Introduction
Osteosarcoma (OS), a type of bone cancer, is one of
the most common and aggressive malignancies in children and
adolescents worldwide (1). The
clinical benefits of existing therapeutic strategies for OS are
unsatisfactory, despite significant developments in diagnostics and
treatments during the last few decades (2,3). The
majority of patients with OS experience recurrence and poor
prognosis following surgical resection or adjuvant chemotherapy
(4). Previous studies have
revealed that the high recurrence rate of OS is primarily due to
chemoresistance to anti-OS therapy (5). Aberrant alterations to certain
proteins and genes are partly responsible for chemoresistance in OS
cells (6). He et al
(1) summarized the major
mechanisms of chemoresistance in OS, including decreased
intracellular drug accumulation, drug inactivation, enhanced DNA
repair, perturbations in signal transduction pathways, apoptosis
and cell cycle-associated gene expression turbulence,
autophagy-associated chemoresistance, microRNA dysregulation and
cancer stem cell-associated drug resistance.
Sphingosine kinases are lipid kinases that catalyze
the production of sphingosine-1-phosphate (S1P) by phosphorylating
sphingosine, a process that regulates cell proliferation, motility,
differentiation, apoptosis and angiogenesis (7). A number of studies have reported a
role for sphingosine kinases in tumor progression, in particular
sphingosine kinase 1 (SphK1) (8–12).
Zhao et al (13) reported
that SphK1 promoted metastasis by activating the S1P/S1P receptor
3/Notch cascade in thyroid carcinoma. Another study reported that
SphK1 inhibited melanoma growth in a mouse model (14). SphK1 has also been reported to be
overexpressed in multiple cancer cell lines, and to be associated
with resistance to chemotherapy (15) and glycolysis promotion (16,17).
Targeting SphK1 has been identified as a promising and effective
anticancer therapeutic strategy for the treatment of multiple types
of cancer, including gastric (18)
and colorectal cancer (19), as
well as nasopharyngeal (20) and
hepatocellular carcinoma (21).
Yao et al (22) reported
that co-administration of doxorubicin and phenoxodiol
synergistically inhibited proliferation both in vivo and
in vitro, which suggested that SphK1-induced chemoresistance
in OS could be reversed by inhibiting the activity of SphK1
(22). However, the precise
molecular mechanisms of SphK1 are not completely understood;
therefore, further investigation is required to identify whether
SphK1 may serve as a potential therapeutic target for OS.
The present study aimed to investigate the potential
role and underlying mechanisms of SphK1 in the chemoresistance and
glycolysis of OS.
Materials and methods
Cell culture
The human OSU2OS, MG63 and SaoS2 cell lines and the
normal human osteoblast hFOB1.19 cell line were purchased from The
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. The cells were maintained in RPMI-1640 medium (HyClone;
Cytiva) supplemented with 10% fetal bovine serum (HyClone; Cytiva)
at 37°C with 5% CO2.
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
Total RNA of cells was isolated using
TRIzol® (Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. The quality and concentration of total
RNA were determined using a NanoDrop spectrophotometer (Thermo
Fisher Scientific, Inc.). Total RNA was reverse transcribed to cDNA
using the PrimeScript™ RT-PCR kit (Takara Bio, Inc.). Subsequently,
qPCR was performed using SYBR green reagent (CoWin Biosciences)
following the manufacturer's protocol. Thermocycling conditions
were: Initial denaturation step at 95°C for 5 min, followed by 40
cycles of 10 sec at 95°C for denaturation and 60 sec at 60°C for
annealing and extension. The primer pairs targeting human SphK1 and
GAPDH were designed using Primer 5.0 (http://www.premierbiosoft.com/primerdesign/index.html),
and the primer specificity was tested using Primer-BLAST
(blast.ncbi.nlm.nih.gov/Blast.cgi). The sequences of the primer
pairs used for qPCR are listed in Table I. mRNA levels were quantified using
the 2−∆∆Cq method (23)
and normalized to the internal reference gene GAPDH.RT-qPCR was
performed in triplicate.
 | Table I.Primers used for reverse
transcription-quantitative PCR. |
Table I.
Primers used for reverse
transcription-quantitative PCR.
|
| Sequence
(5′-3′) |
|---|
|
|
|
|---|
| Gene | Forward | Reverse |
|---|
| GAPDH |
GGTGAAGGTCGGAGTCAACG |
ACCATGTAGTTGAGGTCAATGAAGG |
| SphK1 |
GGAGGAGGCAGAGATAAC |
TTAGCCCATTCACCACTTCA |
Western blotting
Total protein of all cell lines was extracted using
RIPA lysis buffer (Cell Signaling Technology, Inc.). Total protein
was quantified using the Bicinchoninic Acid Protein Assay kit
(Pierce; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol and 20 µg protein samples from each cell
line were separated on 10% SDS-PAGE and transferred to PVDF
membranes. The membranes were blocked with 5% fat free milk for 1 h
at room temperature and subsequently incubated overnight at 4°C
with primary antibodies targeting human GAPDH (cat. no. 5174,
1:1,000, 37 kDa), SphK1 (cat. no. 12071, 1:1,000, 45–60 kDa) and
hypoxia inducible factor-1α (HIF-1α, cat. no. 36169, 1:1,000, 120
kDa); all purchased from Cell Signaling Technology, Inc.).
Following washing with Tris-buffered saline containing 0.1%
Tween-20, the membranes were incubated with HRP-linked anti-rabbit
IgG antibody (Cell Signaling Technology, Inc., cat. no. 7074,
1:1,000) for 1 h at room temperature. Proteins bands were
visualized using the SuperSignal™ West Dura Extended Duration
Chemiluminescence substrate (Thermo Fisher Scientific, Inc.) and
the ChemiDoc™ XRS + system (Bio-Rad Laboratories, Inc.). Blots were
performed in triplicate. GAPDH was used as the loading control.
ImageJ (1.52u; National Institutes of Health) was used to analyze
the gray values of the bands.
Plasmid construction and
transfection
Human SphK1 cDNA was amplified using oligodT primers
and cloned into a pGLV3/H1/GFP + Puro vector (Biovector Science
Lab, Inc.). A pGLV3/H1/GFP + Puro/scramble vector (Biovector
Science Lab, Inc.) with limited homology to any known human
sequences was used as the control. Small interfering (si)RNA
targeting SphK1 (GGCTGAAATCTCCTTCACG) or HIF-1α
(CCGAAUUGAUGGGAUAUGATT) was designed and constructed by Guangzhou
RiboBio Co., Ltd. Cells (5×104/well) were plated into a
6-well plate, cells were transfected with vectors (5,800 bp, 2
µg/well) or siRNAs (5,819 bp, 100 nM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.), according to the manufacturer's protocol. After
culturing for 2 days, the cells were used for the following
experiments.
Detection of glucose uptake, lactate
production and cellular ATP level
The glucose uptake assay was performed using the
Glucose Uptake Assay kit (cat. no. ab136955; Abcam) and the lactate
production assay was performed using the L-Lactate Assay kit (cat.
no. ab65331; Abcam), both according to the manufacturer's protocol.
Cellular ATP levels were measured using the ATP Assay kit (cat. no.
ab83355; Abcam), according to the manufacturer's protocol. All
assays were performed in triplicate.
Survival assay
For the cell survival assay, 4×103
cells/well were plated into a flat bottom 96-well plate with 100 µl
RPMI-1640 medium (HyClone; Cytiva) in triplicate. After 24 h
incubation at 37°C, cells were treated with doxorubicin (0.001–10
µg/ml, cat. no. S1208, Selleck Chemicals) for 72 h at 37°C.
Subsequently, 10 µl Cell Counting Kit-8 reagent following the
manufacturer's protocol (Dojindo Molecular Technologies, Inc.) was
added into each well. The optical density (OD) value was recorded
at a wavelength of 450 nm using a microplate reader.
Statistical analysis
All statistical analyses were performed using SPSS
software (version 21.0; IBM Corp.). Data are presented as the mean
± standard deviation. A Student's unpaired t-test was used for the
comparison of two groups. One-way ANOVA followed by Tukey's post
hoc test was used for the comparison of >2 groups. P<0.05 was
considered to indicate a statistically significant difference.
Results
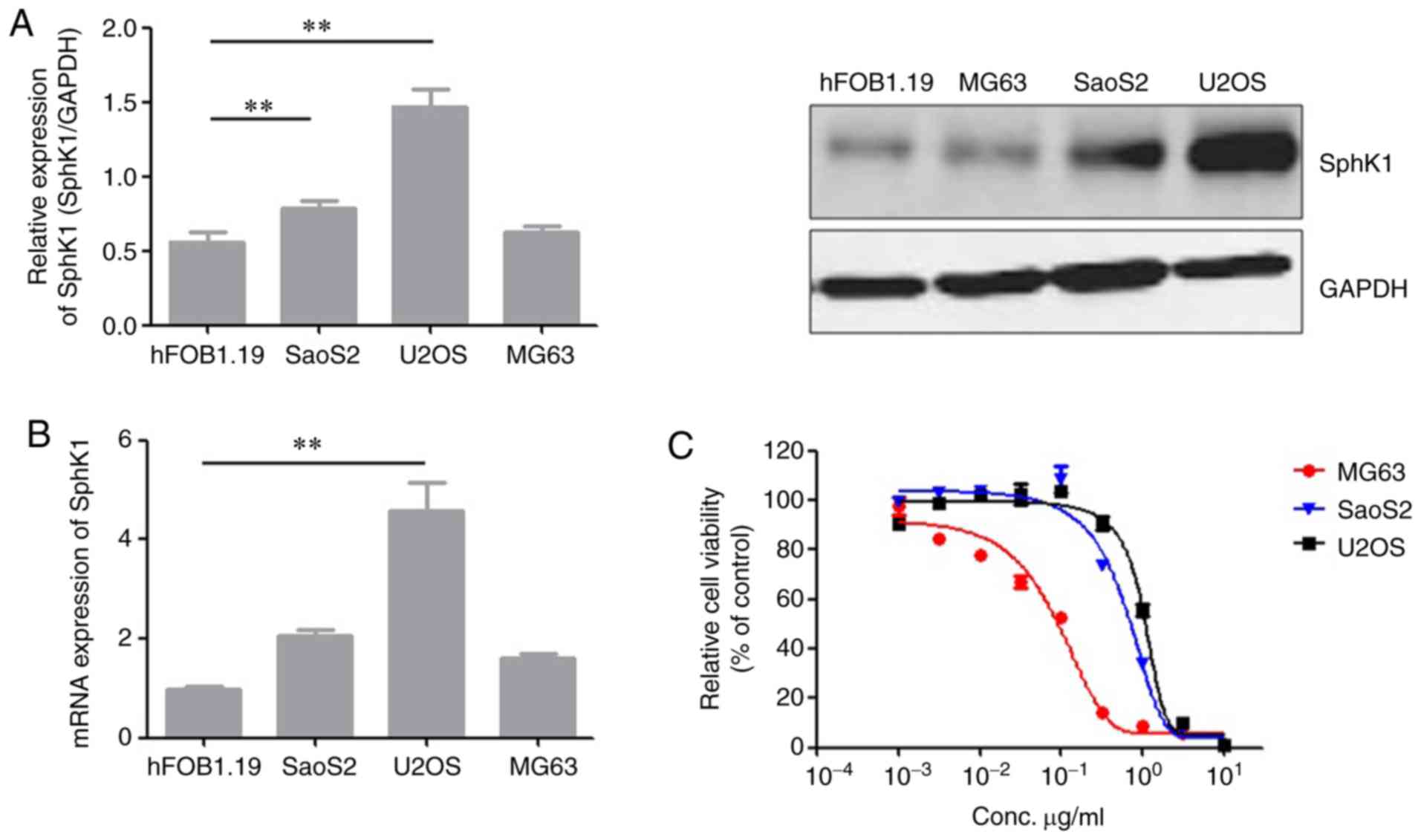
SphK1 is associated with resistance to
doxorubicin in OS cell lines
To investigate the role of SphK1 in resistance to
doxorubicin in OS, the expression of SphK1 in the 3OSU2OS, MG63 and
SaoS2 cell lines and the normal human osteoblast hFOB1.19 cell line
was determined using RT-qPCR and western blotting. The U2OS and
SaoS2 cells exhibited increasedlevels of SphK1 expression compared
with the hFOB1.19 and MG63 cells (Fig.
1A and B). To assess whether the aberrant expression of SphK1
was associated with doxorubicin resistance in lines, a cell
survival assay was performed on cells incubated with increasing
concentrations of doxorubicin (0.001–10 µg/ml) for 72 h. U2OS and
SaoS2 cells exhibited greater resistance to doxorubicin compared
with MG63 cells (Fig. 1C).
Furthermore, these results suggested that there might be a positive
association between SphK1 expression and doxorubicin resistance.
Collectively, the results suggested that increased SphK1 expression
levels were associated with the doxorubicin-resistant phenotype of
OS cell lines.
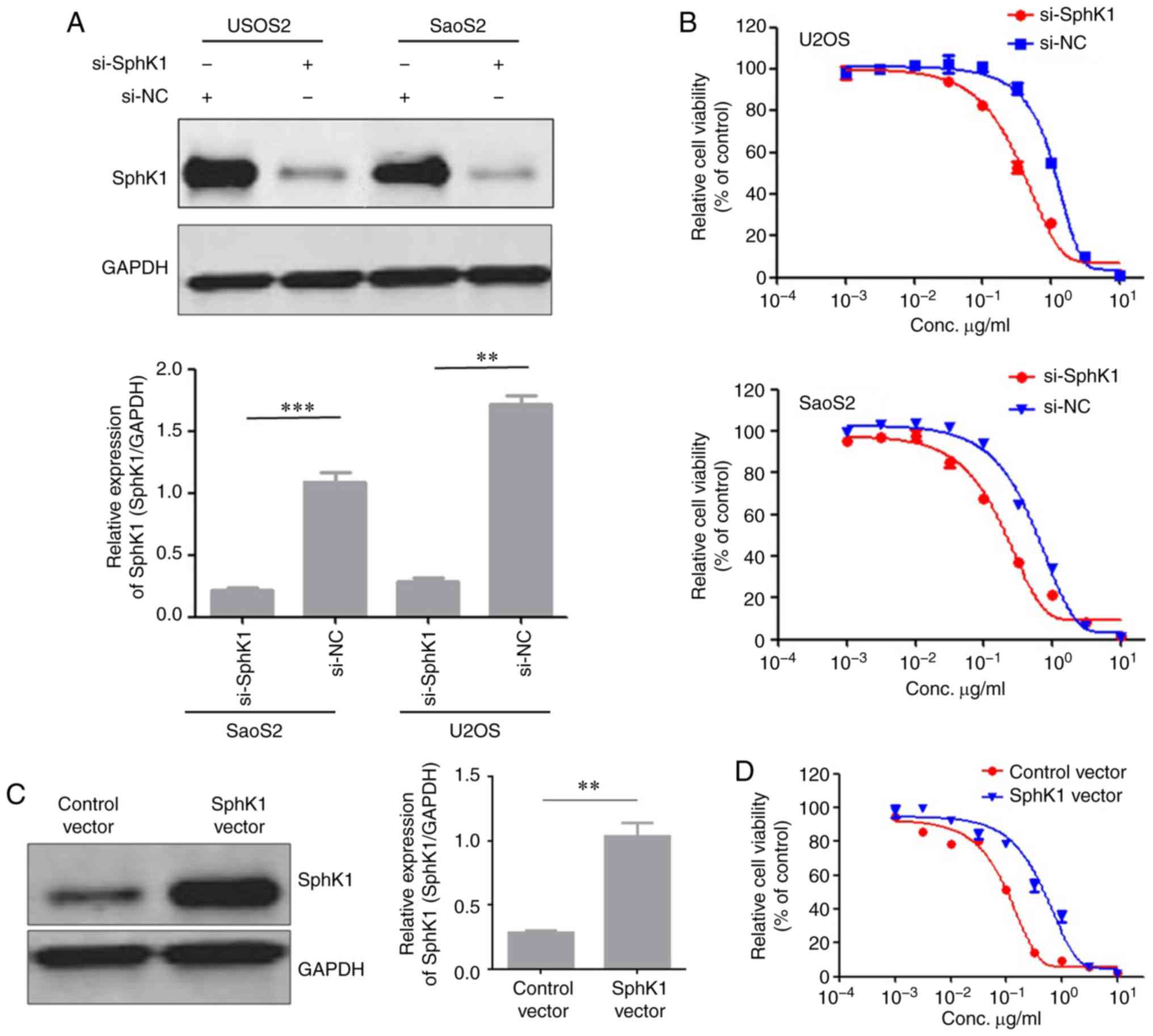
Effects of SphK1 on resistance to
doxorubicin in OS cell lines
To investigate whether SphK1 downregulation in OS
cell lines decreased resistance to doxorubicin, siRNAs were used to
knock down endogenous expression of SphK1 in the U2OS and SaoS2
cells. Successful SphK1 knockdown was confirmed by western blotting
(Fig. 2A). Subsequently, a cell
survival assay was performed to evaluate the effect of SphK1 on
doxorubicin resistance. SphK1 knockdown decreased doxorubicin
resistance in both U2OS and SaoS2 cells compared with the control
cells (Fig. 2B). Based on the
hypothesis that SphK1 was important for maintaining doxorubicin
resistance in OS cell lines, the effect of increasing SphK1
expression on doxorubicin resistance in OS cell lines was
investigated. MG63 cells overexpressing SphK1 were constructed, and
successful transfection was confirmed by western blotting (Fig. 2C). Doxorubicin resistance was
increased in MG63 cells overexpressing SphK1 compared with MG63
control cells (Fig. 2D).
Collectively, the results suggested that SphK1 had a role in the
generation of doxorubicin resistance in OS cell lines.
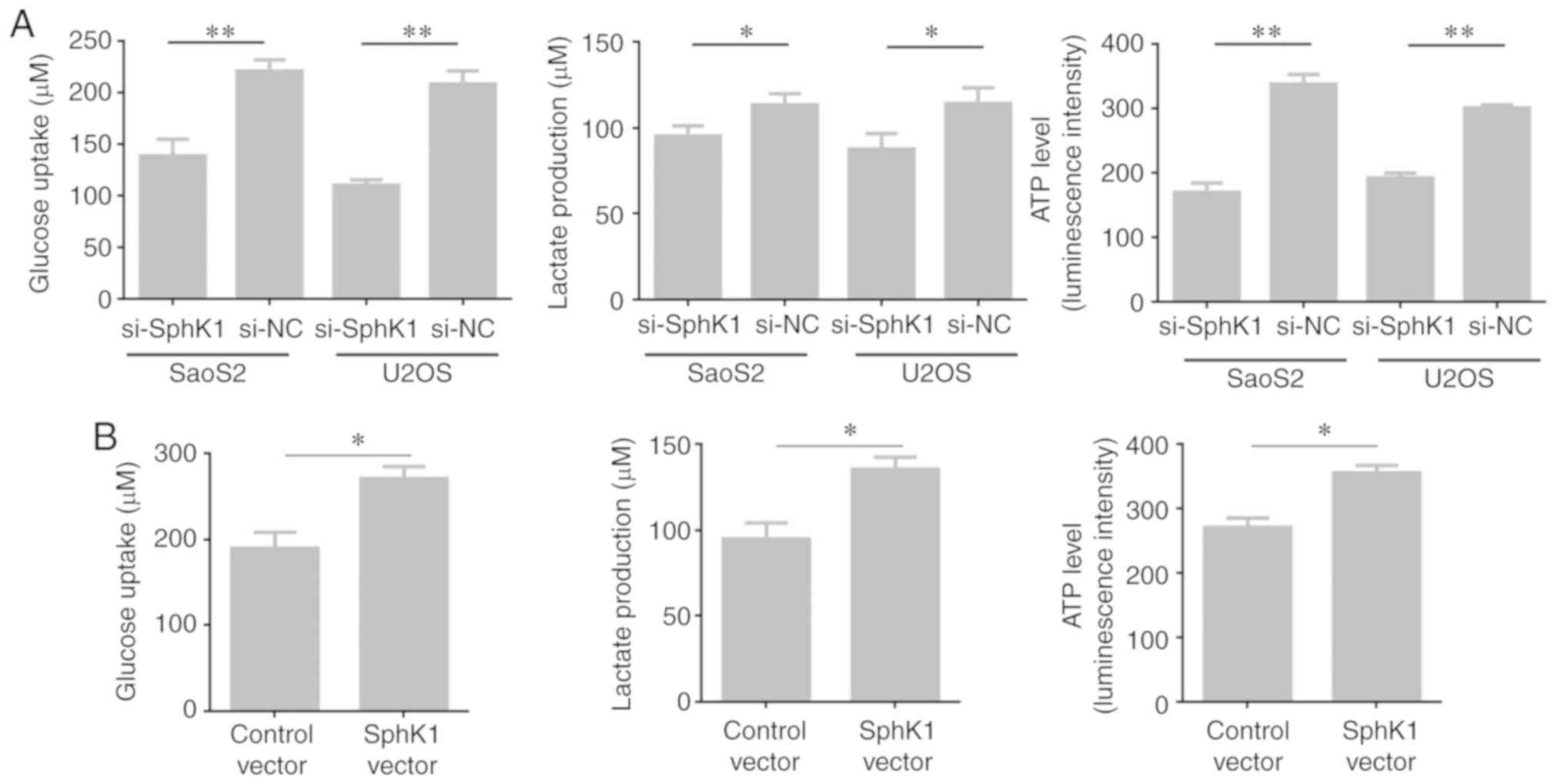
SphK1 contributes to OS cell
glycolysis
To investigate the effect of SphK1 on glycolysis,
glucose uptake, lactate production and cellular ATP levels were
determined. SphK1 knockdown SaoS2 and U2OS cells displayed
significantly decreased glucose uptake, lactate production and
cellular ATP levels compared with the corresponding control cells
(Fig. 3A). By contrast, glucose
uptake, lactate production and cellular ATP levels were
significantly increased in MG63 cells overexpressing SphK1 compared
with the MG63 control cells (Fig.
3B). The results suggested an important role for SphK1 in OS
cell glycolysis.
SphK1-associated effects on glycolysis
and doxorubicin-resistance are mediated by HIF-1α
The underlying mechanisms of the SphK1-associated
effects were investigated. HIF-1α expression was detected in OS
cells by western blotting, and the HIF-1α expression pattern in OS
cells was similar to that of SphK1 (Fig. 4A). SphK1 over expression
significantly incre-ased HIF-1α expression levels in MG63 cells
compared with MG63 cells transfected with control vector (Fig. 4B). Based on the suggestion that
SphK1 may contribute to glycolysis and doxorubicin resistance in
MG63 cells, siRNAs were used to knockdown HIF-1α expression levels
in MG63 and U2OS cells (Fig. 4C).
The survival assay suggested that MG63 cells transfected with
si-HIF-1α were more sensitive to doxorubicin compared with MG63
cells transfected with si-NC (Fig.
4D). Additionally, doxorubicin resistance was decreased in
HIF-1α knockdown U2OS cells compared with U2OS control cells
(Fig. 4E). Furthermore, glucose
uptake, lactate production and cellular ATP levels were decreased
in HIF-1α knockdown MG63 cells compared with MG63 control cells
(Fig. 4F). The decreased glucose
uptake, lactate production and cellular ATP levels were also
observed in U2OS cells transfected with si-HIF-1α compared with
U2OS control cells (Fig. 4G). The
results suggested that the SphK1-mediated effects on glycolysis and
doxorubicin resistance were partially mediated by HIF-1α.
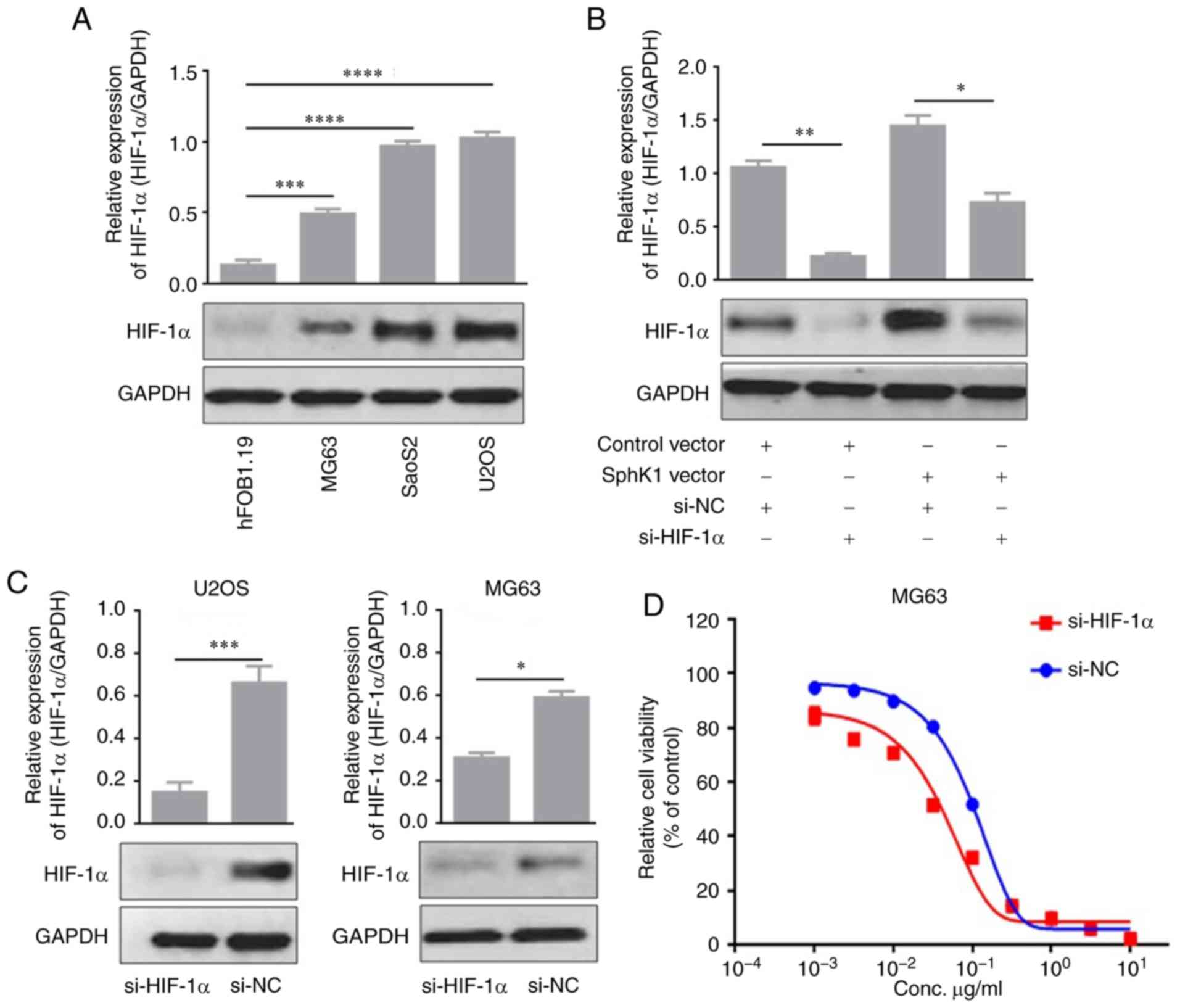 | Figure 4.HIF-1α is involved in SphK1-mediated
effects on glycolysis and doxorubicin resistance. (A) Western
blotting was used to detect HIF-1α expression levels in OS cell
lines. (B) HIF-1α expression was regulated by SphK1 expression in
MG63 cells. (C) HIF-1α was knocked down by siRNA transfection in
MG63 cells and U2OS cells and confirmed by western blotting. The
Cell Counting Kit-8 assay was performed in (D) MG63 cells
transfected with si-HIF-1α to assess cell viability. *P<0.05,
**P<0.01, ***P<0.001 and ****P<0.0001. HIF-1α, hypoxia
inducible factor-1α; SphK1, sphingosine kinase 1; OS, osteosarcoma;
si, small interfering RNA; NC, negative control. HIF-1α is involved
in SphK1-mediated effects on glycolysis and doxorubicin resistance.
The Cell Counting Kit-8 assay was performed in (E) U2OS cells
transfected with si-HIF-1α to assess cell viability. To investigate
glycolysis, glucose uptake, lactate production and cellular ATP
levels were measured in (F) MG63 and (G) U2OS cells transfected
with si-HIF-1α. *P<0.05, **P<0.01 and ***P<0.001. HIF-1α,
hypoxia inducible factor-1α; SphK1, sphingosine kinase 1; OS,
osteosarcoma; si, small interfering RNA; NC, negative control. |
Discussion
In the past 30 years, overall survival time in
patients with OS has significantly improved due to clinical
administration of aggressive chemotherapies (2,3).
However, patients suffering with recurrence or metastasis rarely
benefit from advanced chemotherapy regimens, which is partly due to
chemoresistance to anti-OS agents (2). The development of chemoresistance in
malignancies, including OS, compromises the effectiveness of the
majority of chemotherapeutics.
SphK1 has been frequently reported to function as a
tumor promoter by supporting cancer cell transformation (7). Increased SphK1 expression has been
reported to confer resistance to chemotherapeutic drugs, while
restraining SphK1 may restore or improve sensitivity to
therapeutics (16–18). In the present study, the role of
SphK1 in the chemoresistance of OS cell lines was investigated.
Doxorubicin resistance and SphK1 protein expression levels were
measured in U2OS, SaoS2 and MG63 cells using a survival assay and
western blotting, respectively. The cells that exhibited greater
resistance to doxorubicin also exhibited increased levels of SphK1
expression. Furthermore, a number of previous studies have reported
that SphK1 expression is increased in cancerous tissues compared
with matched non-cancerous tissues (9,10,13,19–24).
To further investigate the effects of SphK1 on doxorubicin
resistance, SphK1 was knocked down in U2OS and SaoS2 cells by siRNA
transfection. SphK1 knockdown in the two OS cell lines decreased
the extent of doxorubicin resistance. SphK1 overexpression in MG63
cells, which endogenously expressed low level SphK1 expression, was
also performed. Similarly, MG63 cells overexpressing SphK1
exhibited increased doxorubicin resistance compared with the MG63
control cells, and the inhibitory effect of doxorubicin on cell
proliferation was also attenuated by increased SphK1 expression
levels. Therefore, the role of SphK1 in OS cell chemoresistance was
established in the OS cell lines. Furthermore, the association
between SphK1 and chemoresistance in other cell lines, including
those with low level endogenous SphK1 expression, requires further
investigation.
In 2006, Bonhoure et al (25) reported that SphK1 contributes to
MDR-associated chemoresistance in acute myeloid leukemia. The role
of SphK1 in chemoresistance has also been investigated in several
different types of cancer, including breast (26), colon (27) and gastroesophageal cancer (28), as well as hepatocellular carcinoma
(12). A study conducted by Wang
and Wu (12) reported that SphK1
expression was associated with poor prognosis and oxaliplatin
resistance in hepatocellular carcinoma (12). Another in vivo study
reported that depletion of SphK1 expression inhibited liver
tumorigenesis in mice treated with diethyl nitrosamine (11). Katsuta et al (26) demonstrated that inhibiting the
activity of SphK1 contributed to doxorubicin-induced cytotoxicity
in breast cancer. He et al (1) summarized the molecular mechanisms of
chemoresistance in OS; however, the detailed molecular mechanisms
by which SphK1 mediates doxorubicin resistance are unknown.
Previous studies (27,29,30)
reported that activation of the SphK1/ERK/p-ERK signaling pathway
in colon cancer cells promoted autophagy, which is one of a number
of mechanisms that have been reported to be responsible for
chemoresistance.
The tumor environment is characterized by low oxygen
levels; therefore, glycolysis is the major source of energy for
rapidly proliferating tumor cells. A number of previous studies
have reported that SphK1 has a role in the glycolysis of cancer and
normal cells (16,31,32).
Cuvillier et al (17) also
reported that SphK1 may serve as a potential therapeutic target for
cancer. Consistently, the present study suggested that increased
levels of SphK1 expression promoted glycolysis in OS cells.
Therefore, further suggesting that SphK1 may serve as a novel
target for the treatment of OS. Subsequently, the underlying
mechanisms of SphK1 were investigated and suggested that HIF-1α
expression was required for SphK1-mediated effects on glycolysis
and doxorubicin resistance in OS cell lines. HIF-1α, as a responder
to hypoxia, has been frequently reported to activate various genes
involved in neoangiogenesis, glycolysis, resistance to therapeutics
and metastasis (33,34). It has also been reported that
HIF-1α upregulates the expression of multidrug resistance genes
(35), and its expression in
breast cancer was significantly associated with P-glycoprotein
expression, a cell membrane protein responsible for the drug efflux
(36,37). Upregulation of HIF-1α in tumor
cells was identified as one of the main mechanisms associated with
doxorubicin resistance (38).
Furthermore, a previous study reported that SphK1 knockdown
prevents the accumulation of HIF-1α in several human cancer cell
lines (including PC-3 and U87), suggesting that SphK1 acts as a
modulator of HIF-1α (39). In
addition, several studies have reported a number of mechanisms that
mediate SphK1-induced doxorubicin resistance. In gastric cancer,
SphK1 expression confers resistance to chemotherapeutic-induced
apoptosis by stimulating the Akt/forkhead box O3a signaling pathway
(18). Additionally, epidermal
growth factor receptor was reported to induce chemoresistance in OS
(40), and a further investigation
reported a relationship between EGFR and SphK1 in resistance to
cetuximab treatment (8).
Collectively, the aforementioned studies and the present study
suggested that SphK1 may serve as a promising therapeutic target
for cancer. However, further investigation into the mechanisms of
SphK1-induced chemoresistance and glycolysis are required to
support the clinical use of SphK1-associated strategies in patients
with OS.
To conclude, the present study suggested that SphK1
participated in the development of doxorubicin resistance and
glycolysis in OS and indicated that HIF-1α may be partially
responsible for SphK1-induced effects. The results of the present
study improved the existing knowledge of the role of SphK1 in OS
and further suggested that SphK1 may serve as a potential
therapeutic target in the disease.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
Both authors were involved in writing the
manuscript. XR mainly provided the conception and design of the
study, and performed the data analysis. CS provided the study
materials. Both authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
He H, Ni J and Huang J: Molecular
mechanisms of chemoresistance in osteosarcoma (Review). Oncol Lett.
7:1352–1362. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Ducreux M, Petersen LN, Ohler L, Bergamo
F, Metges JP, de Groot JW, Wang JY, García Paredes B, Dochy E,
Fiala-Buskies S, et al: Safety and effectiveness of regorafenib in
patients with metastatic colorectal cancer in routine clinical
practice in the prospective, observational CORRELATE study. Eur J
Cancer. 123:146–154. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Morrow JJ and Khanna C: Osteosarcoma
genetics and epigenetics: Emerging biology and candidate therapies.
Crit Rev Oncog. 20:173–197. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Sun K, Gong C, Peng H, Fang H, Zhou J, Li
J, Chen S and Zheng H: High CCL5 expression is associated with
osteosarcoma metastasis and poor prognosis of patients with
osteosarcoma. Mol Med Rep. 16:6953–6957. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
He F, Zhang W, Shen Y, Yu P, Bao Q, Wen J,
Hu C and Qiu S: Effects of resection margins on local recurrence of
osteosarcoma in extremity and pelvis: Systematic review and
meta-analysis. Int J Surg. 36:283–292. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Buljan M, Blattmann P, Aebersold R and
Boutros M: Systematic characterization of pan-cancer mutation
clusters. Mol Syst Biol. 14:e79742018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Heffernan-Stroud LA and Obeid LM:
Sphingosine kinase 1 in cancer. Adv Cancer Res. 117:201–235. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Schiefler C, Piontek G, Doescher J,
Schuettler D, Mißlbeck M, Rudelius M, Haug A, Reiter R, Brockhoff G
and Pickhard A: Inhibition of SphK1 reduces radiation-induced
migration and enhances sensitivity to cetuximab treatment by
affecting the EGFR/SphK1 crosstalk. Oncotarget. 5:9877–9888. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Wang S, Liang Y, Chang W, Hu B and Zhang
Y: Triple negative breast cancer depends on Sphingosine Kinase 1
(SphK1)/Sphingosine-1-Phosphate (S1P)/Sphingosine 1-phosphate
receptor 3 (S1PR3)/notch signaling for metastasis. Med Sci Monit.
24:1912–1923. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hatoum D, Haddadi N, Lin Y, Nassif NT and
McGowan EM: Mammalian sphingosine kinase (SphK) isoenzymes and
isoform expression: Challenges for SphK as an oncotarget.
Oncotarget. 8:36898–36929. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Chen J, Qi Y, Zhao Y, Kaczorowski D,
Couttas TA, Coleman PR, Don AS, Bertolino P, Gamble JR, Vadas MA,
et al: Deletion of sphingosine kinase 1 inhibits liver
tumorigenesis in diethylnitrosamine-treated mice. Oncotarget.
9:15635–15649. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang F and Wu Z: Sphingosine kinase 1
overexpression is associated with poor prognosis and oxaliplatin
resistance in hepatocellular carcinoma. Exp Ther Med. 15:5371–5376.
2018.PubMed/NCBI
|
|
13
|
Zhao Z, Ma J, Hu B, Zhang Y and Wang S:
SPHK1 promotes metastasis of thyroid carcinoma through activation
of the S1P/S1PR3/Notch signaling pathway. Exp Ther Med.
15:5007–5016. 2018.PubMed/NCBI
|
|
14
|
Madhunapantula SV, Hengst J, Gowda R, Fox
TE, Yun JK and Robertson GP: Targeting sphingosine kinase-1 to
inhibit melanoma. Pigment Cell Melanoma Res. 25:259–274. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Gault CR and Obeid LM: Still benched on
its way to the bedside: Sphingosine kinase 1 as an emerging target
in cancer chemotherapy. Crit Rev Biochem Mol Biol. 46:342–351.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sun K, Zhang Y, D'Alessandro A, Nemkov T,
Song A, Wu H, Liu H, Adebiyi M, Huang A, Wen YE, et al:
Sphingosine-1-phosphate promotes erythrocyte glycolysis and oxygen
release for adaptation to high-altitude hypoxia. Nat Commun.
7:120862016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cuvillier O, Ader I, Bouquerel P, Brizuela
L, Gstalder C and Malavaud B: Hypoxia, therapeutic resistance, and
sphingosine 1-phosphate. Adv Cancer Res. 117:117–141. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Xiong H, Wang J, Guan H, Wu J, Xu R, Wang
M, Rong X, Huang K, Huang J, Liao Q, et al: SphK1 confers
resistance to apoptosis in gastric cancer cells by downregulating
Bim via stimulating Akt/FoxO3a signaling. Oncol Rep. 32:1369–1373.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bao Y, Guo Y, Zhang C, Fan F and Yang W:
Sphingosine Kinase 1 and Sphingosine-1-phosphate signaling in
colorectal cancer. Int J Mol Sci. 18:21092017. View Article : Google Scholar
|
|
20
|
Li W, Li J, Wang Y, Zhang K, Li N, Tian Z,
Ni B, Wang H and Ruan Z: Sphingosine kinase 1 is a potential
therapeutic target for nasopharyngeal carcinoma. Oncotarget.
7:80586–80598. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lu PH, Chen MB, Liu YY, Wu MH, Li WT, Wei
MX, Liu CY and Qin SK: Identification of sphingosine kinase 1
(SphK1) as a primary target of icaritin in hepatocellular carcinoma
cells. Oncotarget. 8:22800–22810. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yao C, Wu S, Li D, Ding H, Wang Z, Yang Y,
Yan S and Gu Z: Co-administration phenoxodiol with doxorubicin
synergistically inhibit the activity of sphingosine kinase-1
(SphK1), a potential oncogene of osteosarcoma, to suppress
osteosarcoma cell growth both in vivo and in vitro. Mol Oncol.
6:392–404. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C (T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nazouri AS, Asadpour O, Dabiri S,
Pourseyedi B, Lashkarizadeh MR and Zianalinejad H: High expression
of sphingosine kinase 1 in estrogen and progesterone
receptors-negative breast cancer. Iran J Pathol. 12:218–224. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bonhoure E, Pchejetski D, Aouali N,
Morjani H, Levade T, Kohama T and Cuvillier O: Overcoming
MDR-associated chemoresistance in HL-60 acute myeloid leukemia
cells by targeting sphingosine kinase-1. Leukemia. 20:95–102. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Katsuta E, Yan L, Nagahashi M, Raza A,
Sturgill JL, Lyon DE, Rashid OM, Hait NC and Takabe K: Doxorubicin
effect is enhanced by sphingosine-1-phosphate signaling antagonist
in breast cancer. J Surg Res. 219:202–213. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Xu C, Zhang W, Liu S, Wu W, Qin M and
Huang J: Activation of the SphK1/ERK/p-ERK pathway promotes
autophagy in colon cancer cells. Oncol Lett. 15:9719–9724.
2018.PubMed/NCBI
|
|
28
|
Matula K, Collie-Duguid E, Murray G,
Parikh K, Grabsch H, Tan P, Lalwani S, Garau R, Ong Y, Bain G, et
al: Regulation of cellular sphingosine-1-phosphate by sphingosine
kinase 1 and sphingosine-1-phopshate lyase determines chemotherapy
resistance in gastroesophageal cancer. BMC Cancer. 15:7622015.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Meng Y, Gao R, Ma J, Zhao J, Xu E, Wang C
and Zhou X: MicroRNA-140-5p regulates osteosarcoma chemoresistance
by targeting HMGN5 and autophagy. Sci Rep. 7:4162017. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Kim M, Jung JY, Choi S, Lee H, Morales LD,
Koh JT, Kim SH, Choi YD, Choi C, Slaga TJ, et al: GFRA1 promotes
cisplatin-induced chemoresistance in osteosarcoma by inducing
autophagy. Autophagy. 13:149–168. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Bernacchioni C, Ghini V, Cencetti F,
Japtok L, Donati C, Bruni P and Turano P: NMR metabolomics
highlights sphingosine kinase-1 as a new molecular switch in the
orchestration of aberrant metabolic phenotype in cancer cells. Mol
Oncol. 11:517–533. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Watson DG, Tonelli F, Alossaimi M,
Williamson L, Chan E, Gorshkova I, Berdyshev E, Bittman R, Pyne NJ
and Pyne S: The roles of sphingosine kinases 1 and 2 in regulating
the Warburg effect in prostate cancer cells. Cell Signal.
25:1011–1017. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma Q, Zhang Y, Liu T, Jiang K, Wen Y, Fan
Q and Qiu X: Hypoxia promotes chemotherapy resistance by
down-regulating SKA1 gene expression in human osteosarcoma. Cancer
Biol Ther. 18:177–185. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Sowa T, Menju T, Chen-Yoshikawa TF,
Takahashi K, Nishikawa S, Nakanishi T, Shikuma K, Motoyama H,
Hijiya K, Aoyama A, et al: Hypoxia-inducible factor 1 promotes
chemoresistance of lung cancer by inducing carbonic anhydrase IX
expression. Cancer Med. 6:288–297. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Badowska-Kozakiewicz AM, Sobol M and
Patera J: Expression of multidrug resistance protein P-glycoprotein
in correlation with markers of hypoxia (HIF-1α, EPO, EPO-R) in
invasive breast cancer with metastasis to lymph nodes. Arch Med
Sci. 13:1303–1314. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Xie Y and Zhong DW: AEG-1 is associated
with hypoxia-induced hepatocellular carcinoma chemoresistance via
regulating PI3K/AKT/HIF-1alpha/MDR-1 pathway. Excli J. 15:745–757.
2016.PubMed/NCBI
|
|
37
|
Doublier S, Belisario DC, Polimeni M,
Annaratone L, Riganti C, Allia E, Ghigo D, Bosia A and Sapino A:
HIF-1 activation induces doxorubicin resistance in MCF7 3-D
spheroids via P-glycoprotein expression: A potential model of the
chemo-resistance of invasive micropapillary carcinom of the breast.
BMC Cance. 12:42012. View Article : Google Scholar
|
|
38
|
Cao Y, Eble JM, Moon E, Yuan H, Weitzel
DH, Landon CD, Nien CY, Hanna G, Rich JN, Provenzale JM and
Dewhirst MW: Tumor cells upregulate normoxic HIF-1α in response to
doxorubicin. Cancer Res. 73:6230–6242. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ader I, Brizuela L, Bouquerel P, Malavaud
B and Cuvillier O: Sphingosine kinase 1: A new modulator of hypoxia
inducible factor 1alpha during hypoxia in human cancer cells.
Cancer Res. 68:8635–8642. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Sevelda F, Mayr L, Kubista B, Lötsch D,
van Schoonhoven S, Windhager R, Pirker C, Micksche M and Berger W:
EGFR is not a major driver for osteosarcoma cell growth in vitro
but contributes to starvation and chemotherapy resistance. J Exp
Clin Cancer Res. 34:1342015. View Article : Google Scholar : PubMed/NCBI
|