Introduction
Helicobacter pylori (Hp), a gram-negative
bacterium, thrives in the acidic environment of the stomach and is
the major risk factor for chronic gastric ulcer and gastric cancer.
Hp secretes virulence factors, including cytotoxic-associated gene
A (CagA), vacuolated cytotoxic A (VacA) and urease, which cause DNA
damage and cell vacuolization of gastric mucosa cells, and then
destroy tight junctions between cells and damage the cell barrier
by inducing oxidative stress and activating the inflammatory
response (1,2). In addition to Hp virulence proteins,
it has been suggested that small non-coding RNA may regulate mRNA
stability and protein translation, which finally affect bacterial
growth and reproduction and contribute to virulence.
Bacterial small non-coding RNA (sRNA), 50–500 nt
nucleotides in length, is a type of widespread RNA in prokaryotic
organisms, similar to microRNA (miRNA/miR) in eukaryotes. Through
nucleotide pairing, sRNA is able to target mRNAs and regulate mRNA
stability and protein translation, which finally affects bacterial
growth and reproduction and contributes to virulence. Since 1967,
the first sRNA 6S RNA was isolated from Escherichia coli
(E. coli) and hundreds of sRNAs have been identified in
E. coli and other bacteria (3,4).
However, to date, only 7 sRNAs have been identified in Hp, but
research is still in its infancy (5–9). For
instance, a study has reported that acid-sensing chemotaxis
receptor TlpB, which has a vital role in colonization, was
upregulated in repG (HPnc5490) deletion mutant Hp (6). Hp 5′ureB-sRNA, a cis-encoded
antisense small RNA regulated by the HP0165-HP0166 two-component
system, enhanced the expression of ureAB, consequently protecting
it from an acid environment (7,10).
Another prediction suggested that Hp sRNA NAT-67 was completely
complementary to HP1561 (ceuE), a periplasmic iron-binding protein
that may regulate oxidative stress (8). All of these studies suggested that
sRNAs have critical roles in the growth, colonization and invasion
of Hp. Bioinformatics prediction suggested that there may be
hundreds of sRNAs in Hp (5).
However, the full extent of sRNAs in bacterial chromosomes, as well
as their types and functions, remain elusive. Investigating the
sRNA expression spectrum of HP is crucial to reveal the biological
functions of sRNA and further anti-bacterial therapy.
sRNA has been indicated to be involved in the
response of bacteria to environmental fluctuations, including
oxidative stress and antibiotic drug exposure. The levels of
reactive oxygen species (ROS) were significantly elevated in small
RNAs RyhB-1 and RyhB-2 deletion mutant Salmonella
Typhimurium, proving RyhB to be an anti-oxidative stress factor
(11). Analysis of the small RNA
transcriptional response in multidrug-resistant Staphylococcus
aureus (S. aureus) suggested that 39 sRNAs were
significantly changed by four major classes of antibiotics
(12). Furthermore, sRNA SprX
(alias RsaOR) shaped bacterial resistance to glycopeptides, an
invaluable treatment for methicillin-resistant staphylococcal
infections (13). Azithromycin
inhibits the expression of small RNAs RsmY and RsmZ in
Pseudomonas aeruginosa (P. aeruginosa), hence
affecting quorum sensing and biofilm formation, which may be a
bactericidal mechanism (14). That
is to say that sRNA may contribute to the regulation of sensitivity
to antibiotic exposure and resistance to oxidative stress. It is
well-known that tinidazole (TNZ) is commonly used for HP treatment
by inducing oxidative stress, but the exact mechanism has remained
to be fully elucidated. It was hypothesized that sRNA from HP is
involved in the regulation of oxidative stress induced by TNZ.
In the present study, an HP sRNA profile was
generated to predict the possible functions of sRNA by using
bioinformatics analysis, aiming to explore the role of sRNA in HP
with TNZ exposure.
Materials and methods
Bacterial strain and growth
conditions
The HP stains 26695 and SS1 were obtained from the
American Type Culture Collection. Primary plate cultures of HP were
grown from glycerol stocks on trypticase soy agar (TSA) plates with
5% sheep blood (Thermo Fisher Scientific, Inc.) for 2–3 days in a
microaerobic environment (5% O2, 10% CO2, 85%
N2) at 37°C. The overnight culture of HP26695 on TSA
plates supplemented with 5% sheep blood was suspended in brain
heart infusion medium containing 12% sheep blood to an optical
density of 106 colony-forming units (CFU)/ml with
shaking slowly under microaerobic conditions at 37°C. When
necessary, the following antibiotics were added at the indicated
final concentrations: Vancomycin (50 µg/ml), polymyxin B (10 U/ml),
amphotericin B (125 µg/ml) and trimethoprim (25 µg/ml; all from
Melone Pharmaceutical Co., Ltd.). HP in the logarithmic phase was
treated with TNZ (10−5, 3×10−5,
10−4M) and then subjected to microaerobic conditions at
37°C for 24 h. The logarithmic phase of bacteria is a growth peak,
which is able to avoid the effect of bacterial death on the
results.
RNA preparation
Total RNA was isolated from HP strains using TRIzol
reagent (Takara Biotechnology Co., Ltd.). The concentration and
purity of eluted RNA was determined spectrophotometrically (Beckman
Coulter, Inc.) [optical density at 260 nm (OD260)/OD280 ratio
between 1.8 and 2.2]. The ribosomal RNA was removed prior to deep
sequencing.
Bioinformatics analysis by The Beijing
Genomics Institute
Total RNA from HP26695 was sequenced using an
Illumina HiSeq2000 (Ilumina, Inc.). The reads were spliced based on
overlap relations. According to their genomic locations, tags in
the intergenic region (IGR) and antisense mRNA (AM) were listed as
candidate tags. Among them, candidate sRNAs were defined as sRNAs
whose expression level was >20 reads. The spliced tags were
directly compared with 7 sRNAs with known sequences, since the
database used had not incorporated the known sequences of HP. For
the functional annotation of candidate sRNAs, two methods were
used. Based on sequence similarity, candidate RNAs were subjected
to BLAST analysis against four databases (sRNAMap (http://srnamap.mbc.nctu.edu.tw), sRNATarBase
(http://ccb.bmi.ac.cn), miRBase (http://www.mirbase.org) and SIPHI (http://newbio.cs.wisc.edu). Small RNAs possessed
secondary structure conservation. The secondary structures of
candidate sRNAs were therefore compared using Infernal software
(http://infernal.janelia.org/) and the
Rfam database (http://rfam.janelia.org/) was utilized for family
classification. Subsequently, the secondary structures of candidate
sRNAs were analyzed by RNAfold (http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi),
based on minimum free energy and base-paring probability. In
addition, certain concerned target genes were selected to predict
candidate sRNAs (Fig. S1).
Reverse transcription-quantitative
(RT-q)PCR and product sequencing
In order to detect the expression of mRNAs and
sRNAs, RNA (0.2–0.5 µg) was subjected to RT-PCR using the
PrimeScript reverse transcription reagent kit (Takara Biotechnology
Co., Ltd.) according to the manufacturer's protocol. Quantitative
analysis of the expression of mRNAs and sRNAs was performed using
the ABI 7300 Real-Time PCR system (Thermo Fisher Scientific, Inc.)
with SYBR Premix Ex Taq™ (Takara Biotechnology Co., Ltd.). The
cycling conditions were as follows: Initial incubation at 95°C for
15 sec, followed by 40 cycles of denaturation at 95°C for 5 sec and
annealing at 60°C for 31 sec. 23S rRNA was used as the housekeeping
gene. The primer sequences used are listed in Table I. The 2−ΔΔCq method was
used to analyze the relative changes in gene expression from
RT-qPCR experiments (15). The
products were collected by 2% agarose gel electrophoresis, and
sequencing was performed by Sangon Biotech Co., Ltd. (Shanghai,
China).
 | Table I.Primers used for polymerase chain
reaction. |
Table I.
Primers used for polymerase chain
reaction.
| Nucleotide | Sequence |
|---|
| 5′-ureB-sRNA | Forward:
5′-CTTTAGCATCCATATCCGCATT-3′ |
|
| Reverse:
5′-GACTGGTGGCATTGTCACAATA-3′ |
| NAT-67 | Forward:
5′-ACTTTTTATTGGGCGTCCTC-3′ |
|
| Reverse:
5′-AGAACTGGGTCCACAGCAGA-3′ |
| ceuE | Forward:
5′-CGCAAGAAGTCAAAGTCAAGG-3′ |
|
| Reverse:
5′-CGCCTACAACCCTATTCCAA-3′ |
| 23S rRNA | Forward:
5′-GCTCGTGTCGTGAGATGTTG-3′ |
|
| Reverse:
5′-GATCGTGTAAGTGAGATGG-3′ |
Bacterial density detection
The OD600 was detected using an ultraviolet
spectrophotometer (Beckman Coulter, Inc.). OD600=1 was considered
to indicate a corresponding density for HP of 108
CFU/ml.
Superoxide dismutase (SOD)
activity
The activity of SOD in HP was determined using a
Total Superoxide Dismutase Assay Kit with WST-8 (Beyotime Institute
of Biotechnology). Cell lysates were prepared following various
designated treatments. Assays were performed on 96-well microtiter
plates by incubating 20 µl cell lysate protein per sample in 160 µl
reaction buffer (151 µl SOD detection buffer, 8 µl WST-8 and 1 µl
enzyme solution) and then adding 20 µl starting work buffer (0.5 µl
starting buffer and 19.5 µl SOD detection buffer). Lysates were
incubated at 37°C for 30 min. The OD450 nm were measured using an
ultraviolet spectrophotometer (Beckman Coulter, Inc.). The
procedure was performed according to the manufacturer's
protocol.
ROS determination
ROS in the Hp were visualized using the
dichloro-dihydro-fluorescein diacetate (DCFH-DA) fluorescent probe
(Beyotime Institute of Biotechnology). In brief, HP was prepared
after the respective treatment and then resuspended in DCFH-DA
probe (10 µM) at a density of 107 CFU/ml. The mixture
was incubated at 37°C for 30 min and then washed with PBS buffer
until clear. The ultrasonic probe was immersed into the cell
suspension for ultrasonic crushing. The total ultrasonic time was
~2 min. Finally, the absorbance of cell lysates was measured at 450
nm (Beckman Coulter; DU800).
Statistical analysis
Data were analyzed using SPSS 17.0 (IBM, Corp.).
Values are expressed as the mean ± standard error of the mean.
Analysis of variance followed by the Newman-Student-Keuls test was
used to analyze the results. P<0.05 was considered to indicate a
statistically significant difference.
Results
Construction of the sRNA library of
HP
The concentration of samples used for sequencing was
>150 ng/µl, the ratio of 23S rRNA to 16S rRNA 1.6 was >1.5
and the RNA integrity number was 7.8, which met the requirements
for constructing an sRNA library, according to the Small RNA Sample
Preparation protocol of Illumina HiSeq2000. The 18–150 nt long
fragments were purified after being excised from the gel and then
sequenced by the Illumina HiSeq2000. A total of 592,125 tags were
sequenced, including 84 and 234 that map to AM and the IGR,
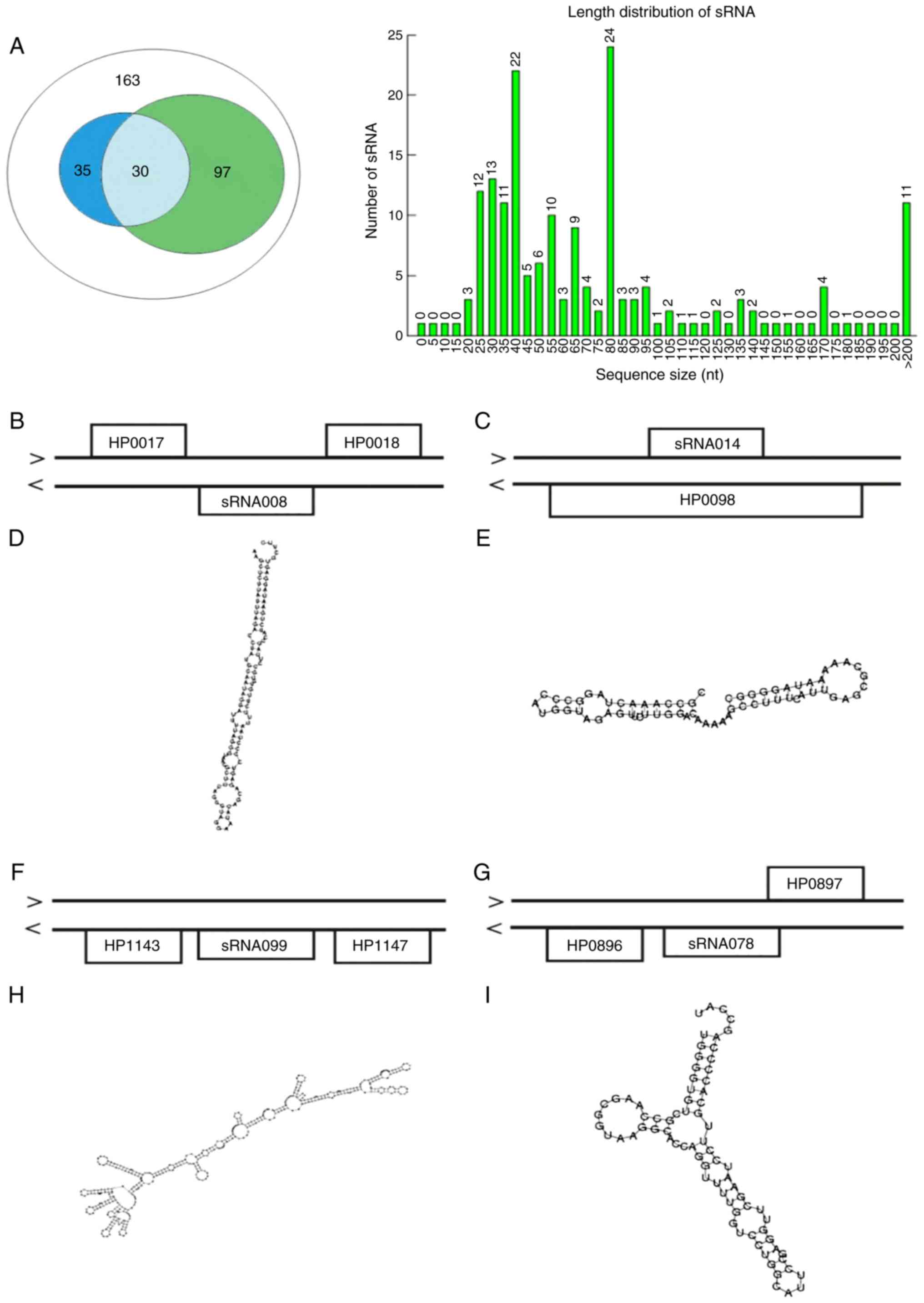
accounting for 4.3 and 11.98%, respectively. Among them, 163 tags
had >20 reads and therefore defined as candidate sRNAs and named
sRNA001-sRNA163. The length distribution of the candidate sRNAs is
illustrated in Fig. 1. A total of
72 candidate sRNAs were <50 nt long, 63 tags were 51–100 nt
long, 11 tags were 101–150 nt long, 6 tags were 151–200 nt long and
11 tags were >200 nt long (Fig.
1A). When mapping the spliced tags to the known sequence, only
5 sRNAs [6S RNA, NAT-67, 5′ureB-sRNA, repG (HPnc5490) and NAT-39]
had matching pieces; the others (IG-443 and IG-524) were
undetected.
Annotation and secondary structure of
candidate sRNAs in HP
Two methods were adopted for candidate sRNA
annotation: Sequence conservation and secondary structure
conservation. As presented in Fig.
1, 35 sRNAs were sequence-similar and 97 were secondary
structure-conservative. Among them, 30 tags were sequence-similar
and secondary structure-conservative. From what has been discussed
above, 61 sRNAs were reported for the first time. RNAfold
(http://rna.tbi.univie.ac.at/cgi-bin/RNAfold.cgi) was
used to predict the secondary structure of candidate sRNAs.
Representative results from Fig.
1B indicate that sRNA008 (112 nt) was encoded at the antisense
strand, which is in opposite orientation to the IGR between HP0017
and HP0018, and sRNA099 (431 nt) was encoded at the antisense
strand in the IGR between HP1143 and HP1147 (Fig. 1F). sRNA014 (66 nt) was encoded at
the sense strand, opposite HP0098 (Fig. 1C). sRNA078 (76 nt) was encoded at
the antisense strand, adjacent to HP0896 and HP0897 (Fig. 1G). Their secondary structures are
presented in Fig. 1.
Target prediction of candidate
sRNAs
IntaRNA was employed to forecast targets of
candidate sRNAs. The strategy of this software is base-paring,
leading to two further predicted targets of one sRNA, and entries
with a free energy of >-22 kcal/mol were filtered out. A total
of 1,247 targets of 79 candidate sRNAs met the requirements.
Therefore, sRNA interactions with genes of interest, including
genes associated with oxidative stress, virulence factors and
chemotaxis, were predicted. The results suggested that sRNAs that
may regulate virulence factors were sRNA057, sRNA126, sRNA055,
sRNA121 and sRNA025 (Fig. 2A).
Chemotaxis-associated genes may be regulated by sRNA057, sRNA090,
sRNA055, sRNA12 and sRNA025 (Fig.
2B). The known sequence NAT-67 and sRNA057 may be associated
with the regulation of oxidative stress (Fig. 2C).
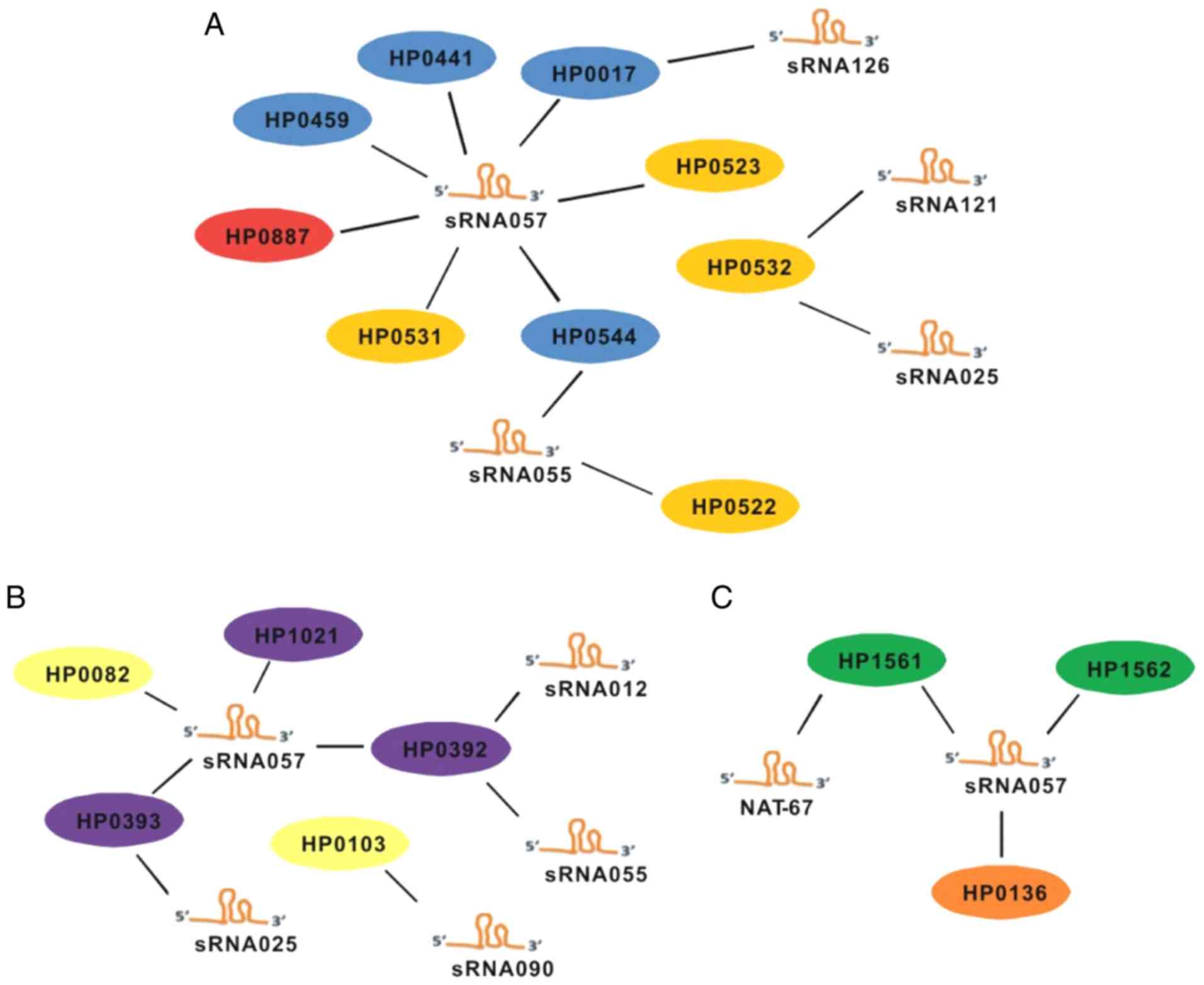 | Figure 2.Target prediction of candidate sRNAs.
Targets associated with (A) virulence factors, (B) chemotaxis and
(C) oxidative stress are provided. Blue, type IV secretion
system-associated genes; yellow, cag-pathogenicity island protein;
red, vacuolating cytotoxin-associated genes; purple, chemotaxis
protein-encoding genes; pale yellow, methyl-accepting chemotaxis
protein-associated genes; green, ceuE; orange, peroxidase. Hp,
Helicobacter pylori; sRNA, small non-coding RNA. |
Verification of NAT-67 and
5′ureB-sRNA
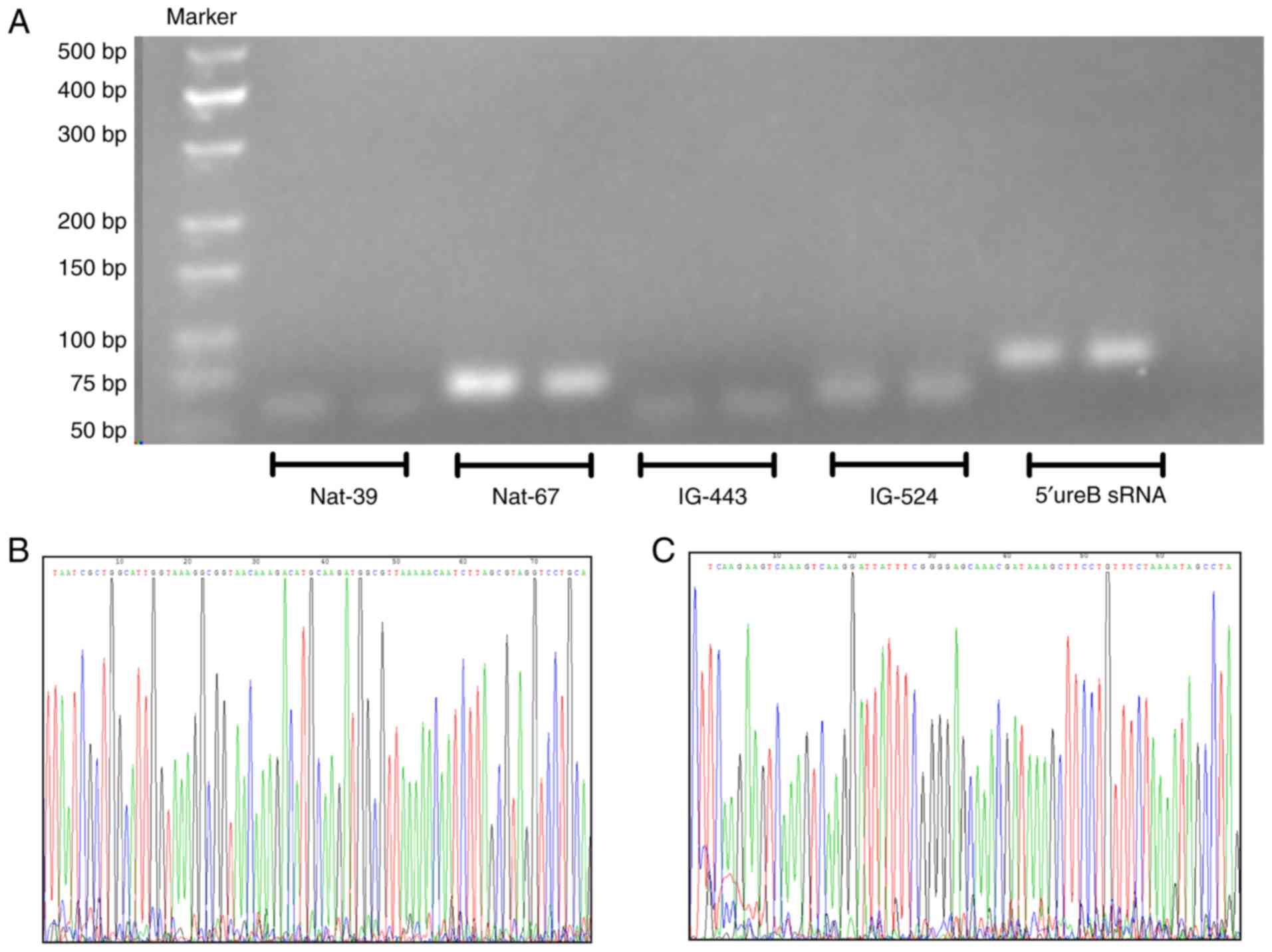
In order to verify the existence of HP RNAs, RT-qPCR
primers for NAT-67 and 5′ureB-sRNA were designed. The DNA fragment
was directly electrophoresed and sequenced. The results (Fig. 3) were consistent with previous
reports (5,8).
TNZ inhibits bacterial viability and
downregulates NAT-67 and 5′ureB-sRNA
TNZ is used for HP treatment by inducing oxidative
stress and cell death but the exact mechanism has remained to be
fully elucidated. Furthermore, it has been demonstrated that
certain sRNAs, including RyhB and RybA, are involved in the
regulation of oxidative stress (11). Consequently, the association
between TNZ-induced cell death and sRNAs was investigated. Under
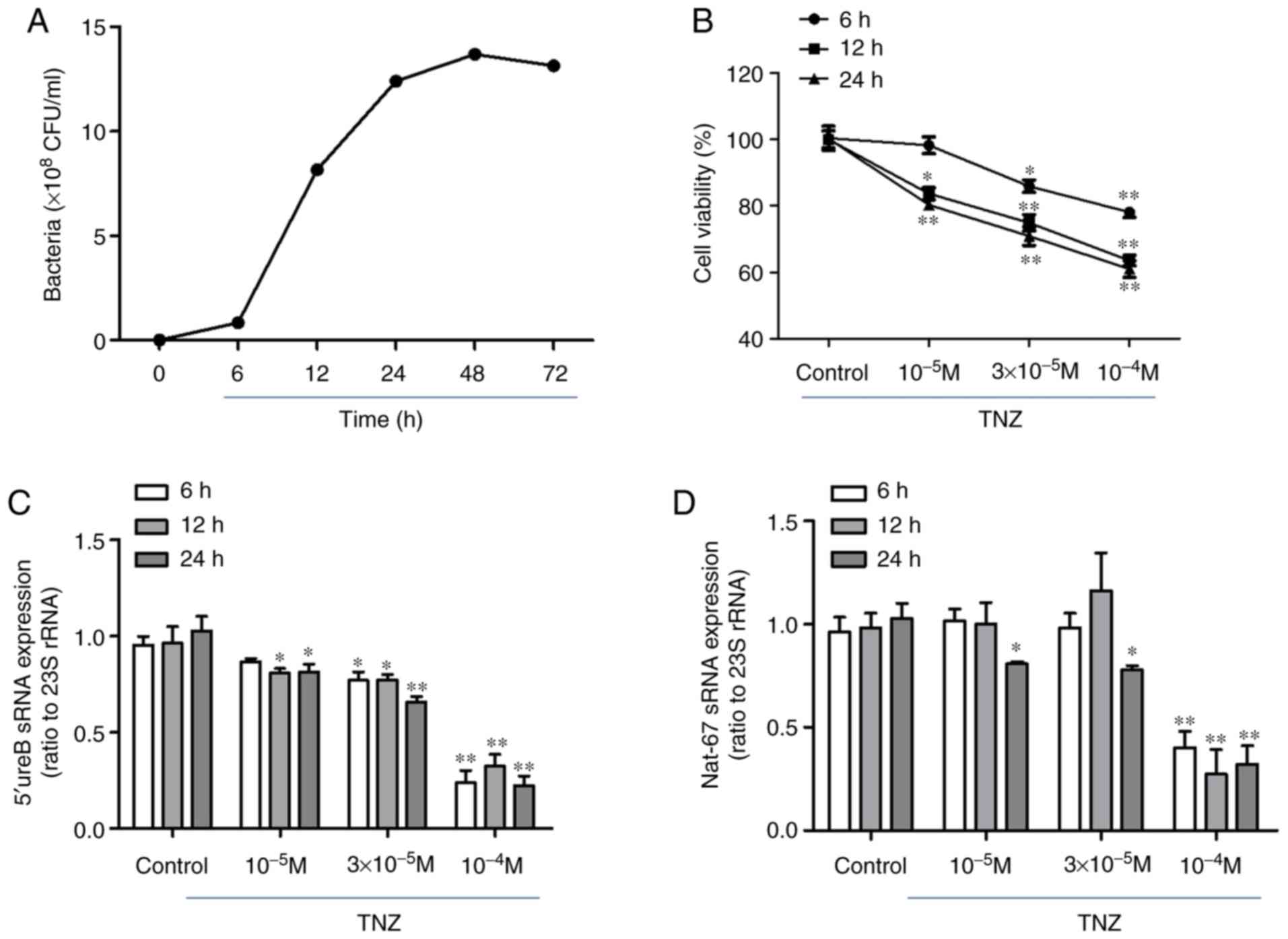
liquid culture conditions, the growth cycle of HP was ~48 h
(Fig. 4A). TNZ was added at the
middle of the logarithmic phase (10 h), since, during this phase,
sRNAs begin to be expressed and the response of bacteria to
environmental stimuli is at its most sensitive. The growth of HP
was significantly suppressed following treatment with TNZ,
particularly at 12 h (Fig. 4B). Of
note, the expression of NAT-67 and 5′ureB-sRNA was significantly
reduced (Fig. 4C and D).
TNZ downregulates ceuE and induces
oxidative stress
It was reported that NAT-67 may be the regulator of
iron-binding protein ceuE, which may participate in the regulation
of oxidative stress. The expression of ceuE and oxidative stress in
TNZ-treated Hp were therefore observed. It was indicated that TNZ
significantly decreased ceuE expression and SOD activity, while
increasing ROS levels (Fig. 4E-G).
These results suggested that the induction of cell death by TNZ may
be achieved through the downregulation of NAT-67 and induction of
oxidative stress.
Discussion
The significance of sRNA (50–500 nt in length) in
the regulation of bacteria is becoming increasingly apparent. These
molecules are involved in the regulation of a wide range of
physiological responses, cellular metabolism and reactions to
environmental stimuli (e.g., pH or temperature shifts), ultimately
contributing to survival and virulence (16). It has been demonstrated that sRNA
is involved in the response of bacteria to environmental
fluctuations, including oxidative stress and antibiotic drug
exposure. For instance, azithromycin inhibits the expression of
small RNAs RsmY and RsmZ in P. aeruginosa, which may be a
bactericidal mechanism (14).
Hp, which specifically colonizes the gastric
epithelium, is the dominant pathogenic factor for chronic gastric
ulcer and gastric cancer. Recent studies have indicated that sRNAs
were crucial to the growth, colonization and pathogenicity of Hp.
For instance, nucleotide pairing revealed that Hp sRNA NAT-67 was
completely complementary to periplasmic iron-binding protein HP1561
(ceuE), suggesting that NAT-67 may be a regulator in iron
homeostasis and oxidative stress (8). 5′ureB-sRNA, a cis-encoded antisense
small RNA regulated by the HP0165-HP0166 two-component system in
Hp, enhanced expression of ureB, consequently protecting it from
acid (7,10). Furthermore, 5′ureB knockout in Hp
markedly inhibited the inflammatory response of gastric mucosa
epithelial cells (17). However,
at present, the full extent of sRNAs in Hp chromosomes remains
elusive. While bioinformatics prediction indicated that hundreds of
sRNAs may exist in Hp, only 7 sRNAs had a known nucleotide sequence
(5,8–9). In
the present study, the sRNA expression spectrum was reported for
the first time. The results provided 163 candidate sRNAs in Hp.
Among the 7 known sRNAs, 5, including 6S RNA, NAT-67, 5′ureB-sRNA,
repG (HPnc5490) and NAT-39, matched the sequence in the present
study, while IG-443 and IG-524 were undetected. A possible reason
for this is that sRNA varies with time and environment.
Furthermore, RNAfold was used for secondary structure and IntaRNA
for target prediction. Based on base-paring, the so-called
predicted targets with a free energy of below −22 kcal/mol were
filtered out. In order to obtain a more specific result, the
interactions between sRNAs and genes of interest, including genes
associated with oxidative stress, virulence factors and chemotaxis,
were predicted. It was indicated that several sRNAs, including
NAT-67, are highly correlated with these genes. Bioinformatics
analysis of HP is momentous and may facilitate future research on
sRNA and understanding the pathogenic mechanism of HP.
In the present study, bioinformatics analysis and
gene sequencing were combined to determine gene location, sequence
similarity and secondary structure. However, this approach had its
limitations in predicting series of sRNAs. First, the sRNAs
screened in the present study were located in IGRs and AM, but some
were encoded by open reading frames and untranslated regions
(UTRs), and therefore, certain tags may have been missed. In
addition, it is required to analyze the initiator and terminator,
as the candidate sRNAs were short clips. However, the major
limitation is the reliability and repeatability of the
bioinformatics analysis, the results of which require to be
confirmed. In the present study, RT-qPCR and sequencing were
performed. The northern blot and gene chip would be the choices in
further studies.
NAT-67 is involved in regulating ceuE (HP1561) and
ceuE is associated with HP growth (9). For this reason, the influence of TNZ,
a frequently-used clinical drug for HP infection, on sRNA
expression and bacterial growth was assessed. TNZ suppressed Hp
growth while downregulating NAT-67 and ceuE. However, the effects
of sRNA regulation depend on the binding site of target mRNAs. If
the binding site contains a ribosome binding site, it prevents 30S
ribosomes from binding to mRNA, which suppresses transcription or
degrades mRNA (18). On the other
hand, if the binding site of mRNA forms a specific secondary
structure, combination with sRNA results in hairpin opening, so
that mRNA stabilizes and/or promotes translation (4). E. coli sRNA RyhB binding to
the 5′-UTR of cirA increased mRNA stability and translation
(19). S. aureus sRNA FasX
combining with a virulence-associated gene also enhanced its
activity (20). These results
supported that TNZ exerts its antibacterial effect by
downregulating NAT-67 and then inhibiting ceuE.
HP1561 (ceuE), a periplasmic iron-binding protein
that binds and transports iron into cell, has an important role in
iron homeostasis. NAT-67 (90 nt) was encoded in the IGR between
gene ftsW and tsaA, with perfect complementarity to HP1561 (ceuE).
Iron is the key ion for survival and growth. Hp expresses only a
single SOD, the iron-cofactor one, known as SODB (21). Therefore, a decrease in the
cellular iron concentration of HP leads to a decrease in SOD
activity and increase in oxidative stress. The present study
indicated that TNZ exposure restrained NAT-67 and ceuE expression,
accompanied by a SOD activity decrease and ROS increase. These
results provided novel experimental evidence with regards to the
antimicrobial mechanism of TNZ.
It is now accepted that miRNAs have crucial
regulatory roles in almost all pathological processes by regulating
cell growth, proliferation, differentiation and apoptosis. Recent
results have suggested that miRNAs not only execute functions
within the original cells but may also be initiatively secreted to
regulate adjacent or distant cells functions, and are even
transmitted from one species to another, facilitating crosstalk,
communication or signal interference between them (22–24).
For example, Rhodovulum sulfidophilum bacterial
extracellular RNAs have been reported in the literature (25). Nucleic acids were detected in
Lactobacillus culture filtrate and the major group were RNAs
(including sRNAs) (26). Studies
have reported that sequence-similar miRNAs may coordinately
regulate one target mRNA and thus, microRNA-mediated miRNA
regulation may be a novel model (27). For instance, cardiac miR-378 and
miR-499 regulated miR-143 (28–29).
It may therefore be speculated that bacteria-secreted sRNAs may be
a novel pathogenic factor.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (grant nos. 81703592 to JD and 81573486
to YJL). It was also supported by the Natural Science Foundation of
Hunan province (grant no. 2019JJ50934) and the Open Sharing Fund
for the Large-scale Instruments and Equipments of Central South
University.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JD, XL and YL designed the present study; JD and WZ
performed the experiments; JD and WZ analyzed the data and wrote
this manuscript. JD and WZ contributed to the manuscript equally.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
Hp
|
Helicobacter pylori
|
|
TNZ
|
tinidazole
|
|
sRNA
|
small non-coding RNA
|
|
ROS
|
reactive oxygen species
|
|
SOD
|
superoxide dismutase
|
|
CagA
|
cytotoxic-associated gene A
|
|
E. coli
|
Escherichia coli
|
|
VacA
|
vacuolated cytotoxic A
|
|
S. aureus
|
Staphylococcus aureus
|
|
AM
|
antisense mRNA
|
|
P. aeruginosa
|
Pseudomonas aeruginosa
|
|
IGR
|
intergenic region
|
References
|
1
|
Salama NR, Hartung ML and Müller A: Life
in the human stomach: Persistence strategies of the bacterial
pathogen Helicobacter pylori. Nat Rev Microbiol. 11:385–399.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Posselt G, Backert S and Wessler S: The
functional interplay of Helicobacter pylori factors with
gastric epithelial cells induces a multi-step process in
pathogenesis. Cell Commun Signal. 11:772013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wassarman KM: 6S RNA: A small RNA
regulator of transcription. Curr Opin Microbiol. 10:164–168. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lalaouna D, Simoneau-Roy M, Lafontaine D
and Massé E: Regulatory RNAs and target mRNA decay in prokaryotes.
Biochim Biophys Acta. 1829:742–747. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sharma CM, Hoffmann S, Darfeuille F,
Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller
J, Reinhardt R, et al: The primary transcriptome of the major human
pathogen Helicobacter pylori. Nature. 464:250–255. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pernitzsch SR, Tirier SM, Beier D and
Sharma CM: A variable homopolymeric G-repeat defines small
RNA-mediated posttranscriptional regulation of a chemotaxis
receptor in Helicobacter pylori. Proc Natl Acad Sci USA.
111:E501–E510. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wen Y, Feng J and Sachs G: Helicobacter
pylori 5′ureB-sRNA, a cis-encoded antisense small RNA,
negatively regulates ureAB expression by transcription termination.
J Bacteriol. 195:444–452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xiao B, Li W, Guo G, Li BS, Liu Z, Tang B,
Mao XH and Zou QM: Screening and identification of natural
antisense transcripts in Helicobacter pylori by a novel
approach based on RNase I protection assay. Mol Biol Rep.
36:1853–1858. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiao B, Li W, Guo G, Li B, Liu Z, Jia K,
Guo Y, Mao X and Zou Q: Identification of small noncoding RNAs in
Helicobacter pylori by a bioinformatics based approach. Curr
Microbiol. 58:258–263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yi W, Jing F, Scott DR, Marcus EA and
Sachs G: A cis-encoded antisense small RNA regulated by the
HP0165-HP0166 two-component system controls expression of ureB in
Helicobacter pylori. J bacterial. 193:40–51. 2011.
View Article : Google Scholar
|
|
11
|
Calderón IL, Morales EH, Collao B,
Calderón PF, Chahuán CA, Acuña LG, Gil F and Saavedra CP: Role of
salmonella typhimurium small RNAs RyhB-1 and RyhB-2 in the
oxidative stress response. Res Microbiol. 165:30–40. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Howden BP, Beaume M, Harrison PF,
Hernandez D, Schrenzel J, Seemann T, Francois P and Stinear TP:
Analysis of the small RNA transcriptional response in
multidrug-resistant staphylococcus aureus after antimicrobial
exposure. Antimicrob Agents Chemother. 57:3864–3874. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eyraud A, Tattevin P, Chabelskaya S and
Felden B: A small RNA controls a protein regulator involved in
antibiotic resistance in staphylococcus aureus. Nucleic Acids Res.
42:4892–4905. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pérez-Martínez I and Haas D: Azithromycin
inhibits expression of the GacA-dependent small RNAs RsmY and RsmZ
in pseudomonas aeruginosa. Antimicrob Agents Chemother.
55:3399–3405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wassarman KM: Small RNAs in bacteria:
Diverse regulators of gene expression in response to environmental
changes. Cell. 109:141–144. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Perrais M, Rousseaux C, Ducourouble MP,
Courcol R, Vincent P, Jonckheere N and Van Seuningen I:
Helicobacter pylori urease and flagellin alter mucin gene
expression in human gastric cancer cells. Gastric Cancer.
17:235–246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Desnoyers G, Bouchard MP and Massé E: New
insights into small RNA-dependent translational regulation in
prokaryotes. Trends Genet. 29:92–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Salvail H, Caron MP, Bélanger J and Massé
E: Antagonistic functions between the RNA chaperone Hfq and an sRNA
regulate sensitivity to the antibiotic colicin. EMBO J.
32:2764–2778. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ramirez-Peña E, Treviño J, Liu Z, Perez N
and Sumby P: The group A Streptococcus small regulatory RNA
FasX enhances streptokinase activity by increasing the stability of
the ska mRNA transcript. Mol Microbiol. 78:1332–1347. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tsugawa H, Suzuki H, Satoh K, Hirata K,
Matsuzaki J, Saito Y, Suematsu M and Hibi T: Two amino acids
mutation of ferric uptake regulator determines Helicobacter
pylori resistance to metronidazole. Antioxid Redox Signal.
14:15–23. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang H, Zen K, Zhang J, Zhang CY and Chen
X: New roles for microRNAs in cross-species communication. RNA
Biol. 10:367–370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Scott RB, Christopher N, Xie F, Wood JR
and Zempleni J: MicroRNAs are absorbed in biologically meaningful
amounts from nutritionally relevant doses of cow milk and affect
gene expression in peripheral blood mononuclear cells, HEK-293
kidney cell cultures, and mouse livers. J Nutr. 144:1495–1500.
2013.
|
|
24
|
Zhou Z, Li X, Liu J, Dong L, Chen Q, Liu
J, Kong H, Zhang Q, Qi X, Hou D, et al: Honeysuckle-encoded
atypical microRNA2911 directly targets influenza A viruses. Cell
Res. 25:39–49. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li J, Zhang Y, Li D, Liu Y, Chu D, Jiang
X, Hou D, Zen K and Zhang CY: Small non-coding RNAs transfer
through mammalian placenta and directly regulate fetal gene
expression. Protein Cell. 6:391–396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ando T, Suzuki H, Nishimura S, Tanaka T,
Hiraishi A and Kikuchi Y: Characterization of extracellular RNAs
produced by the marine photosynthetic bacterium
rhodovulumsulfidophilum. J Biochem. 139:805–811. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li T, Ling ZX and Wen S: Analysis of
nucleic acids in lactobacillus culture filtrate. Chin J Microecol.
23:104–106. 2011.
|
|
28
|
Guo L, Sun B, Wu Q, Yang S and Chen F:
miRNA-miRNA interaction implicates for potential mutual regulatory
pattern. Gene. 511:187–194. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matkovich SJ, Hu Y and Dorn GW II:
Regulation of cardiac microRNAs by cardiac microRNAs. Circ Res.
113:62–71. 2013. View Article : Google Scholar : PubMed/NCBI
|