Introduction
Apolipoprotein L (APOL) belongs to the high-density
lipoprotein (HDL) family and is present in a number of species,
including all mammals (1);
however, its role remains poorly understood. The human APOL family
consists of six genes clustered on chromosome 22 (2). Human APOL1 was first identified and
characterized as one of the proteins associated with a subset of
HDL particles (3). Its lipid
binding capability was later demonstrated in vitro and is
therefore considered to be involved in lipid transport and
metabolism (4). Its toxicity
against Trypanosoma brucei brucei was later demonstrated to
involve a protein domain that can form anionic pores in lipid
membranes (5). More recent studies
have suggested novel actions for APOL1 in programmed cell death
(PCD), more specifically autophagy. These actions have been shown
to be associated with the BH3 domain, a Bcl2 homology domain; BH3
domain is ubiquitously expressed in regulators of programmed cell
death in the Bcl2 and BH3-only families (6,7).
The network of these regulators is complex,
containing eight BH3-only and at least 20 Bcl2 family members in
humans, and is known to be involved in numerous types of cell
death. As this machinery remains much simpler in yeast, involving
only one Bcl2 family member, yBH3 (8), the present study used the yeast
Saccharomyces cerevisiae as a model organism in order to
investigate APOL1 function. In the present study, a yeast model for
APOL1 expression was developed and used to investigate APOL1
function. The present study revealed its mitochondrial localization
and interference with mitochondrial integrity, which had
deleterious effects on yeast proliferation; the effect was observed
when the cells were obliged to undergo respiration in a medium
containing glycerol.
Materials and methods
Yeast strain and growth
conditions
The S. cerevisiae strains used in the present
study were isogenic to the wild type Σ1278b strain: 23344c
(ura3, lab collection: Laboratory of Membrane Transport
Biology, IBMM, Université Libre de Bruxelles) and 27061b
(ura3 and trp1, lab collection: Laboratory of
Membrane Transport Biology, IBMM, Université Libre de Bruxelles)
(9,10). Cells were grown in a minimum
buffered (pH 6.1) medium prepared as described previously (11) with 3% galactose (MGal), glucose
(MGlu) or glycerol (MGly) as the carbon source or in non-inducible
medium raffinose (MRaf) and glutamate as a nitrogen source,
supplemented with vitamins and minerals (11). Yeasts were grown at 29°C.
Plasmids and mutagenesis
For APOL1 expression, the human APOL1 was cloned
into the centromeric p416Gal.1 (HA) (12) yeast expression plasmid under the
control of a GAL1 promoter by homologous recombination. Yeast cells
transformed with empty p416Gal.1 (HA) vector were used as control.
The primers used are listed in Table
I. The APOL with the mutated BH3 domain was amplified using PCR
with Phusion™ High-Fidelity DNA Polymerase (Thermo
Fisher Scientific, Inc.) and the APOL1-∆BH3F/R oligonucleotides
(Table I) and introduced in the
APOL1-pGal.1 vector. The reaction mixtures were processed with an
initial denaturation period at 98°C for 30 sec, followed by a
three-step PCR program for 30 cycles that consisted of 98°C for 10
sec, 70°C for 20 sec and 72°C for 25 sec, prior to a final
extension step at 72°C for 10 min.
 | Table I.Primers used for construction of
plasmids. |
Table I.
Primers used for construction of
plasmids.
| Oligonucleotides or
primers | Sequence,
5′-3′ | Purpose |
|---|
| APOL1-F |
GTTAATATACCTCTATACTTTAACGTCAAGGAGAAAAAACTATAGGTACCTAGATGGAGGGAGCTGCTTTGC3 | Plasmid
construction |
| APOL1-R |
CAGCACCGGCTGCTCCTGCTCCTGCTCCTGCTCCTGCTCCCTCGAGCAGTTCTTGGTCCGCC | Plasmid
construction |
| APOL1-∆BH3-F |
TTGTGGACCTTCCTTCTTATGTTATCCTCAAGC | Plasmid
construction |
| APOL1-∆BH3-R |
ATAAGAAGGCTCAAGGTCCACAAAGGCACCAC | Plasmid
construction |
The mutant APOL1-∆BH3 (BH3 domain deletion) was
sequenced by Beckman Coulter (Illumina HiSeq) to verify the
introduction of the desired substitution. pYX232-mtGFP (TRP1),
encoding green fluorescent protein (GFP) fused to the mitochondrial
pre-sequence subunit 9 of the F0-ATPase (mt-GFP) under
the control of the constitutive triosephosphate isomerase promoter
was used to monitor mitochondrial structure and morphology.
pYX232-mtGFP was kindly provided by Professor Benedikt Westermann
(Universität Bayreuth, Germany) (13).
Western blot analysis
Total protein extracts were performed as previously
described (14). Aliquots of 1 ml
at an optical density (OD) of 0.2 (107 cells/ml; λ=660
nm) from cells at the exponential growth phase were harvested by
centrifugation at room temperature for 5 min at 3,500 × g. The cell
pellet was suspended in 500 µl water and the cells were lysed with
50 µl of 2 M NaOH for 10 min. The proteins were precipitated with
50 µl of 50% trichloroacetic acid and collected by centrifugation
at 4°C for 5 min at 12,000 × g. The pellet was suspended with gel
loading buffer containing 4% (w/v) SDS, 100 mM Tris pH 6.8,
β-mercaptoethanol 2% (v/v), 20% (v/v) glycerol and blue
bromophenol. Samples were heat-treated at 37°C for 10 min. For the
western blotting analysis, equal protein amounts (~20 µg) were
loaded onto a 8% SDS-PAGE. After transfer to nitrocellulose
membranes, HA-tagged APOL1 was probed with anti-HA (1:10,000, cat.
no. 26183, Molecular Probes; Thermo Fisher Scientific, Inc.), while
Sc-Dpm1, Sc-Pma and Sc-porin were probed with anti-Dpm1 (1:5,000,
cat. no. A-6429), anti-Pma1 (1:10,000, cat. no. MA1-91567) and
anti-porin (1:5,000, cat. no. 459500; all from Invitrogen; Thermo
Fisher Scientific, Inc.). Primary antibodies were detected with
horseradish peroxidase-conjugated secondary antibodies
anti-rabbit-IgG (1:1,000, cat. no. NA934; Cytiva) and
anti-mouse-IgG (1:5,000, cat. no. NA931; Cytiva) for 1 h at room
temperature) followed by measurement of chemo-luminescence
(Lumi-LightPLUS, Roche Diagnostics). The western blot
experiments in the present study analyzed membrane proteins;
accordingly, Pma1 was used as a suitable control (15).
Growth curve and clonogenic assay
For growth in liquid medium, yeast were grown
overnight at 29°C in MGlu or MGal medium and diluted to an OD
(λ=660 nm) of 0.2 in MGlu and MGal, respectively. OD was measured
repeatedly over 9 days. In order to test the proliferation of cells
grown on agar plates, yeast previously grown in liquid MGlu or MGal
were spotted at a final concentration of 103 cells on
MGlu or MGal solid media. In parallel, aliquots were collected for
immunostaining for the detection of APOL1/APOL1-∆BH3 expression in
inducible conditions.
Reactive oxygen species (ROS)
assessment
Cells were cultured in MGal overnight at 29°C and
then diluted in fresh media to an OD (λ=660 nm) of 0.2. Cell
aliquots were collected, washed twice in PBS and re-suspended at
1×107 cells/ml in 1 ml of 2.5 µg/ml dihydroethidium
(DHE) (37291, Sigma-Aldrich) in PBS and incubated for 15 min in the
dark at room temperature. Then, cells were washed with 1 ml PBS and
analyzed via flow cytometry. Flow cytometric analysis was performed
on a Canto II (BD Biosciences) and results were analyzed with the
FlowJo program (V10.5.0, FlowJo LLC).
Tetramethylrhodamine, ethyl ester
(TMRE) staining
Yeast strain cell cultures were collected at OD=0.2
(λ=660 nm) of the exponential phase in MGal medium. Cells were
incubated with 50 pM TMRE (TMRE-Mitochondrial Membrane Assay Kit,
ab113852, Abcam) for 10 min at 4°C, washed twice with PBS 1X, then
cells were harvested, centrifuged (1,600 × g for 4 min at 4°C) and
suspended (~1×106 cells ml−1) in PBS, in
order to be analyzed by flow cytometry, according to the
manufacturer's protocol, on a Canto II (BD Biosciences).
Carbonilcyanide p-triflouromethoxyphenylhydrazone (FCCP,
ab120081, Abcam) treatment, an uncoupling agent, was used as a
positive control, as it is able to completely depolarize the
mitochondrial outer membrane. It was added to cell cultures at a
final concentration of 20 µM 20 min prior to the incubation with
TMRE.
Fluorescence microscopy for yeast
organelles
For vacuole staining, cells were collected at OD=0.2
(λ=660 nm) of the exponential phase in MGAl growth medium and
resuspended in 1 ml of pre-heated medium and 80 nM FM4-64 (T3166,
Invitrogen, Thermo Fisher Scientific, Inc.). Cells were incubated
at 29°C for 15 min, centrifuged (5,000 × g at room temperature for
5 min), collected and resuspended in 5 ml of medium and incubated
at 29°C for 120 min. For fixed (1.1 ml of 37% formaldehyde
incubated at 29°C for 30 min) or live cells, mt-GFP was monitored
at a wavelength of 510 nm (magnification, ×63 and ×100).
Immunofluorescence assay
Yeast strains were cultured at 29°C overnight in
MGal media until they reached the log growth phase. At OD=0.2
(λ=660 nm), cell aliquots of 9 ml were fixed with 1.1 ml of
formaldehyde 37% and incubated at 29°C for 30 min. Cells were
collected, washed and resuspended in Buffer B [Sorbitol (1 M),
K2HPO4, 3H2O (1 M) and
KH2PO4 (1 M) at pH 7.5]. A total of 5 µl of
lyticase (10,000 u/ml) and 2 µl of β-mercaptoethanol were added.
After 30 min incubation at 29°C, cells were collected, washed and
suspended in 1 ml of PBS 1X. A total of 20 µl of yeast culture was
seeded on 10-well slides (MP Biomedicals, LLC.). Cells were stained
for APOL1/APOL1-∆BH3 expression levels with mouse anti-HA (1:100,
cat. no. 26183, Molecular Probes; Thermo Fisher Scientific, Inc.)
overnight at room temperature in a humidified chamber. A series of
PBS washes preceded the secondary anti-mouse antibody (1:1,500,
A-11001; Invitrogen; Thermo Fisher Scientific, Inc.) incubation for
1 h at room temperature in a humid chamber. DAPI (cat. no. D9542;
Sigma Aldrich) was used to stain the nuclei of yeast cells. The
revelation of APOL1/APOL1-∆BH3 expression was assessed using a
Zeiss inverted fluorescence microscope (magnification, ×63 and
×100).
Mitochondrial isolation
The procedure was performed as previously described
(16). Yeast strains were grown at
29°C for 16 h, collected by centrifugation (3,000 × g for 5 min at
room temperature) and then washed twice with distilled water. Cells
were suspended in dithiothreitol (DTT) buffer (100 mM Tris, pH 9.4,
10 mM DTT) and incubated at 29°C for 20 min with gentle shaking.
After removing the supernatant by centrifugation at 3,000 × g for 5
min at room temperature, cells were resuspended in Zymolyase buffer
(1.2 M sorbitol buffer containing 1 mg zymolyase 20T per gram of
cells). Following incubation at 29°C for 30 min with gentle
shaking, cells were centrifuged at 2,200 × g for 8 min and
resuspended in homogenization buffer [0.6 M sorbitol, 1 mM EDTA, 10
mM Tris, 0.2% (w/v) BSA pH 6.0]. These cells were centrifuged at
2,200 × g for 8 min at 4°C and then homogenized 15 times with a
tight pestle in a glass homogenizer. The unbroken cells were then
pelleted at 1,500 × g for 5 min at 4°C. The resulting supernatant
was collected and centrifuged first at 3,000 × g for 5 min at 4°C
and again at 12,000 × g for 15 min at 4°C. After repeating
centrifugation (3,000 × g for 5 min and 12,000 × g for 15 min at
4°C) and resuspension of this pellet to yield pure mitochondria,
the mitochondrial pellet was finally resuspended in SEM buffer (10
mM MOPS, 250 mM sucrose, 1 mM EDTA pH 7.4). Highly pure enriched
mitochondria fraction was subjected to a sucrose gradient
purification. After centrifugation of 134,000 × g for 1 h at 4°C,
an oxidized band was extracted and pelleted again at 10,000 × g for
30 min at 4°C. Western blot analyses were performed to examine
preparation of the mitochondrial fraction as detailed above.
Index of respiratory competence assay
(IRC)
Exponentially MGal-growing cells at a final density
of 103 cells were spread onto MGly agar plates or MGal
in parallel, incubated at 29°C for three days. At the end of the
incubation, cells were observed and counted manually, and. the IRC
was calculated as colony number observed on MGly plates divided by
the number of colonies on MGal plates.
Statistical analysis
PRISM (version 6.0 GraphPad Prism software, Inc.)
was used to perform statistical data analysis and plot the graphs.
Data are presented as the mean ± standard error of the mean (SEM).
A minimum of three experiments were performed for statistical
analysis. One-way ANOVA followed by Tukey's post hoc test and
Student's t-test were used to determine statistically significant
differences between the different experimental conditions.
P<0.001 was considered to indicate a statistically significant
difference.
Results
APOL1 does not inhibit yeast
proliferation in fermentable media
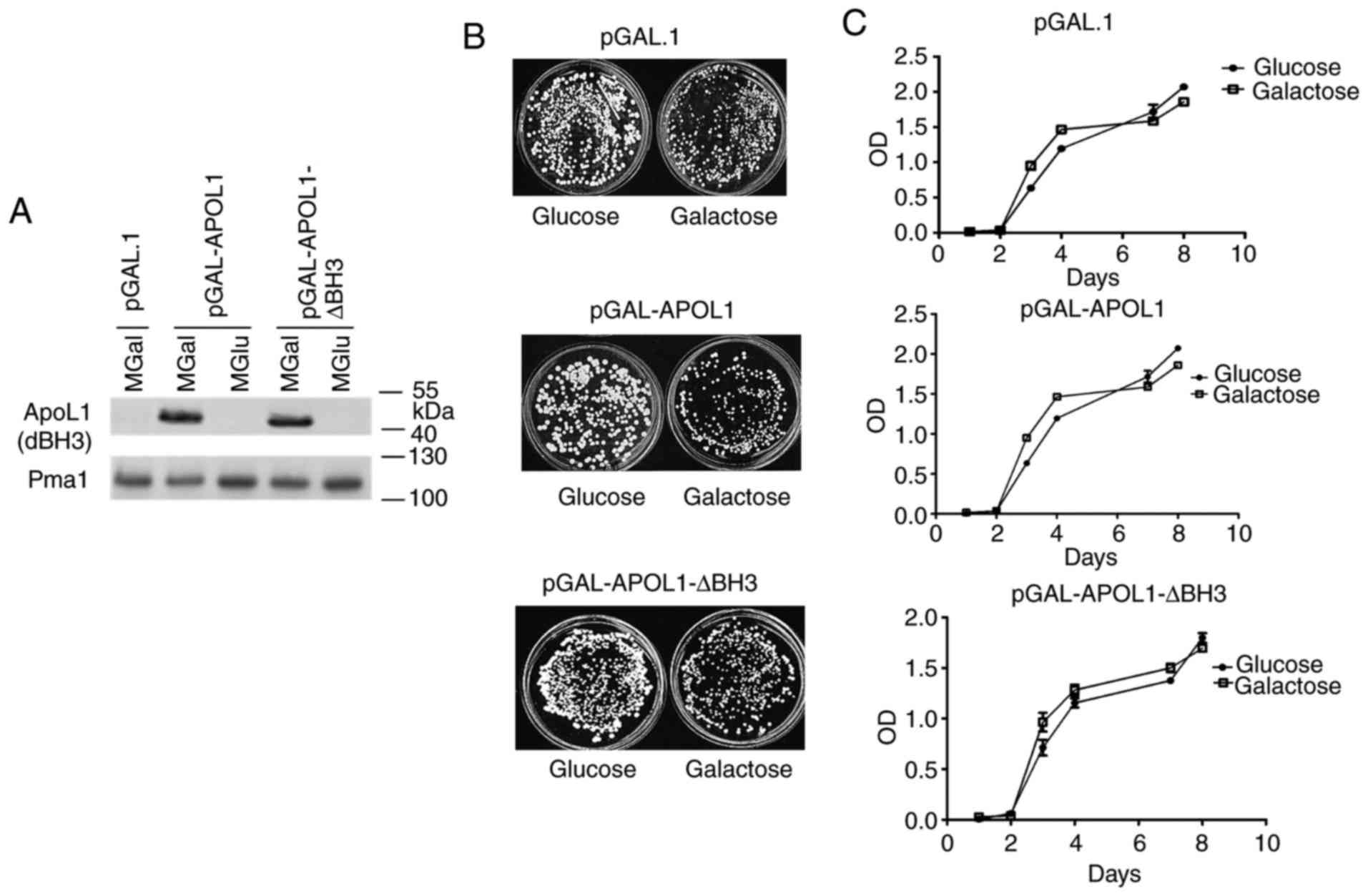
In order to investigate the action of human APOL1 in
yeast, a sequence covering the whole APOL1 open reading frame was
inserted in the yeast vector (pGAL1) allowing expression induction
by galactose and repression by glucose. As the BH3 domain is one of
the primary features of APOL1 and has been associated with the
pro-apoptotic function of Bcl2 family proteins, the activity of the
BH3-deleted version of APOL1 (∆BH3) was tested. Cells were cultured
in a medium containing glucose (MGlu-repressive conditions,
negative control) or galactose (MGal-induced conditions) as carbon
sources. In order to test cell proliferation, cells expressing
APOL1 variants and control cells were plated on MGlu and MGal. No
significant differences were observed in the number of colonies
between cells expressing or not expressing APOL1 (Fig. 1B). As expected, no significant
difference was observed between the different cells on MGlu, the
repressive medium. Similarly, Fig.
1C shows no significant differences in the proliferation of
cells expressing or not expressing an APOL1 variant on liquid MGal
or MGlu media although its expression was properly induced in the
MGal medium (Fig. 1A). Thus, APOL1
expression levels do not influence cell proliferation in these
conditions. In the presence of glucose or galactose, yeast cells
primarily undergo anaerobic fermentation, generating ROS. In order
to monitor ROS generation in induced and non-induced conditions,
cells were stained with DHE detecting superoxide molecules and
analyzed by flow cytometry in the present study. As presented in
Fig. 1D and quantified in Fig. 1E, this analysis did not detect any
significant difference in the fraction of the cell population
producing ROS between the strains expressing or not APOL1.
APOL1 inhibits yeast proliferation in
non-fermentable media
In the presence of a fermentable carbon source such
as glucose or galactose, yeast cells preferentially generate energy
by undergoing fermentation, and can grow normally with a minimal
level of mitochondrial respiration, generating ethanol as the end
product of fermentation. When fermentable carbon sources become
limiting, genes required for respiration are induced, and ATP is
generated by metabolizing non-fermentable carbon sources such as
glycerol, ethanol or lactate in the mitochondria (17).
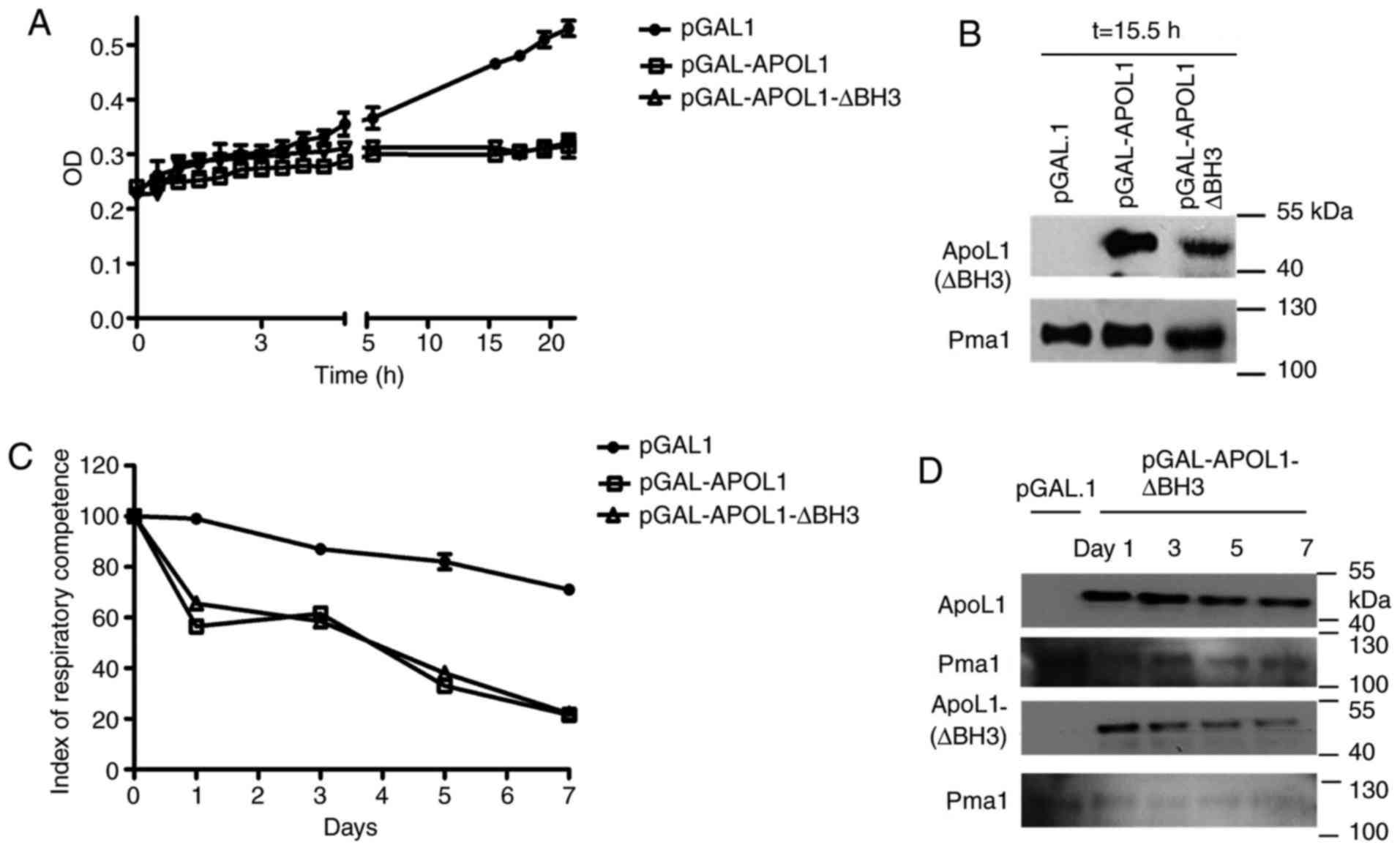
In order to determine whether human APOL1 could
interfere with mitochondrial function, cells initially grown on
MGal to induce APOL1 expression were transferred to an MGly medium,
containing glycerol, a non-fermentable carbon source. As presented
in Fig. 2A, yeast proliferation
was inhibited in MGly media when APOL1 was induced (Fig. 2B) compared with the control
culture. This indicated that APOL1 was able to inhibit yeast
proliferation in conditions in which aerobic respiration is
required, indicating that this protein could interfere with
mitochondrial function.
As mitochondria are primarily involved in
respiration, the present study assessed the IRC in cells expressing
or not APOL1 variants. This index measures the percentage of viable
respiration-competent cells and reflects the fraction of cells that
can grow on non-fermentable (glycerol) carbon sources compared with
the fraction of cells growing on fermentable (galactose) carbon
sources (18). At the same time,
the expression levels of the induced gene were verified (Fig. 2D). As presented in Fig. 2C, while the IRC value was 100% at
the beginning of the experiment for all three strains, the IRC
value started decreasing at day 1 only in the cells expressing
APOL1 and APOL1 ∆BH3. This decrease continued, reaching almost 20%
after 7 days, while IRC remained higher than 70% for the control
strain. The absence of the BH3 domain in APOL1 ∆BH3 expressing
cells had no effect on the number of viable respiration-competent
cells.
APOL1 depolarizes the mitochondrial
membrane
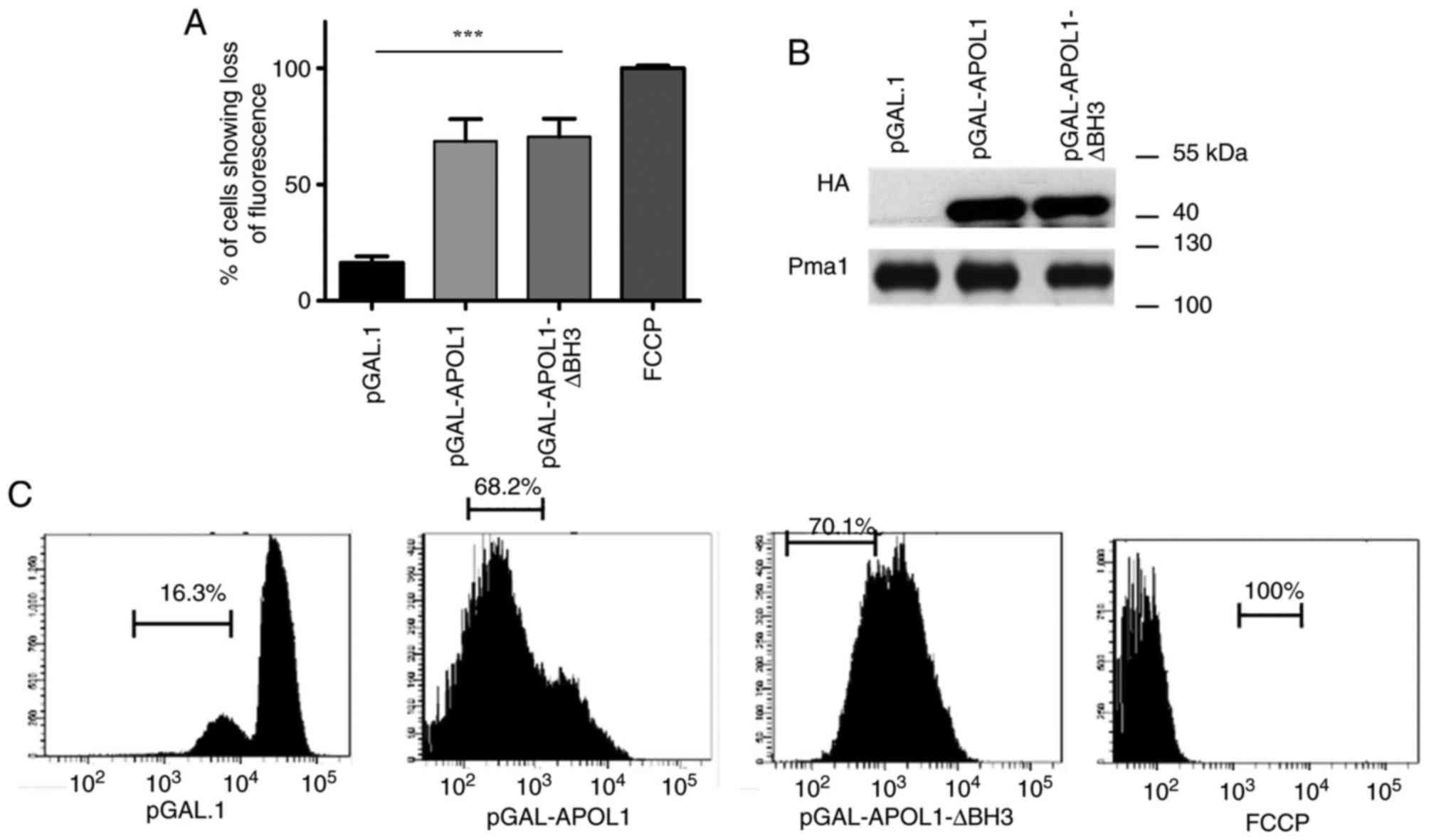
As a result of the cell proliferation inhibition
observed on MGly media and the drop in respiratory index, the
present study investigated whether the mitochondrial membrane
potential (∆ψm) of yeast is affected by APOL1 induction.
Maintenance of the membrane potential is key for mitochondrial
functions (19). Thus, cells
expressing or not expressing native APOL1 or APOL1-∆BH3 on a
fermentable carbon source (MGal), were incubated with TMRE, a
chemical dye able to accumulate in the mitochondria depending on
membrane polarization (20). The
positive control, which included treatment with FCCP (an ionophore
that uncouples the electron transport chain from ATP production),
completely prevented TMRE staining. As presented in Fig. 3, cells expressing APOL1, whether
native or ∆BH3, showed a loss of membrane potential. TMRE
uptake into the mitochondria was decreased by 68 and 70% in cells
expressing native APOL1and ∆BH3 APOL1 respectively
(P<0.0001). This suggests that APOL1 expression levels promote
loss of mitochondrial membrane potential and that this ability does
not require the BH3 domain.
APOL1 is found in the mitochondrial
fraction
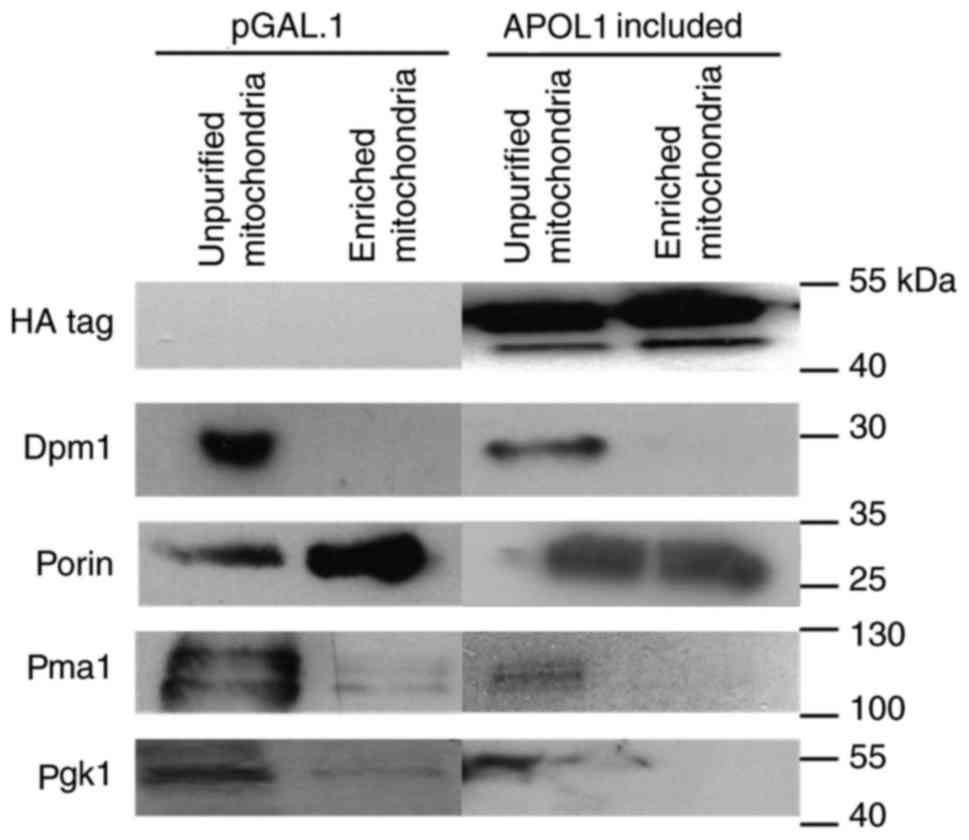
As APOL1 induction provoked mitochondrial membrane
depolarization, the present study investigated a potential direct
APOL1 interaction within the yeast mitochondria. In order to assess
the subcellular localization of APOL1 in yeast, APOL1-expressing
cells were grown on galactose medium for 24 h and mitochondria
enrichment was performed using these cells. A fraction of HA-APOL1
was consistently found in the mitochondrial-enriched fraction
(enriched mitochondria) as validated by the detection of the outer
membrane mitochondrial channel-forming protein porin in this
fraction (Fig. 4).
APOL1 induces mitochondrial
morphological alterations in yeast cells
As APOL1 localizes in the mitochondria and
interferes with proper function, the present study investigated
whether their morphology was also affected. Morphology was
visualized using mt-GFP, a mitochondria-targeted GFP protein. Cells
co-expressing native APOL1 or APOL1-∆BH3 with mt-GFP were grown in
MGal. In control cells not expressing APOL1, a normal branched
tubular mitochondrial network located below the cell cortex
(21) was observed. In contrast,
in yeast cells expressing APOL1, the mitochondrial network was
altered; the branched network disappeared and fluorescent patches
appeared predominantly at the periphery of some of the cells; the
mitochondrial signal in APOL1-expressing cells concentrated in
fewer mitochondria that were larger compared with those in control
cells and mt-GFP was not detectable in certain APOL1-expressing
cells (Fig. 5A).
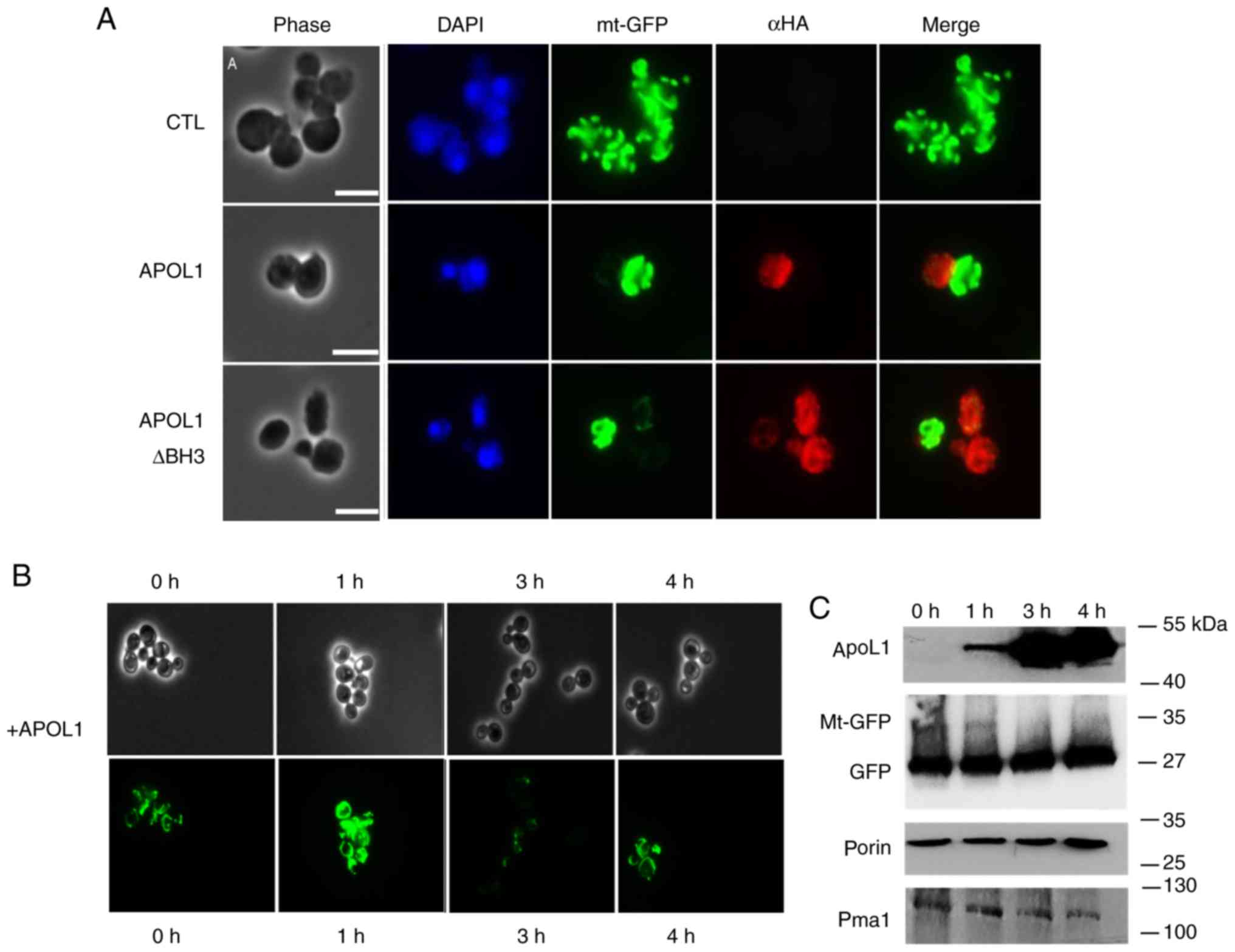 | Figure 5.APOL1 expression levels and
mitochondrial alterations. (A) APOL1 expression levels modified
mitochondrial morphology as visualized by a mitochondrial marker.
Cells were transformed by the plasmid pYX232-mtGFP, which expresses
GFP fused to a mitochondrial matrix targeting sequence (mtGFP)
alone (CTL) or together with plasmids expressing wild type APOL1 or
BH3-deleted APOL1. Aliquots from cultures in MGal growth medium
were then taken at OD 0.2 and treated with an anti-HA antibody
(red), and stained with DAPI (blue). Magnification, ×100. (B) Cells
transformed by pYX232-mtGFP (with a constitutive promoter) and
APOL1 expression vector were induced by galactose added to MRaf
growth medium for the indicated times [time-dependent kinetics (0,
1, 3 and 4 h)]. (C) Immunoblot analysis of cell extracts from (B)
demonstrated the expression levels of APOL1 (anti-HA), porin (Sc
anti-porin) and GFP (anti-GFP). Scale bar, 5 µm. Magnification,
×63. APOL1, apolipoprotein L 1; GFP, green fluorescent protein;
MGal, galactose-containing medium; OD, optical density; MRaf,
raffinose-containing medium. |
In order to test if this morphological change was
accompanied by a change in mitochondrial content, cells were grown
on non-inducible and non-repressive medium (raffinose) and APOL1
was induced in a time-dependent manner by the addition of galactose
to the growth medium. Using western blotting, the present study
observed the stability of two mitochondrial components after
inducing APOL1 for 4 h in the cells. Fig. 5C shows that HA-APOL1 (used as a
control) was immunodetectable during the 4 h of induction. Porin, a
mitochondrial outer membrane protein, remained stable during the 4
h induction. A decrease in the mitochondrial mt-GFP signal (35 KDa)
was observed.
In order to further analyze these different
structural patterns, the present study used a fluorescence
microscopy assay during APOL1 induction kinetics. Cells were grown
in MRaf and galactose was added at time (T)=0 to induce HA-APOL1
expression. At different times after the induction, cell aliquots
were collected for mitochondrial visualization using mt-GFP as a
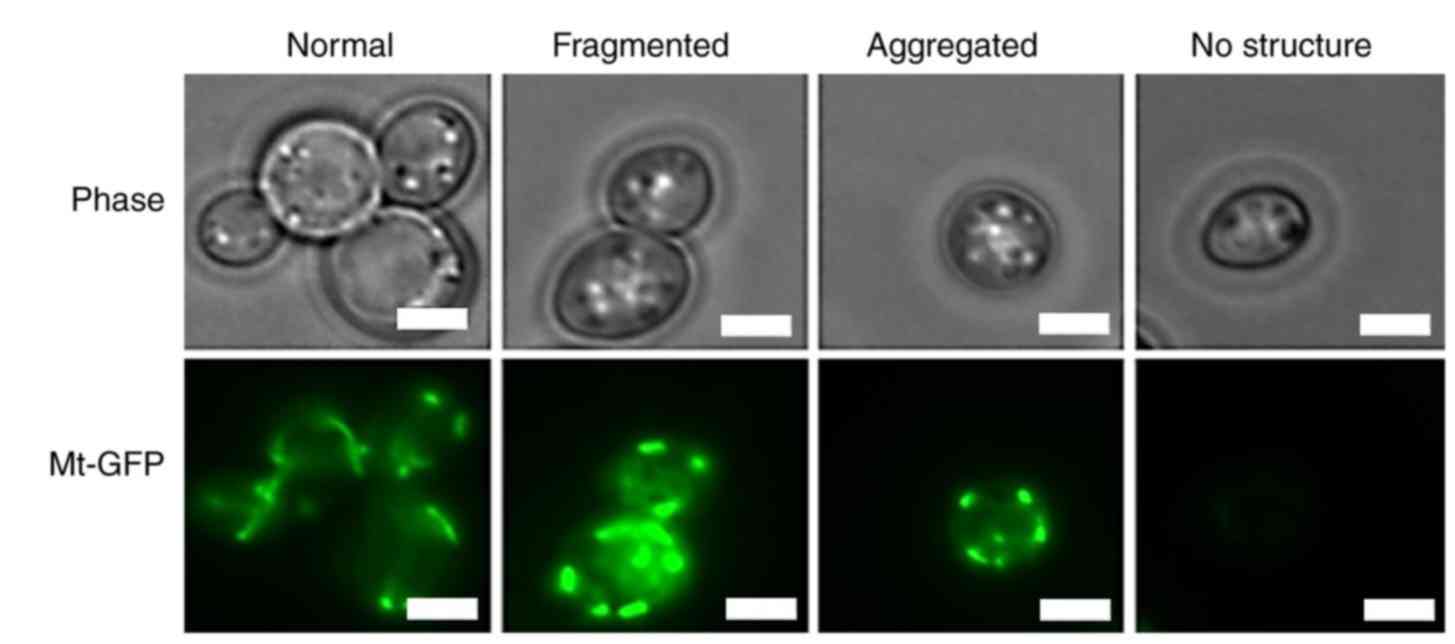
marker and for immunoblot analysis. The present study identified
different categories of the mitochondrial morphological structures
that are consistent with what has been previously described
(22). At T=0, 92% of the cells
contained the typical tubular shaped mitochondria (Fig. 6 and Table II). Mitochondria remained tubular
in 90% of the cells at (T)=0. Following 1 h of APOL1 induction,
67.2% still contained a normal mitochondrial structure while 5% had
fragmented but widespread mitochondria, 12.9% showed mitochondria
aggregated to one side of the cell, and 14.6% of cells lost the
mitochondrial signal. In the control cells, mitochondria remained
tubular throughout the experiment, while only 26.0% of
APOL1-expressing cells displayed normal tubular mitochondria
following 4 h of induction. At T=4, a notable proportion of the
cell population lacked detectable mitochondrial structure (42.5%).
This suggested that prolonged APOL1 expression levels may
progressively modify mitochondrial structure.
 | Table II.Cells were counted on the basis of
different mitochondrial morphology: Mean count (%) of yeast cells
presenting the mitochondrial morphologies illustrated in Figure 6 at the indicated times. At least
100 cells were examined at each time point. |
Table II.
Cells were counted on the basis of
different mitochondrial morphology: Mean count (%) of yeast cells
presenting the mitochondrial morphologies illustrated in Figure 6 at the indicated times. At least
100 cells were examined at each time point.
| +APOL1 | Time | Normal % | Fragmented % | Aggregated % | No structure % |
|---|
|
| T=0 h | 92 | – | – | 8 |
|
| T=1 h |
67.2 |
5.1 | 12.9 |
14.6 |
|
| T=3 h |
42.9 | 6 | 10.1 |
40.9 |
|
| T=4 h | 26 |
12.2 | 19.1 |
42.5 |
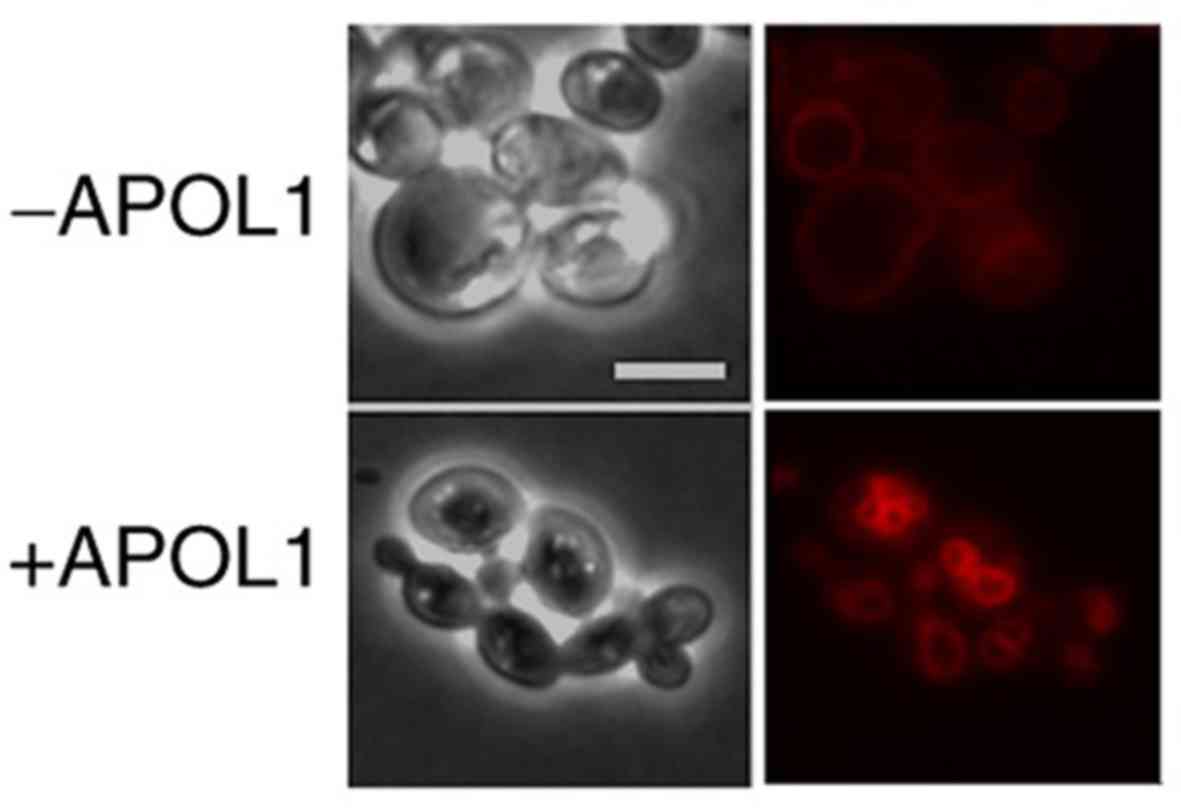
As APOL1 uptake by African trypanosomes promotes
lysosome swelling (23), the
present study also observed the behavior of lysosome-like
organelles, namely the vacuole, following APOL1 induction in yeast
cells. Yeast cells were grown on MGal medium and stained with the
fluorescent dye FM4-64, which accumulated at the vacuolar membrane.
As presented in Fig. 7,
APOL1-expressing cells exhibited alterations in vacuolar
morphology, showing a vacuole fragmented into smaller vesicles
compared with the control cells, which possessed a single, regular
and large vacuole. These smaller vesicles restained as intensely as
the single wild type vacuole.
Discussion
Previous studies have suggested an association
between certain APOL family members and PCD of neutrophils in
chronically ill patients and murine dendritic cells (24,25).
As PCD is much simpler in yeast than in mammals, the present study
used yeast to investigate the molecular action of APOL1. The APOL
family is conserved in evolution as members are found in fish, but
they are also divergent between mammals (4). For instance, APOL1 is specific to
humans (26). The present study
demonstrated that ectopic expression of human APOL1 in S.
cerevisiae is associated with disruption of mitochondrial
function, dissipation of the mitochondrial membrane potential and
alteration of mitochondrial and vacuolar morphology. The present
study further showed that APOL1 alters yeast respiratory function.
APOL1 induction did not have any effect on cell proliferation in
the presence of fermentable carbon source although it affected
yeast proliferation in non-fermentable media, which was associated
with mitochondrial membrane depolarization. As this was also
associated with a drop in respiratory index, this proliferation
defect may be ascribed to a loss of mitochondrial functionality
that is only detrimental if fermentation is inhibited. Likewise,
mutations that affect the mitochondria are known to affect yeast
respiration capacity in MGly media (27). The association between loss of
membrane potential and APOL1 induction may be the result of direct
APOL1 action on the mitochondrial membrane, similar to the
mitochondrial pore forming capability of the Bax and Bak members of
the Bcl2 family. This pore-forming capacity is associated with
multimerization mediated partly by the BH3 domain, a peptide
sequence conserved in APOL1 (28).
The observation that an APOL1 deleted on the BH3 domain was still
able to interfere with mitochondrial function is in disagreement
with this finding. On the contrary, the present study demonstrated
that, in yeast, APOL1 localized at the mitochondria. This
observation is in line with recent findings regarding the
localization of APOL1 to structures surrounding the mitochondria in
human podocyte cell lines where it also triggers a decrease in
mitochondrial respiration (29).
This suggests that the molecular associations required for this
specific mitochondrial localization are conserved between mammalian
and yeast mitochondria. APOL1 intracellular function may regulate
the outer mitochondrial membrane potential in certain
circumstances. This is suggested by the fact that APOL1 can induce
lysosomal and mitochondrial membrane permeabilization in the
distant Trypanosoma brucei eukaryote (22). Ongoing studies are attempting to
elucidate the molecular requirements using a battery of mutants
affecting different components of mitochondrial metabolism and cell
death. The effects of APOL1 also seem to extend to the vacuole. It
is not clear how APOL1 exerts this effect but it is known that Bax
expressed in yeast cells causes vacuoles to fragment. Vacuole
fragmentation is also induced by certain mutants, such as Vph1p
(partial deletions of the V0 subunit of V-ATPase); Vph1p affects
vacuole acidification (30). This
APOL1 effect in yeast is similar to that previously found in
trypanosomes; APOL1 lyses these protozoans, which involves
localization in organelles, lysosome swelling and mitochondrial
depolarization (31). This effect
is also consistent with previous discoveries regarding the
consequences of APOL1 high-risk variants (G1 and G2) on podocytes
in people with chronic kidney diseases. It has been shown that
APOL1 G1 and G2 variants were implicated in promoting cell damage
by mediating endoplasmic reticulum stress, compromising lysosomal
membrane permeability and disrupting trafficking processes
including vacuole acidification (32,33).
The yeast model provides insights into the mode of action of
mammalian APOL1; it suggests that APOL1 interacts with the
mitochondrial and vacuolar membranes, causing alteration of
mitochondrial and vacuolar morphology, disruption of their
functions (primarily by depolarization of the mitochondrial
membrane) and the disruption of trafficking processes that may
involve changes in vacuolar pH. The present study further showed
that APOL1 alters yeast respiratory function, which is followed by
a proliferation arrest when yeast cells are forced to use
mitochondrial respiration as a unique source of energy. A combined
genetics and biochemistry approach may identify the mechanisms that
underlie the function of APOL1 and its role in mammalian cells. For
example, APOL1 may be involved when mammalian cells shift from
mitochondrial metabolism to glycolysis for energy production, a
phenomenon encountered in numerous types of cancers (the Warburg
effect), or during an immune reaction when T lymphocytes are
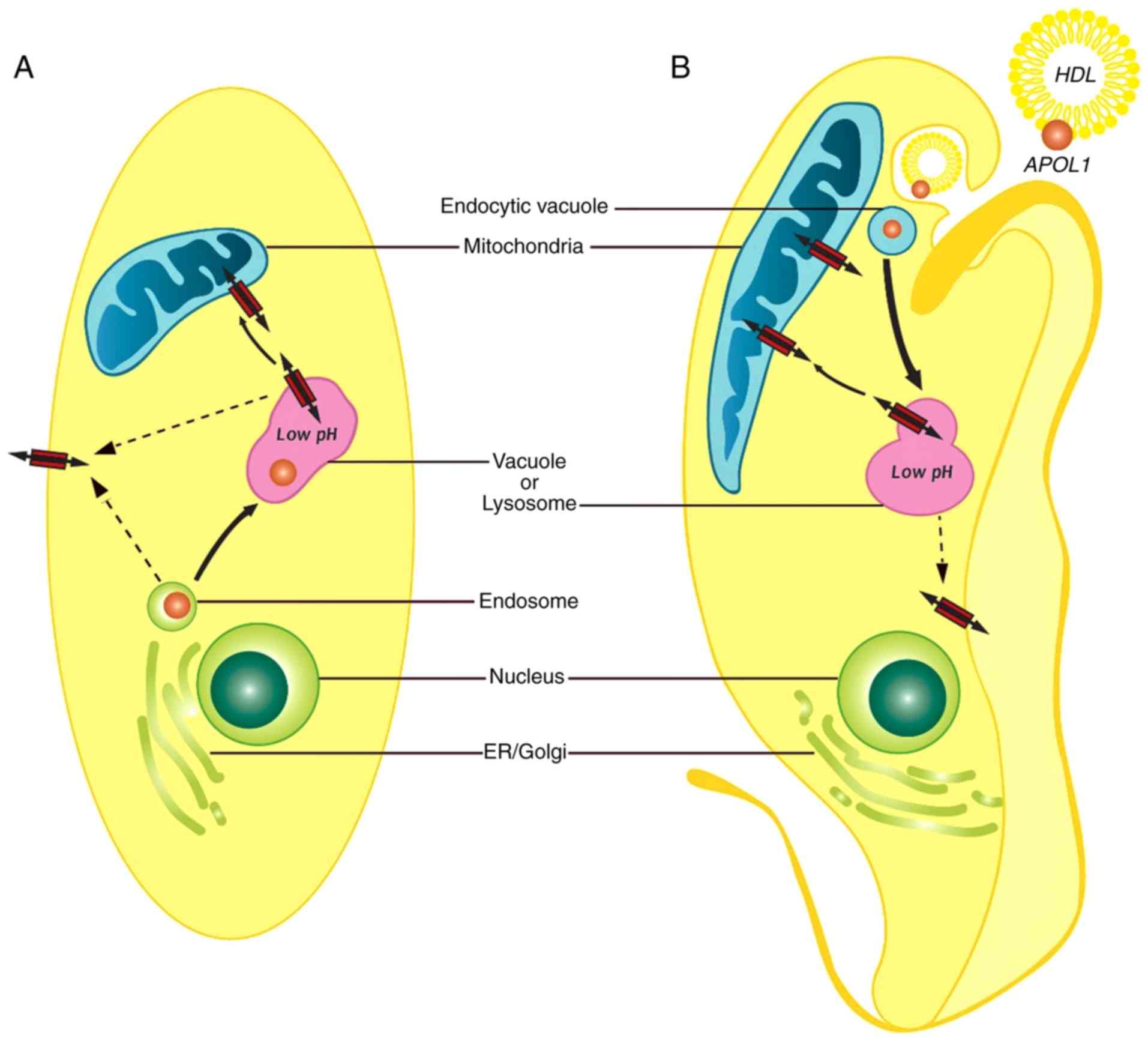
actively proliferating. A model of how APOL1 induces cell death is
outlined in Fig. 8. In yeast and
human kidney cells, APOL1 is produced intracellularly by normal
biosynthetic pathways. APOL1 may cause changes in mitochondrial
membrane potential and vacuolar acidification, ultimately resulting
in cell hypertrophy and death (34,35).
In trypanosomes, the process of trypanolysis starts with the uptake
of HDL particles containing APOL1 by the parasite. As acidity
increase within the endocytic compartments within the cell, APOL1
localizes to the lysosomal membrane causing lysosomal membrane
permeabilization and osmotic swelling. In addition, acidified APOL1
can be transported to the mitochondrial membrane causing
depolarization of the mitochondria. The two processes result in
cell lysis and death (31).
Although not examined in the present study, APOL1 may be inserted
into plasma membranes of both trypanosomes and human kidney cells
(36).
Acknowledgements
Not applicable.
Funding
The present study has been funded by research
programs of the FNRS-FRS (Belgian National Fund for Scientific
Research), Azm and Saadé Association (grant no. 110617) and AUF
(Agence Universitaire de la Francophonie; grant no. 140909).
Additional funding was provided by the Bureau of International
Relations (grant no. bric98156) of the Université Libre de
Bruxelles (ULB).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
MC, JD, MB and PP were involved in performing the
experiments and analyzing the results. MC drafted the original
manuscript. JD and LV wrote the manuscript. JD, MB, BB, AMM, LV and
RK reviewed the manuscript. JD, LV and RK edited the manuscript.
AMM and LV developed the methodology. LV and BB conceptualized the
study. RK interpreted the data. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
LV is Director of Research at the FNRS. AMM is
Senior Research Associate at the FNRS and WELBIO investigator. MB
is a scientific research worker supported by WELBIO.
References
|
1
|
Page NM, Butlin DJ, Lomthaisong K and
Lowry PJ: The human apolipoprotein L gene cluster: Identification,
classification, and sites of distribution. Genomics. 74:71–78.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Monajemi H, Fontijn RD, Pannekoek H and
Horrevoets AJ: The apolipoprotein L gene cluster has emerged
recently in evolution and is expressed in human vascular tissue.
Genomics. 79:539–546. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Duchateau PN, Pullinger CR, Orellana RE,
Kunitake ST, Naya-Vigne J, O'Connor PM, Malloy MJ and Kane JP:
Apolipoprotein L, a new human high density lipoprotein
apolipoprotein expressed by the pancreas. Identification, cloning,
characterization, and plasma distribution of apolipoprotein L. J
Biol Chem. 272:25576–25582. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Vanhollebeke B and Pays E: The function of
apolipoproteins L. Cell Mol Life Sci. 63:1937–1944. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Thomson R and Finkelstein A: Human
trypanolytic factor APOL1 forms pH-gated cation-selective channels
in planar lipid bilayers: Relevance to trypanosome lysis. Proc Natl
Acad Sci USA. 112:2894–2899. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Youle RJ and Strasser A: The BCL-2 protein
family: Opposing activities that mediate cell death. Nat Rev Mol
Cell Biol. 9:47–59. 2008. View
Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhaorigetu S, Wan G, Kaini R, Jiang Z and
Hu CA: ApoL1, a BH3-only lipid-binding protein, induces autophagic
cell death. Autophagy. 4:1079–1082. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Büttner S, Ruli D, Vögtle FN, Galluzzi L,
Moitzi B, Eisenberg T, Kepp O, Habernig L, Carmona-Gutierrez D,
Rockenfeller P, et al: A yeast BH3-only protein mediates the
mitochondrial pathway of apoptosis. EMBO J. 30:2779–2792. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Bechet J, Greenson M and Wiame JM:
Mutations affecting the repressibility of arginine biosynthetic
enzymes in Saccharomyces cerevisiae. Eur J Biochem.
12:31–39. 1970. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Springael JY, De Craene JO and André B:
The yeast Npi1/Rsp5 ubiquitin ligase lacking its N-terminal C2
domain is competent for ubiquitination but not for subsequent
endocytosis of the Gap1 permease. Biochem Biophys Res Commun.
257:561–566. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Jacobs P, Jauniaux JC and Grenson M: A
cis-dominant regulatory mutation linked to the argB-argC gene
cluster in Saccharomyces cerevisiae. J Mol Biol.
139:691–704. 1980. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Mumberg D, Müller R and Funk M:
Regulatable promoters of Saccharomyces cerevisiae: Comparison of
transcriptional activity and their use for heterologous expression.
Nucleic Acids Res. 22:5767–5768. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Westermann B and Neupert W:
Mitochondria-targeted green fluorescent proteins: Convenient tools
for the study of organelle biogenesis in Saccharomyces
cerevisiae. Yeast. 16:1421–1427. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Volland C, Urban-Grimal D, Géraud G and
Haguenauer-Tsapis R: Endocytosis and degradation of the yeast
uracil permease under adverse conditions. J Biol Chem.
269:9833–9841. 1994.PubMed/NCBI
|
|
15
|
Boeckstaens M, Merhi A, Llinares E, Van
Vooren P, Springael JY, Wintjens R and Marini AM: Identification of
a novel regulatory mechanism of nutrient transport controlled by
TORC1-Npr1-Amu1/Par32. PLoS Genet. 11:e10053822015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gregg C, Kyryakov P and Titorenko VI:
Purification of mitochondria from yeast cells. J Vis Exp.
30:e14172009.
|
|
17
|
Piskur J, Rozpedowska E, Polakova S,
Merico A and Compagno C: How did Saccharomyces evolve to
become a good brewer? Trends Genet. 22:183–186. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Parrella E and Longo VD: The chronological
life span of Saccharomyces cerevisiae to study mitochondrial
dysfunction and disease. Methods. 46:256–262. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Crowley LC, Christensen ME and Waterhouse
NJ: Measuring mitochondrial transmembrane potential by TMRE
staining. Cold Spring Harb Protoc 2016. 2016. View Article : Google Scholar
|
|
20
|
Hoffmann HP and Avers CJ: Mitochondrion of
yeast: Ultrastructural evidence for one giant, branched organelle
per cell. Science. 181:749–751. 1973. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Pevala V, Kolarov J and Polčic P:
Alterations in mitochondrial morphology of Schizosaccharomyces
pombe induced by cell-death promoting agents. Folia Microbiol
(Praha). 52:381–390. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Vanwalleghem G, Fontaine F, Lecordier L,
Tebabi P, Klewe K, Nolan DP, Yamaryo-Botté Y, Botté C, Kremer A,
Burkard GS, et al: Coupling of lysosomal and mitochondrial membrane
permeabilization in trypanolysis by APOL1. Nat Commun. 6:80782015.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zamzami N, Marchetti P, Castedo M,
Decaudin D, Macho A, Hirsch T, Susin SA, Petit PX, Mignotte B and
Kroemer G: Sequential reduction of mitochondrial transmembrane
potential and generation of reactive oxygen species in early
programmed cell death. J Exp Med. 182:367–377. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Akl I, Lelubre C, Uzureau P, Piagnerelli
M, Biston P, Rousseau A, Badran B, Fayyad-Kazan H, Ezedine M,
Vincent JL, et al: Apolipoprotein L expression correlates with
neutrophil cell death in critically ill patients. Shock.
47:111–118. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Uzureau S, Coquerelle C, Vermeiren C,
Uzureau P, Van Acker A, Pilotte L, Monteyne D, Acolty V,
Vanhollebeke B, Van den Eynde B, et al: Apolipoproteins L control
cell death triggered by TLR3/TRIF signaling in dendritic cells. Eur
J Immunol. 46:1854–1866. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Francis BR, White KH and Thorsness PE:
Mutations in the Atp1p and Atp3p subunits of yeast ATP synthase
differentially affect respiration and fermentation in
Saccharomyces cerevisiae. J Bioenerg Biomembr. 39:127–144.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Granado D, Müller D, Krausel V,
Kruzel-Davila E, Schuberth C, Eschborn M, Wedlich-Söldner R,
Skorecki K, Pavenstädt H, Michgehl U and Weide T: Intracellular
APOL1 risk variants cause cytotoxicity accompanied by energy
depletion. J Am Soc Nephrol. 28:3227–3238. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Baars TL, Petri S, Peters C and Mayer A:
Role of the V-ATPase in regulation of the vacuolar fission-fusion
equilibrium. Mol Biol Cell. 18:3873–3882. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kruzel-Davila E, Shemer R, Ofir A,
Bavli-Kertselli I, Darlyuk-Saadon I, Oren-Giladi P, Wasser WG,
Magen D, Zaknoun E, Schuldiner M, et al: APOL1-mediated cell injury
involves disruption of conserved trafficking processes. J Am Soc
Nephrol. 28:1117–1130. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lan X, Jhaveri A, Cheng K, Wen H, Saleem
MA, Mathieson PW, Mikulak J, Aviram S, Malhotra A, Skorecki K and
Singhal PC: APOL1 risk variants enhance podocyte necrosis through
compromising lysosomal membrane permeability. Am J Physiol Renal
Physiol. 307:F326–F336. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Vanhamme L, Paturiaux-Hanocq F, Poelvoorde
P, Nolan DP, Lins L, Van Den Abbeele J, Pays A, Tebabi P, Van Xong
H, Jacquet A, et al: Apolipoprotein L-I is the trypanosome lytic
factor of human serum. Nature. 422:83–87. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wen H, Kumar V, Lan X, Shoshtari SSM, Eng
JM, Zhou X, Wang F, Wang H, Skorecki K, Xing G, et al: APOL1 risk
variants cause podocytes injury through enhancing endoplasmic
reticulum stress. Biosci Rep. 38:BSR201717132018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Greene AS and Hajduk SL: Trypanosome lytic
factor-1 initiates oxidation stimulated osmotic lysis of
Trypanosoma brucei brucei. J Biol Chem. 291:3063–3075. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Cheng D, Weckerle A, Yu Y, Ma L, Zhu X,
Murea M, Freedman BI, Parks JS and Shelness GS: Biogenesis and
cytotoxicity of APOL1 renal risk variant proteins in hepatocytes
and hepatoma cells. J Lipid Res. 56:1583–1593. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Olabisi OA, Zhang JY, VerPlank L, Zahler
N, DiBartolo S III, Heneghan JF, Schlöndorff JS, Suh JH, Yan P,
Alper SL, et al: APOL1 kidney disease risk variants cause
cytotoxicity by depleting cellular potassium and inducing
stress-activated protein kinases. Proc Natl Acad Sci USA.
113:830–837. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Limou S, Dummer PD, Nelson GW, Kopp JB and
Winkler CA: APOL1 toxin, innate immunity, and kidney injury. Kidney
Int. 88:28–34. 2015. View Article : Google Scholar : PubMed/NCBI
|