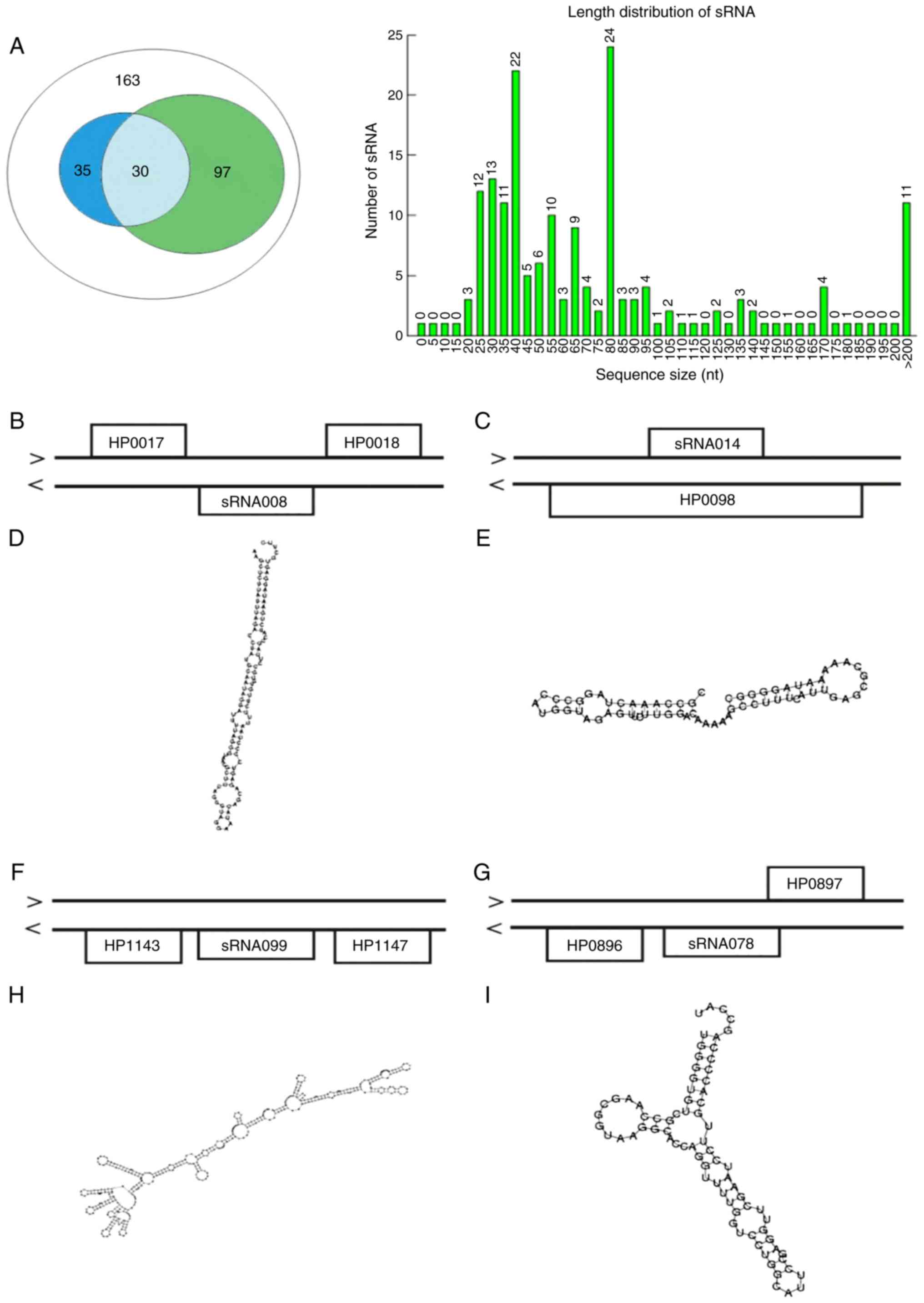
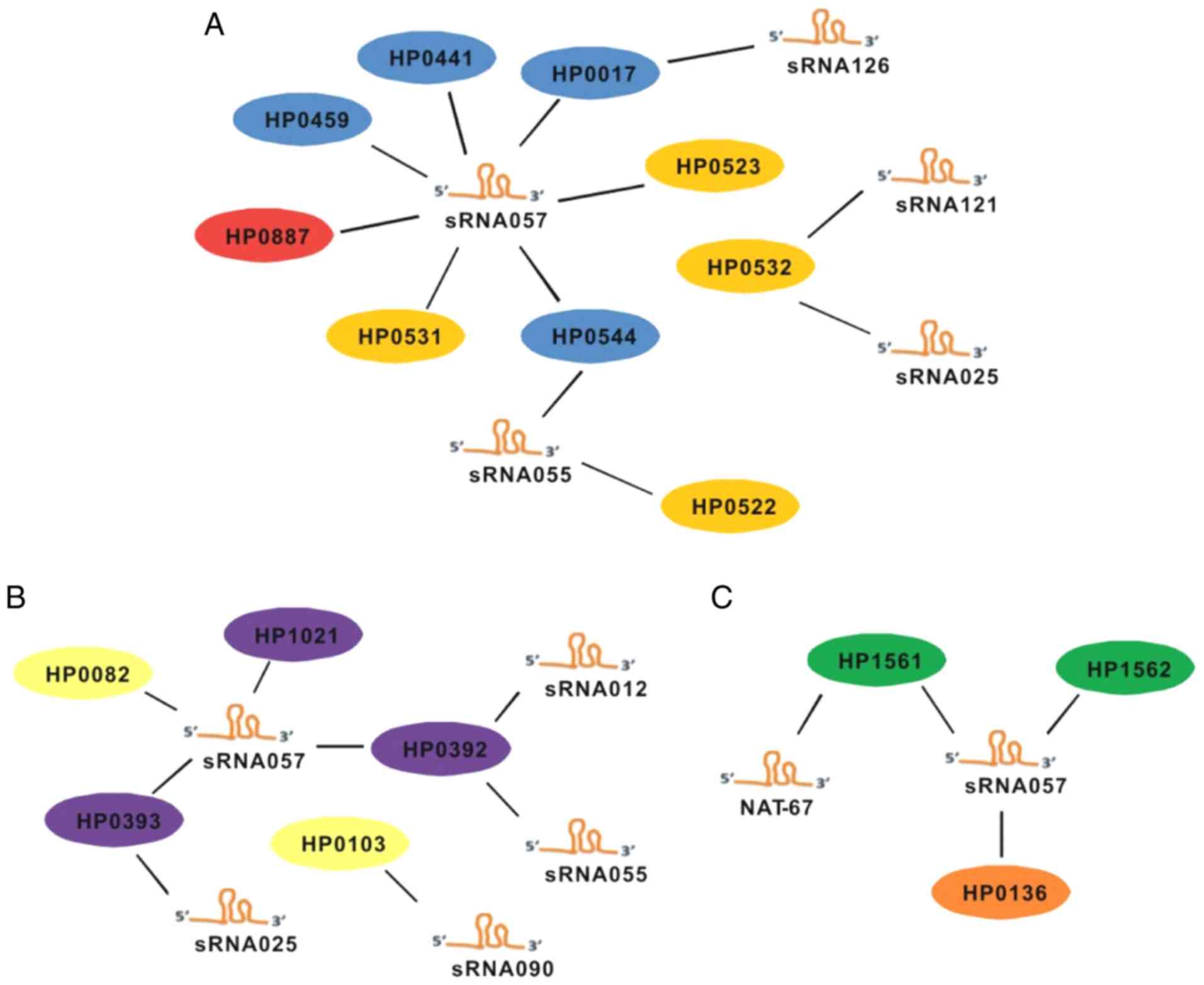
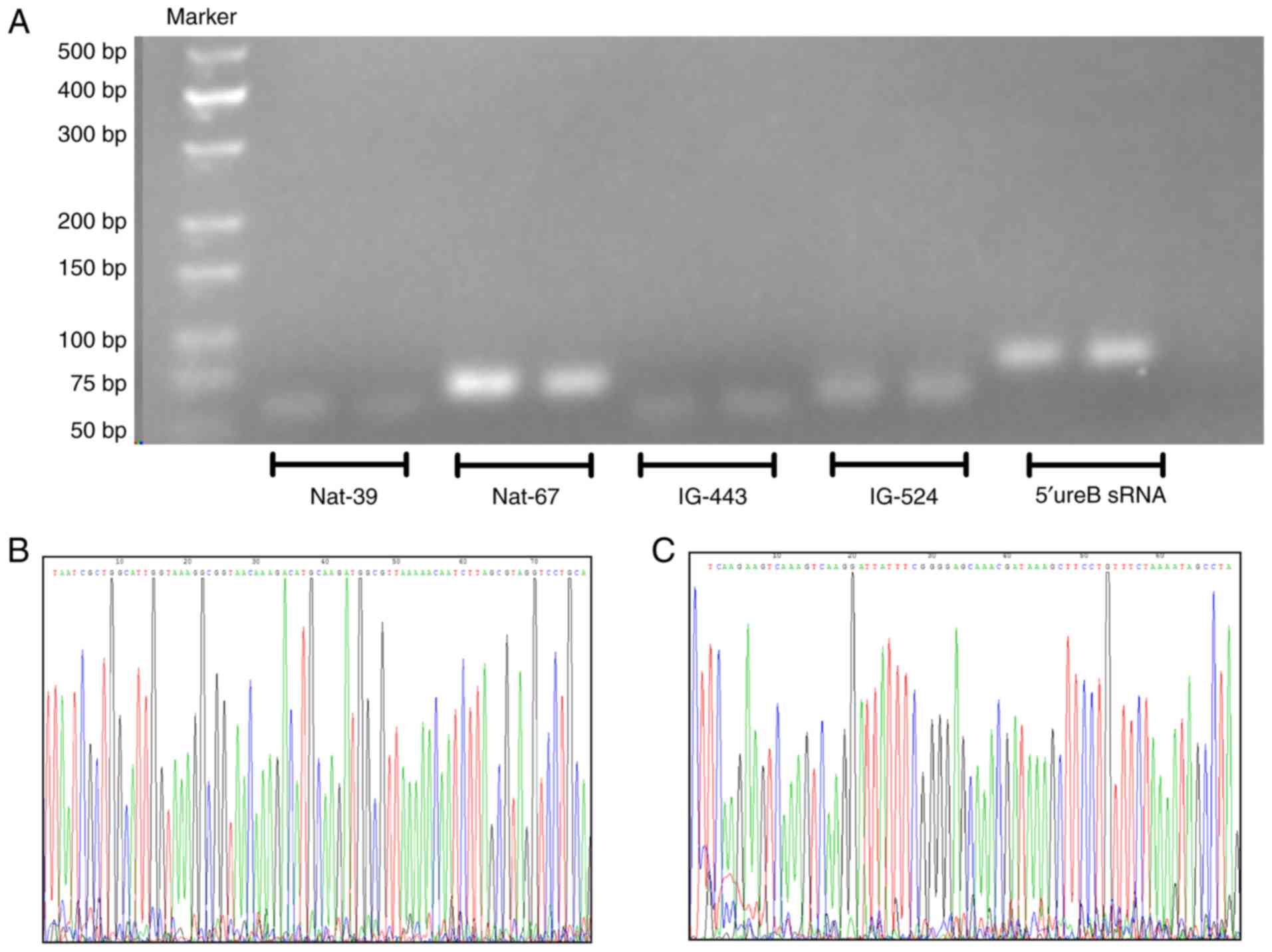
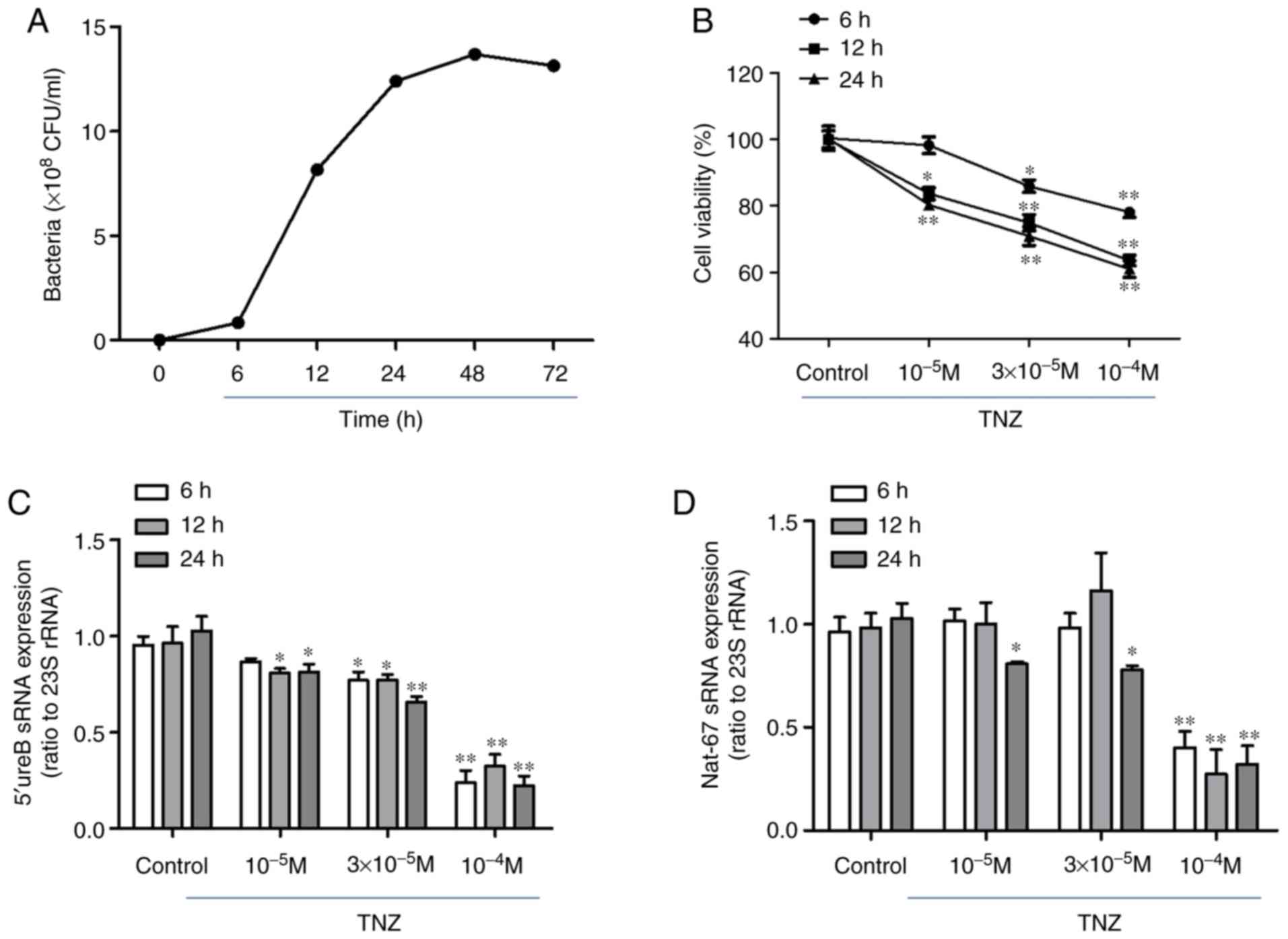
|
1
|
Salama NR, Hartung ML and Müller A: Life
in the human stomach: Persistence strategies of the bacterial
pathogen Helicobacter pylori. Nat Rev Microbiol. 11:385–399.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Posselt G, Backert S and Wessler S: The
functional interplay of Helicobacter pylori factors with
gastric epithelial cells induces a multi-step process in
pathogenesis. Cell Commun Signal. 11:772013. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Wassarman KM: 6S RNA: A small RNA
regulator of transcription. Curr Opin Microbiol. 10:164–168. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Lalaouna D, Simoneau-Roy M, Lafontaine D
and Massé E: Regulatory RNAs and target mRNA decay in prokaryotes.
Biochim Biophys Acta. 1829:742–747. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sharma CM, Hoffmann S, Darfeuille F,
Reignier J, Findeiss S, Sittka A, Chabas S, Reiche K, Hackermüller
J, Reinhardt R, et al: The primary transcriptome of the major human
pathogen Helicobacter pylori. Nature. 464:250–255. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pernitzsch SR, Tirier SM, Beier D and
Sharma CM: A variable homopolymeric G-repeat defines small
RNA-mediated posttranscriptional regulation of a chemotaxis
receptor in Helicobacter pylori. Proc Natl Acad Sci USA.
111:E501–E510. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wen Y, Feng J and Sachs G: Helicobacter
pylori 5′ureB-sRNA, a cis-encoded antisense small RNA,
negatively regulates ureAB expression by transcription termination.
J Bacteriol. 195:444–452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Xiao B, Li W, Guo G, Li BS, Liu Z, Tang B,
Mao XH and Zou QM: Screening and identification of natural
antisense transcripts in Helicobacter pylori by a novel
approach based on RNase I protection assay. Mol Biol Rep.
36:1853–1858. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xiao B, Li W, Guo G, Li B, Liu Z, Jia K,
Guo Y, Mao X and Zou Q: Identification of small noncoding RNAs in
Helicobacter pylori by a bioinformatics based approach. Curr
Microbiol. 58:258–263. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yi W, Jing F, Scott DR, Marcus EA and
Sachs G: A cis-encoded antisense small RNA regulated by the
HP0165-HP0166 two-component system controls expression of ureB in
Helicobacter pylori. J bacterial. 193:40–51. 2011.
View Article : Google Scholar
|
|
11
|
Calderón IL, Morales EH, Collao B,
Calderón PF, Chahuán CA, Acuña LG, Gil F and Saavedra CP: Role of
salmonella typhimurium small RNAs RyhB-1 and RyhB-2 in the
oxidative stress response. Res Microbiol. 165:30–40. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Howden BP, Beaume M, Harrison PF,
Hernandez D, Schrenzel J, Seemann T, Francois P and Stinear TP:
Analysis of the small RNA transcriptional response in
multidrug-resistant staphylococcus aureus after antimicrobial
exposure. Antimicrob Agents Chemother. 57:3864–3874. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Eyraud A, Tattevin P, Chabelskaya S and
Felden B: A small RNA controls a protein regulator involved in
antibiotic resistance in staphylococcus aureus. Nucleic Acids Res.
42:4892–4905. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Pérez-Martínez I and Haas D: Azithromycin
inhibits expression of the GacA-dependent small RNAs RsmY and RsmZ
in pseudomonas aeruginosa. Antimicrob Agents Chemother.
55:3399–3405. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wassarman KM: Small RNAs in bacteria:
Diverse regulators of gene expression in response to environmental
changes. Cell. 109:141–144. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Perrais M, Rousseaux C, Ducourouble MP,
Courcol R, Vincent P, Jonckheere N and Van Seuningen I:
Helicobacter pylori urease and flagellin alter mucin gene
expression in human gastric cancer cells. Gastric Cancer.
17:235–246. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Desnoyers G, Bouchard MP and Massé E: New
insights into small RNA-dependent translational regulation in
prokaryotes. Trends Genet. 29:92–98. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Salvail H, Caron MP, Bélanger J and Massé
E: Antagonistic functions between the RNA chaperone Hfq and an sRNA
regulate sensitivity to the antibiotic colicin. EMBO J.
32:2764–2778. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ramirez-Peña E, Treviño J, Liu Z, Perez N
and Sumby P: The group A Streptococcus small regulatory RNA
FasX enhances streptokinase activity by increasing the stability of
the ska mRNA transcript. Mol Microbiol. 78:1332–1347. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Tsugawa H, Suzuki H, Satoh K, Hirata K,
Matsuzaki J, Saito Y, Suematsu M and Hibi T: Two amino acids
mutation of ferric uptake regulator determines Helicobacter
pylori resistance to metronidazole. Antioxid Redox Signal.
14:15–23. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Liang H, Zen K, Zhang J, Zhang CY and Chen
X: New roles for microRNAs in cross-species communication. RNA
Biol. 10:367–370. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Scott RB, Christopher N, Xie F, Wood JR
and Zempleni J: MicroRNAs are absorbed in biologically meaningful
amounts from nutritionally relevant doses of cow milk and affect
gene expression in peripheral blood mononuclear cells, HEK-293
kidney cell cultures, and mouse livers. J Nutr. 144:1495–1500.
2013.
|
|
24
|
Zhou Z, Li X, Liu J, Dong L, Chen Q, Liu
J, Kong H, Zhang Q, Qi X, Hou D, et al: Honeysuckle-encoded
atypical microRNA2911 directly targets influenza A viruses. Cell
Res. 25:39–49. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li J, Zhang Y, Li D, Liu Y, Chu D, Jiang
X, Hou D, Zen K and Zhang CY: Small non-coding RNAs transfer
through mammalian placenta and directly regulate fetal gene
expression. Protein Cell. 6:391–396. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ando T, Suzuki H, Nishimura S, Tanaka T,
Hiraishi A and Kikuchi Y: Characterization of extracellular RNAs
produced by the marine photosynthetic bacterium
rhodovulumsulfidophilum. J Biochem. 139:805–811. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li T, Ling ZX and Wen S: Analysis of
nucleic acids in lactobacillus culture filtrate. Chin J Microecol.
23:104–106. 2011.
|
|
28
|
Guo L, Sun B, Wu Q, Yang S and Chen F:
miRNA-miRNA interaction implicates for potential mutual regulatory
pattern. Gene. 511:187–194. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Matkovich SJ, Hu Y and Dorn GW II:
Regulation of cardiac microRNAs by cardiac microRNAs. Circ Res.
113:62–71. 2013. View Article : Google Scholar : PubMed/NCBI
|