Introduction
Molecules obtained from medicinal plants have
traditionally been used by people to maintain good health.
According to the World Health Organization, >80% of the world's
population use traditional medicinal plants to improve their health
(1). A previous study has
suggested that medicinal plants contain chemical components and
exhibit biological activities that are useful for physiological
processes including antioxidant, anti-inflammation and anti-cancer
activity in the body (2). The most
widely used of these plant bioactive components are alkaloids,
flavonoids and phenolic compounds (3,4). Due
to their aromatic structure, phenolic compounds can act as electron
donors, and this antioxidant potential can relatively reduce
reactive oxygen species (ROS) levels (5). DNA is under constant stress resulting
from environmental factors or cellular metabolism (6). ROS cause DNA damage through oxidative
stress, thereby playing an important role in tumor initiation
(7). In response to DNA damage,
cells activate complex signaling networks to either repair the
damage or promote cell death (8).
Oxidative DNA damage has been implicated in the pathogenesis of
several diseases including cancer, inflammation, and heart disease
(9). Thus, it is important for
organisms to suppress the excessive generation of ROS.
ROS-mediated DNA damage influences cell survival
through the modification of histone H2AX (10–14).
The tumor suppressor p53 regulates baseline and DNA
damage-inducible levels of ROS (15–17).
p53 prevents DNA damage through an activated phosphatidylinositol
3-kinase-related kinase (PI3K) that plays a pivotal role in
activating DNA repair proteins (18). Phosphorylated H2AX (γ-H2AX) serves
an important role in the DNA damage response to oxidative stress by
recruiting genes involved in DNA repair (19). Biological processes that eliminate
ROS are important in the DNA damage response and in preventing the
progression of numerous diseases (20).
Abeliophyllum distichum Nakai is a deciduous
flowering plant belonging to the Oleaceae family. A.
distichum is a monotypic genus and has been regarded as one of
the important plant resources for research on its genetic property
and bioactivity (21).
Additionally, it is known to contain some glycosides including
acteoside, isoacteoside, rutin and hirsutrin (22). Acteoside exhibits several
pharmacological properties such as anti-hepatotoxic (23), anti-inflammatory (24), antinociceptive (25) and antioxidant activities (26–28).
It also improves sexual function (29). However, to the best of our
knowledge, whether acteoside from A. distichum (AAD) can
prevent oxidative DNA damage remains unknown. Therefore, the aim of
the present study was to determine whether AAD could exert a
protective effect over oxidative DNA damage.
Materials and methods
Plant material and extraction
A. distichum Nakai [voucher no.
Park1001(ANH)] was collected from the Misun-Hyang Theme Park. An
extract from air-dried A. distichum was obtained by
immersion in 80% methanol for 3 days and a using a sonicator (40
KHz; 700 W; Hwashin Technology Co., Ltd.). The methanol-soluble
extracts were filtered and concentrated using an N-1110S vacuum
evaporator (Eyela). The extracts were sequentially fractionated
three times with 1:1 ratio of petroleum ether, 1:1 ratio of
chloroform and 1:1 ratio of ethyl acetate. The ethyl acetate
fraction from A. distichum was harvested and refrigerated at
4°C until use.
Isolation and purification
Acteoside was isolated from the ethyl acetate
fraction (500 g) of A. distichum using an Isolera™ Spektra
Accelerated Chromatographic Isolation system at 25°C with SNAP
KP-SIL (100 g; 170×40 mm) and SNAP Ultra (25 g; 80 × 35 mm)
cartridges (both from Biotage AB). The mobile phases consisted of
chloroform (solvent A) and methanol (solvent B). The flow rate was
kept at 50 ml/min for a total run volume of 1,600 ml.
The system was run with a gradient program: 0–32
min, 6–50% B and the peaks of interest were monitored at 335 nm by
a photodiode array (PDA) detector with SNAP KP-SIL cartridges.
Subsequently, using the same mobile phases, the flow rate was kept
constant at 75 ml/min for a total run volume of 600 ml. Then the
system was run with a gradient program: 0–32 min, 6–50% B and the
peaks of interest were monitored at 335 nm by a PDA detector using
SNAP Ultra cartridges. Lastly, 2.5 g AAD was further analyzed by
high-performance liquid chromatography (HPLC)-PDA and liquid
chromatography-mass spectrometry (LC-MS) for further analysis and
identification.
Identification of acteoside by HPLC
analysis
AAD (1 mg/ml) was analyzed in a Waters 2695 HPLC
system equipped with Waters 2996 PDA using an Xbridge-C18 column
(250×4.6 mm; 5 µm; all from Waters Corporation). The binary mobile
phase consisted of acetonitrile (solvent A) and water containing 1%
acetic acid (solvent B). Both solvents were filtered through a
0.45-µm filter prior to use. The flow rate was kept constant at 1.0
ml/min for a total run time of 40 min at 25°C. The system was run
with a gradient program: i) 0–5 min, 10–15% A: 90–85% B; ii) 5–15
min, 15% A: 85% B; iii) 15–40 min, 15–40% A: 85–60% B. The sample
injection volume was 10 µl. The peaks of interest were monitored at
335 nm by a PDA and compared with an acteoside standard
(V4015-10MG; Sigma-Aldrich; Merck KGaA).
Identification of acteoside by
LC-MS
LC-MS analysis was performed using a Nexera X2 Ultra
high-performance liquid chromatograph/mass selective detector at
25°C, equipped with an LCMS-8050 quadrupole mass spectrometer (all
from Shimadzu Corporation). The liquid chromatographic system
consisted of a binary pump, on-line vacuum degasser and
thermostatic column compartment, connected in-line to the mass
spectrometer. Data acquisition and analysis were carried out using
Shimadzu LabSolutions 5.0 (Shimadzu Corporation). Samples (3 µl)
were injected into the HPLC system using a binary gradient of
deionized water (solvent A) and acetonitrile (solvent B) and
isocratically eluted in 50% solvent B for 1 min at a flow rate of
0.3 ml/min. The mass spectrometer was fitted to an atmospheric
pressure electrospray ionization source operated in negative ion
mode. The electrospray capillary voltage was set to 4.0 kV, with a
nebulizing gas (10.7 bar) flow rate of 3 l/min and a drying gas
temperature of 300°C. MS data were acquired in the Scan mode (mass
range m/z 300–800) and the Product ion scan mode.
1,1-Diphenyl-2-picryl hydrazyl (DPPH)
radical scavenging activity
DPPH radical scavenging activity was measured as
previously described (30), with
minor modifications. A 300 µM DPPH solution (Sigma-Aldrich; Merck
KGaA) in 95% alcohol was prepared. The solution was adjusted to an
absorbance value of 1.00 at 515 nm. A 40-µl volume of sample was
mixed with 760 µl DDPH solution, then incubated for 20 min in the
dark at 25°C. L-ascorbic acid was used as a positive control.
Absorbance was measured at 515 nm using an ultraviolet (UV)/visible
spectrophotometer (Xma-3000PC; Human Corporation). The radical
scavenging activity was measured as a decrease in the absorbance of
DPPH and calculated using the following formula:
DPPH radical scavenging activity (%) =
[1-(ASample-ABlank)/AControl] ×100
where ‘ASample’ = Absorbance values of DPPH radicals
following treatment with sample, ‘ABlank’ = Absorbance
values of ethanol and ‘AControl’ = Absorbance values of
DPPH radicals.
2,2′-Azino-bis
(3-ethylbenzothiazoline-6-sulfonic acid) diammonium salt (ABTS)
radical scavenging activity
ABTS radical scavenging activity was measured as
previously described (31), with
some modifications. A 7.4 mM ABTS and 2.6 mM potassium persulfate
solution (both from Sigma-Aldrich; Merck KGaA) in distilled water
was prepared 24 h prior to use. The solution was adjusted to an
absorbance value of 0.70 at 732 nm. A 40 µl volume of sample was
mixed with 760 µl ABTS solution, then incubated for 20 min in the
dark at 25°C. L-ascorbic acid was used as a positive control.
Absorbance was measured using a UV/visible spectrophotometer at 732
nm. The radical scavenging activity was measured as a decrease in
the absorbance of ABTS and calculated using the following formula:
ABTS radical scavenging activity (%) =
[1-(ASample-ABlank)/AControl] ×100
where ‘ASample’ = Absorbance values of ABTS radicals
after treatment with sample, ‘ABlank’ = Absorbance
values of H2O and ‘AControl’ = Absorbance
values of ABTS radicals.
Inhibitory effect of AAD on oxidative
DNA damage using ΦX-174 RF I plasmid DNA
The inhibitory effect on oxidative DNA damage was
measured as previously described (32). Conversion of supercoiled plasmid
DNA to open circular forms has been used as an index of DNA damage
(32). In the absence of
OH− radicals and Fe2+ ions, plasmid DNA
mainly exists in a supercoiled form. However, in the presence of
OH− radicals and Fe2+, plasmid DNA is
cleaved, converting its supercoiled form into open-circular forms.
AAD (0–200 µM) was pre-incubated with 5 mM FeCl2 or 0.4
mM FeSO4 + 0.4 mM H2O2 for 15 min
at 25°C. The ΦX-174 RF I plasmid DNA (50 µg/ml; Promega
Corporation) was then added to the mixtures. A solution containing
50% glycerol (v/v), 40 mM EDTA, and 0.05% bromophenol blue
(Sigma-Aldrich; Merck KGaA) was added to stop the reaction.
Reaction mixtures were analyzed by agarose gel electrophoresis on
1% gels with DNA SafeStain (Lamda Biotech). DNA in the gel was
visualized using the Chemi-Doc imaging system (Bio-Rad
Laboratories, Inc.) controlled by Image Lab software 6.0 (Bio-Rad
Laboratories, Inc.).
Cell culture
The murine skin fibroblast cell line NIH 3T3 was
purchased from American Type Culture Collection. Cells were
cultured under sterile conditions at 37°C in a humid incubator with
5% of CO2, in Dulbecco's modified Eagle's medium
supplemented with 10% bovine calf serum (Biowest), 1% antibiotics
(penicillin/streptomycin; HyClone; Cytiva) and 0.2% anti-mycoplasma
agents (Plasmocin prophylactic; InvivoGen). The oxidative damage
was induced by 150 µM FeSO4 + 600 µM
H2O2 for the cell viability, western
blotting, and PCR assays.
Cell viability
Cells (1.0×104 cells/ml) were treated
with AAD (0–200 µg/ml) and incubated at 37°C for 24 h. After 24 h,
cells were treated with 10 µl alamarBlue® cell viability
reagent (Thermo Fisher Scientific, Inc.). After 2 h, cell viability
was measured using a UV/Visible spectrophotometer at 570 nm.
Immunoblotting
Cells were lysed using radioimmunoprecipitation
assay buffer (Thermo Fisher Scientific, Inc.) supplemented with a
protease inhibitor cocktail (Sigma-Aldrich; Merck KGaA). The
protein concentration in the sample was determined using the
Bradford protein assay (Bio-Rad Laboratories, Inc.). Proteins (20
µg) were separated by SDS-PAGE on 10% gels, then transferred to a
PVDF (Bio-Rad Laboratories, Inc.). The membrane was blocked for 1 h
at 20°C with 5% bovine serum albumin (BSA; Biosesang) in TBS
containing 1% Tween-20 (TBS-T), then incubated with primary
antibodies in 3% BSA overnight at 4°C. The following primary
antibodies were used at 1:1,000 (all from Abcam): i) Anti-γH2AX
(Ser139) polyclonal antibody (cat. no. ab11174); ii) anti-H2AX
polyclonal antibody (cat. no. ab11175); iii)
anti-phosphorylated-p53 (p-p53; Ser15) polyclonal antibody (cat.
no. ab1431); iv) anti-p53 polyclonal antibody (cat. no. ab131442);
and v) anti-GAPDH monoclonal antibody (cat. no. ab8245). After
washing with TBS-T, membranes were incubated with anti-rabbit IgG
monoclonal antibody (cat. no. 18-8816-33; Rockland Immunochemicals
Inc.; 1:5,000) or anti-mouse IgG monoclonal antibody (cat. no.
18-8817-33; Rockland Immunochemicals Inc.; 1:5,000) horseradish
peroxidase-conjugated secondary antibodies for 1 h. Immunoreactive
bands were visualized using an ECL western blotting substrate and
the Chemi-Doc imaging system (both from Bio-Rad Laboratories,
Inc.). Bands were analyzed using the ImageJ software v1.51K
(National Institutes of Health).
Reverse transcription
(RT)-semi-quantitative PCR (qPCR)
Total RNA was extracted from NIH 3T3 cells using the
NucleoSpin™ RNA Plus kit (Macherey-Nagel; Thermo Fisher Scientific,
Inc.). cDNA was synthesized using ReverTra Ace-α-™ (Toyobo Life
Science), according to the manufacturer's protocol. qPCR was
carried out using Quick Taq® HS Dye Mix (Toyobo Life
Science). Amplification was initiated at 94°C for 2 min, followed
by 30 amplification cycles at 94°C for 30 sec (denaturation), then
54.2°C for 30 sec (annealing/extension) using Thermal cycler
(T100™; Bio-Rad Laboratories, Inc.). Primer sequences using Primer
3 program 0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/) are presented in
Table I. DNA density was
visualized by 2% agarose gel (A1200; Biosesang) with 1% DNA
SafeStain (C138; Lamda Biotech Corporation). Transcription levels
were normalized to GAPDH. Density was analyzed using the ImageJ
software 1.51K (National Institutes of Health).
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| A,
RT-semi-qPCR |
|---|
|
|---|
| Gene | Forward primer
sequence (5–3) | Reverse primer
sequence (5–3) |
|---|
| H2AX |
TTGCTTCAGCTTGGTGCTTAG |
AACTGGTATGAGGCCAGCAAC |
| p53 |
CGGATAGTATTTCACCCTCAAGATCCG |
AGCCCTGCTGTCTCCAGACTC |
| GAPDH |
AACTTTGGCATTGTGGAAGG |
ATGCAGGGATGATGTTCTGG |
|
| B,
RT-qPCR |
|
| Gene | Forward primer
sequence (5–3) | Reverse primer
sequence (5–3) |
|
| H2AX |
GCGTCTTTGCTTCAGCTTG |
CACACTGGCCTACGAATGG |
| p53 |
GATGTTCCGGGAGCTGAAT |
GCCCCACTTTCTTGACCAT |
| GAPDH |
CCTCCAAGGAGTAAGAAACC |
CTAGGCCCCTCCTGTTATTA |
RT-qPCR analysis
cDNA samples were generated using the aforementioned
method. qPCR was carried out using the Quanti Tect®
SYBR-Green PCR kit (Qiagen GmbH) on a 7500 Real-Time PCR system
(Applied Biosystems; Thermo Fisher Scientific, Inc.). Primers were
designed using the Primer 3 program 0.4.0 (http://bioinfo.ut.ee/primer3-0.4.0/; Table I). Thermocycling conditions
consisted of a 15-min initial denaturation at 95°C, followed by 40
amplification cycles at 94°C for 15 sec (denaturation), 60°C for 30
sec (annealing) and 72°C for 30 sec (extension). Transcription
levels were normalized to GAPDH. The 2−ΔΔCq formula was
used to analyze mRNA expression levels, where ΔΔCq =
(Cqtarget - CqGAPDH)sample -
(Cqtarget - CqGAPDH)control
(33). Data were analyzed using
the ABI 7500 Software 2.3 (Applied Biosystems; Thermo Fisher
Scientific, Inc.).
Immunofluorescence staining
γ-H2AX and p-p53 are markers for DNA damage and
repair (34). NIH 3T3 cells were
treated with AAD (0, 100 µM) and oxidative damage (150 µM
FeSO4 + 600 µM H2O2) at 37°C for
24 h. Following treatment, samples were prepared on sterilized
glass coverslips (BD Biosciences). Cells were then fixed with 4%
paraformaldehyde for 15 min at 20°C, then blocked for 1 h with 3%
BSA (Biosesang) and 0.1% Triton X-100 for permeabilization at 20°C.
Cells were incubated with anti-γ-H2AX (cat. no. ab11174; 1:500) and
anti-p-p53 (cat. no. ab1431; 1:500) overnight at 4°C, then with
Alexa Fluor 488 (cat. no. ab150077; 1:500) and Alexa Fluor
568-conjugated secondary antibodies (cat. no. ab175471; 1:500; All
antibodies from Abcam) for an additional 1 h at 20°C in the dark.
Nuclei were stained using DAPI (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C. Slides were mounted and images captured
using a CKX53 fluorescence microscope (magnification, ×400; Olympus
Corporation).
Statistical analysis
All data were analyzed using GraphPad Prism 5.0
(GraphPad Software, Inc.) and are presented as the mean ± standard
deviation. All experiments were performed ≥3 times. Data of DPPH
and ABTS radical scavenging activity were analyzed using paired
Student's t-test. Other data were analyzed by One-way ANOVA
followed by Tukey's post hoc test were used to compare the means
across multiple groups. P<0.05 was considered to indicate a
statistically significant difference.
Results
Isolation, purification and
identification of AAD
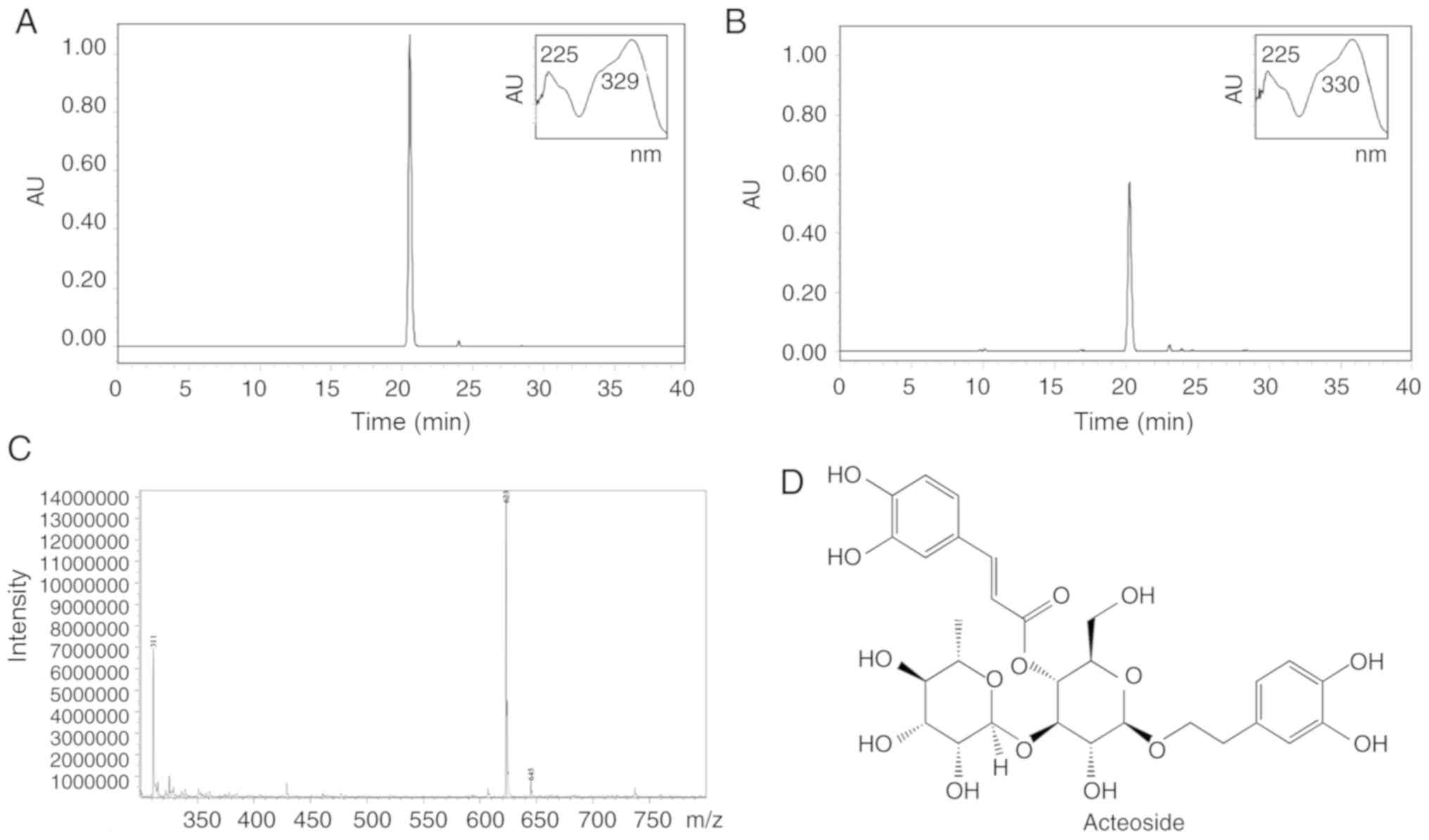
Acteoside standard and AAD extract were analyzed
using HPLC-PDA. The standard (Fig.
1A) and the sample (Fig. 1B)
displayed similar retention times (20.3 min) and UV spectra. The
LC-MS data (Fig. 1C) also
exhibited 623 m/z (M-H−). Based on these observations,
the AAD sample was identified as an acteoside (Fig. 1D). The purity of AAD was >95%,
based on a calibration curve (y = 16089× − 93989, R2 =
0.9971).
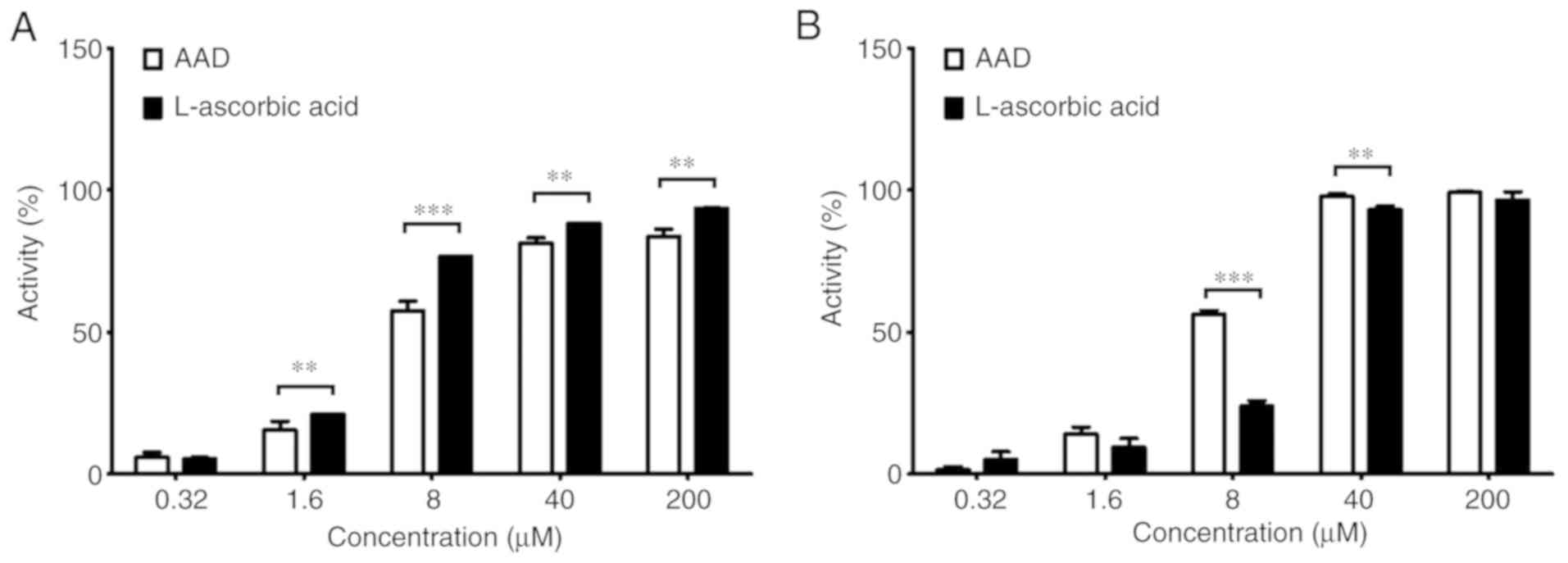
Antioxidant activity of AAD
AAD eliminated DPPH radicals in a dose-dependent
manner (Fig. 2A). DPPH radicals
were scavenged at all AAD concentrations, except 0.32 µM, although
not to the same extent as with L-ascorbic acid. The half-maximal
inhibitory concentration (IC50) values of AAD and
L-ascorbic acid were 8.81 and 5.08 µg/ml, respectively. Similarly,
AAD also eliminated ABTS radicals in a dose-dependent manner
(Fig. 2B). The IC50
values of AAD and L-ascorbic acid were 6.47 and 10.49 µg/ml,
respectively. Furthermore, at concentrations of 8 and 40 µM, the
scavenging activity of AAD was significantly higher compared with
L-ascorbic acid. These results indicated that AAD effectively
scavenged DPPH and ABTS radicals in vitro.
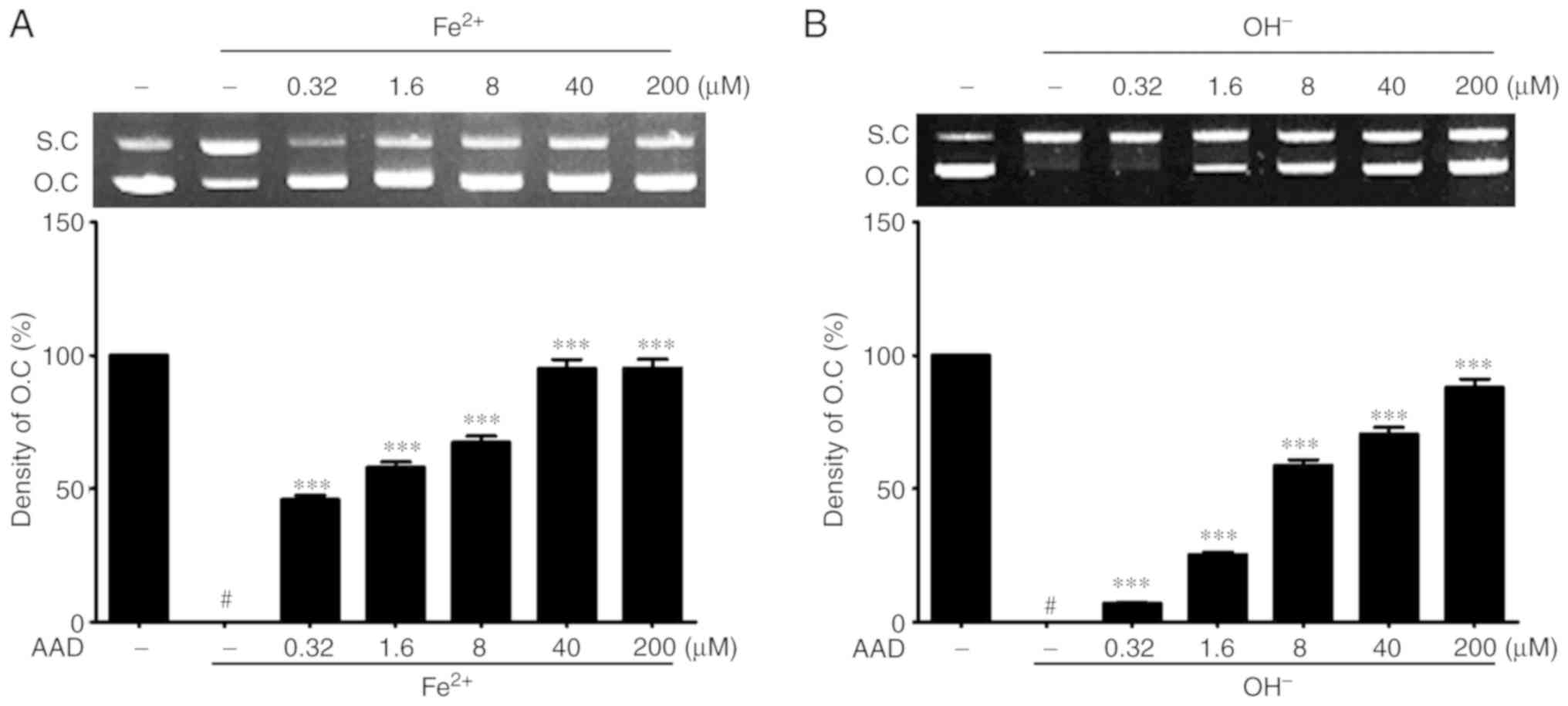
Protective effect against oxidative
stress-induced DNA damage of AAD
The protective effects of AAD on oxidative DNA
damage were evaluated in a DNA cleavage assay using ΦX-174 RF I
DNA. The untreated control group (lane 1) was used as a positive
control and assigned an open circular (OC) DNA band density value
of 100%. In lane 2, the groups receiving Fe2+ or
OH− without AAD treatment were used as a negative
control and given a value of 0%.
AAD inhibited Fe2+-induced oxidative DNA
damage in a dose-dependent manner, as suggested by the significant
OC band density increase at all concentrations, compared with the
negative control (47.14±1.64% at 0.32 µM AAD vs. 97.66±3.41% at 200
µM; Fig. 3A). Additionally, AAD
also significantly protected plasmid DNA from OH- radicals-induced
oxidative damage in a dose-dependent manner (7.09±0.25% at 0.32 µM
AAD vs. 88.05±3.09% at 200 µM; Fig.
3B). These results suggested that AAD could protect plasmid DNA
from oxidative damage.
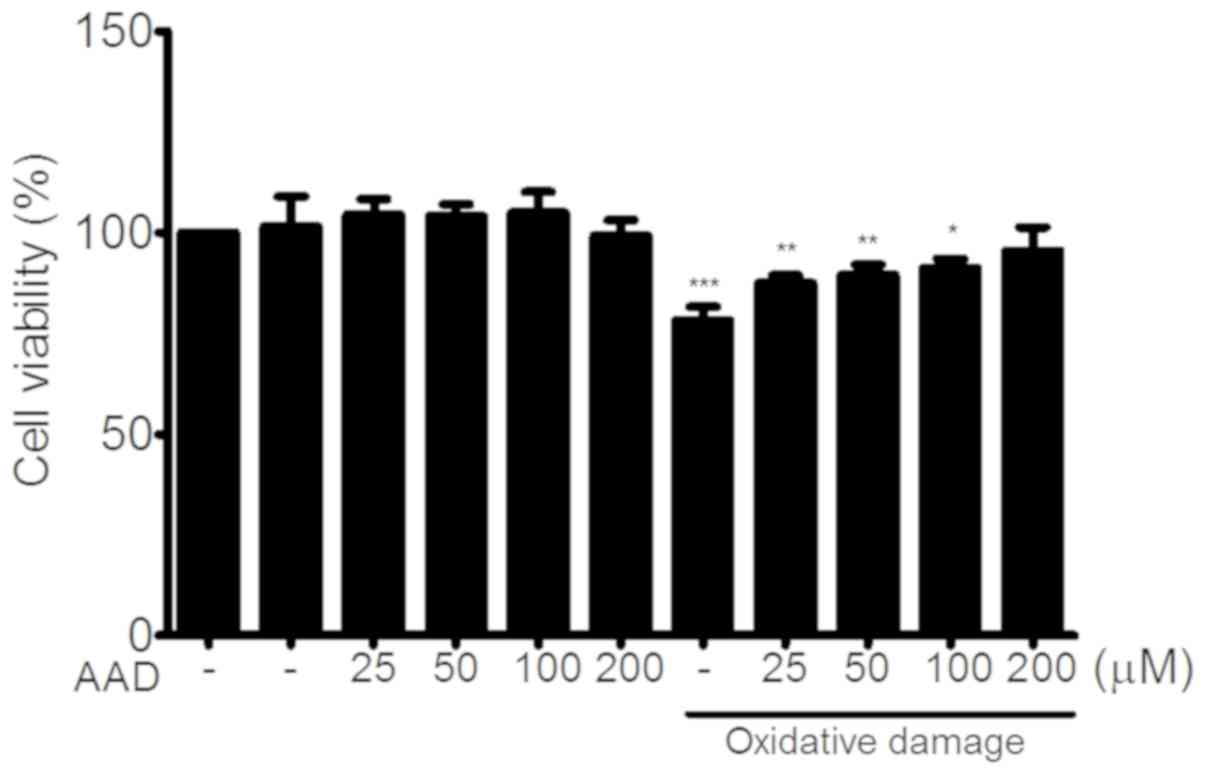
Cytotoxicity of AAD in NIH 3T3
cells
To determine whether AAD had any cytotoxic effect,
NIH 3T3 cells were cultured in AAD concentrations ranging between
25 and 200 µM, and cell viability was then assessed following 24-h
incubation. AAD had no effect on cell viability. In oxidative
damage conditions, AAD increased cell viability compared with
untreated cells (Fig. 4).
AAD protects NIH 3T3 cells against
oxidative DNA damage
Western blotting assays suggested that, in the
absence of AAD, oxidative damage significantly increased
phosphorylation of p-53 and H2AX in NIH 3T3 cells. However, under
oxidative damage conditions, the levels of p-p53 and γ-H2AX were
significantly reduced by treatment with AAD. This reduction in
p-p53 and γ-H2AX was dose-dependent (Fig. 5A and B). Additionally, gene
expression levels of p53 and H2AX followed the same pattern, as
indicated by RT-semi-qPCR (Fig. 5C and
D) and RT-qPCR (Fig. 5E).
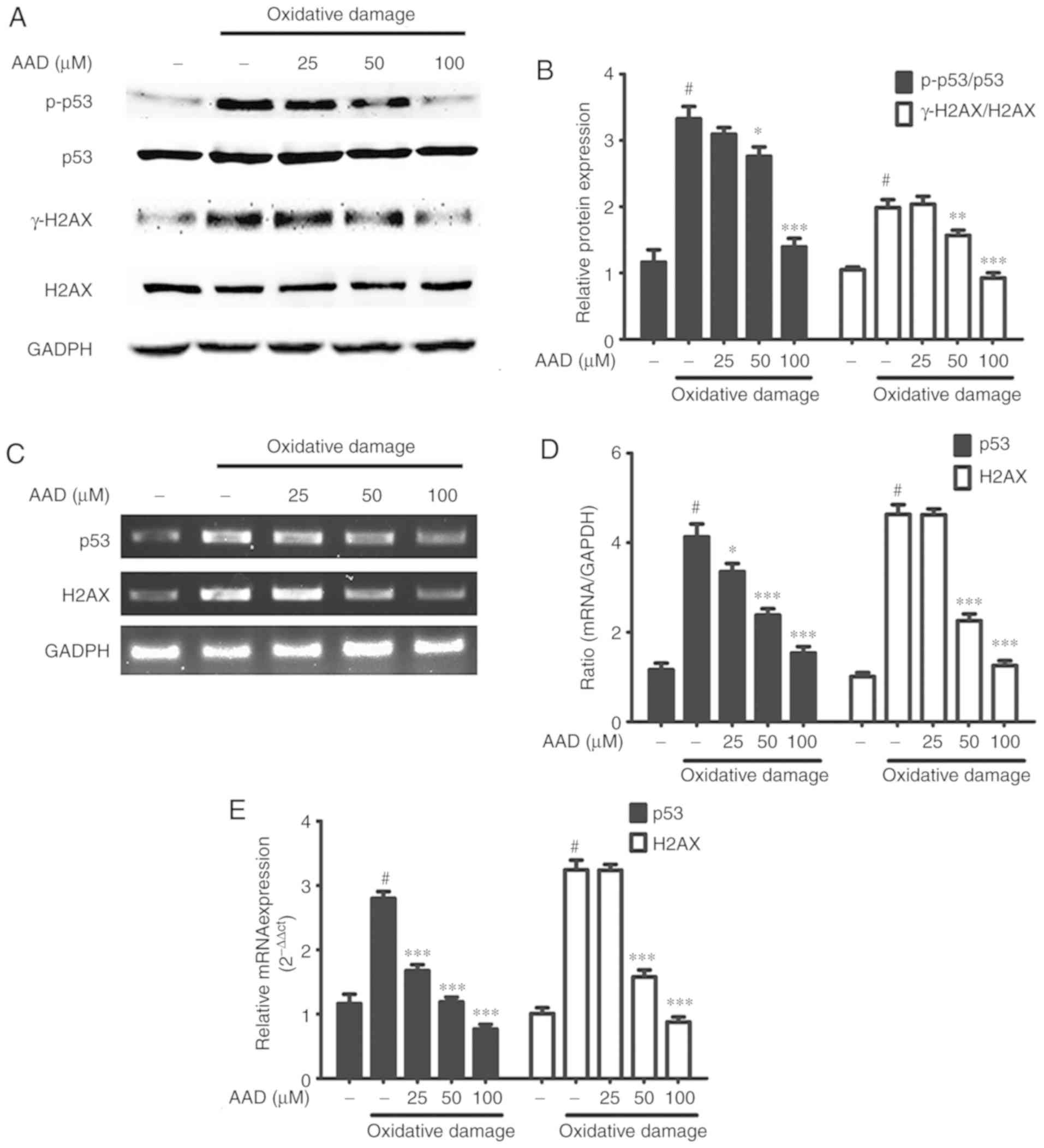 | Figure 5.AAD protects NIH 3T3 cells from
oxidative DNA damage. (A) Western blots of p-p53, p53, γ-H2AX, H2AX
and GAPDH in NIH 3T3 cells treated with AAD (25–100 µM) in the
presence of oxidative damage (150 µM FeSO4 + 600 µM
H2O2) for 24 h. (B) Densitometry of p-p53,
p53, γ-H2AX and H2AX in NIH 3T3 cells. (C) DNA gel electrophoresis
of p53 and H2AX genes in NIH 3T3 cells treated with AAD, and
oxidative damage for 24 h. (D) Semi-quantitative analysis of p53
and H2AX gene expression in NIH 3T3 cells. (E) Reverse
transcription-quantitative PCR analysis of p53 and H2AX expression
in NIH 3T3 cells. Data are presented as the mean ± standard
deviation of ≥3 independent experiments; #P<0.05 vs.
untreated group; *P<0.05, **P<0.01, ***P<0.001 vs.
oxidative damage group without AAD (the second bar). AAD, acteoside
from Abeliophyllum distichum; H2AX, H2A histone family
member X; p/γ, phosphorylated. |
Immunofluorescence staining of p-p53 and γ-H2AX was
also performed (Fig. 6). The
levels of p-p53 and γ-H2AX increased following oxidative damage.
However, p-p53 and γ-H2AX levels were reduced in the presence of
AAD compared with untreated oxidative damaged cells. These results
suggested that AAD could regulate the expression of p-p53 and
γ-H2AX.
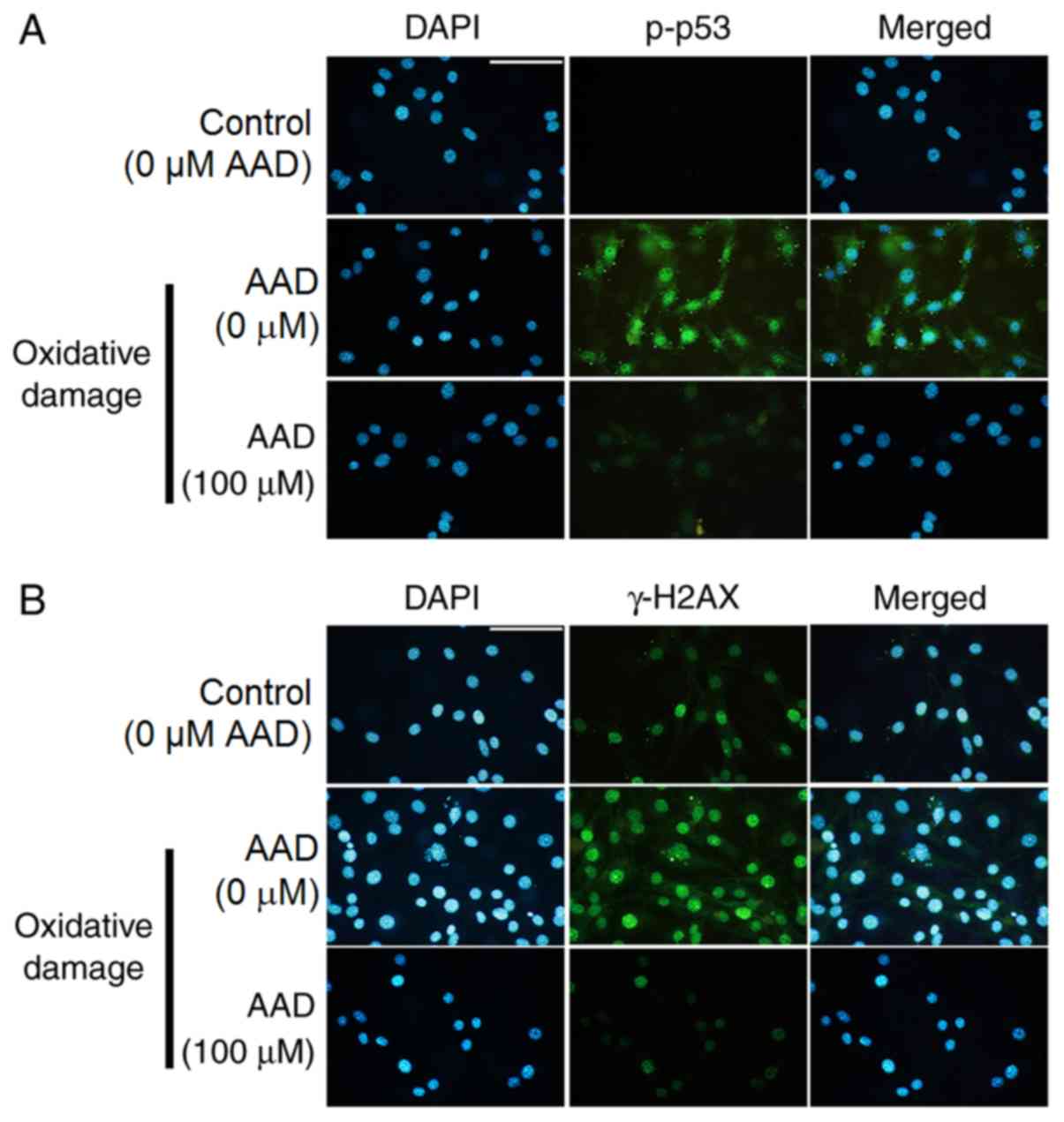 | Figure 6.AAD regulates p-p53 and γ-H2AX in NIH
3T3 cells. (A) Fluorescence microscopy shows expression of
anti-p-p53 antibody in NIH 3T3 cells treated for 24 h with 150 µM
FeSO4 and 600 µM H2O2 (oxidative
damage) in the presence or absence of 100 µM AAD. Green, p-p53;
Blue, DAPI. Scale bar, 40 µm. (B) Fluorescence microscopy shows
expression of anti-γ-H2AX antibody in NIH 3T3 cells treated for 24
h with 150 µM FeSO4 and 600 µM
H2O2 (oxidative damage) in the presence or
absence of 100 µM AAD. Green, γ-H2AX; Blue, DAPI. Scale bar, 40 µm.
Control (0 µM AAD; no oxidative damage); AAD, acteoside from
Abeliophyllum distichum; H2AX, H2A histone family member X;
p/γ, phosphorylated; DAPI, 4,6-diamidino-2-phenylindole. |
Discussion
Due to the rarity of A. distichum Nakai,
previous studies have focused on its ecological characterization,
genomic sequencing and biological properties (35–40).
Bremer et al (41)
demonstrated that a number of plants in the Oleaceae family, such
as the Abeliophyllum, Forsythia and Jasminum genera,
contain acteoside. Although the potential bioactivities of
acteoside may have important uses in industry and research, AAD
remains poorly characterized.
Several plant-sourced compounds, such as vitamin C,
phenolics and flavonoids, can act as natural antioxidants by
reacting with free radicals, chelating catalytic metals and
scavenging oxygen (42,43). Antioxidants suppress oxygen,
electrons and hydrogen atoms that are generated during
intracellular metabolism. They also protect cells against harmful
effects of ROS and associated oxidative stress generated in aerobic
organisms (44). For these
reasons, numerous studies on the regulation of ROS and the role of
natural antioxidants derived from plants have been conducted
(45–47).
In the present study, HPLC and LC-MS analyses were
carried out to characterize an AAD extract. The AAD sample had the
same retention time and a similar UV spectrum compared with a
standard acteoside. Additionally, based on LC-MS analysis, the AAD
peak was observed at 623 m/z (M-H−), which coincided
with the molecular weight of acteoside (48). Consequently, based on HPLC-PDA and
LC-MS data, the AAD extract used in this study was identified as
acteoside. Additionally, the effects of the antioxidant properties
of AAD on oxidative DNA damage were also examined. Our previous
studies suggested that A. distichum had potential
antioxidant activities (49,50).
DPPH and ABTS radicals are the most common and most stable
chromogens used to estimate antioxidant activity of biological
materials. Furthermore, DPPH radical scavenging and ABTS radical
cation decolorization assays can both be used to evaluate
antioxidant activities in a relatively short time (51,52).
Oxidation of ABTS with potassium persulfate generates
ABTS•+ radical cations with the transfer of one
electron. Under prolonged oxidative conditions, these radicals can
generate di-cation ABTS2+ radicals (53,54).
In the present study, AAD scavenged both DPPH and ABTS
radicals.
DNA damage leads to an irreversible covalent
modification of the DNA molecule. This can occur through single- or
double-strand breaks (DSBs) in the DNA sugar-phosphate backbone,
disruption of the or N-glycosidic bonds linking nucleobases to the
sugar, as well as chemical modification of nucleobase residues
(55,56). Oxidative DNA damage represents one
of the most frequent consequences of exposure to exogenous
environmental stimuli or endogenous genotoxic agents. These
reactions are associated with the activity of ROS, such as hydroxyl
radicals, superoxides, peroxides or single oxygen (57,58).
For instance, the oxidative DNA damage produced by the Fenton
reaction plays a major role in the aging process, as well as
several diseases, including Alzheimer's disease, cancer and
multiple sclerosis (59–62). In the present study, the protective
effects of AAD against oxidative DNA damage were evaluated using
non-cellular and cellular models. A ΦX-174 RF I plasmid DNA
cleavage assay was conducted using OH radicals and Fe2+
ions. Hydrogen peroxide itself cannot directly oxidize DNA
molecules, but can react with transition metals, such as
Fe2+ to form OH− radicals. These
OH− radicals attack DNA structures, leading to sugar
fragmentation, strand scission and base adducts (63). The Fe2+ ion is an
essential transition metal element in humans. In the present study,
AAD decreased oxidative DNA damage in a dose-dependent manner.
Therefore, due to its scavenging capacity, AAD may prevent cell
damage caused by OH- radicals and Fe2+ ions. This type
of DNA damage is one of the most genotoxic types occurring in
cells, either resulting in cell cycle arrest and DNA repair, or
elimination of the injured cells. Appropriate cellular responses to
DNA DSBs are important for tumor suppression and maintaining
genetic stability (64,65).
One of the first cellular responses to the
introduction of DSBs is the phosphorylation of H2AX, a sensitive
marker for DNA DSBs. DSBs induce phosphorylation of H2AX at its
C-terminal serine residues (Ser136 and Ser139) (66). The phosphorylated formed of H2AX,
called γ-H2AX, is detectable at the sites of the damage following
the introduction of DSBs (67,68).
Therefore, γ-H2AX plays a distinct role in the DNA damage response.
Additionally, the p53 pathway is also a key effector of the DNA
damage response and is activated by several stimuli that induce DNA
lesions (69). The expression of
phosphorylated p53 (p-p53) may represent a negative regulator of
the cellular response to DNA damage (70). In the present study, AAD treatment
reduced the phosphorylation of γ-H2AX and p-p53 in NIH 3T3 cells
under oxidative damage conditions. Moreover, the expression levels
of H2AX and p53 were also reduced by AAD treatment. Although
several other possible mechanisms may also explain these
observations, the present study identified a relationship between
the antioxidant properties of AAD and inhibition of DNA damage. In
conclusion, the present study demonstrated that AAD displayed
antioxidant properties that could protect DNA against oxidative
damage.
Acknowledgements
Not applicable.
Funding
This research was supported by The Basic Science
Research Program through The National Research Foundation of Korea
funded by The Ministry of Education (grant no.
NRF-2016R1D1A1B03934869).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
TWJ and JHP drafted the manuscript and performed the
experiments. TWJ and JSC performed the experiments and data
interpretation. TWJ contributed materials and analytical tools and
helped in data analysis. TWJ, JSC and JHP designed the experiments,
provided critical suggestions for the manuscript, and reviewed and
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Akerele O: Nature's medicinal bounty:
Don't throw it away. World Health Forum. 14:390–395.
1993.PubMed/NCBI
|
|
2
|
Omale J and Okafor PN: Comparative
antioxidant capacity, membrane stabilization, polyphenol
composition and cytotoxicity of the leaf and stem of Cissus
multistriata. Afr J Biotechnol. 7:172008.
|
|
3
|
Hill AF: Economic Botany. McGraw-Hill.
(New York, NY). 5601952.
|
|
4
|
Edeoga HO, Okwu D and Mbaebie B:
Phytochemical constituents of some Nigerian medicinal plants. Afr J
Biotechnol. 4:685–688. 2005. View Article : Google Scholar
|
|
5
|
Valko M, Morris H and Cronin MT: Metals,
toxicity and oxidative stress. Curr Med Chem. 12:1161–1208. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Schärer OD: Chemistry and biology of DNA
repair. Angew Chem Int Ed Engl. 42:2946–2974. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Han HM, Kwon YS and Kim MJ: Antioxidant
and antiproliferative activity of extracts from water chestnut
(Trapa japonica Flerow). Hanguk Yakyong Changmul Hakhoe Chi.
24:14–20. 2016. View Article : Google Scholar
|
|
8
|
Roos WP, Thomas AD and Kaina B: DNA damage
and the balance between survival and death in cancer biology. Nat
Rev Cancer. 16:20–33. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Gilgun-Sherki Y, Rosenbaum Z, Melamed E
and Offen D: Antioxidant therapy in acute central nervous system
injury: Current state. Pharmacol Rev. 54:271–284. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hamanaka RB and Chandel NS: Mitochondrial
reactive oxygen species regulate cellular signaling and dictate
biological outcomes. Trends Biochem Sci. 35:505–513. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Simon HU, Haj-Yehia A and Levi-Schaffer F:
Role of reactive oxygen species (ROS) in apoptosis induction.
Apoptosis. 5:415–418. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fernandez-Capetillo O, Lee A, Nussenzweig
M and Nussenzweig A: H2AX: The histone guardian of the genome. DNA
Repair (Amst). 3:959–967. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stucki M, Clapperton JA, Mohammad D, Yaffe
MB, Smerdon SJ and Jackson SP: MDC1 directly binds phosphorylated
histone H2AX to regulate cellular responses to DNA double-strand
breaks. Cell. 123:1213–1226. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cook PJ, Ju BG, Telese F, Wang X, Glass CK
and Rosenfeld MG: Tyrosine dephosphorylation of H2AX modulates
apoptosis and survival decisions. Nature. 458:591–596. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu B, Chen Y and St Clair DK: ROS and
p53: A versatile partnership. Free Radic Biol Med. 44:1529–1535.
2008. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Polyak K, Xia Y, Zweier JL, Kinzler KW and
Vogelstein B: A model for p53-induced apoptosis. Nature.
389:300–305. 1997. View
Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sablina AA, Budanov AV, Ilyinskaya GV,
Agapova LS, Kravchenko JE and Chumakov PM: The antioxidant function
of the p53 tumor suppressor. Nat Med. 11:1306–1313. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shibata A and Jeggo PA: DNA double-strand
break repair in a cellular context. Clin Oncol (R Coll Radiol).
26:243–249. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Paull TT, Rogakou EP, Yamazaki V,
Kirchgessner CU, Gellert M and Bonner WM: A critical role for
histone H2AX in recruitment of repair factors to nuclear foci after
DNA damage. Curr Biol. 10:886–895. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ji K, Jang NY and Kim YT: Isolation of
lactic acid bacteria showing antioxidative and probiotic activities
from kimchi and infant feces. J Microbiol Biotechnol. 25:1568–1577.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Nakai T: Genus novum Oleacearum in Corea
media inventum. Shokubutsugaku Zasshi. 33:153–154. 1919. View Article : Google Scholar
|
|
22
|
Oh H, Kang DG, Kwon TO, Jang KK, Chai KY,
Yun YG, Chung HT and Lee HS: Four glycosides from the leaves of
Abeliophyllum distichum with inhibitory effects on
angiotensin converting enzyme. Phytother Res. 17:811–813. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xiong Q, Hase K, Tezuka Y, Tani T, Namba T
and Kadota S: Hepatoprotective activity of phenylethanoids from
Cistanche deserticola. Planta Med. 64:120–125. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Schlesier K, Harwat M, Böhm V and Bitsch
R: Assessment of antioxidant activity by using different in vitro
methods. Free Radic Res. 36:177–187. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xie J: Effect of ethanolic extract of
Cistanche deserticola on the contents of monoamine
neurotransmitters in rat brain. Chin Tradit Herbal Drugs.
24:417–419. 1993.
|
|
26
|
He ZD, Lau KM, Xu H-X, Li PC and Pui-Hay
But P: Antioxidant activity of phenylethanoid glycosides from
Brandisia hancei. J Ethnopharmacol. 71:483–486. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Wang PF, Zheng R, Liu ZM and Jia Z:
Protection of phenylpropanoid glycosides from Pedicularis
against oxidative hemolysis in vitro. Planta Med. 59:315–317. 1993.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Xiong Q, Kadota S, Tani T and Namba T:
Antioxidative effects of phenylethanoids from Cistanche
deserticola. Biol Pharm Bull. 19:1580–1585. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zong G, He W, Wu G, Chen M, Shen X and Shi
M: Comparisons between Cistanche deserticola Y.C. Ma and
C. tubulosa (Schenk) Wight on Some Pharmacological Actions.
Zhongguo Zhong Yao Za Zhi. 21:436–437. 1996.(In Chinese).
PubMed/NCBI
|
|
30
|
Bondet V, Brand-Williams W and Berset C:
Kinetics and mechanisms of antioxidant activity using the DPPH.
free radical method. Lebensm Wiss Technol. 30:609–615. 1997.
View Article : Google Scholar
|
|
31
|
van den Berg R, Haenen GR, van den Berg H
and Bast A: Applicability of an improved Trolox equivalent
antioxidant capacity (TEAC) assay for evaluation of antioxidant
capacity measurements of mixtures. Food Chem. 66:511–517. 1999.
View Article : Google Scholar
|
|
32
|
Jung Y and Surh Y: Oxidative DNA damage
and cytotoxicity induced by copper-stimulated redox cycling of
salsolinol, a neurotoxic tetrahydroisoquinoline alkaloid. Free
Radic Biol Med. 30:1407–1417. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2−ΔΔCT method. Methods. 25:402–408. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Otterbein LE, Hedblom A, Harris C,
Csizmadia E, Gallo D and Wegiel B: Heme oxygenase-1 and carbon
monoxide modulate DNA repair through ataxia-telangiectasia mutated
(ATM) protein. Proc Natl Acad Sci USA. 108:14491–14496. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Kang U, Chang CS and Kim YS: Genetic
structure and conservation considerations of rare endemic
Abeliophyllum distichum Nakai (Oleaceae) in Korea. J Plant
Res. 113:127–138. 2000. View Article : Google Scholar
|
|
36
|
Kim NY and Lee HY: Effect of antioxidant
and skin whitening of ethanol extracts from ultrasonic pretreated
Abeliophyllum distichum Nakai. Hanguk Yakyong Changmul
Hakhoe Chi. 23:155–160. 2015. View Article : Google Scholar
|
|
37
|
Kim HW, Lee HL, Lee DK and Kim KJ:
Complete plastid genome sequences of Abeliophyllum distichum
Nakai (Oleaceae), a Korea endemic genus. Mitochondrial DNA B
Resour. 1:596–598. 2016. View Article : Google Scholar
|
|
38
|
Park J, Kim Y, Xi H, Jang T and Park JH:
The complete chloroplast genome of Abeliophyllum distichum
Nakai (Oleaceae), cultivar Ok Hwang 1ho: Insights of cultivar
specific variations of A. distichum. Mitochondrial DNA B
Resour. 4:1640–1642. 2019. View Article : Google Scholar
|
|
39
|
Min J, Kim Y, Xi H, Jang T, Kim G, Park J
and Park JH: The complete chloroplast genome of a new candidate
cultivar, Sang Jae, of Abeliophyllum distichum Nakai
(Oleaceae): Initial step of A. distichum intraspecies
variations atlas. Mitochondrial DNA B Resour. 4:3716–3718. 2019.
View Article : Google Scholar
|
|
40
|
Park J, Min J, Kim Y, Xi H, Kwon W, Jang
T, Kim G and Park JH: The complete chloroplast genome of a new
candidate cultivar, Dae Ryun, of Abeliophyllum distichum
Nakai (Oleaceae). Mitochondrial DNA B Resour. 4:3713–3715. 2019.
View Article : Google Scholar
|
|
41
|
Bremer B, Bremer K, Heidari N, Erixon P,
Olmstead RG, Anderberg AA, Källersjö M and Barkhordarian E:
Phylogenetics of asterids based on 3 coding and 3 non-coding
chloroplast DNA markers and the utility of non-coding DNA at higher
taxonomic levels. Mol Phylogenet Evol. 24:274–301. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Pisoschi AM, Cheregi MC and Danet AF:
Total antioxidant capacity of some commercial fruit juices:
Electrochemical and spectrophotometrical approaches. Molecules.
14:480–493. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Prasad KN, Chew LY, Khoo HE, Kong KW,
Azlan A and Ismail A: Antioxidant capacities of peel, pulp, and
seed fractions of Canarium odontophyllum Miq. fruit. J
Biomed Biotechnol. 2010:8713792010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sies H: Strategies of antioxidant defense.
Eur J Biochem. 215:213–219. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Devasagayam TP, Tilak JC, Boloor KK, Sane
KS, Ghaskadbi SS and Lele RD: Free radicals and antioxidants in
human health: Current status and future prospects. J Assoc
Physicians India. 52:794–804. 2004.PubMed/NCBI
|
|
46
|
Alfadda AA and Sallam RM: Reactive oxygen
species in health and disease. J Biomed Biotechnol.
2012:9364862012. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Asif M: Chemistry and antioxidant activity
of plants containing some phenolic compounds. Chem Int. 1:35–52.
2015.
|
|
48
|
Blazics B, Alberti Á, Kursinszki L, Kéry
Á, Béni S and Tölgyesi L: Identification and LC-MS-MS determination
of acteoside, the main antioxidant compound of Euphrasia
rostkoviana, using the isolated target analyte as external
standard. J Chromatogr Sci. 49:203–208. 2011. View Article : Google Scholar
|
|
49
|
Ahn J and Park JH: Effects of
Abeliophyllum distichum Nakai flower extracts on
antioxidative activities and inhibition of DNA damage. Korean J
Plant Resour. 26:355–361. 2013. View Article : Google Scholar
|
|
50
|
Park JH: Antioxidant activities and
inhibitory effect on oxidative DNA damage of extracts from
Abeliophylli distichi Folium. Korea J Herbology. 26:95–99.
2011.
|
|
51
|
Sieniawska E, Baj T and Głowniak K:
Influence of the preliminary sample preparation on the tannins
content in the extracts obtained from Mutellina purpurea.
Ann UMCS Sect. 1500:47–54. 2010.
|
|
52
|
Luís Â, Domingues F, Gil C and Duarte AP:
Antioxidant activity of extracts of Portuguese shrubs:
Pterospartum tridentatum, Cytisus scoparius and Erica spp. J
Med Plants Res. 3:886–893. 2009.
|
|
53
|
Venkatasubramanian L and Maruthamuthu P:
Kinetics and mechanism of formation and decay of
2,2-azinobis-(3-ethylbenzothiazole-6-sulphonate) radical cation in
aqueous solution by inorganic peroxides. Int J Chem Kinet.
21:399–421. 1989. View Article : Google Scholar
|
|
54
|
Branchi B, Galli C and Gentili P: Kinetics
of oxidation of benzyl alcohols by the dication and radical cation
of ABTS. Comparison with laccase-ABTS oxidations: An apparent
paradox. Org Biomol Chem. 3:2604–2614. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Gates KS: An overview of chemical
processes that damage cellular DNA: Spontaneous hydrolysis,
alkylation, and reactions with radicals. Chem Res Toxicol.
22:1747–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Sancar A and Sancar GB: DNA repair
enzymes. Annu Rev Biochem. 57:29–67. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Cowan JA: Chemical nucleases. Curr Opin
Chem Biol. 5:634–642. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Vacek J, Mozga T, Cahová K, Pivoňková H
and Fojta M: Electrochemical sensing of chromium-induced DNA
damage: DNA strand breakage by intermediates of chromium (VI)
electrochemical reduction. Electroanalysis. Int J Devoted Fundam
Pract Asp Electroanalysis. 19:2093–2102. 2007.
|
|
59
|
Kanvah S and Schuster GB: One-electron
oxidation of DNA: The effect of replacement of cytosine with
5-methylcytosine on long-distance radical cation transport and
reaction. J Am Chem Soc. 126:7341–7344. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Demple B and Harrison L: Repair of
oxidative damage to DNA: Enzymology and biology. Annu Rev Biochem.
63:915–948. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Poulsen HE, Prieme H and Loft S: Role of
oxidative DNA damage in cancer initiation and promotion. Eur J
Cancer Prev. 7:9–16. 1998.PubMed/NCBI
|
|
62
|
Hasty P and Vijg J: Aging. Genomic
priorities in aging. Science. 296:1250–1251. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Hutchinson F: Chemical changes induced in
DNA by ionizing radiation. Prog Nucleic Acid Res Mol Biol.
32:115–154. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Bennett CB, Lewis AL, Baldwin KK and
Resnick MA: Lethality induced by a single site-specific
double-strand break in a dispensable yeast plasmid. Proc Natl Acad
Sci USA. 90:5613–5617. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Mills KD, Ferguson DO and Alt FW: The role
of DNA breaks in genomic instability and tumorigenesis. Immunol
Rev. 194:77–95. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Rogakou EP, Pilch DR, Orr AH, Ivanova VS
and Bonner WM: DNA double-stranded breaks induce histone H2AX
phosphorylation on serine 139. J Biol Chem. 273:5858–5868. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Celeste A, Fernandez-Capetillo O, Kruhlak
MJ, Pilch DR, Staudt DW, Lee A, Bonner RF, Bonner WM and
Nussenzweig A: Histone H2AX phosphorylation is dispensable for the
initial recognition of DNA breaks. Nat Cell Biol. 5:675–679. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Sedelnikova OA, Pilch DR, Redon C and
Bonner WM: Histone H2AX in DNA damage and repair. Cancer Biol Ther.
2:233–235. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Phillips ER and McKinnon PJ: DNA
double-strand break repair and development. Oncogene. 26:7799–7808.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Kastan MB, Onyekwere O, Sidransky D,
Vogelstein B and Craig RW: Participation of p53 protein in the
cellular response to DNA damage. Cancer Res. 51:6304–6311.
1991.PubMed/NCBI
|