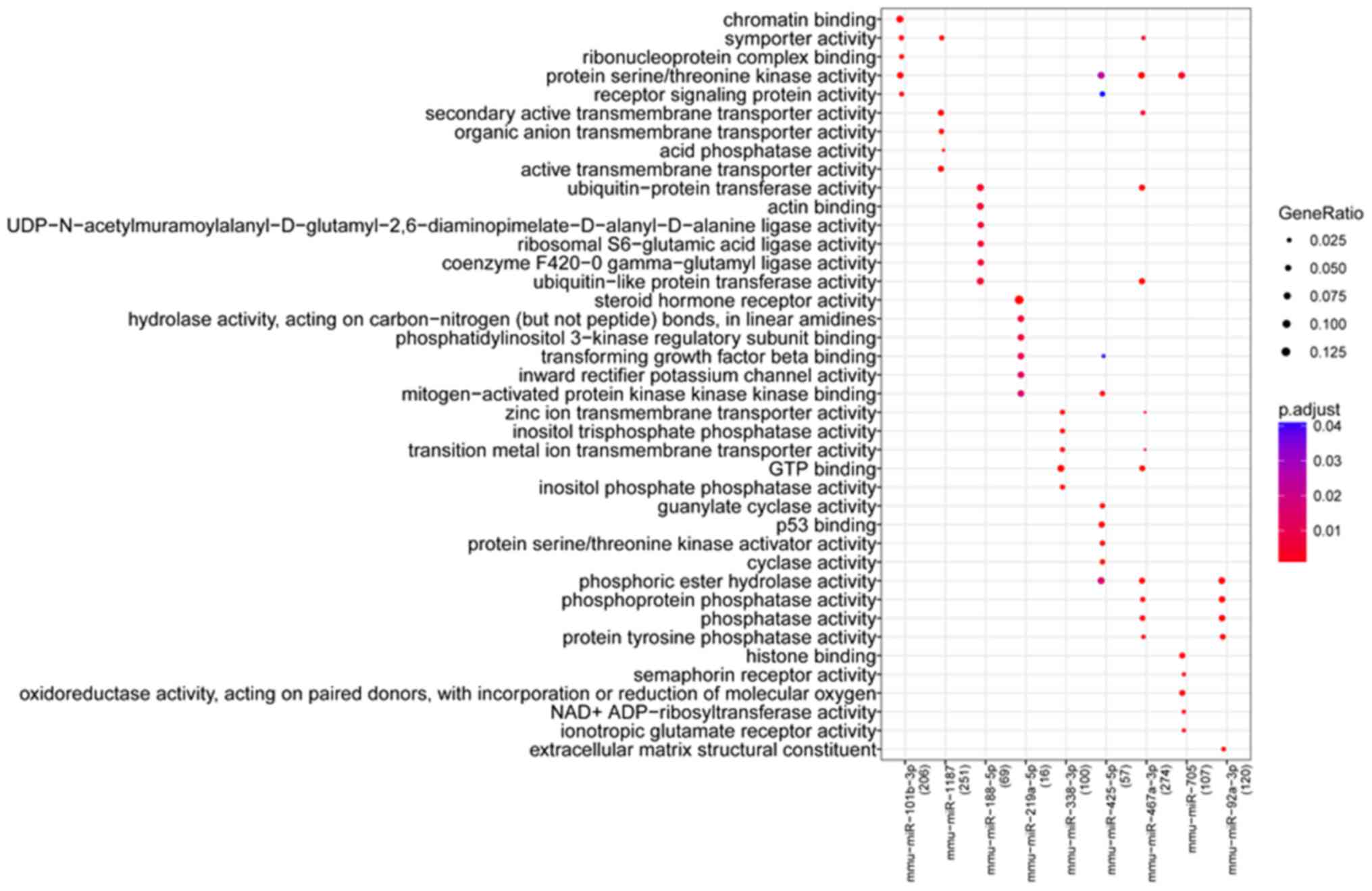
biological function and pathways of the nine miRNAs were assessed
by GO and pathway enrichment analysis. Results identified 190
biological process terms and 27 pathways significantly enriched by
the nine miRNAs. Results showed that the biological functions and
pathways closely related with miRNAs were relatively different. The
top five GO function terms (ranked by P-value) of each miRNA are
listed in Fig. 3. miR-101b-3p was
closely related with ‘chromatin binding’ and ‘protein
serine/threonine kinase activity’. miR-1187 was significantly
enriched in ‘secondary active transmembrane transporter activity’,
‘organic anion transmembrane transporter activity’ and ‘active
transmembrane transporter activity’. miR-219a-5p was involved in
‘steroid hormone receptor activity’, ‘phosphatidylinositol 3-kinase
regulatory subunit binding’ and ‘transforming growth factor-β
binding’. miR-467a-3p and miR-92a-3p were closely related with
phosphatase activity.
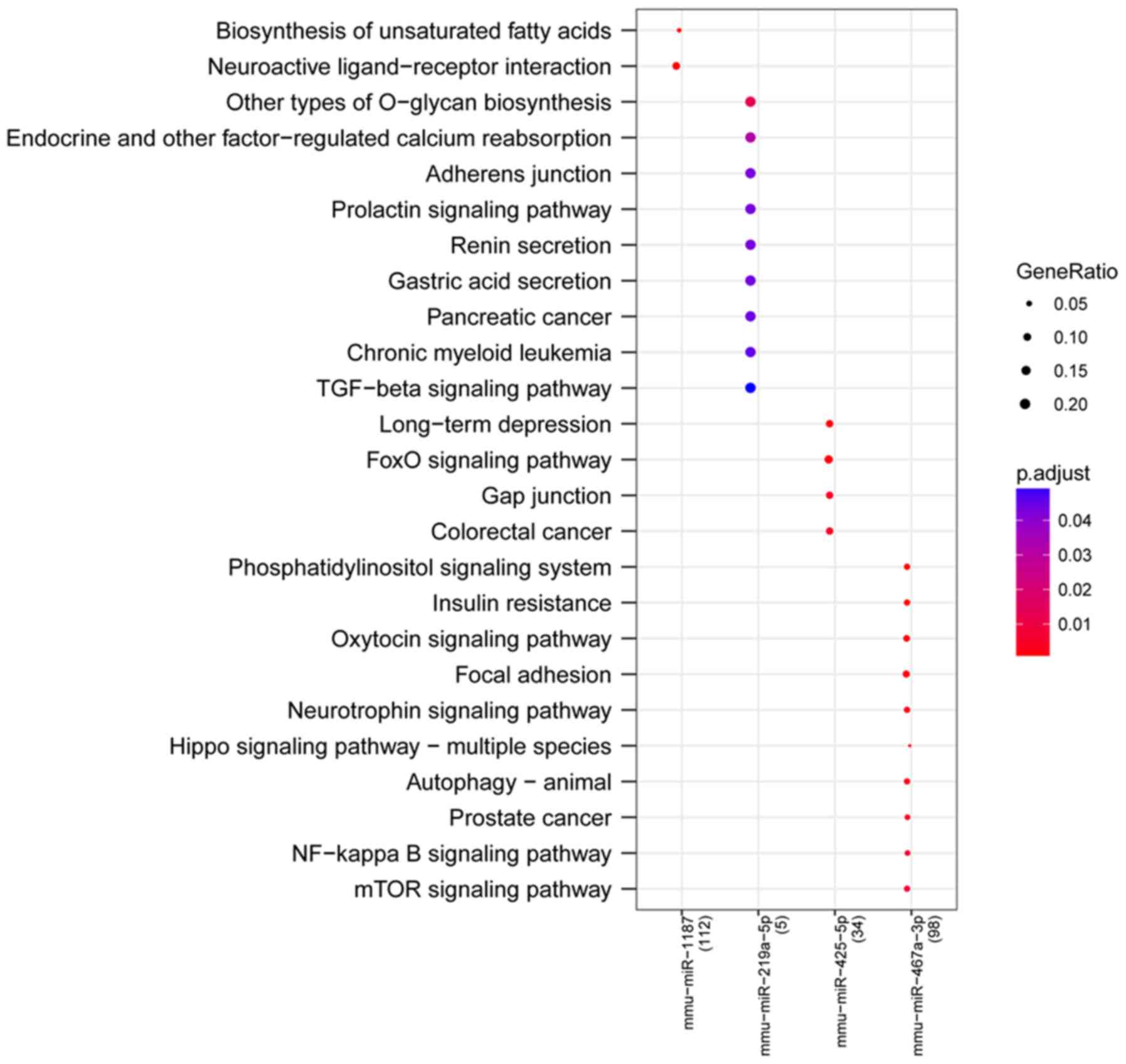
Pathways that were particularly affected by four
miRNAs (miR-1187, miR-219a-5p, miR-425-5p and miR-467a-3p) were
screened out by KEGG signaling pathway enrichment analysis. The
most significantly affected pathways for miRNAs included
‘biosynthesis of unsaturated fatty acids’ (miR-1187), ‘neuroactive
ligand-receptor interaction’ (miR-1187), ‘other types of O-glycan
biosynthesis’ (miR-219a-5p), ‘long-term depression’ (miR-425-5p),
‘FoxO signaling pathway’ (miR-425-5p), ‘phosphatidylinositol
signaling system’ (miR-467a-3p), ‘neurotrophin signaling pathway’
(miR-467a-3p), ‘Hippo signaling pathway-multiple species’ and
‘NF-κB signaling pathway’ (miR-467a-3p; Fig. 4).
miRNA-target gene regulatory
network
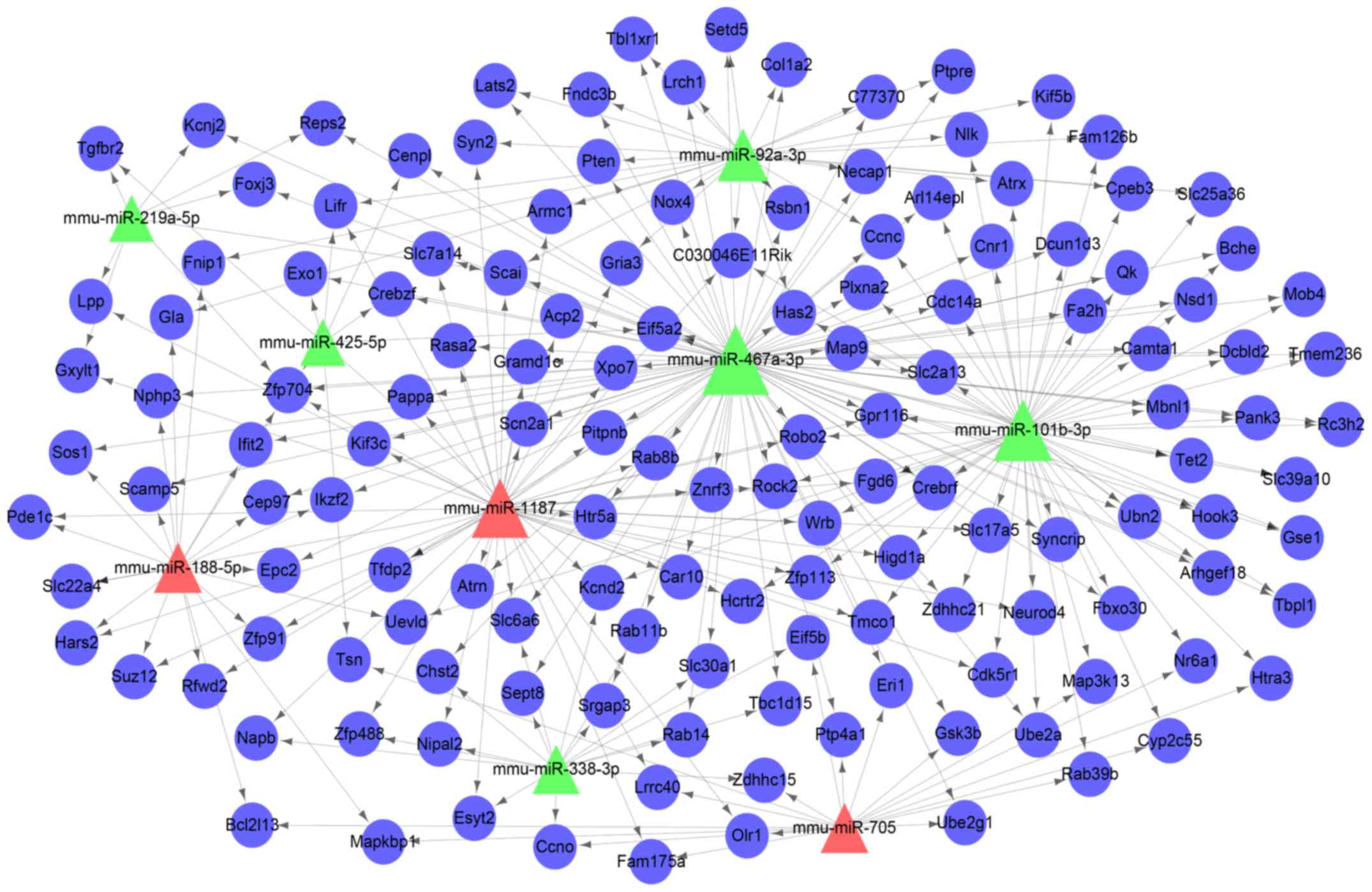
A miRNA regulatory network containing the nine
miRNAs and 141 genes was constructed (Fig. 5). The top 10 nodes with the highest
degrees were listed in Table II.
The hub nodes included miR-467a-3p (degree=89), miR-101b-3p
(degree=59), miR-1187 (degree=51) and Zfp704 (degree=5).
 | Table II.Top 10 nodes in the miR regulatory
network. |
Table II.
Top 10 nodes in the miR regulatory
network.
| Name | Degree | Regulated |
|---|
| miR-467a-3p | 89 | Down |
| miR-101b-3p | 59 | Down |
| miR-1187 | 51 | Up |
| miR-92a-3p | 28 | Down |
| miR-188-5p | 19 | Up |
| miR-705 | 18 | Up |
| miR-338-3p | 16 | Down |
| miR-425-5p | 9 | Down |
| miR-219a-5p | 8 | Down |
| Zfp704 | 5 | / |
Discussion
The alterations of the miRNA expression profile in
the hippocampus tissues of mice exposed to sevoflurane may provide
novel insight to understand the molecular mechanisms of memory and
learning impairments resulting from altered development of the
brain and allow opportunities to develop new therapeutic management
strategies. The aim of this paper was to identify differentially
expressed miRNAs in hippocampus tissues between mice exposed to
sevoflurane and controls, and elucidate the molecular mechanisms
that govern memory impairment in sevoflurane-exposed mice. The
results identified 18 differentially expressed miRNAs. After
function and pathway enrichment analysis, the target genes of
differentially expressed miRNAs were found to be significantly
enriched in the ‘Wnt signaling pathway’.
A previous study also showed that the dysregulated
miRNAs were closely related with Wnt signaling pathways underlying
sevoflurane-induced neurotoxicity in the development of mice brains
(31), which was consistent with
the present results. Ye et al (31) predicted the gene targets for miRNAs
with TargetScan, miRanda and PicTar. In the present study, the
target genes for dysregulated miRNAs were predicted by miRWalk 2.0
combined with miRanda, miRDB, PITA, RNA22, RNAhybrid and TargetScan
databases, which enhanced the accuracy of significant pathway
identification and indicated the importance of the Wnt signaling
pathway in the impaired cognition resulting from exposure to
sevoflurane.
The Wnt signaling pathway is involved in various
biological processes, such as cell proliferation, tissue
regeneration, stem cell renewal and axon guidance (15,32,33).
It is reported that the Wnt signaling pathway is involved with ~3%
of differentially expressed transcripts that are prominent in the
brain and spinal cord after spinal cord injury (SCI) in lampreys
(34). Previous work has suggested
an essential role for Wnt signaling in developing and adult brains
(35). Blocking Wnt signaling has
been shown to inhibit functional recovery following SCI (34), which indicates the significant role
of Wnt signaling in neural repair and regeneration of the
peripheral nervous system. In addition, synapse degeneration is
closely associated with cognitive impairment and deficits in
learning and memory. Evidence suggests that synapse degeneration,
as an early event in neurodegenerative disease, is linked with Wnt
signaling deficiency (36). The
Wnt signaling pathway has been proposed as a therapeutic target for
neuronal circuit recovery following synapse degeneration. A
previous study also showed that reactivation of the Wnt signaling
pathway improves neuroblast formation and neural function in the
brain after focal cerebral ischemia in mice (37). Additionally, the Wnt signaling
cascade is involved in the neuronal differentiation of human
non-neural tissue-derived stem cells (35). Wnt signaling pathways are
implicated in the differentiation of neural stem cells in human
brain development (38). Taken
together, these findings indicated a significant role of the Wnt
signaling pathway in mediating cognitive disorders in brains
exposed to sevoflurane.
The miRNA target gene regulatory network illustrated
that miR-467a-3p (degree=89), miR-101b-3p (degree=59), and miR-1187
(degree=51) were hub nodes with multiple connections with target
genes, which suggested a regulatory role for these miRNAs. The
upregulated miR-188-5p (degree=19) was found to be another
significant node in the miRNA regulatory network. A recent study
suggested that miR-188-3p was upregulated in sevoflurane-treated
mice and involved in sevoflurane-induced cognitive dysfunction
(39), which was consistent with
the present results. miR-188-5p is an alternative mature body of
miR-188 and, to our knowledge, has not yet been reported to be
dysregulated following sevoflurane exposure. It has been reported
that miRNA-188-3p targeting mouse double minute 2 plays a
significant role in sevoflurane-induced apoptosis pathways
(39). The key role of miR-188-5p
in miRNA-target gene regulatory networks may provide new insight
into further gene targets in cognitive impairment induced by
sevoflurane.
A recent study showed that overexpression of
miR-467a-3p inhibits the neural differentiation of mouse embryonic
stem cells (ESCs) (40). Neural
stem cells differentiated from ESCs are suggested to be involved in
cognition impairment-related diseases in humans (41,42).
miR-467a-3p has also previously been found to be involved in the
apoptosis of vascular smooth muscle cells (43). The present study showed that
miR-467a-3p was differentially expressed in the hippocampus tissues
of mice exposed to sevoflurane, which indicated that the neural
differentiation and proliferation were dysregulated upon
sevoflurane exposure. In addition, the pathway enrichment analysis
conducted in this study showed that miR-467a-3p was closely related
with the neurotrophin signaling pathway. Neurotrophic factors are
implicated in the development and maintenance of the nervous system
(44). The dysregulation of
neurotrophin signaling has been found to be associated with
neurodegeneration in Alzheimer's disease (44). Taken together, these data suggested
that miR-467a-3p may play a significant regulatory role in the
maintenance of neuron function.
Furthermore, miR-1187 has been found to be a novel
miRNA in the inhibition of osteoblast differentiation (45). miR-101b-3p has been found to be
enriched in hepatocytes and is markedly upregulated following
hepatocyte damage (46). Although
evidence of the role of miR-1187 and miR-101b-3p in the regulation
of plasticity in the hippocampus is lacking, the present study
showed that miR-1187 was closely associated with ‘neuroactive
ligand-receptor interaction’ and miR-101b-3p was closely related
with ‘receptor signaling protein activity’. The differential
expression of miR-1187 and miR-101b-3p may impact neuroactive
signaling interactions in brains exposed to sevoflurane.
In conclusion, the present findings suggested that
the Wnt signaling pathway is involved in mediating cognitive
disorder upon exposure to sevoflurane. miR-467a-3p may play a
significant regulatory role in the maintenance of neuron function.
miR-1187 and miR-101b-3p may be implicated in the regulation of
neuroactive signaling interactions. The miRNAs and their related
pathways may be important therapeutic targets to prevent
sevoflurane-induced memory and learning disorders.
Acknowledgements
Not applicable.
Funding
This study was supported by Zhejiang Medical and
Health Science and Technology Plan (grant no. 2014KYA159), Zhejiang
Traditional Chinese Medicine Science and Technology Plan (grant no.
2019ZA047) and Natural Science Foundation of Zhejiang Province
(grant no. LY20H150002).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
HS and ZL conceived and designed the study; HH and
XX acquired the data and analyzed and interpreted the data; TT
performed the statistical analysis; HS drafted the manuscript; and
ZL revised the manuscript for important intellectual content. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Approval was obtained from Animal Care and Ethics
Committee of Zhejiang Chinese Medical University and all the animal
procedures were performed according to the institution's ethical
standards.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang SQ, Fang F, Xue ZG, Cang J and Zhang
XG: Neonatal sevoflurane anesthesia induces long-term memory
impairment and decreases hippocampal PSD-95 expression without
neuronal loss. Eur Rev Med Pharmacol Sci. 17:941–950.
2013.PubMed/NCBI
|
|
2
|
Xiao H, Liu B, Chen Y and Zhang J:
Learning, memory and synaptic plasticity in hippocampus in rats
exposed to sevoflurane. Int J Dev Neurosci. 48:38–49. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kamal A, Ramakers G, Gispen WH and
Biessels GJ: Hyperinsulinemia in rats causes impairment of spatial
memory and learning with defects in hippocampal synaptic plasticity
by involvement of postsynaptic mechanisms. Exp Brain Res.
226:45–51. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Liu XS, Xue QS, Zeng QW, Li Q, Liu J, Feng
XM and Yu BW: Sevoflurane impairs memory consolidation in rats,
possibly through inhibiting phosphorylation of glycogen synthase
kinase-3β in the hippocampus. Neurobiol Learn Mem. 94:461–467.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu X, Song X, Yuan T, He J, Wang X and
Wang Q: Effects of calpain on sevoflurane-induced aged rats
hippocampal neuronal apoptosis. Aging Clin Exp Res. 28:633–639.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ge X, Zhang Y, Zuo Y, Israr M, Li B, Yu P,
Gao G, Chang YZ and Shi Z: Transcriptomic analysis reveals the
molecular mechanism of Alzheimer-related neuropathology induced by
sevoflurane in mice. J Cell Biochem. 120:17555–17565. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Liu Y, Gao M, Ma L, Zhang L and Pan N:
Sevoflurane alters the expression of receptors and enzymes involved
in Aβ clearance in rats. Acta Anaesthesiol Scand. 57:903–910. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Li Y, Liu L, Tian Y and Zhang J: Rapamycin
improves sevoflurane-induced cognitive dysfunction in aged rats by
mediating autophagy through the TLR4/MyD88/NF-κB signaling pathway.
Mol Med Rep. 20:3085–3094. 2019.PubMed/NCBI
|
|
9
|
Xu B, Hsu PK, Karayiorgou M and Gogos JA:
MicroRNA dysregulation in neuropsychiatric disorders and cognitive
dysfunction. Neurobiol Dis. 46:291–301. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Maffioletti E, Tardito D, Gennarelli M and
Bocchio-Chiavetto L: Micro spies from the brain to the periphery:
New clues from studies on microRNAs in neuropsychiatric disorders.
Front Cell Neurosci. 8:752014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Veltri A, Lang C and Lien WH: Concise
review: Wnt Signaling pathways in skin development and epidermal
stem cells. Stem Cells. 36:22–35. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Amjadi-Moheb F and Akhavan-Niaki H: Wnt
signaling pathway in osteoporosis: Epigenetic regulation,
interaction with other signaling pathways, and therapeutic
promises. J Cell Physiol. 234:14641–14650. 2019. View Article : Google Scholar
|
|
13
|
Clevers H and Nusse R: Wnt/β-catenin
signaling and disease. Cell. 149:1192–1205. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Mahmood S, Bhatti A, Syed NA and John P:
The microRNA regulatory network: A far-reaching approach to the
regulate the Wnt signaling pathway in number of diseases. J Recept
Signal Transduct Res. 36:310–318. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Shih J, May LD, Gonzalez HE, Lee EW, Alvi
RS, Sall JW, Rau V, Bickler PE, Lalchandani GR, Yusupova M, et al:
Delayed environmental enrichment reverses sevoflurane-induced
memory impairment in rats. Anesthesiology. 116:586–602. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ishikawa M, Tanaka S, Arai M, Genda Y and
Sakamoto A: Differences in microRNA changes of healthy rat liver
between sevoflurane and propofol anesthesia. Anesthesiology.
117:1245–1252. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Su D, Zhao Y, Wang B, Xu H, Li W, Chen J
and Wang X: Isoflurane-induced spatial memory impairment in mice is
prevented by the acetylcholinesterase inhibitor donepezil. PLoS
One. 6:e276322011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Gautier L, Cope L, Bolstad BM and Irizarry
RA: Affy-analysis of affymetrix GeneChip data at the probe level.
Bioinformatics. 20:307–315. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bolstad BM, Irizarry RA, Astrand M and
Speed TP: A comparison of normalization methods for high density
oligonucleotide array data based on variance and bias.
Bioinformatics. 19:185–193. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Irizarry RA, Hobbs B, Collin F,
Beazer-Barclay YD, Antonellis KJ, Scherf U and Speed TP:
Exploration, normalization, and summaries of high density
oligonucleotide array probe level data. Biostatistics. 4:249–264.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ritchie ME, Phipson B, Wu DI, Hu Y, Law
CW, Shi W and Smyth GK: Limma powers differential expression
analyses for RNA-sequencing and microarray studies. Nucleic Acids
Res. 43:e472015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu J, Zhu M, Lv M, Wu X, Zhang X, Zhang Y,
Li J and Zhang Q: Characterization of a five-microRNA signature as
a prognostic biomarker for esophageal squamous cell carcinoma. Sci
Rep. 9:198472019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhang S, Liu W, Liu X, Qi J and Deng C:
Biomarkers identification for acute myocardial infarction detection
via weighted gene co-expression network analysis. Medicine
(Baltimore). 96:e83752017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Breuer K, Foroushani AK, Laird MR, Chen C,
Sribnaia A, Lo R, Winsor GL, Hancock REW, Brinkman FSL and Lynn DJ:
InnateDB: Systems biology of innate immunity and beyond-recent
updates and continuing curation. Nucleic Acids Res. 41((Database
issue)): D1228–D1233. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dweep H and Gretz N: miRWalk2.0: A
comprehensive atlas of microRNA-target interactions. Nat Methods.
12:697. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ashburner M, Ball CA, Blake JA, Botstein
D, Butler H, Cherry JM, Davis AP, Dolinski K, Dwight SS, Eppig JT,
et al: Gene ontology: Tool for the unification of biology. The gene
ontology consortium. Nat Genet. 25:25–29. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Kanehisa M and Goto S: KEGG: Kyoto
encyclopaedia of genes and genomes. Nucleic Acids Res. 28:27–30.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Yu G, Wang LG, Han Y and He QY:
clusterProfiler: An R package for comparing biological themes among
gene clusters. OMICS. 16:284–287. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Shannon P, Markiel A, Ozier O, Baliga NS,
Wang JT, Ramage D, Amin N, Schwikowski B and Ideker T: Cytoscape: A
software environment for integrated models of biomolecular
interaction networks. Genome Res. 13:2498–2504. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Jaksik R, Polańska J, Herok R and
Rzeszowska-Wolny J: Calculation of reliable transcript levels of
annotated genes on the basis of multiple probe-sets in affymetrix
microarrays. Acta Biochim Pol. 56:271–277. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Ye J, Zhang Z, Wang Y, Chen C, Xu X, Yu H
and Peng M: Altered hippocampal microRNA expression profiles in
neonatal rats caused by sevoflurane anesthesia: MicroRNA profiling
and bioinformatics target analysis. Exp Ther Med. 12:1299–1310.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Kyritsis N, Kizil C, Zocher S, Kroehne V,
Kaslin J, Freudenreich D, Iltzsche A and Brand M: Acute
inflammation initiates the regenerative response in the adult
zebrafish brain. Science. 338:1353–1356. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
MacDonald BT, Tamai K and He X:
Wnt/β-catenin signaling: Components, mechanisms, and diseases. Dev
Cell. 17:9–26. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Herman PE, Papatheodorou A, Bryant SA,
Waterbury CKM, Herdy JR, Arcese AA, Buxbaum JD, Smith JJ, Morgan JR
and Bloom O: Highly conserved molecular pathways, including Wnt
signaling, promote functional recovery from spinal cord injury in
lampreys. Sci Rep. 8:7422018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
L'Episcopo F, Tirolo C, Caniglia S, Testa
N, Morale MC, Serapide MF, Pluchino S and Marchetti B: Targeting
Wnt signaling at the neuroimmune interface for dopaminergic
neuroprotection/repair in parkinson's disease. J Mol Cell Biol.
6:13–26. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Marzo A, Galli S, Lopes D, McLeod F,
Podpolny M, Segovia-Roldan M, Ciani L, Purro S, Cacucci F, Gibb A
and Salinas PC: Reversal of synapse degeneration by restoring Wnt
signaling in the adult hippocampus. Curr Biol. 26:2551–2561. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qiu CW, Liu ZY, Hou K, Liu SY, Hu YX,
Zhang L, Zhang FL, Lv KY, Kang Q, Hu WX, et al: Wip1 knockout
inhibits neurogenesis by affecting the Wnt/β-catenin signaling
pathway in focal cerebral ischemia in mice. Exp Neurol. 309:44–53.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Vangipuram SD and Lyman WD: Ethanol
affects differentiation-related pathways and suppresses Wnt
signaling protein expression in human neural stem cells. Alcohol
Clin Exp Res. 36:788–797. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang L, Zheng M, Wu S and Niu Z:
MicroRNA-188-3p is involved in sevoflurane anesthesia-induced
neuroapoptosis by targeting MDM2. Mol Med Rep. 17:4229–4236.
2018.PubMed/NCBI
|
|
40
|
Zhang L, Xue Z, Yan J, Wang J, Liu Q and
Jiang H: LncRNA Riken-201 and Riken-203 modulates neural
development by regulating the Sox6 through sequestering miRNAs.
Cell Prolif. 52:e125732019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
De Filippis L, Zalfa C and Ferrari D:
Neural stem cells and human induced pluripotent stem cells to model
rare CNS diseases. CNS Neurol Disord Drug Targets. 16:915–926.
2017.PubMed/NCBI
|
|
42
|
Sugaya K and Vaidya M: Stem cell therapies
for neurodegenerative diseases. Adv Exp Med Biol. 1056:61–84. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Cui R, Ye S, Zhong J, Liu L, Li S, Lin X,
Yuan L and Yi L: MicroRNA-494 inhibits apoptosis of murine vascular
smooth muscle cells in vitro. Mol Med Rep. 19:4457–4467.
2019.PubMed/NCBI
|
|
44
|
Chen XQ, Sawa M and Mobley WC:
Dysregulation of neurotrophin signaling in the pathogenesis of
alzheimer disease and of alzheimer disease in down syndrome. Free
Radic Biol Med. 114:52–61. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
John AA, Prakash R, Kureel J and Singh D:
Identification of novel microRNA inhibiting actin cytoskeletal
rearrangement thereby suppressing osteoblast differentiation. J Mol
Med (Berl). 96:427–444. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Takeuchi M, Oda S, Tsuneyama K and Yokoi
T: Comprehensive analysis of serum microRNAs in hepatic sinusoidal
obstruction syndrome (SOS) in rats: Implication as early phase
biomarkers for SOS. Arch Toxicol. 92:2947–2962. 2018. View Article : Google Scholar : PubMed/NCBI
|