Introduction
Ossification of the ligamentum flavum (OLF) is one
of the degenerative changes to the ligamentum flavum (LF) that
occur with aging (1). Since the
first report of OLF in 1920, OLF has been investigated mainly in
Asian countries (2). In China and
Japan, the prevalence of OLF is 3.8% (3) and 6.2% (4), respectively. OLF arises from the
lateral capsular portion and exhibits continuity with the bony
laminae (5). Clinical
manifestations of OLF include regional thoracic stiffness and pain,
which usually progresses to myelopathy (6). The diagnosis of OLF is often missed
or delayed due to its slow progression and the co-occurrence of
other spinal diseases (5,6), which results in a heavy burden to
both the patients and society.
OLF is a heterotopic ossification that occurs in the
ligament (5,6). OLF usually starts from the
degeneration and hypertrophy of the elastic fibrous tissues, which
results in the production of fibrocartilage-like cells and the
proliferation of chondrocytes (7).
Subsequently, chondrocytes are calcified, and with the growth of
small vessels into the LF tissues, mature bone structures are
formed (7). However, the etiology
of OLF is not fully understood. Multiple factors have been
implicated in the development of OLF, including genetics (8), systematic hormones (9), inflammatory markers (10) and mechanical stressors (11). Moreover, OLF is closely related to
disturbance of the local mechanical microenvironment of the spine
(12). The reduced stability of
the middle and lower thoracic joints (T) due to long-term
rotational motions has been indicated the anatomical basis of OLF
(13). A previous study reported
T9-T12 are the common segments affected by OLF, suggesting that
increases in local stress caused by longitudinal axial instability
of the middle and lower thoracic spine may be the biomechanical
basis of OLF (14).
In rats, after periodic mechanical stress
stimulation, LF of the caudal vertebrae shows similar pathology to
that found in humans with OLF: An extensive proliferation of the
chondrocyte-like cells and ectopic cartilage formation near the
insertion point of the LF (15).
In addition, Cai et al (16) found that the expression levels of
the ossification markers osteopontin (OPN) and
sex-determining region Y-box protein 9 (SOX9) are
significantly upregulated after stress stimulation. In contrast,
normal ligamentum cells did not show this significant upregulation
in expression levels (16).
Therefore, periodic mechanical stress can facilitate the
progression of OLF at the cellular level.
Indian hedgehog (IHH), a member of the
secretory morphogen signaling family, is a secreted molecule that
regulates cells via concentration gradients (17). The IHH signaling pathway is key for
bone growth and development (18).
IHH is one of the essential signaling pathways mediating
heterotopic ossification of the extremities (19). Furthermore, the IHH pathway can
directly or indirectly promote the proliferation and hypertrophy of
the chondrocytes, induce the expression of parathyroid
hormone-related peptide in cells surrounding the chondrocytes,
facilitate the differentiation of resting chondrocytes into
proliferating ones and induce differentiation of deep
perichondrocytes into osteoblasts (20,21).
In an IHH−/− mouse model, no deep
perichondrocytes differentiation into osteoblasts is observed
(22). Regard et al
(23), found that the IHH pathway
independently causes heterotopic ossification of the extremities;
furthermore, inhibition of the IHH pathway significantly reduced
the degree of heterotopic ossification. Thus, previous studies have
indicated that the IHH signaling pathway may play a role in the
process of bone development and heterotopic ossification. However,
whether the IHH signaling pathway mediates osteogenic
differentiation during ossification of LF requires further
investigation.
Currently, there is no effective medication to
prevent or delay the progress of OLF (5). Once ossification causes oppression to
the spinal cord, the only treatment solution for OLF is surgery
(5). However, OLF progression does
not decrease or stop, despite this surgical intervention (24–26).
Therefore, it is necessary to investigate the signaling pathways
underlying the progression of OLF in order to understand the
pathogenesis of OLF. The aim of the present study was to examine
the involvement of the IHH signaling pathway in the development of
OLF at the cellular and tissue levels, by simulating the in
vivo stress environment of the LF. The present results will
help in the understanding of the mechanisms underlying the
development of OLF, and provide evidence for potential targets in
novel therapeutic strategies.
Materials and methods
Patient specimens
The Ethics Committee from The Second Military
Medical University approved the present study. Participants
provided written informed consent prior to specimen collection. The
diagnosis of OLF was confirmed by clinical symptoms and
radiological examinations. Patients were included if they received
posterior open decompressive laminectomy between January 2016 and
January 2019 at Changzheng Hospital, Second Military Medical
University. A total of 18 LF tissue samples (male patients, 10;
female patients, 8; mean age, 61.2 years; age range, 52–73 years)
from patients with OLF were obtained, of which 10 samples were
harvested for cell culture. The remaining eight samples were used
for histology. The non-ossified LF samples from 12 patients were
used as controls (male patients, 7; female patients, 5; mean age,
56.2 years; age range, 42–68 years), of which four samples were
harvested for cell culture. The remaining eight samples were used
for histology. Patients in the control group underwent posterior
surgical procedures for disc herniation (n=7) and fractures (n=5).
Thus, eight samples from the OLF group and eight samples from the
control group were used for the tissue experiments. Whole pieces of
ligaments were isolated and harvested after removing the lamina
during the surgery. Patients who had congenital bone diseases or
musculoligamentous tissue abnormalities were excluded.
Cell culture
A tissue explant method (16) was used to obtain the cultured LF
cells. The LF and OLF tissues were obtained aseptically during
surgery. For OLF tissues, the ossified areas were separated and
removed under a microscope to avoid contamination with osteogenic
cells. The LF and OLF ligaments were digested in 0.25% trypsin,
followed by 250 U/ml type I collagenase (Gibco; Thermo Fisher
Scientific, Inc.). Subsequently, the fragments were washed with PBS
and cultured at 37°C with 5% CO2 in a 10-cm dish with
DMEM: Nutrient Mixture F-12 (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.)
and 100 µg/ml penicillin/streptomycin. The cultures were left
undisturbed for 2 days and then replaced with fresh medium. The
cells, which were obtained from the explants, were treated with
0.25% trypsin containing 0.02% EDTA for 1–2 min at 37°C,
re-suspended and cultured for further passages. Cells at passage
three were used for subsequent experiments.
Procedures of cyclic stretch
application
A Flexcell FX-5000 strain unit (Flexcell
International Corporation) was used in this study, with procedures
similar to a previous study (16).
Cells were cultured (5×105 cells/well) in a flexible
bottomed polystyrene plate (6 wells) with type I collagen (0.15
mg/ml) coated at the bottom (Flex I, Bioflex Plates; Flexcell
International Corporation). After cell attachment, cyclic stretch
was applied to the cells at 37°C for different durations (0, 6, 12
or 24 h). Based on a previous study (16), the stretch parameter was set at a
maximum 15% elongation. In the present study, the cultured cells
were subjected to 15% elongation for 10 sec and then relaxation for
the next 10 sec. In pathway inhibition tests, subconfluent cells
were pre-treated with 0.5 µM Cyclopamine (Cpn; Sigma-Aldrich; Merck
KGaA) for 2 h at 37°C and subsequently subjected to cyclic stretch
for 24 h at 37°C.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from OLF and LF samples or
cultured OLF cells using TRIzol® reagent (Invitrogen;
Thermo Fisher Scientific, Inc.), according to the manufacturer's
instructions. RNA concentration was then measured and converted to
cDNA using a PrimerScript RT kit (Takara Bio, Inc.), according to
the manufacturer's instructions. The mRNA expression levels of the
osteogenic genes Runt-related transcription factor 2
(RUNX2), Osterix, alkaline phosphatase (ALP)
and osteocalcin (OCN), and the IHH signaling pathway genes
IHH, Smoothened (SMO), GLI family zinc finger 1
(GLI1), GLI2 and GLI3 were quantified using
SYBR Premix Ex Taq™ II (Takara Bio, Inc.). GAPDH was used as
an endogenous control gene. The following qPCR conditions were
used: Initial denaturation at 95°C for 10 min; followed by 40
cycles of 95°C for 30 sec; and final extension at 60°C for 1 min.
The relative expression levels of each gene were calculated by the
2−ΔΔCq method (27).
Primer sequences used for qPCR are presented in Table I.
 | Table I.Primers used for reverse
transcription-quantitative PCR. |
Table I.
Primers used for reverse
transcription-quantitative PCR.
| Target | Primer
sequence | Genbank accession
no. |
|---|
| GAPDH | F:
5′-AGGTCGGTGTGAACGGATTTG-3′ | NM_008084 |
|
| R:
5′-TGTAGACCATGTAGTTGAGGTCA-3′ |
|
| RUNX2 | F:
5′-ATGCTTCATTCGCCTCACAAA-3′ | NM_001146038 |
|
| R:
5′-GCACTCACTGACTCGGTTGG-3′ |
|
| Osterix | F:
5′-ATGGCGTCCTCTCTGCTTG-3′ | AF477981.1 |
|
| R:
5′-TGAAAGGTCAGCGTATGGCTT-3′ |
|
| ALP | F:
5′-CCAACTCTTTTGTGCCAGAGA-3′ | NM_007431 |
|
| R:
5′-GGCTACATTGGTGTTGAGCTTTT-3′ |
|
| OCN | F:
5′-CTGACCTCACAGATCCCAAGC-3′ | X04142 |
|
| R:
5′-TGGTCTGATAGCTCGTCACAAG-3′ |
|
| IHH | F:
5′-CGGCTTTGACTGGGTGTATT-3′ | KR710697.1 |
|
| R:
5′-AAAATGAGCACATCGCTGAA-3′ |
|
| SMO | F:
5′-CTGTCCTGCGTCATCATCTTT-3′ | NM_005631.5 |
|
| R:
5′-CCACAGCAAGGATTGCCAC-3′ |
|
| GLI1 | F:
5′-AAGCGTGAGCCTGAATCTGT-3′ | BC013000.2 |
|
| R:
5′-CAGCATGTACTGGGCTTTGA-3′ |
|
| GLI2 | F:
5′-CGACACCAGGAAGGAAGGTA-3′ | BC172434.1 |
|
| R:
5′-TGCACAGAACGGAGGTAGTG-3′ |
|
| GLI3 | F:
5′-CTTTGCAAGCCAGGAGAAAC-3′ | BC113616.1 |
|
| R:
5′-TTGTTGGACTGTGTGCCATT-3′ |
|
Western blot analysis
Protein samples were extracted from the OLF cells
using RIPA lysis buffer (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's instructions. Detailed
procedures were described previously (28). The bicinchoninic acid protein assay
(Pierce; Thermo Fisher Scientific, Inc.) was used to measure the
protein concentration. The protein samples (~60 µg/lane) were
separated by electrophoresis using 10% separating gel and 5%
stacking gel, and then transferred onto PVDF membranes. After
blocking with skimmed milk in 5% Tris-buffered saline with 0.05%
Tween-20 for 1 h at room temperature, the membranes were
subsequently incubated overnight at 4°C with primary antibodies
targeted against: RUNX2 (cat. no. ab23981; Abcam; 1:1,000), Osterix
(cat. no. ab22552; Abcam; 1:500), ALP (cat. no. ab67228; Abcam;
1:500), OCN (cat. no. ab93876; Abcam; 1:500), IHH (cat. no.
ab39634; Abcam; 1:1,000), SMO (cat. no. ab113438; Abcam, 1:500) and
GAPDH (cat. no. ab181602; Abcam; 1:5,000). The next day, membranes
were incubated with a horseradish peroxide (HRP)-conjugated
secondary antibody (cat. no. ab205718; Abcam; 1:2,000) for 1 h at
room temperature and detected with electrochemiluminescence (GE
Healthcare). Protein expression was quantified using Image J
software (v1.51; National Institutes of Health) with GAPDH as the
loading control.
Immunohistochemical analysis
Immunohistochemistry was performed in OLF and LF
tissues following previously described (28). OLF and LF samples were fixed with
4% formaldehyde for 24 h at room temperature, embedded in paraffin
and cut at 4-µm thickness. The sections underwent antigen retrieval
at 120°C for 10 min in a citrate solution (10 mmol/l; pH 6.0).
After blocking with 1% bovine serum albumin (Gibco; Thermo Fisher
Scientific, Inc.) for 37°C at 15 min, the sections were
subsequently incubated overnight at 4°C with the following primary
antibodies: Rabbit polyclonal antibody against RUNX2 (cat. no.
ab23981; Abcam; 1:500), Osterix (cat. no. ab22552; Abcam; 1:200),
ALP (cat. no. ab224335; Abcam; 1:400), OCN (cat. no. ab93876;
Abcam; 1:200), IHH (cat. no. ab39634; Abcam; 1:200) and SMO (cat.
no. ab113438; Abcam; 1:100). The next day, the sections were
incubated with the HRP-conjugated secondary antibody (cat. no.
ab205718; Abcam; 1:2,000) for 1 h at room temperature, followed by
incubation with 3,3′-diaminobenzidine solution for 2–5 min at room
temperature and counterstaining with hematoxylin at room
temperature for 30 sec-1 min. Stained cells were identified using a
ZEISS light microscope (magnification, ×400; ZEISS Axio Imager A2;
Carl Zeiss AG). Within each section, 200 cells were counted at
random and the proportion of immunopositive cells was calculated
(29). For quantitative analysis,
images from three different sections of each sample were analyzed
using Image J software (v1.51; National Institutes of Health).
Measurement of ALP activity and
histochemical staining
ALP activity was quantitatively measured using an
LabAssay ALP kit (cat. no. 291-58601; Wako Pure Chemical
Industries, Ltd.) and histochemical staining was performed using a
ALP color development kit (cat. no. C3206; Beyotime Institute of
Biotechnology) as previously described (30). Cell pellets were obtained by
centrifugation at 3,000 × g for 5 min at 4°C. The total cellular
protein was collected from the cultured cells by lysing cell
pellets with RIPA lysis buffer (Invitrogen; Thermo Fisher
Scientific, Inc.). The concentration of the protein was measured
using the BCA method (Pierce; Thermo Fisher Scientific, Inc.). The
protein solution was then analyzed for ALP activity using the ALP
kit. The absorbance at 450 nm was measured using Spectra Max 250
spectrophotometer. The ALP-specific activity was determined using a
standard curve and normalized to the protein concentration (U/mg).
For histochemical staining, the cultured cells were washed with PBS
and then fixed in 4% formaldehyde for 10 min at room temperature.
The cells were stained with ALP reagent at 37°C for 30 min, and
ALP-positive cells were stained blue. Stained cells were identified
using a ZEISS light microscope (magnification, ×400; ZEISS Axio
Imager A2; Carl Zeiss AG). In total, five random fields in each
well were used for the analysis. ALP staining intensities were
applied to evaluate the ability of osteogenic differentiation in
OLF cells.
Statistical analysis
Statistical analyses were performed using SPSS v19.0
(SPSS, Inc.). The experiments were repeated three times. Data are
presented as the mean ± SD. The results were analyzed with a
Student's t-test, or one-way ANOVA followed by Tukey's post hoc
test or Dunnett's test where appropriate. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression of osteogenic genes in
OLF
RUNX2, ALP, Osterix and OCN are
important genes that define the osteogenesis process (31,32).
In the present study, the mRNA and protein expression levels of
RUNX2, Osterix, ALP and OCN were compared between the
LF and OLF groups (Fig. 1).
RT-qPCR and immunohistochemical analyses indicated that the OLF
group had higher mRNA and protein expression levels of RUNX2
compared with the LF group. Moreover, the OLF group had
significantly higher mRNA and protein expression levels of
Osterix, ALP and OCN compared with the LF group
(P<0.01). Therefore, the present results suggested that the
differentiation of fibroblasts into osteoblasts plays a role in OLF
development.
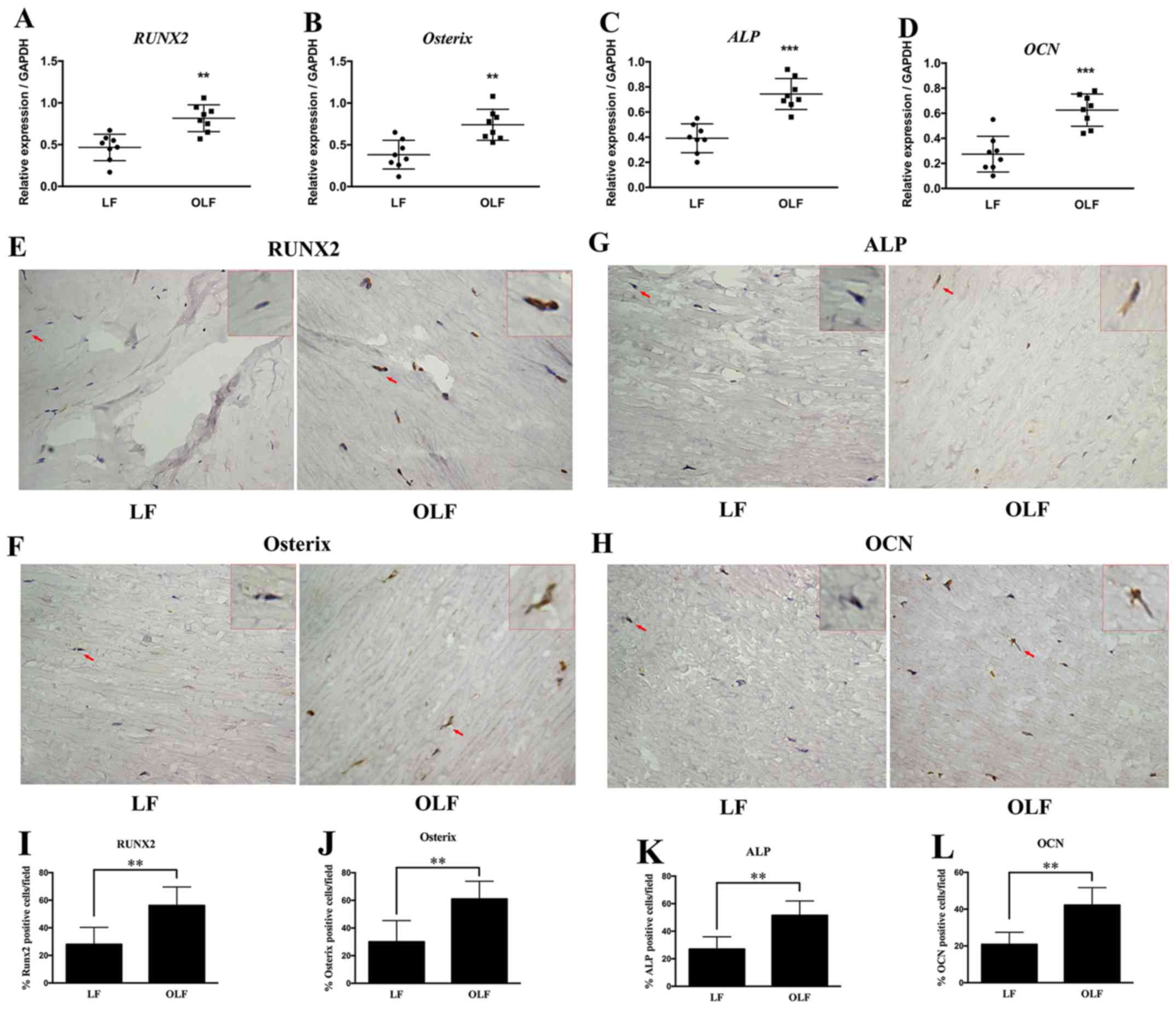 | Figure 1.Expression of RUNX2, Osterix,
ALP and OCN in LF and OLF tissues. Reverse
transcription-quantitative PCR results for the mRNA expression
levels of (A) RUNX2, (B) Osterix, (C) ALP and
(D) OCN in the LF and OLF groups. Immunohistochemistry
images of (E) RUNX2, (F) Osterix, (G) ALP and
(H) OCN. Quantification of the protein expression levels of
(I) RUNX2, (J) Osterix, (K) ALP and (L)
OCN in the LF group and OLF group. Magnification, ×400. The
red arrows represent the typical cells, which were magnified and
shown in the upper right-hand corner (magnification, × 1,600). N=8.
**P<0.01, ***P<0.001 vs. LF samples. OLF, ossification of
ligamentum flavum; LF, ligamentum flavum; RUNX2,
Runt-related transcription factor 2; ALP, alkaline
phosphatase; OCN, osteocalcin. |
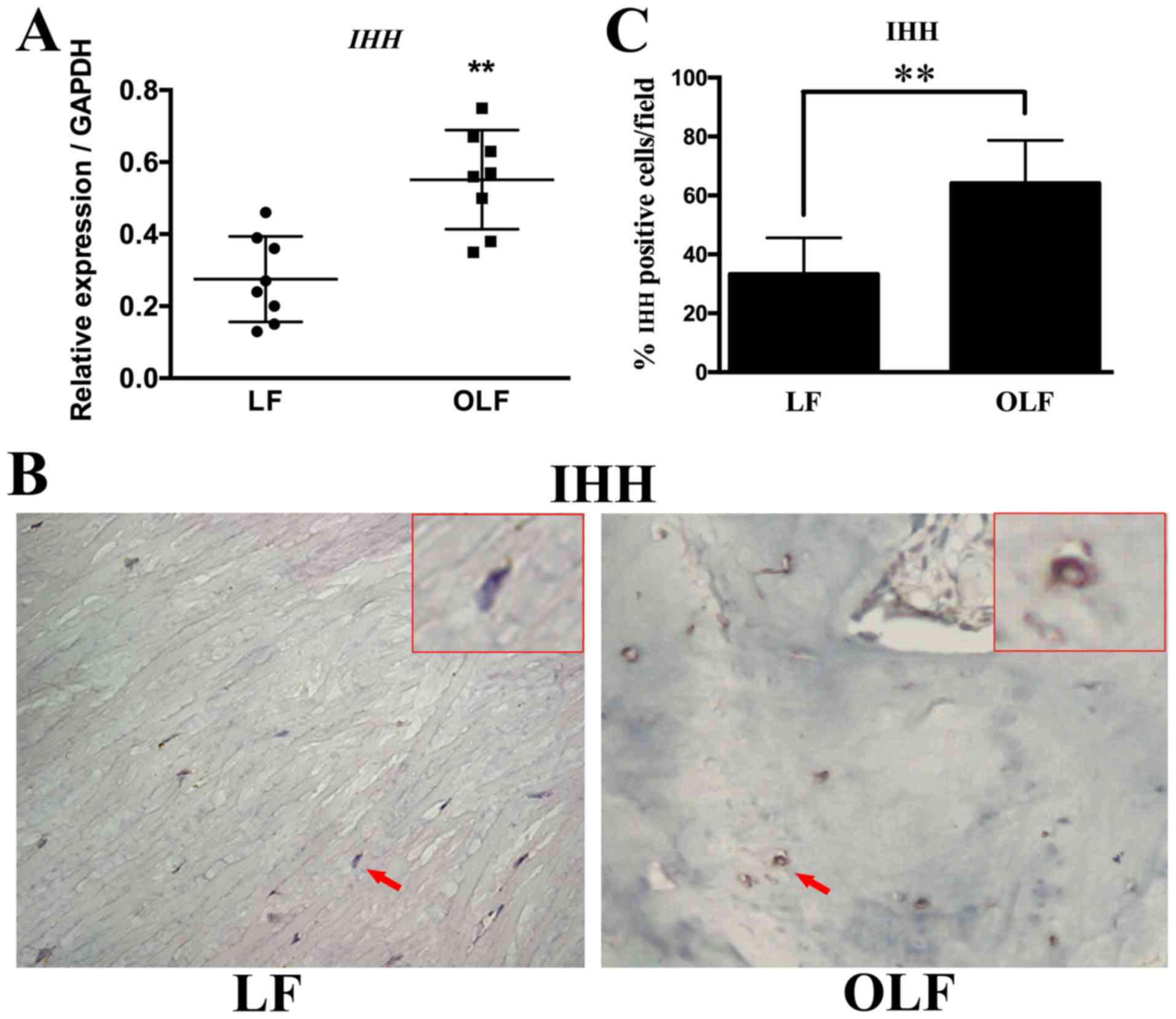
Expression of IHH signaling genes in
OLF
The mRNA and protein expression levels of IHH
in both groups are shown in Fig.
2. The RT-qPCR and immunohistochemical results revealed that
the OLF group had significantly higher mRNA and protein expression
levels of IHH compared with the LF group (P<0.01). Thus,
IHH may be involved in the osteogenesis of OLF.
Effects of cyclic stretch on
osteogenic differentiation of LF and OLF cells
The mRNA expression levels of osteogenic genes and
IHH signaling genes in OLF and LF cells are shown in Fig. 3. It was demonstrated that
application of cyclic stretch to OLF cells resulted in significant
increases in mRNA expression levels of RUNX2, Osterix, ALP
and OCN, and the IHH signaling genes IHH and
SMO at 24 h. Moreover, in LF cells the application of cyclic
tensile strain significantly increased the mRNA expression level of
ALP, but not of RUNX2, Osterix, OCN, IHH and
SMO (Fig. 3).
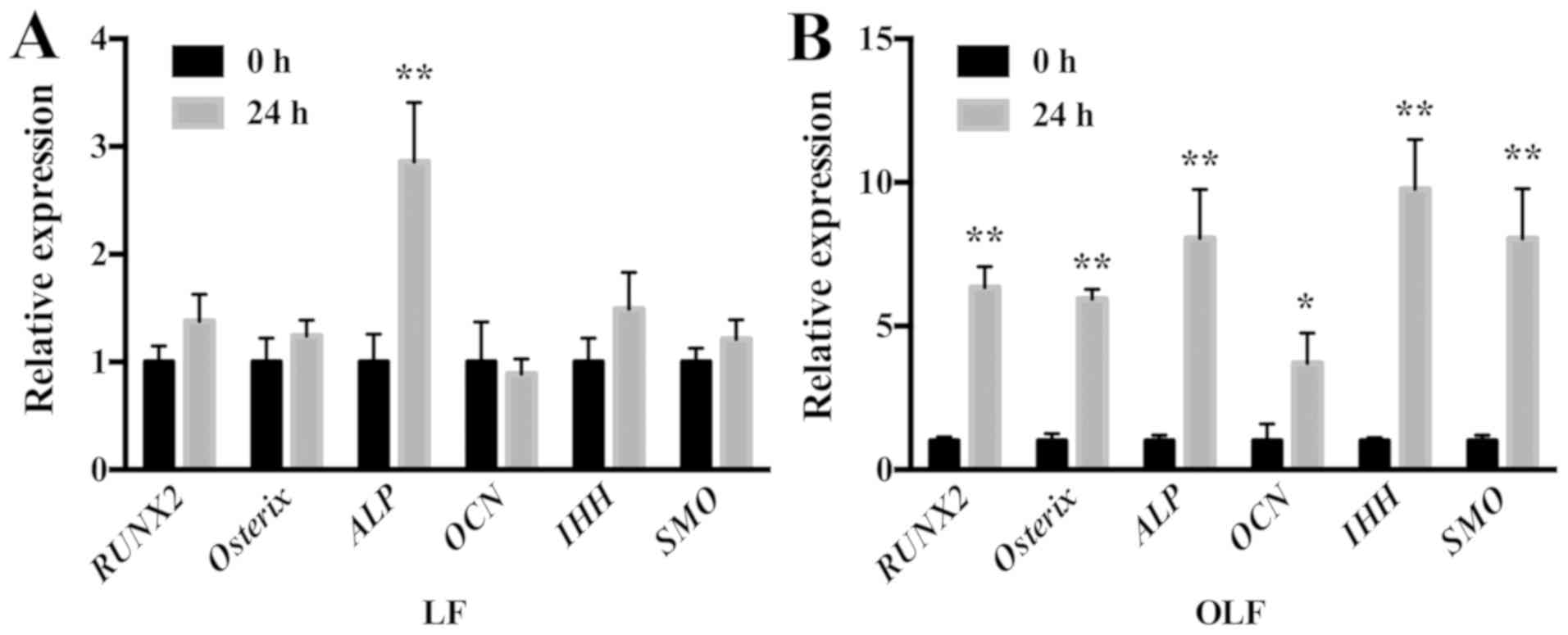 | Figure 3.Expression level of osteogenic genes
and IHH signaling genes in LF and OLF cells after cyclic
stretch. (A) Reverse transcription-quantitative PCR results showed
that in LF cells, cyclic stretch increased the ALP expression
level, while other gene expression levels did not change. (B)
Relative mRNA expression levels of RUNX2, Osterix, ALP,
OCN, IHH and SMO significantly increased after the
application of cyclic stretch in OLF cells. *P<0.05, **P<0.01
vs. 0 h. Data are presented as the mean ± SD. N=3. OLF,
ossification of ligamentum flavum; LF, ligamentum flavum;
RUNX2, Runt-related transcription factor 2; ALP, alkaline
phosphatase; OCN, osteocalcin; IHH, Indian
hedgehog. |
To further examine the effect of mechanical stress
on the osteogenesis of OLF cells, the expression levels of
osteogenic genes were compared after OLF cells were treated with or
without cyclic stretch for 6, 12 and 24 h. It was found that cyclic
stretch caused a significant elevation in the mRNA expression
levels of RUNX2, Osterix, ALP and OCN at all time
points (Fig. 4A-D). Furthermore,
cyclic stretch was determined to significantly increase the protein
expression levels of RUNX2, Osterix and ALP at 6, 12
and 24 h (Fig. 4E-G). In addition,
it was demonstrated that cyclic stretch caused a significant
increase in the protein expression level of OCN at 12 and 24
h (Fig. 4H). ALP activity was also
significantly enhanced at 12 and 24 h, with the greatest increase
observed at 24 h (Fig. 4I and J).
Collectively, the present results indicated that mechanical stress
induced the osteogenesis process during the development of OLF.
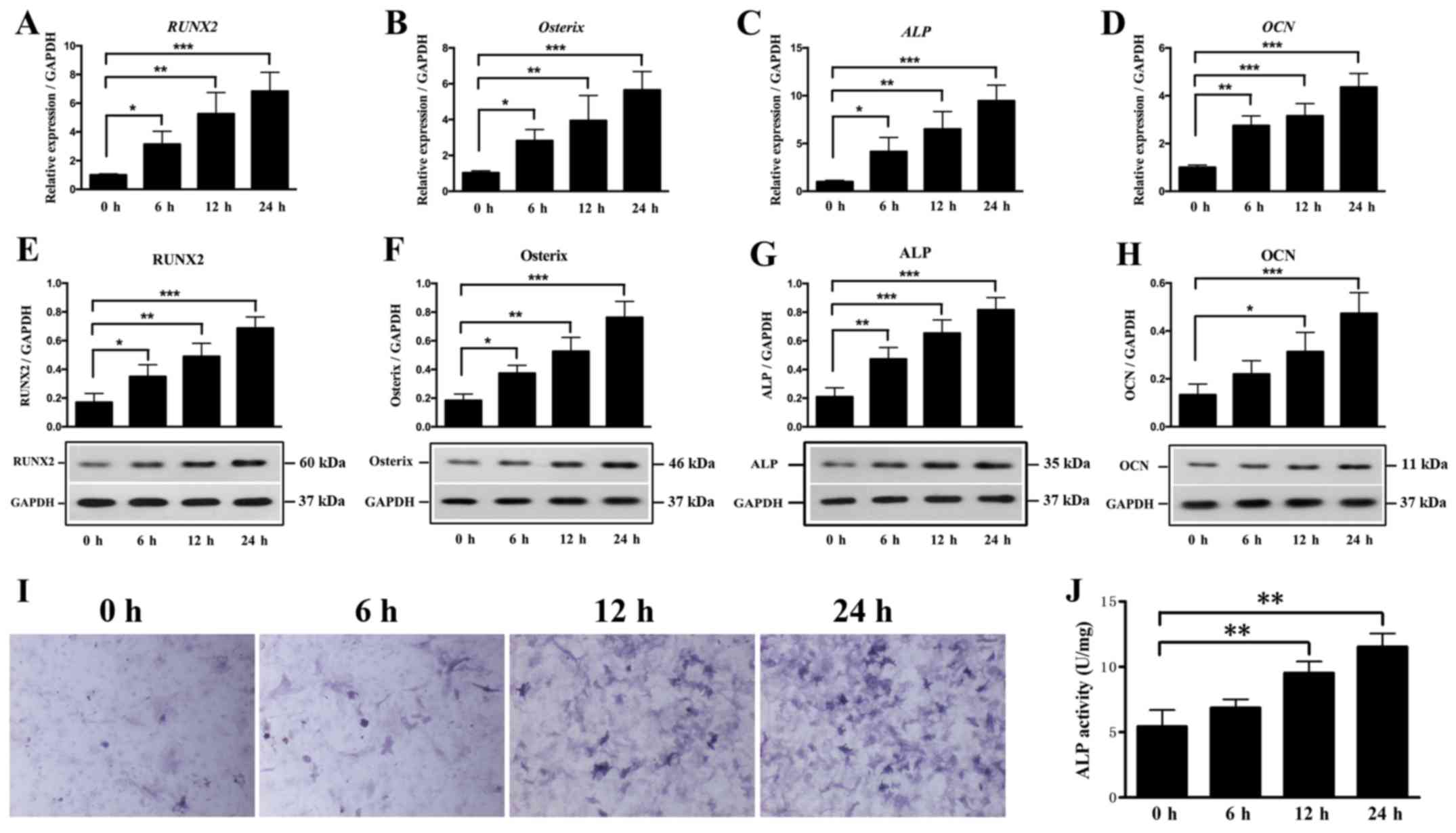 | Figure 4.Effect of cyclic stretch on
osteogenic genes and ALP activity. Expression levels of (A)
RUNX2, (B) Osterix, (C) ALP and (D) OCN
in OLF cells were detected after cyclic stretch treatment for 0, 6,
12 and 24 h. Protein expression levels of (E) RUNX2, (F)
Osterix, (G) ALP and (H) OCN after cyclic
stretch. (I) Representative images of ALP staining.
Magnification, ×400. (J) Cyclic stretch increased ALP
activity in OLF cells. *P<0.05, **P<0.01, ***P<0.001. Data
are presented as the mean ± SD. N=3. OLF, ossification of
ligamentum flavum; RUNX2, Runt-related transcription factor
2; ALP, alkaline phosphatase; OCN, osteocalcin. |
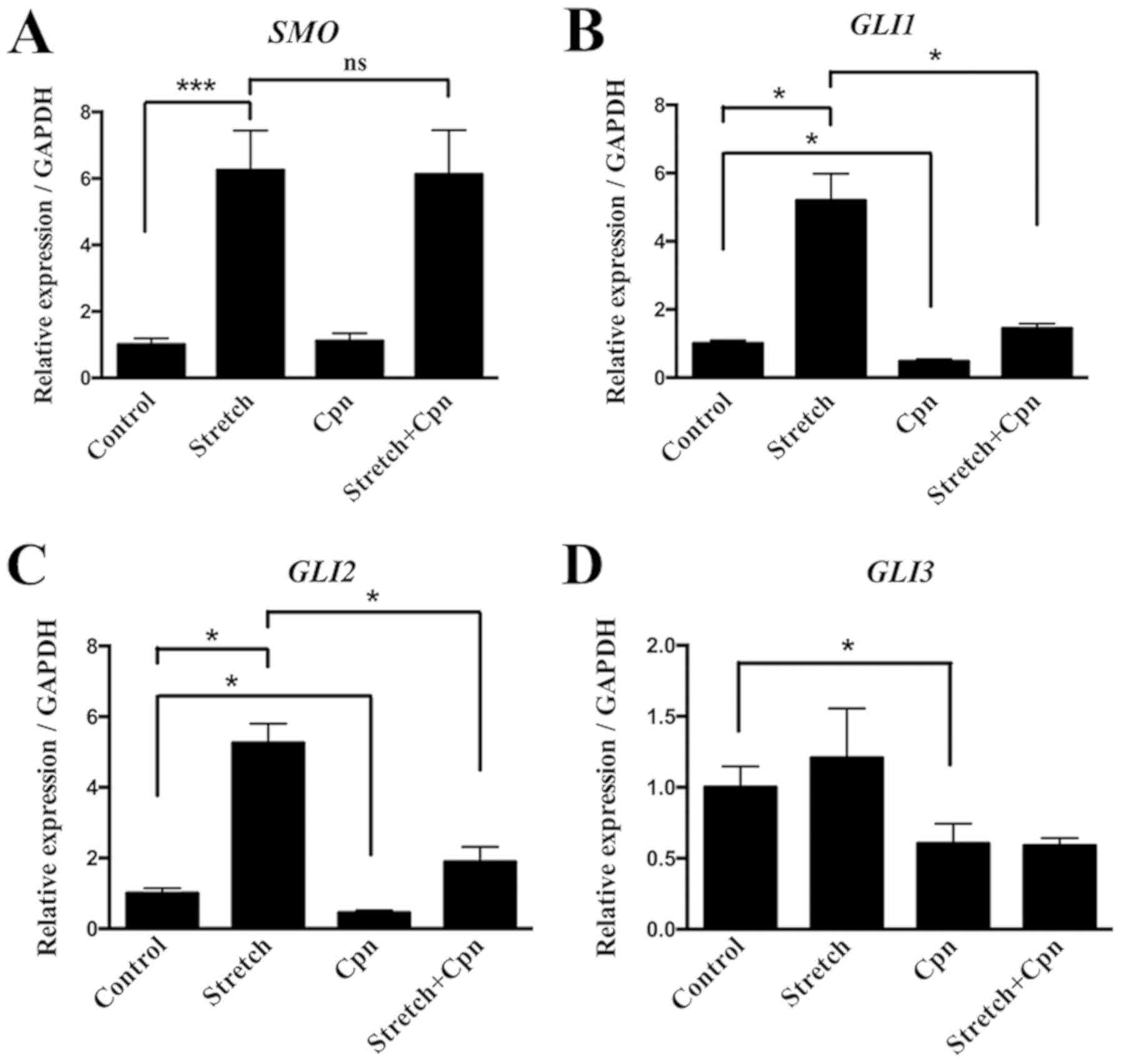
Effects of cyclic stretch on the IHH
signaling pathway
The IHH signaling pathway, which includes IHH,
SMO and GLI, is an important pathway in the osteogenic
differentiation (33). To further
examine the effect of local stress on the IHH signaling pathway and
osteogenesis in OLF, cyclic stretch was applied to OLF cells for 6,
12 and 24 h. The relative ratios of IHH, SMO and GLI
expression levels in OLF cells are shown in Fig. 5. It was found that cyclic stretch
significantly increased the mRNA and protein expression level of
IHH at 6, 12 and 24 h (Fig. 5A
and B). Moreover, the mRNA and protein expression levels of
SMO were significantly increased at all time points
(Fig. 5C and D). Cyclic stretch
also significantly upregulated the expression level of GLI1
at 6, 12 and 24 h, and the mRNA expression level of GLI2 at
12 and 24 h (Fig. 5E and F);
however, no significant effect on GLI3 expression level was
detected (Fig. 5G). Therefore, the
present results suggested that local stress could promote the
expression of osteogenic markers by regulating the IHH signaling
pathway in OLF cells.
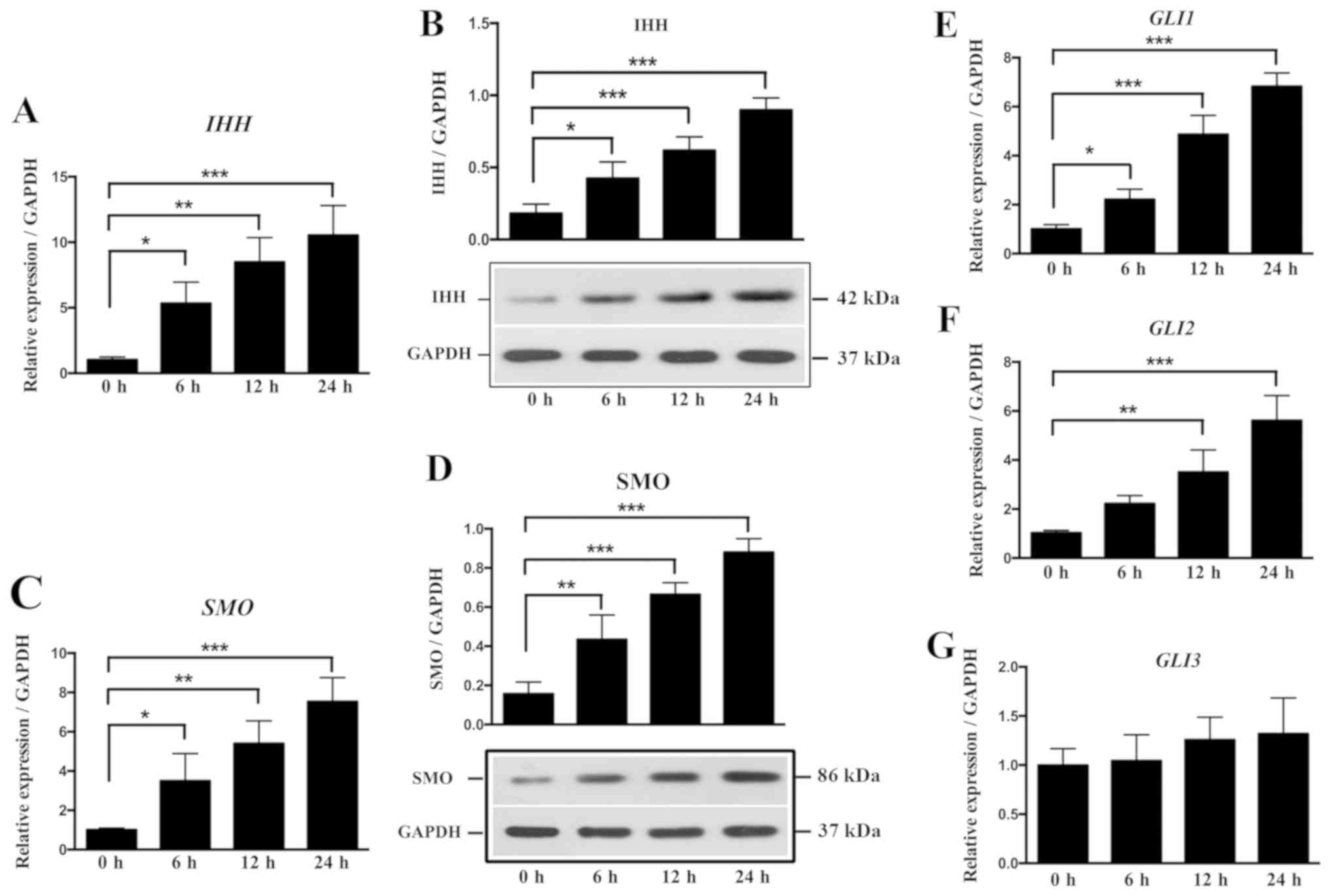 | Figure 5.Effect of cyclic stretch on the
expression of molecules involved in the IHH signaling
pathway in OLF cells. Cyclic stretch of OLF cells resulted in
significant increases in the (A) mRNA and (B) protein expression
levels of IHH. Cyclic stretch of OLF cells resulted in
significant increases in the (C) mRNA and (D) protein expression
levels of SMO at 6, 12 and 24 h. mRNA expression levels of
(E) GLI1 and (F) GLI2 were significantly increased after cyclic
stretch. (G) No significant effect was observed on the expression
level of GLI3. *P<0.05, **P<0.01, ***P<0.001. Data are
presented as the mean ± SD. N=3. OLF, ossification of ligamentum
flavum; RUNX2, Runt-related transcription factor 2;
ALP, alkaline phosphatase; OCN, osteocalcin;
IHH, Indian hedgehog; SMO, Smoothened; GLI, GLI
family zinc finger. |
IHH signaling is involved in
osteogenesis induced by cyclic stretch
Cyclopamine (Cpn) can downregulate the IHH signaling
pathway by binding to SMO directly, which is a key component
in IHH signaling (34). To examine
whether cyclic stretch can directly affect the expression levels of
RUNX2, Osterix, ALP and OCN via the IHH signaling
pathway, OLF cells were treated with cyclic stretch alone or both
cyclic stretch and Cpn. Fig. 6
showed the expression levels of RUNX2, Osterix, ALP and
OCN after Cpn treatment. It was found that the expression
levels of all genes were significantly decreased in the Cpn +
cyclic stretch group compared with the cyclic stretch alone group
(Fig. 6A-H). Furthermore, ALP
activity was reduced with Cpn treatment compared with cyclic
stretch alone (Fig. 6I and J). It
was demonstrated that cells treated with cyclic stretch showed a
significant increase in the expression levels of the downstream IHH
signaling molecules SMO, GLI1 and GLI2. However,
cyclic stretch did not significantly alter the expression of
GLI3 compared with the control group. Moreover, Cpn alone
decreased the expression of GLI1, GLI2 and GLI3
compared with the control group. The effect of cyclic stretch on
the expression of GLI1 and GLI2 was decreased by the
addition of Cpn (Fig. 7). Thus,
the present results provided evidence of the role of the IHH
signaling pathway in the pathogenesis of OLF.
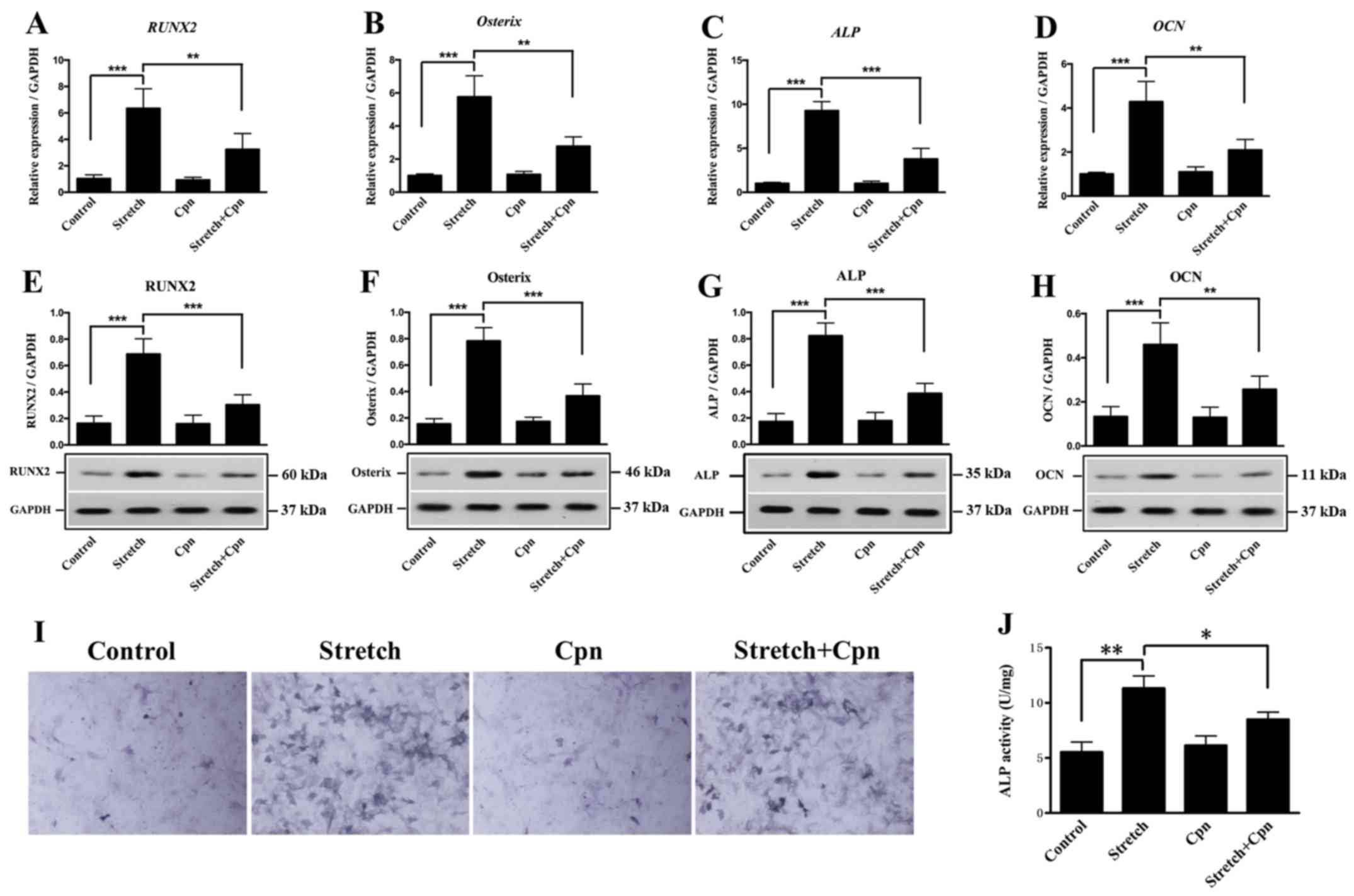 | Figure 6.Effect of the Indian hedgehog
signaling pathway inhibitor Cpn on the expression levels of
RUNX2, Osterix, ALP and OCN in OLF cells. mRNA
expression levels of (A) RUNX2, (B) Osterix, (C)
ALP and (D) OCN. Protein expression levels of (E)
RUNX2, (F) Osterix, (G) ALP (H) and OCN
were significantly decreased in the stretch + Cpn group compared
with cyclic stretch alone. (I) Representative images of ALP
staining. Magnification, ×400. (J) ALP activity was
significantly reduced in the Cpn + stretch group compared with
cyclic stretch alone. *P<0.05, **P<0.01, ***P<0.001. Data
are presented as the mean ± SD. N=3. OLF, ossification of
ligamentum flavum; RUNX2, Runt-related transcription factor
2; ALP, alkaline phosphatase; OCN, osteocalcin; Cpn,
cyclopamine. |
Discussion
OLF in East Asian populations is more common
compared with individuals from other regions, and is characterized
by pathological heterotopic ossification in the LF (5,6).
Various genetic, mechanical and inflammatory factors are involved
in the development OLF (35), but
the molecular mechanistic pathways remain to be elucidated. IHH
signaling is an essential signaling pathway mediating heterotopic
ossification of the extremities (19). To the best of our knowledge, the
present study is one of the first to examined whether IHH signaling
mediates osteogenic differentiation during OLF. It was found that
the expression levels of osteoblastic markers and IHH were
higher in OLF tissues compared with in LF tissues. Moreover, cyclic
stretch elevated the expression levels of osteogenic
differentiation markers in the LF cells of patients with OLF.
Furthermore, it was demonstrated that osteogenic induction was
attenuated by inhibiting IHH signaling. Collectively, the present
results suggested that the IHH signaling pathway may play a role in
the pathogenesis of OLF.
The development of OLF involves chondrogenic and
osteogenic differentiation of the fibroblasts into osteoblasts
(7). Therefore, examining
osteogenic differentiation of the LF may help to understand the
pathogenesis of OLF. Previous studies have identified higher
expression levels of osteogenesis-related genes in the OLF cells
and tissues (16,36). Using RNA sequencing, Yang et
al (36), examined gene
expression profiles of LF cells in both immature and mature
ossification groups. Moreover, Yang et al (36) found that 42 osteogenesis-related
factors were differentially expressed, of which the expression
levels of SOX11, RUNX2, Osterix and secreted phosphoprotein
1 were increased. Cai et al (16), performed immunohistochemistry in
the decalcified paraffin OLF sections, and found that β-catenin and
SOX9 were immunopositive in premature chondrocytes. In addition,
Cai et al (16) identified
that the expression of RUNX2 and OPN was
significantly increased in hypertrophic chondrocytes surrounding
the calcification front. According to a previous in vitro
study (37), cultured OLF cells
have higher mRNA expression levels of β-catenin, RUNX2, SOX9
and OPN compared with non-OLF cells. In line with these
previous results, the present study found that the expression
levels of the osteoblastic markers RUNX2, Osterix, ALP and
OCN were higher in OLF tissues compared with LF tissues.
Thus, the process of fibroblasts differentiating into osteoblasts
may be an important physiopathological mechanism in OLF.
Mechanical and genetic factors may facilitate the
development of OLF (5). Previous
studies have demonstrated that the cyclic stretch could initiate
and promote the ossification process by modulating the osteogenic
differentiation of the spinal ligament cells (15,38).
The present study applied cyclic stretch to OLF cells, and found
that both ALP activity and the expression levels of RUNX2,
Osterix, ALP and OCN were elevated as the duration of
the stretch force was increased; these results were in line with
previous results (11,39). Ning et al (39), showed that cyclic mechanical
application produces higher ALP activity and expression levels of
ALP, BMP2, OPN and Osterix in the multiple-level OLF
group compared with the single-level group. Furthermore, a previous
in vitro study revealed that mechanical stress significantly
increases the mRNA expression levels of RUNX2, ALP and
OCN in OLF cells compared with the control group (11).
The present results indicated that cyclic stretch
increased the expression levels of IHH, SMO, GLI1 and
GLI2 genes, which are involved in the IHH signaling pathway,
at different time points. Furthermore, the present study blocked
the IHH signaling pathway by treating OLF cells with Cpn, which is
a potent inhibitor of the IHH signaling pathway (40). It was found that the impact of
cyclic stretch on the expression levels of osteogenic genes was
significantly attenuated after Cpn treatment. Thus, the present
results suggested that cyclic stretch may affect osteogenic
differentiation of OLF cells via the IHH signaling pathway. IHH
signaling plays a critical role in prenatal endochondral bone
formation and limb development, as well as postnatal bone formation
(18,41). Therefore, the complete ablation of
IHH signaling can lead to the closure of the growth plate and
immature bone growth (18,41). IHH signaling may also participate
in the bone repair process after a fracture (42). Furthermore, abnormal bone
development and ectopic bone formation due to overexpression of the
IHH signaling have been previously reported (22,43).
In the present study, the expression of IHH, SMO, GLI1 and
GLI2 was upregulated by cyclic stretch, suggesting the IHH
signaling pathway was involved in the pathogenesis of OLF. By
contrast, it has been shown that the upregulation of patched 1
(PTC1) can decrease IHH signaling function (44). Moreover, it has been suggested that
several IHH molecules can interact with the increased PTC1,
resulting in reduced SMO release (45). However, the present study did not
identify any changes in the expression level of GLI3 under
cyclic stretch conditions. Thus, GLI3 may not play a
critical role in the regulation of osteogenic differentiation of
OLF cells.
There are certain limitations to the present study.
First, only the IHH signaling pathway was investigated, and other
signaling pathways are likely involved in the differentiation of
OLF cells. A previous study (16)
demonstrated that cyclic stretch significantly elevated the
expression levels of β-catenin, RUNX2, SOX9 and OPN,
which indicates that the β-catenin signaling pathway may be
involved in the pathogenesis of OLF. Second, the present study was
conducted in vitro. Therefore, in vivo OLF models are
required to assess the therapeutic effect of inhibiting IHH
signaling on the progression of OLF. In addition, only the effects
of mechanical stress on osteogenic differentiation were
investigated, and other types of stress, such as compression and
shear force, were not examined. Thus, further studies are required
to investigate the specific mechanisms and therapeutic targets for
OLF.
In conclusion, the present results suggested that
the expression levels of osteogenic markers and IHH signaling genes
were higher in OLF cells. Moreover, inhibition of the IHH signaling
pathway reduced the expression levels of osteogenic markers. Thus,
the IHH signaling pathway may be involved in the progression of
OLF. If cell differentiation can be regulated by modulating this
pathway, the homeostatic balance of cell differentiation could be
maintained and the ossification of spinal ligaments may be
prevented..
Acknowledgements
Not applicable.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant nos. 81501917 and
81802120) and the Shanghai Sailing Program (grant no.
18YF1423100).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
RG and CS performed experiments, analyzed data and
prepared the manuscript. CY and YZ analyzed and interpreted the
data, and prepared the manuscript. XC and XZ designed experiments,
analyzed data and finalized the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study was approved by the Ethics Committee of
Second Military Medical University, and the experimental procedures
were performed in accordance with the standard guidelines. Informed
consent was obtained from the patients or relatives for the
collection of LF during surgery, and the use of these samples for
scientific research.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
IHH
|
Indian hedgehog
|
|
OLF
|
ossification of ligamentum flavum
|
|
RUNX2
|
Runt-related transcription factor
2
|
|
ALP
|
alkaline phosphatase
|
|
OCN
|
osteocalcin
|
|
LF
|
ligamentum flavum
|
|
Cpn
|
cyclopamine
|
|
OPN
|
osteopontin
|
|
SOX9
|
sex-determining region Y-box protein
9
|
|
BCA
|
bicinchoninic acid
|
References
|
1
|
Nouri A, Tetreault L, Singh A, Karadimas
SK and Fehlings MG: Degenerative cervical myelopathy: Epidemiology,
genetics, and pathogenesis. Spine. 40:E675–E693. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Takahashi T, Hanakita J and Minami M:
Pathophysiology of calcification and ossification of the ligamentum
flavum in the cervical spine. Neurosurg Clin N Am. 29:47–54. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Guo JJ, Luk KDK, Karppinen J, Yang H and
Cheung KMC: Prevalence, distribution, and morphology of
ossification of the ligamentum flavum: A population study of one
thousand seven hundred thirty-six magnetic resonance imaging scans.
Spine. 35:51–56. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Kawaguchi Y, Yasuda T, Seki S, Nakano M,
Kanamori M, Sumi S and Kimura T: Variables affecting postsurgical
prognosis of thoracic myelopathy caused by ossification of the
ligamentum flavum. Spine J. 13:1095–1107. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ahn DK, Lee S, Moon SH, Boo KH, Chang BK
and Lee JI: Ossification of the ligamentum flavum. Asian Spine J.
8:89–96. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hirabayashi S: Ossification of the
ligamentum flavum. Spine Surg Relat Res. 1:158–163. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yayama T, Uchida K, Kobayashi S, Kokubo Y,
Sato R, Nakajima H, Takamura T, Bangirana A, Itoh H and Baba H:
Thoracic ossification of the human ligamentum flavum:
Histopathological and immunohistochemical findings around the
ossified lesion. J Neurosurg Spine. 7:184–193. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Qu X, Chen Z, Fan D, Xiang S, Sun C, Zeng
Y, Li W, Guo Z, Qi Q, Zhong W, et al: Two novel BMP-2 variants
identified in patients with thoracic ossification of the ligamentum
flavum. Eur J Hum Genet. 25:565–571. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li H, Jiang LS and Dai LY: Hormones and
growth factors in the pathogenesis of spinal ligament ossification.
Eur Spine J. 16:1075–1084. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhang C, Chen Z, Meng X, Li M, Zhang L and
Huang A: The involvement and possible mechanism of pro-inflammatory
tumor necrosis factor alpha (TNF-α) in thoracic ossification of the
ligamentum flavum. PLoS One. 12:e01789862017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Shunzhi Y, Zhonghai L and Ning Y:
Mechanical stress affects the osteogenic differentiation of human
ligamentum flavum cells via the BMP-Smad1 signaling pathway. Mol
Med Rep. 16:7692–7698. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Edwards DS and Clasper JC: Heterotopic
ossification: A systematic review. J R Army Med Corps. 161:315–321.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kłosiński M, Skrzat J, Walocha J and Mizia
E: Contemporary views on the ossification of the ligamenta flava.
Ortop Traumatol Rehabil. 14:495–503. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Gao R, Yuan W, Yang L, Shi G and Jia L:
Clinical features and surgical outcomes of patients with thoracic
myelopathy caused by multilevel ossification of the ligamentum
flavum. Spine J. 13:1032–1038. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Tsukamoto N, Maeda T, Miura H, Jingushi S,
Hosokawa A, Harimaya K, Higaki H, Kurata K and Iwamoto Y:
Repetitive tensile stress to rat caudal vertebrae inducing
cartilage formation in the spinal ligaments: A possible role of
mechanical stress in the development of ossification of the spinal
ligaments. J Neurosurg Spine. 5:234–242. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Cai HX, Yayama T, Uchida K, Nakajima H,
Sugita D, Guerrero AR, Yoshida A and Baba H: Cyclic tensile strain
facilitates the ossification of ligamentum flavum through β-catenin
signaling pathway: In vitro analysis. Spine. 37:E639–E646. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gallet A: Hedgehog morphogen: From
secretion to reception. Trends Cell Biol. 21:238–246. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang J, Andre P, Ye L and Yang YZ: The
Hedgehog signalling pathway in bone formation. Int J Oral Sci.
7:73–79. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kan C, Chen L, Hu Y, Ding N, Lu H, Li Y,
Kessler JA and Kan L: Conserved signaling pathways underlying
heterotopic ossification. Bone. 109:43–48. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Minina E, Wenzel HM, Kreschel C, Karp S,
Gaffield W, McMahon AP and Vortkamp A: BMP and Ihh/PTHrP signaling
interact to coordinate chondrocyte proliferation and
differentiation. Development. 128:4523–4534. 2001.PubMed/NCBI
|
|
21
|
Wongdee K, Thonapan N, Saengamnart W,
Krishnamra N and Charoenphandhu N: Bromocriptine modulates the
expression of PTHrP receptor, Indian hedgehog, and Runx2 proteins
in the growth plate of lactating rats. Mol Cell Biochem.
381:191–199. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
St-Jacques B, Hammerschmidt M and McMahon
AP: Indian hedgehog signaling regulates proliferation and
differentiation of chondrocytes and is essential for bone
formation. Genes Dev. 13:2072–2086. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Regard JB, Malhotra D, Gvozdenovic-Jeremic
J, Josey M, Chen M, Weinstein LS, Lu J, Shore EM, Kaplan FS and
Yang Y: Activation of Hedgehog signaling by loss of GNAS causes
heterotopic ossification. Nat Med. 19:1505–1512. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Sugita S, Chikuda H, Takeshita K, Seichi A
and Tanaka S: Progression of ossification of the posterior
longitudinal ligament of the thoracic spine following posterior
decompression and stabilization. J Neurosurg Spine. 21:773–777.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ando K, Imagama S, Ito Z, Kobayashi K,
Ukai J, Muramoto A, Shinjo R, Matsumoto T, Nakashima H and Ishiguro
N: Progressive relapse of ligamentum flavum ossification following
decompressive surgery. Asian Spine J. 8:835–839. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Tokuhashi Y, Ajiro Y and Umezawa N: A
patient with two re-surgeries for delayed myelopathy due to
progression of ossification of the posterior longitudinal ligaments
after cervical laminoplasty. Spine. 34:E101–E105. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Shi C, Wu H, Du D, Im HJ, Zhang Y, Hu B,
Chen H, Wang X, Liu Y, Cao P, et al: Nicotinamide
phosphoribosyltransferase inhibitor APO866 prevents IL-1β-induced
human nucleus pulposus cell degeneration via autophagy. Cell
Physiol Biochem. 49:2463–2482. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Hu B, Shi C, Xu C, Cao P, Tian Y, Zhang Y,
Deng L, Chen H and Yuan W: Heme oxygenase-1 attenuates IL-1β
induced alteration of anabolic and catabolic activities in
intervertebral disc degeneration. Sci Rep. 6:211902016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Du D, Zhou Z, Zhu L, Hu X, Lu J, Shi C,
Chen F and Chen A: TNF-α suppresses osteogenic differentiation of
MSCs by accelerating P2Y2 receptor in estrogen-deficiency induced
osteoporosis. Bone. 117:161–170. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Krane SM: Identifying genes that regulate
bone remodeling as potential therapeutic targets. J Exp Med.
201:841–843. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Karsenty G: The genetic transformation of
bone biology. Genes Dev. 13:3037–3051. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Shimoyama A, Wada M, Ikeda F, Hata K,
Matsubara T, Nifuji A, Noda M, Amano K, Yamaguchi A, Nishimura R,
et al: Ihh/Gli2 signaling promotes osteoblast differentiation by
regulating Runx2 expression and function. Mol Biol Cell.
18:2411–2418. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Gould A and Missailidis S: Targeting the
hedgehog pathway: The development of cyclopamine and the
development of anti-cancer drugs targeting the hedgehog pathway.
Mini Rev Med Chem. 11:200–213. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Uchida K, Yayama T, Cai HX, Nakajima H,
Sugita D, Guerrero AR, Kobayashi S, Yoshida A, Chen KB and Baba H:
Ossification process involving the human thoracic ligamentum
flavum: Role of transcription factors. Arthritis Res Ther.
13:R1442011. View
Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yang X, Chen Z, Meng X, Sun C, Li M, Shu
L, Fan D, Fan T, Huang AY and Zhang C: Angiopoietin-2 promotes
osteogenic differentiation of thoracic ligamentum flavum cells via
modulating the Notch signaling pathway. PLoS One. 13:e02093002018.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Park JO, Lee BH, Kang YM, Kim TH, Yoon JY,
Kim H, Kwon UH, Lee KI, Lee HM and Moon SH: Inflammatory cytokines
induce fibrosis and ossification of human ligamentum flavum cells.
J Spinal Disord Tech. 26:E6–E12. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zuscik MJ, Hilton MJ, Zhang X, Chen D and
O'Keefe RJ: Regulation of chondrogenesis and chondrocyte
differentiation by stress. J Clin Invest. 118:429–438. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ning S, Chen Z, Fan D, Sun C, Zhang C,
Zeng Y, Li W, Hou X, Qu X, Ma Y, et al: Genetic differences in
osteogenic differentiation potency in the thoracic ossification of
the ligamentum flavum under cyclic mechanical stress. Int J Mol
Med. 39:135–143. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Incardona JP, Gaffield W, Kapur RP and
Roelink H: The teratogenic Veratrum alkaloid cyclopamine inhibits
sonic hedgehog signal transduction. Development. 125:3553–3562.
1998.PubMed/NCBI
|
|
41
|
Ohba S: Hedgehog Signaling in endochondral
ossification. J Dev Biol. 4:E202016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Shi Y, He G, Lee WC, McKenzie JA, Silva MJ
and Long F: Gli1 identifies osteogenic progenitors for bone
formation and fracture repair. Nat Commun. 8:20432017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Day TF and Yang Y: Wnt and hedgehog
signaling pathways in bone development. J Bone Joint Surg Am. 90
(Suppl 1):19–24. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Quijada L, Callejo A, Torroja C and
Guerrero I: The patched receptor. In: Hedgehog-Gli Signaling in
Human Disease. Boston, MA: Ruiz i Altaba A: Springer, US; pp.
23–33. 2006
|
|
45
|
Salem O, Wang HT, Alaseem AM, Ciobanu O,
Hadjab I, Gawri R, Antoniou J and Mwale F: Naproxen affects
osteogenesis of human mesenchymal stem cells via regulation of
Indian hedgehog signaling molecules. Arthritis Res Ther.
16:R1522014. View
Article : Google Scholar : PubMed/NCBI
|