Introduction
MicroRNAs (miRNAs/miRs) are a class of endogenous
small single-stranded non-coding RNAs that have post-transcription
regulatory functions in eukaryotes (1). Ranging from 20 to 25 nucleotides in
size, miRNAs recognize the 3′ untranslational region (3′UTR) of
their target mRNAs through base pairing, and suppress the
translation or promote the degradation of target mRNAs via the
formation of RNA-induced silencing complexes (2–4).
miRNAs are involved in a variety of physiological processes
including cell proliferation, differentiation, apoptosis, energy
metabolism, and various pathological conditions such as cardiac
remodeling, B cell development, diabetes and allergic inflammation
(5–10).
Accumulating studies have demonstrated that miRNAs
are also actively involved in regulating chondrogenesis and
cartilage development (11). For
example, miR-410 (12), miR-29b
(13) and miR-218 (14) have been identified to regulate
chondrogenic differentiation of bone marrow mesenchymal stem cells.
miR-1 is expressed in muscle tissues and is involved in the
regulation of proliferation and differentiation of muscle tissues
through targeting histone deacetylase 4, as well as the activation
of oxidative metabolism during muscle cell differentiation through
the miR-1/133a-myocyte-specific enhancer factor 2A-Δ like
non-canonical notch ligand 1-iodothyronine deiodinase 3 axis
(15,16). Knockout of skeletal muscle-specific
miRNAs-1–2 in vivo resulted in the death of 50% of mice due
to cardiac morphological abnormalities, electrical conduction and
cell cycle disorders (17). Our
previous study demonstrated that miR-1 is highly expressed in the
hypertrophic zone of growth plate cartilage, and regulates
chondrocyte phenotypes during growth plate development (18). However, the roles of miR-1 in
regulating matrix synthesis and chondrocyte proliferation and
differentiation have not been extensively investigated.
The Hedgehog genes were originally identified during
the study of the gene mutations in Drosophila melanogaster
(19). One subtype of secretory
Hedgehog proteins, Indian hedgehog (Ihh), is expressed
predominantly in mammalian prehypertrophic chondrocytes (20,21).
Ihh plays an important role in bone development and maintains bone
balance before and after birth. Activation of Ihh has been reported
to promote chondrocyte hypertrophy in human osteoarthritic
cartilage (22) and cultured
chicken chondrocytes (23).
Previous studies suggest that Ihh plays an important regulatory
role in the growth and development of articular cartilage (24–26),
but whether it is regulated by miRNAs is unclear.
In the present study, mouse primary chondrocytes
were isolated and miR-1 levels were altered via the transfection of
a miR-1-specific miRNA mimic and inhibitor in chondrocytes. The
expression of matrix synthesis associated molecules collagen
(Col)-II and aggrecan (AGG), and chondrocyte differentiation
markers Col-X and matrix metallopeptidase (MMP)-13 were evaluated
upon miR-1 overexpression and inhibition in chondrocytes.
Importantly, this study demonstrated that miR-1 promotes cartilage
matrix synthesis and regulates the chondrocyte differentiation by
the post-transcriptional suppression of the Ihh gene.
Materials and methods
miRNA mimic, inhibitor and small
interfering (si)RNA oligonucleotides (oligos)
The miR-1 mimic, corresponding negative control
mimic (ConmiR), the miR-1 inhibitor (Anti-miR-1), control miRNA
inhibitor (Control), and the siRNA oligos were purchased from
Shanghai GenePharma Co., Ltd. The miRNA-1 mimics were
double-stranded siRNA oligos. The sense strand of miRNA-1 mimic
(5′-UGGAAUGUAAAGAAGUAUGUAU-3′) consisted of 21 bases, and the
antisense strand was complementary to the sense chain. The miR-1
inhibitor consisted of RNA oligos of 21 bases fully complementary
to the target sequences and modified with 2′ oxygen methyl. The
siRNA oligo for knockdown of Ihh (siIhh) was designed and
synthesized by Shanghai GenePharma Co., Ltd., and the sequences
were as follows: sense, 5′-CCUUCAGUGAUGUGCUUAUTT-3′.
Isolation and culture of primary
chondrocytes
C57BL/6 mice of specific-pathogen-free-grade (male,
6–8 weeks old) were purchased and maintained in the Animal
Experimental Center of Shanxi Medical University. A total of 10
mice were maintained in a specific pathogen-free (SPF) ‘barrier’
facility and housed under 25°C and humidity and alternating 12-h
light and dark cycles. The mice received SPF mouse food and were
provided with sterile drinking water ad libitum. All
experiments involving the use of animals in this study were
approved by the Ethics Committee of Shanxi Medical University. The
isolation and culture of murine primary chondrocytes were conducted
as previously reported (27).
Briefly, mouse thoraxes were isolated and digested in PBS
supplemented with 3 mg/ml collagenase D (Roche Diagnostics GmbH)
for 90 min at 37°C until soft tissues were peeled clean. Then, the
tissues were further digested with fresh digestion medium
containing 3 mg/ml collagenase D at 37°C for an additional 4 h with
shaking. Chondrocytes were harvested after centrifugation of the
suspension at 1,000 × g, 4°C for 10 min, and cultured in Dulbecco's
modified Eagle's medium/Nutrient Mixture F-12 (DMEM/F-12; 1:1 ratio
mixture) medium supplemented with 10%, 100 U/ml of penicillin and
100 µg/ml of streptomycin (all purchased from Gibco; Thermo Fisher
Scientific, Inc.). Chondrocytes were grown in a humidified
atmosphere with 5% CO2 at 37°C, and the medium was
changed every other day.
Chondrocyte transfection
Chondrocytes at passages 2–4 were used for
transfection of the miR-1 mimic, inhibitor and siRNA. Transfection
experiments were performed with Lipofectamine® 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) following the
manufacturer' s protocols. Briefly, chondrocytes were seeded at
3×105/well in 6-well plates. The miR-1 mimic, and its
inhibitor, and siRNA oligos against Ihh (40 pM) were resuspended in
GenMutt buffer (Promega Corp.) and mixed with 10 µl of
Lipofectamine 2000 reagent for transfection of cells in a single
well. At 24 or 48 h after the transfection, the cells were
harvested for RNA isolation, western blotting and
immunofluorescence.
Cell proliferation assay
Cell proliferation was detected by
5-ethynyl-2′-deoxyuridine (EdU; Cell Light EdU Apollo 567 In
Vitro Imaging Kit; RiboBio) labeling of cultured cells
according to a previous study (13). After permeabilization with 0.5%
Triton X-100 in PBS for 10 min and washing with PBS for 3 times,
the cells were subsequently stained with Apollo and DAPI at room
temperature for 1 h. The cells were observed immediately after
staining under a fluorescence microscope (DM6 B; Leica Microsystems
GmbH) under ×40 magnification, and the percentages of positively
stained cells (red) were calculated using Image Lab software 5.1
(Bio-Rad Laboratories, Inc.). A total of ~300 cells in each group
were counted, and three independent experiments were performed.
ELISA
The levels of Ihh (cat. no. SED116Mu; Wuhan USCN
Business Co., Ltd.), MMP-13 (cat. no. SEA099Mu; Wuhan USCN Business
Co., Ltd.), tissue inhibitor of metalloproteinases-1 (TIMP-1) (cat.
no. SEA552Mu; Wuhan USCN Business Co., Ltd.), glycosaminoglycan
(GAG) (cat. no. EK3456; Signalway Antibody, Ltd.) and Col-X (cat.
no. SEC156Mu; Wuhan USCN Business Co., Ltd.) in cell culture
supernatants were determined using ELISA. Chondrocytes seeded in
24-well plates at a density of 8,000 cells/well in DMEM/F-12 medium
were cultured overnight, and then separately transfected with the
miR-1 mimic, the miR-1 mimic control, the miR-1 inhibitor and the
miR-1 inhibitor control. ELISAs were performed according to the
manufacturer' s protocols. All samples were normalized according to
the total protein levels and run in duplicate, and the average
value was calculated.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was performed as previously described
(18). Total RNA was isolated from
in vitro cultured chondrocytes using the RNeasy Mini kit
(Qiagen GmbH). Total RNA (1 µg) was reverse transcribed to
complementary DNA (cDNA) using the iScript™ cDNA Synthesis kit
(Bio-Rad Laboratories, Inc.). The cDNA (40 ng) was used to quantify
the expression of the target genes by RT-qPCR, using the QuantiTect
SYBR®-Green PCR kit (Qiagen GmbH) with the DNA Engine
Opticon® 2 CFD-3220 Continuous Fluorescence Detector (MJ
Research Inc.). The internal controls were 18S and U6 (cat. no.
218300; Qiagen GmbH) for mRNA and miRNA, respectively (U6 primer
sequence not commercially available). The stem-loop primers for
miR-1 were designed and purchased from Qiagen GmbH. The mRNA
expression changes of genes, including Ihh, GLI family zinc finger
(Gli)-1, smoothened, frizzled class receptor (Smo), AGG, Col-II,
Col-X, Sox-9, parathyroid hormone-like hormone (PTHrP), Gli-2 and
Gli-3, were quantified as described in our previous publications
(22,28). The thermocycling conditions were as
follows: Pre-incubation of samples at 95°C for 5 min, then 40
cycles of denaturation at 95°C for 10 sec, annealing at 55°C for 30
sec, and extension at 72°C for 30 sec. The sequences of primers
used in this study are listed in Table
I. Relative transcript levels were calculated by the
2−ΔΔCq method (29).
 | Table I.Sequences of PCR primers used in this
study. |
Table I.
Sequences of PCR primers used in this
study.
| Gene | Primer sequence
(5–3) |
|---|
| Ihh | F:
CCACTTCCGGGCCACATTTG |
|
| R:
GGCCACCACATCCTCCACCA |
| Gli-1 | F:
GGTCCGGATGCCCACGTGAC |
|
| R:
TCCCGCTTGGGCTCCACTGT |
| Gli-2 | F:
CATGGTATCCCTAGCTCCTC |
|
| R:
GATGGCATCAAAGTCAATCT |
| Gli-3 | F:
CATGAACAGCCCTTTAAGAC |
|
| R:
TCATATGTGAGGTAGCACCA |
| Smo | F:
CTCCTACTTCCACCTGCTCAC |
|
| R:
CAAAACAAATCCCACTCACAGA |
| PTHrP | F:
CAACCAGCCCACCAGAGGA |
|
| R:
GGCGGCTGAGACCCTCCA |
| Col-X | F:
GCCAGGAAAGCTGCCCCACG |
|
| R:
GAGGTCCGGTTGGGCCTGGT |
| MMP-13 | F:
GGACCTTCTGGTCTTCTGGC |
|
| R:
GGATGCTTAGGGTTGGGGTC |
| Col-II | F:
AAGGGACACCGAGGTTTCACTGG |
|
| R:
GGGCCTGTTTCTCCTGAGCGT |
|
Aggrecan | F:
CAGTGGGATGCAGGCTGGCT |
|
| R:
CCTCCGGCACTCGTTGGCTG |
| Sox9 | F:
CGTGGACATCGGTGAACTGA |
|
| R:
GGTGGCAAGTATTGGTCAAACTC |
| 18s | F:
CGGCTACCACATCCAAGGAA |
|
| R:
GCTGGAATTACCGAGGCT |
Western blotting
Total proteins were obtained after lysing in
vitro-cultured chondrocytes with RIPA lysis buffer (Beyotime
Institute of Biotechnology). Total protein was quantified using a
bicinchoninic acid assay kit (cat. no. 23225; Pierce; Thermo Fisher
Scientific, Inc.) and 30 µg protein/lane was separated on a 10%
SDS-PAGE gel and then transferred to a PVDF membrane (Beijing
Solarbio Science & Technology Co., Ltd.). Non-specific binding
was blocked with 5% non-fat milk in Tris-buffered saline plus 0.1%
TBST (cat. no. T1081; Beijing Solarbio Science & Technology
Co., Ltd) at 25°C for 2 h, and then the membranes were incubated
with the primary antibodies (purchased from Abcam) against Ihh
(1:1,000; cat. no. ab52919), Gli-1 (1:50; cat. no. ab49314), Smo
(1:100; cat. no. ab236465), Col-II (1:5,000; cat. no. ab185430),
Col-X (1:300; cat. no. ab58632), and MMP-13 (1:3,000; cat. no.
ab39012) overnight at 4°C. β-actin (1:1,000; cat. no. ab8227;
Abcam) was used as the loading control. After washing with TBST,
the immobilized primary antibodies were incubated with a
horseradish peroxidase-conjugated secondary goat anti-rabbit IgG
antibody (1:2,000; cat. no. ab205718; Abcam) for 1 h at 25°C and
visualized using the ECL kit (Thermo Fisher Scientific, Inc.).
Finally, the blots were analyzed quantitatively using Image Lab
software (version 5.1; Bio-Rad Laboratories, Inc.) as previously
described (30).
Immunofluorescence assay
Chondrocytes cultured in vitro were rinsed in
PBS three times and fixed with 4% formaldehyde in PBS for 15 min at
room temperature. After blocking non-specific binding with 5%
normal goat serum (Sigma-Aldrich; Merck KGaA) at 25°C for 1 h,
cells were then incubated with antibody against Col-I (1:200; cat.
no. ab6308; Abcam), Col-II (1:200; cat. no. ab34712; Abcam) or Ihh
(1:1,000; cat. no. ab52919; Abcam) in PBS for 8 h at 4°C. After
washing with PBS 3 times at room temperature, the cells were
incubated with FITC-conjugated goat anti-rabbit IgG H&L
antibody (1:1,000; cat. no. ab6717; Abcam) in PBS at 25°C for 1 h.
After another three washes with PBS, the samples were incubated
with 10 µg/ml DAPI (Beyotime Institute of Biotechnology) for 5 min
at room temperature. After the final round of three washes, the
samples were mounted on slides and examined using a confocal
microscope (Nikon Eclipse 80i; Nikon Corporation) under ×40
magnification.
Dual-luciferase reporter assay
The online tools including TargetScan (http://www.targetscan.org) and miRanda (microrna.org)
were utilized to identify target genes for miR-1. Sequence analysis
indicated that miR-1 is conserved across mammalian species (miRBas
accession no. MIMAT0000416; microrna.sanger.ac.uk/sequences/) and
there are two miR-1 target sites in the 3′UTR of Ihh in different
species (CATTCCAT and ATGACCTTCCC).
The dual-luciferase assay was employed to determine
the effect of miR-1 on controlling luciferase expression, which was
linked with and regulated by the 3′UTR of human Ihh gene. Three
luciferase reporter plasmids containing wild-type (WT) or mutated
miR-1 seed sites within the 3′UTR sequence of human Ihh gene were
purchased from Sangon Biotech Co., Ltd. The 293T cells were
co-transfected with a combination of 200 ng of the
wild-type-Ihh-3′UTR-Luc reporter plasmid (WT 3′UTR of human Ihh
gene), or plasmid containing the 3′UTR of Ihh muta1 (miR-1 binding
site 1 mutation) or the 3′UTR of Ihh muta2 (miR-1 binding site 2
mutation), or the 3′UTR of Ihh muta1 and muta2, with miR-1 mimic or
control miR-NC (20 nM), and a Renilla plasmid (SV40
promoter; Sangon Biotech Co., Ltd.) using Lipofectamine 2000
reagent (Invitrogen; Thermo Fisher Scientific, Inc). After
transfection for 48 h, firefly luciferase activity was determined
and adjusted by Renilla luminescence using the assay kit
according to the manufacturer's instructions (Promega Corporation).
Each experiment was repeated four times.
Statistical analysis
All the experiments were repeated at least three
times and the results are expressed as the means ± SD. Statistical
analyses were performed using SPSS (version 19.0; IBM Corp.).
Independent sample t-test was used to compare the data from two
different groups. The data from multiple groups were analyzed by
one-way ANOVA followed by the Turkey-Kramer multiple comparisons
tests. Two-way ANOVA was used to compare the time-dependent changes
in GAG. Statistical significance was set at P<0.05.
Results
miR-1 promotes the proliferation of
mouse thorax chondrocytes
First, mouse thorax chondrocytes were isolated, and
the expression of matrix components Col-I and Col-II were measured
by immunofluorescence. As shown in Fig. 1A, Col-I expression was not
detected, while Col-II was abundantly expressed in the chondrocytes
(Fig. 2C). To determine the impact
of miR-1 expression on chondrocyte proliferation, the miR-1 mimic
or inhibitor (anti-miR-1) was transfected into these chondrocytes
to alter miR-1 expression. As validated by RT-qPCR, transfection of
the miR-1 mimic significantly increased miR-1 transcription, while
anti-miR-1 significantly decreased its expression, compared with
the corresponding controls (Fig.
1B). EdU staining at 48 h post-transfection demonstrated that
miR-1 levels were positively associated with chondrocyte
proliferation (Fig. 1C). The miR-1
mimic almost doubled the percentage of EdU-positive chondrocytes,
whereas the anti-miR-1 reduced the percentage of EdU-stained
chondrocytes by ~50% (Fig. 1D).
Moreover, the association between miR-1 level and cell
proliferation in mouse thorax chondrocytes was further validated
using the CCK-8 cell proliferation assay (Fig. 1E). Furthermore, in chondrocytes at
24 h after transfection of the miR-1 mimic, or inhibitor, or
controls, the mRNA levels of the matrix enzyme Sox-9, which is also
a marker of chondrocyte proliferation, were associated with miR-1
expression levels (Fig. 1F).
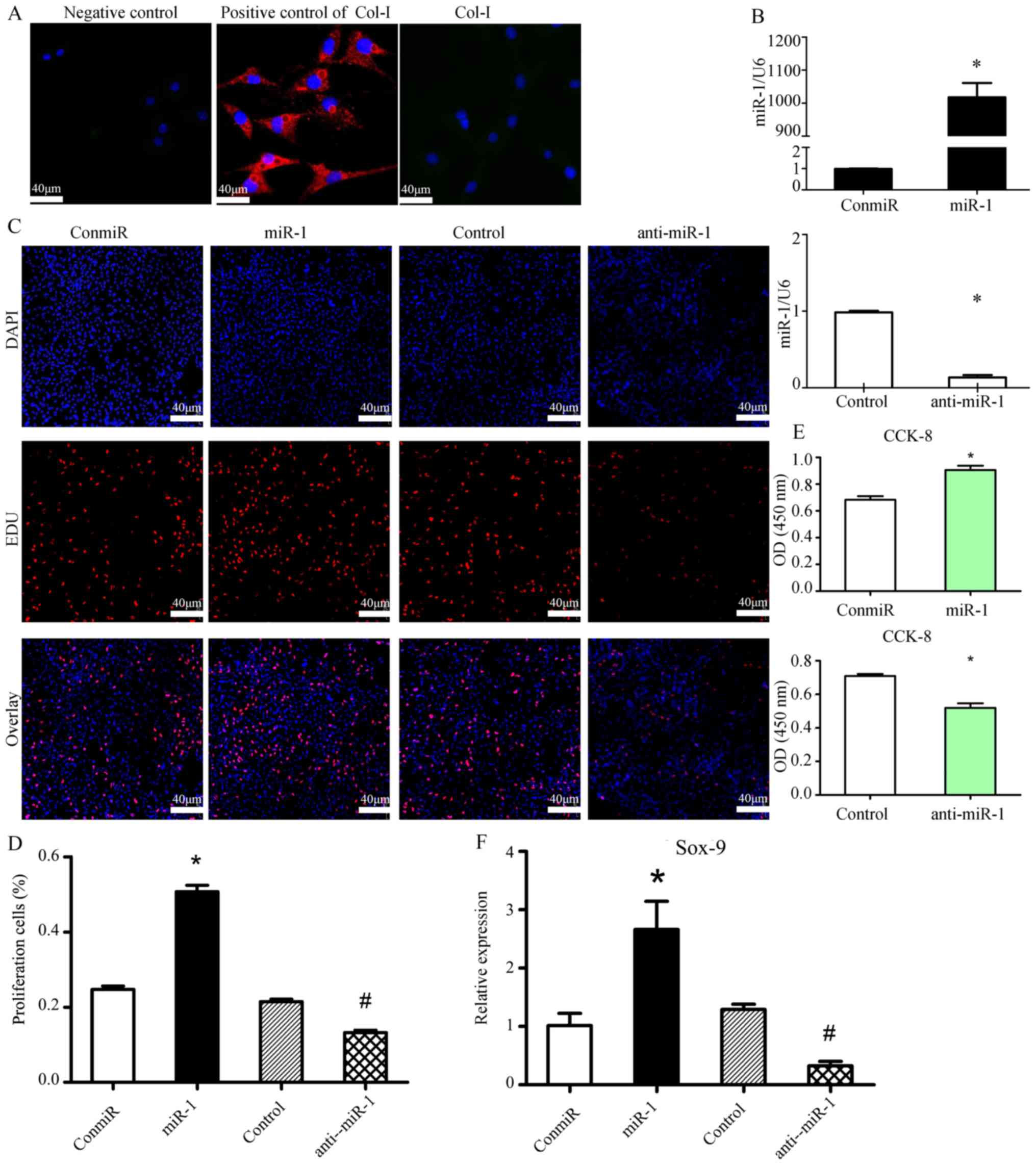 | Figure 1.miR-1 promotes the proliferation of
mouse thorax chondrocytes. (A) Expression of Col-I in mouse thorax
chondrocytes was assessed using immunofluorescence staining. Mouse
fibroblasts were used as the positive control for Col-I staining;
magnification, ×40; scale bars, 40 µm. (B) RT-qPCR results showed
that transfection of the miR-1 mimic (40 pM) increased miR-1
levels, whereas transfection of the anti-miR-1 decreased its
expression, compared with the transfection of a negative control
mimic (ConmiR) or a control miRNA inhibitor (Control) in the mouse
sterna chondrocytes at 24 h post-transfection. n=3 for each group;
*P<0.05 vs. ConmiR or Control. (C and D) Cell growth was
measured by EdU cell proliferation staining 48 h after mouse sterna
chondrocytes were transfected with the miR-1 mimic (miR-1) or
negative control mimic (ConmiR), and miR-1 inhibitor (anti-miR-1)
or a control miRNA inhibitor (Control) at 120 nM, respectively; (C)
images show the staining for EdU and DAPI, and (D) bar graph data
summarizes the percentage of EdU-proliferating cells in 5 view
fields per group; miR-1 stimulates chondrocyte proliferation. Scale
bars, 40 µm; *P<0.05 vs. ConmiR; #P<0.05 vs.
Control. (E) Transfection of miR-1 enhanced mouse thorax
chondrocyte proliferation, while transfection of anti-miR-1
inhibited proliferation, as measured by the CCK-8 cell
proliferation assay. n=5 for each group; *P<0.05 vs. ConmiR or
Control. (F) miR-1 increased Sox-9 mRNA levels, a marker for
chondrocyte proliferation. Sox-9 mRNA levels in mouse sterna
chondrocytes transfected with the miR-1 mimic (miR-1), or control
miRNA mimic (conmiR), and inhibitor (anti-miR-1) or control miRNA
inhibitor (Control) were quantified by RT-qPCR at 24 h post
transfection. n=3 for each group; *P<0.05 vs. ConmiR;
#P<0.05 vs. Control. Col, collagen; Sox9, SRY-box
transcription factor 9; RT-qPCR, reverse transcription-quantitative
PCR; miR/miRNA, microRNA. |
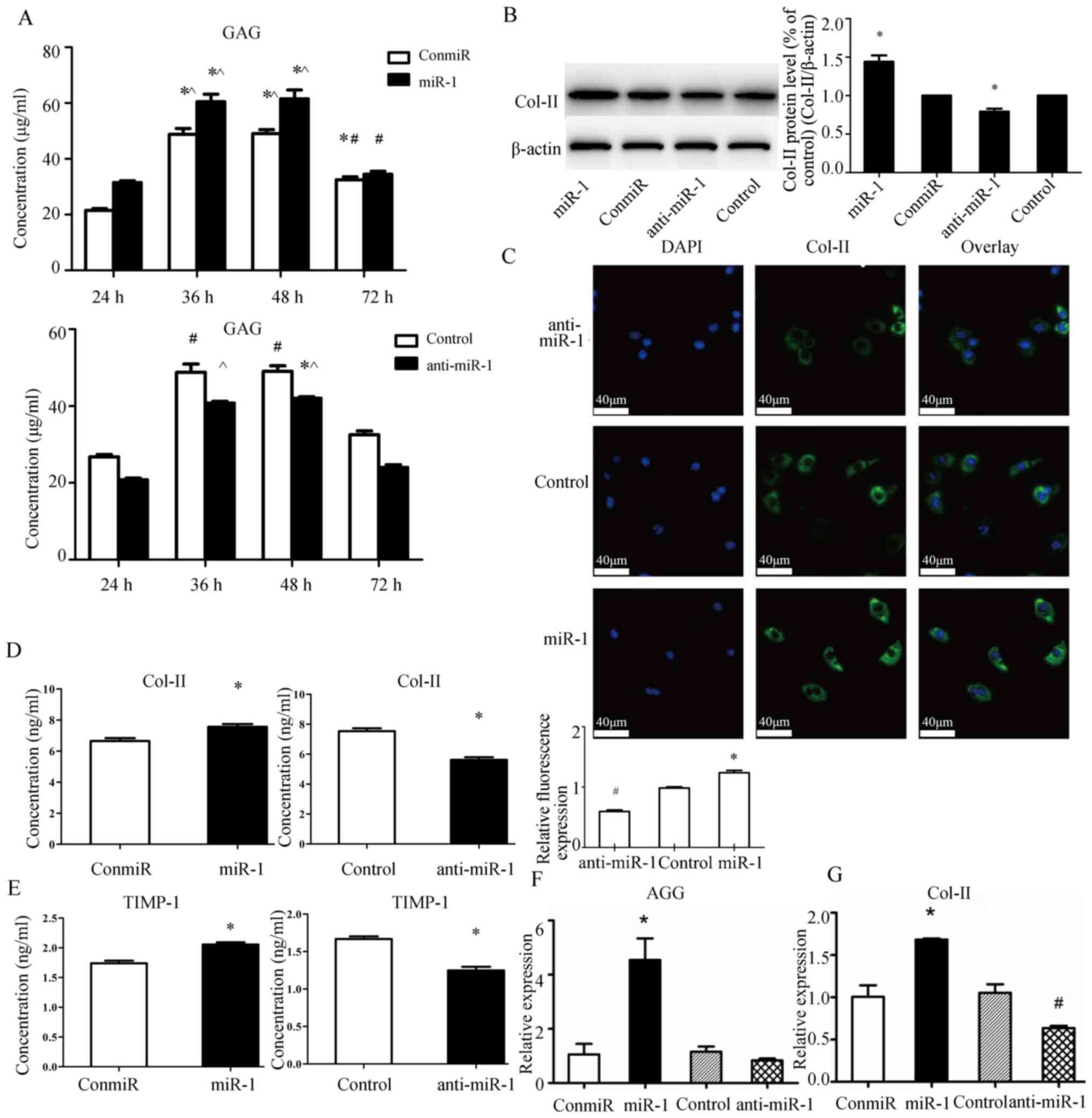
miR-1 increases the expression of
matrix synthesis associated molecules Col-II and AGG in mouse
thorax chondrocytes
Next, the impact of miR-1 expression on matrix
synthesis was examined by measuring GAG, Col-II and TIMP1 protein
levels in cell culture supernatants using ELISA, and Col-II protein
levels using western blotting. Whereas overexpression of miR-1
increased GAG levels in supernatant at 24, 36 and 48 h
post-transfection, suppression of miR-1 by anti-miR-1 significantly
decreased GAG levels (Fig. 2A).
Immunofluorescence staining and western blotting also indicated an
association between miR-1 expression and Col-II protein expression
(Fig. 2B and C). ELISA results
demonstrated that a miR-1 mimic promoted the secretion of Col-II
(Fig. 2D) and TIMP1 (Fig. 2E) proteins from the chondrocytes at
48 h after transfection, whereas transfection of anti-miR-1 reduced
expression levels of these proteins in cell culture supernatants.
Moreover, miR-1 promoted transcription of AGG and Col-II, as the
chondrocytes transfected with the miR-1 mimic had significantly
higher mRNA levels of AGG and Col-II, whereas transfection of
anti-miR-1 reduced AGG and Col-II transcript levels (Fig. 2F and G).
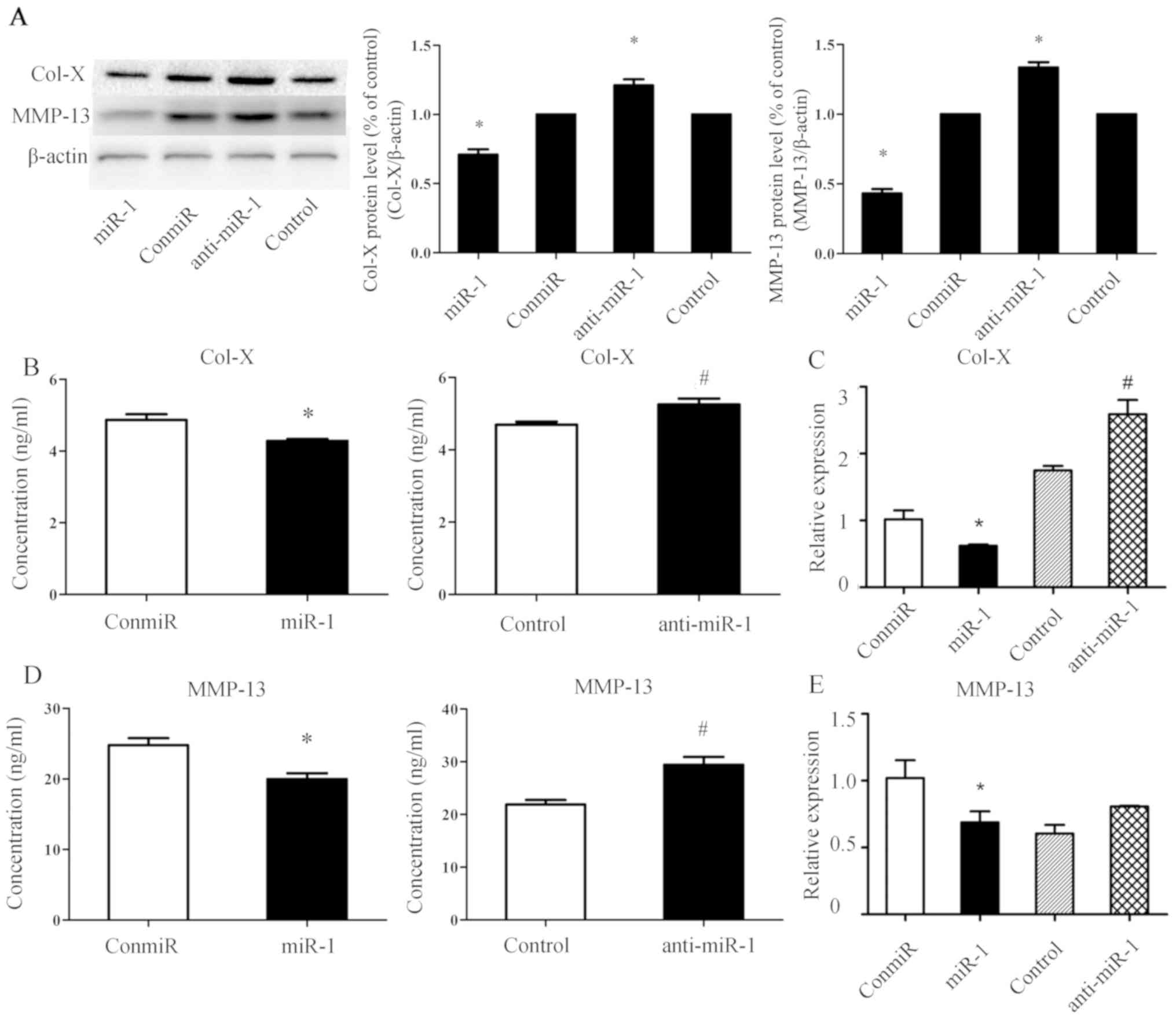
miR-1 downregulates the expression of
chondrocyte differentiation associated molecules Col-X and MMP-13
in mouse thorax chondrocytes
The impact of miR-1 expression on chondrocyte
differentiation was then evaluated by quantifying protein and mRNA
levels of chondrocyte differentiation related molecules, Col-X and
MMP-13. Western blotting and ELISA results indicated that Col-X
(Fig. 3A and B) and MMP-13
(Fig. 3A and D) protein levels
decreased at 48 h after transfection of the miR-1 mimic and
increased after transfection of anti-miR-1. In addition, as
revealed by RT-qPCR assays, mRNA levels of Col-X (Fig. 3B) decreased at 24 h after
transfection of the miR-1 mimic but increased after transfection of
anti-miR-1. Therefore, an association between the expression of
miR-1 and hypertrophy-associated molecules Col-X and MMP-13 was
observed in mouse thorax chondrocytes.
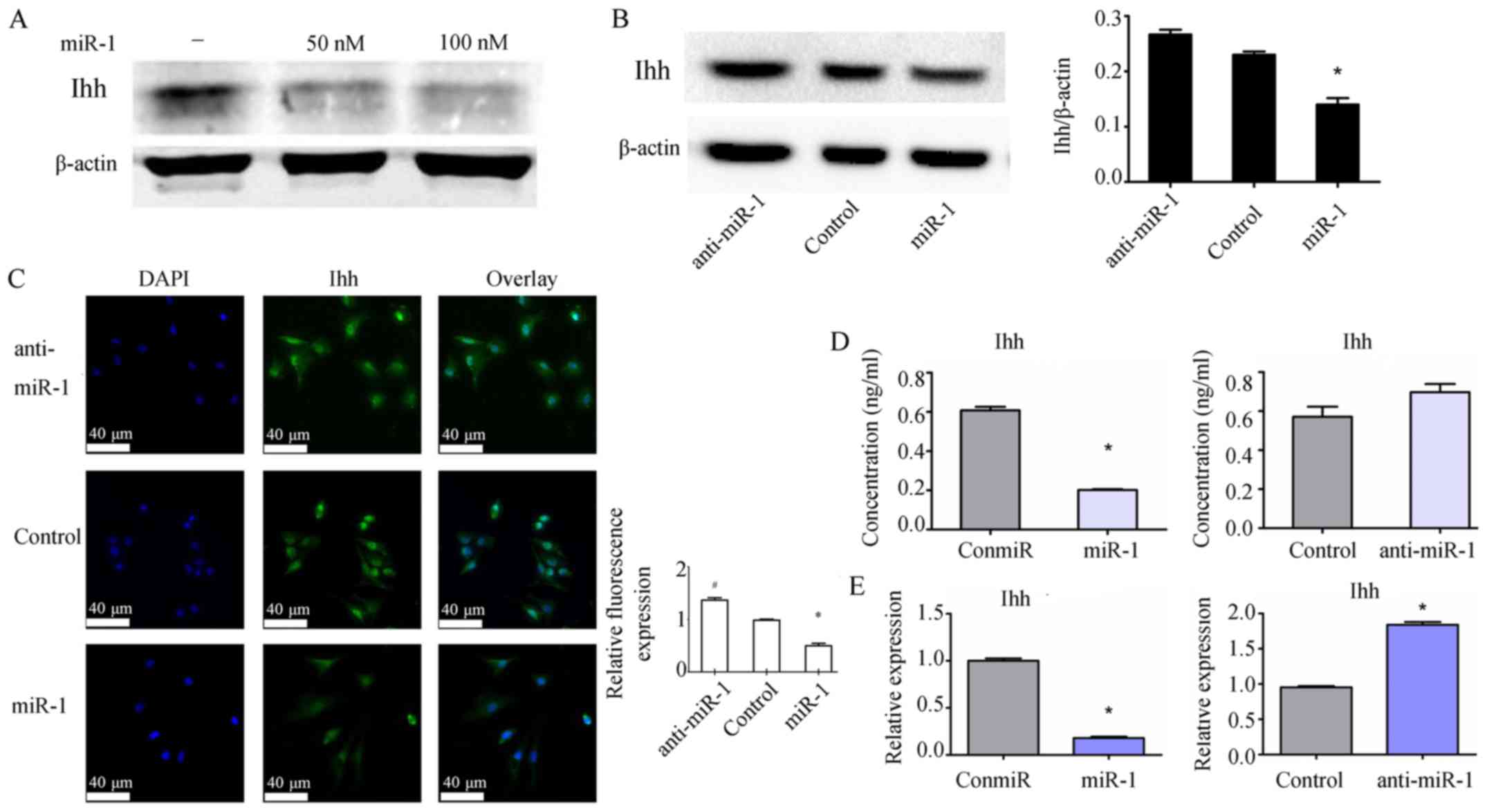
miR-1 suppresses the expression of Ihh
in mouse thorax chondrocytes
To determine the potential interaction between miR-1
function and Hedgehog signaling pathway in chondrocytes, the levels
of Ihh protein and mRNA in mouse chondrocytes was quantified after
transfection of the miR-1 mimic and anti-miR-1. At 48 h after
transfection, the miR-1 mimic inhibited the expression of Ihh
protein in the chondrocytes in a dose-dependent manner (Fig. 4A). Consistently, whereas the miR-1
mimic inhibited the production of Ihh protein, anti-miR-1 increased
Ihh protein expression as revealed by western blotting (Fig. 4B). Immunofluorescence staining also
demonstrated a role of miR-1 in downregulating Ihh expression
(Fig. 4C). Moreover, the protein
levels of Ihh in cell culture supernatant decreased at 48 h after
transfection of the miR-1 mimic (Fig.
4D). Furthermore, similar findings were observed on Ihh mRNA
levels at 24 h after transfection of the miR-1 mimic and anti-miR-1
in mouse thorax chondrocytes (Fig.
4E).
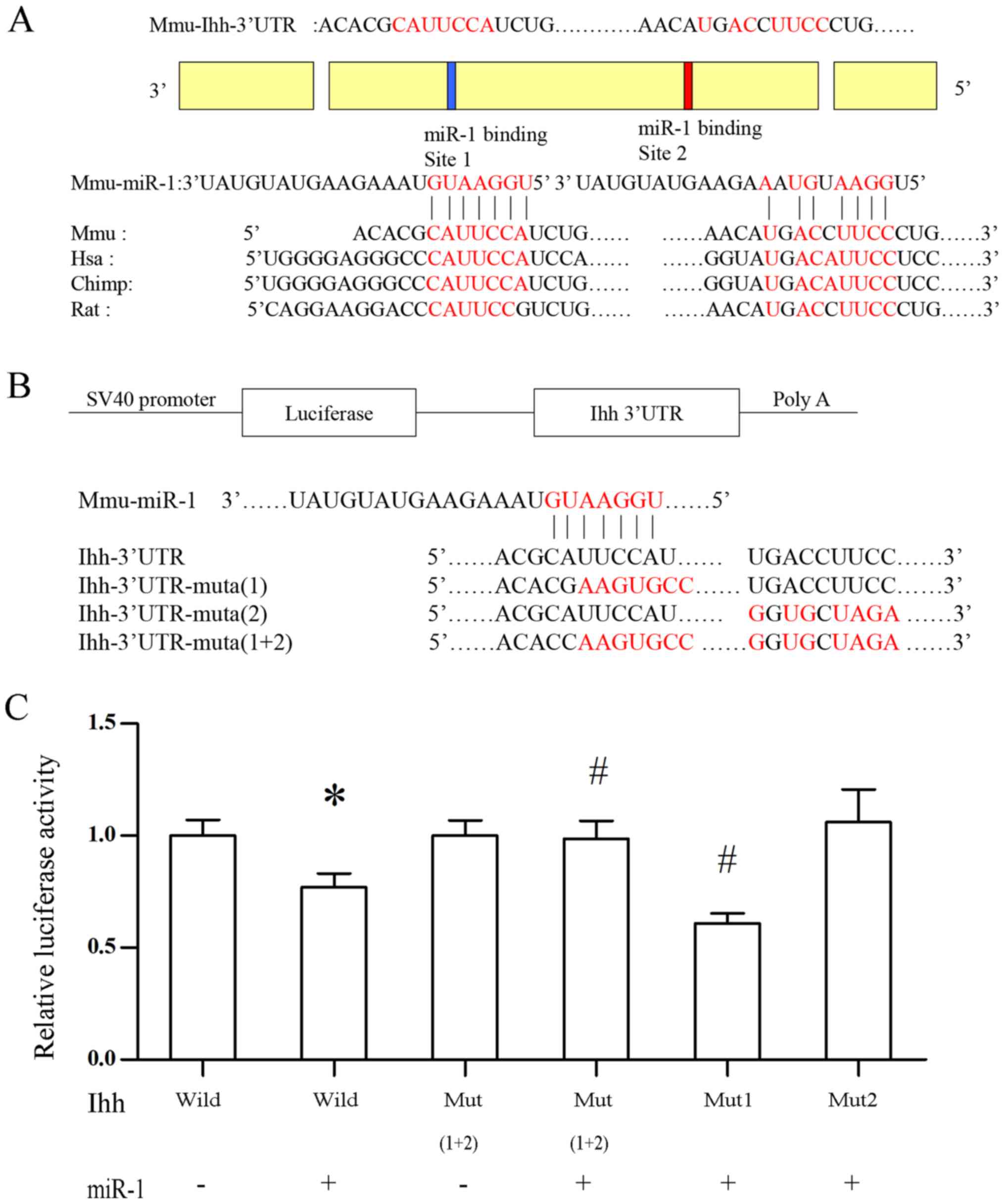
Post-transcriptional regulation of Ihh
by miR-1 is dependent on the miR-1 binding site in the 3′UTR of Ihh
gene
Since miR-1 regulates Ihh mRNA levels, the potential
post-transcriptional inactivation of Ihh by miR-1 was explored.
Sequence analysis indicated that miR-1 is conserved in different
mammalian species, and there are two putative seed regions of miR-1
in the 3′UTR of the Ihh gene (positions: 5′-271-278-3′ and
5′-490-498-3′), and the homologous binding sites in Ihh gene among
human, mouse, chimpanzee and rat were identified (Fig. 5A). To confirm the contribution of
these miR-1 binding sequences in downregulating the Ihh mRNA
levels, vectors were constructed that linked the coding sequence of
the luciferase gene and the 3′UTR of human Ihh gene. In these
vectors, the 3′UTR of human Ihh gene included either two WT miR-1
seed regions, one mutated miR-1 seed region, or two mutated seed
regions. Luciferase reporter assays indicated that a miR-1 mimic
suppressed luciferase activity of the WT Ihh-3′UTR reporter
containing two binding sites, and this suppression was abolished
when expressing the Ihh-3′UTR reporter containing mutations of two
putative miR-1 binding sites (mutated-1 and 2) or a single mutation
of the miR-1 putative binding site 2 (mutated-2; Fig. 5C). These results suggested that the
miR-1 binding site 2 contributes to the suppression of Ihh
expression by miR-1.
miR-1 decreases the expression of Ihh
downstream molecules in the Hedgehog signaling pathway in mouse
thorax chondrocytes
To further validate the post-transcriptional
regulation of miR-1 on the Ihh gene, the impact of altered miR-1
expression on known genes downstream of Ihh in the Hedgehog
signaling pathway was examined in mouse thorax chondrocytes.
Overexpression of miR-1 with the miR-1 mimic decreased the mRNA
levels of Gli-1, Gli-2, Gli-3, Smo and PTHrP, while knockdown of
miR-1 by anti-miR-1 transfection significantly increased the
expression of these Ihh downstream genes, compared with the control
miRNAs (Fig. 6A). To determine
whether the regulation of these genes required Ihh, the mRNA levels
of Gli-1, Gli-2, Gli-3, Smo and PTHrP was quantified in mouse
chondrocytes with knockdown of miR-1 (anti-miR-1) and knockdown of
both miR-1 (anti-miR-1) and Ihh (siIhh; Fig. 6B). Results of RT-qPCR assays
demonstrated that anti-miR-1 significantly increased Gli-1, Gli-2,
Gli-3, Smo and PTHrP mRNAs, and knockdown of Ihh decreased Smo and
Gli-1 expressions (Fig. 6B), but
knockdown of Ihh completely abrogated the effects of anti-miR-1 on
mRNA levels of these genes (Fig.
6C). Moreover, western blotting results showed that the Gli-1
and Smo protein levels were significantly increased in chondrocytes
transfected with anti-miR-1, and Ihh knockdown abolished the effect
of anti-miR-1 (Fig. 6D).
Collectively, these data indicated that miR-1 regulated the
expression of downstream genes in the Hedgehog signaling pathway
via the post-transcriptional inactivation of Ihh.
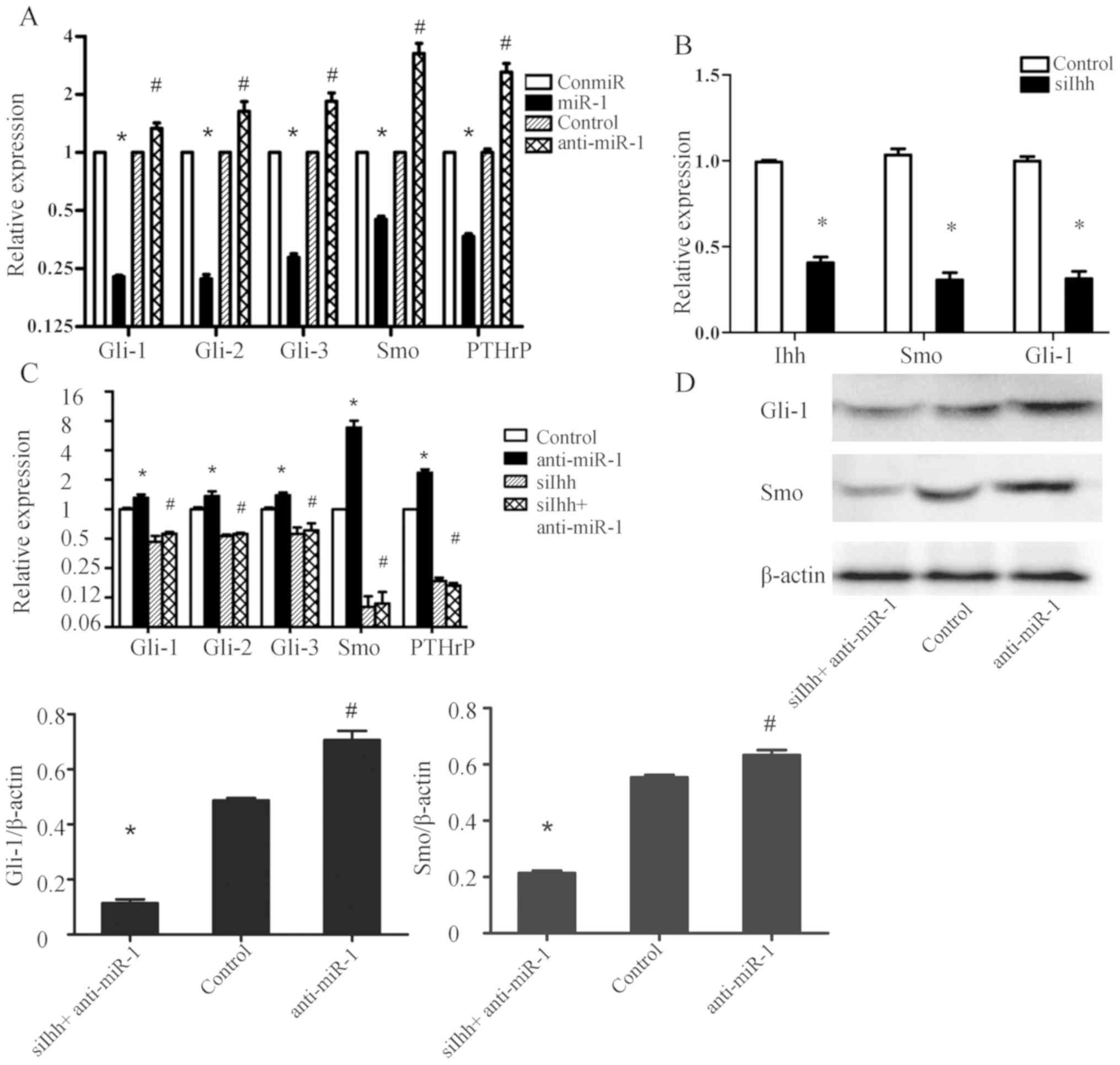 | Figure 6.miR-1 decreases the expression of Ihh
downstream molecules in the Hedgehog signaling pathway in mouse
thorax chondrocytes. (A) Mouse thorax chondrocytes with the
indicated transfection were subjected to RT-qPCR in order to
analyze mRNA levels of Gli-1, Gli-2, Gli-3, Smo and PTHrP genes at
24 h post-transfection. n=6 for each group; *P<0.05 vs. ConmiR
group; #P<0.05 vs. Control group. (B) Knockdown
efficiency of siRNA against mouse Ihh in the chondrocytes and the
impact of Ihh knockdown on the expression of Ihh downstream genes
Smo and Gli-1 were determined by RT-qPCR at 24 h post-siRNA
transfection. n=6 for each group. *P<0.05 vs. Control. (C) Mouse
thorax chondrocytes with indicated treatments were subjected to
RT-qPCR for Gli-1, Gli-2, Gli-3, Smo and PTHrP at 24 h post-siRNA
transfection. Control, cells transfected with the control miRNA;
Anti-miR-1, cells transfected with the miR-1 inhibitor; siIhh,
cells transfected with the mouse Ihh-targeted siRNA oligos; siIhh +
Anti-miR-1, cells simultaneously transfected the mouse Ihh-targeted
siRNA oligos and anti-miR-1. n=6 for each group. *P<0.05 vs.
Control group; #P<0.05 vs. siIhh group. (D) Protein
expression of Gli-1 and Smo were assessed by western blot assays.
The representative images are from one of at least three different
experiments with similar results. The histograms show the
densitometric analysis of Gli-1 and Smo expression, with
normalization to the β-actin levels. n=3 for each group; *P<0.05
vs. Control group; #P<0.05 anti-miR-1 vs. Control
group. miR/miRNA, microRNA; Ihh, Indian hedgehog signaling
molecule; RT-qPCR, reverse transcription-quantitative PCR; Gli, GLI
family zinc finger; Smo, smoothened, frizzled class receptor;
PTHrP, parathyroid hormone-like hormone; siRNA, short interfering
RNA. |
Discussion
The signaling pathways regulating chondrocyte
hypertrophy are not completely understood. Significant changes in
the extracellular matrix have been observed after hypertrophy, such
as downregulation of Col-II, and upregulation of MMP-13 and Col-X
(31). When Ihh expression was
suppressed, the expression of Gli-1, Gli-2, type X collagen and
MMP-13 was decreased and the expression of AGG and type II collagen
was increased (32). However, the
mechanism by which miRNAs are involved in the regulation of
extracellular matrix metabolism during chondrocyte hypertrophy is
not fully understood. miR-1 plays an important role in muscle cell
proliferation and differentiation (15,16).
The effects of miR-1 on the growth and development of growth plate
have been systematically studied by our group (18). In the present study it was shown
that miR-1 enhanced the expression of the matrix
synthesis-associated molecules Col-II, GAG, TIMP1 and AGG, and
inhibited the expression of chondrocyte differentiation markers
Col-X and MMP-13 in mouse thorax chondrocytes. Notably, it was
demonstrated that miR-1 post-transcriptionally suppressed Ihh
expression via the targeting of one of the putative binding sites
in the 3′UTR of the Ihh gene, and regulated Hedgehog signaling
downstream genes.
Our previous results showed that miR-1 was expressed
in the articular cartilage and growth plate in addition to muscle
tissues of various species (18).
Expression levels of miR-1 in different regions of chicken tibia
growth plates were different as revealed by various experimental
methods. In particular, the pre-hypertrophic and hypertrophic zones
had higher miR-1 expression than proliferative zone, which implies
a correlation between miR-1 expression levels and Ihh (18,24).
Additionally, miR-1 has been identified as an miRNA involved in the
regulation of chondrocyte phenotypes during the late stages of
differentiation, and its expression was most repressed upon
hypertrophic differentiation (33). Consistent with those findings, the
current study demonstrated that miR-1 promoted chondrocyte
proliferation. Increased EdU-positive cells and higher OD values in
the CCK-8 assay were observed when miR-1 was overexpressed in
chondrocytes by the miR-1 mimic transfection, whereas knockdown of
miR-1 by inhibitor transfection significantly reduced chondrocyte
proliferation. Moreover, overexpression of miR-1 promoted the mRNA
expression of Sox-9, a specific marker of chondrocyte proliferation
(34), which further substantiates
the proliferative role of miR-1 in mouse primary chondrocytes.
Overexpression of miR-1 increased GAG levels in the supernatant at
24, 36 and 48 h post-transfection, and the reason for a lack of a
significant change of GAG at 72 h upon miR-1 post-transfection may
be due to the transient transfection and expression of miR-1. The
increased expression of Col-II and AGG in mouse thorax chondrocytes
supported the hypothesis that miR-1 promotes matrix synthesis
metabolism.
Ihh is a secretory protein that belongs to the
Hedgehog signaling family. It is mainly expressed in mammalian
pre-hypertrophy chondrocytes, and regulates the growth and
differentiation of growth plate chondrocytes by regulating their
hypertrophy (20,21,24).
Western blotting results demonstrated that overexpression of miR-1
inhibited Ihh protein expression, whereas inhibition of miR-1
displayed the opposite effect. The negative correlation between
miR-1 and Ihh expression is consistent with the notion that miRNAs
control the translation and/or mRNA degradation of target genes
(9). As the IHH-PTHrP axis
participates in the maintenance of articular cartilage and
regulates the proliferation and differentiation of articular
chondrocytes (24), the present
data suggested that inhibition of Ihh and the Hedgehog signaling
pathway contributed to the role of miR-1 in stimulating chondrocyte
proliferation.
It has been demonstrated that miRNAs act via the
3′UTRs of targeted transcripts (9). Sequence analysis indicated that miR-1
is conserved in mammalian species (miRBas accession no.
MIMAT0000416; microrna.sanger.ac.uk/sequences/) and homologous
binding sites in the Ihh gene between multiple species were
identified. Within the Ihh 3′UTR two miR-1 binding sites were
identified. The present data demonstrated that the miR-1 mimic
inhibited luciferase activity that was linked with the
post-transcriptional suppression of miR-1, when the luciferase
reporter plasmid harboring the two WT binding sites of Ihh 3′UTR
was used. Thus, a decrease of luciferase activity by miR-1
expression indicated the post-transcriptional degradation of Ihh
mRNA. The mutagenesis study with single and combined mutations of
the miR-1 binding sites further revealed that the binding region 2
in the 3′UTR of the Ihh mRNA was required for the miR-1
actions.
This report provided evidence that miR-1 is a
critical mediator in regulating chondrocyte proliferation, which is
associated with Ihh suppression via the targeting the 3′UTR of Ihh
mRNA by miR-1. However, the role of miR-1 in regulating Ihh
expression seems to be species-dependent and tissue-dependent. In
primary chicken embryonic chondrocytes, the mRNA levels of Ihh and
Col-X can be significantly increased at 24 h after the transfection
of the miR-1 mimic (18).
Therefore, miR-1 was proposed to induce chondrocyte
differentiation. On the contrary, the present study revealed that
miR-1 downregulated the expression of Ihh and Col-X in cultured
mouse thorax chondrocytes. miR-1 seems to differentially regulate
the expression of AGG (33), the
major cartilaginous proteoglycan gene, in mouse and human or
chicken chondrocytes. Transfection of human chondrocytic HCS-2/8
cells, and chicken normal chondrocytes with miR-1 reduced the
expression of AGG, whereas overexpression of miR-1 in culture mouse
chondrocytes significantly increased AGG mRNA. Therefore, the
specific roles of miR-1 in regulating chondrogenesis under the
context of species, development stages, and tissues need to be
further investigated.
In summary, the present data indicated that miR-1 is
able to regulate cell proliferation and the expression of matrix
synthesis and chondrocyte differentiation associated molecules in
mouse thorax chondrocytes. miR-1 post-transcriptionally suppressed
Ihh expression by binding to the 3′UTR of the Ihh gene, and
inhibiting downstream Hedgehog signaling. These findings may
provide new insights into the molecular mechanisms of miR-1 in
regulating chondrocyte phenotypes and suggest the potential use of
a miR-1 mimic to treat chondrocyte hypertrophy.
Acknowledgements
Not applicable.
Funding
This study was supported by grants from the National
Natural Science Foundation of China (grant no. 81601949),
International Cooperation Projects in Shanxi Province (grant no.
201803D421066), Research Project Supported by Shanxi Scholarship
Council of China (grant no. 2016-118), Supported by Scientific and
Technological Innovation Programs of Higher Education Institutions
in Shanxi (grant no. 20161119), Innovation and Entrepreneurship
Training Program for College Students in Shanxi Province (grant no.
2018185), Scientific and Technological Innovation Programs of
Shanxi Medical University (grant no. 01201509).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
TC, XC, JL and ZH performed the experiments,
contributed to data analysis and wrote the manuscript. PH, CW and
BL analyzed the data. XW, LW and PL conceptualized the study
design, contributed to data analysis and revision of the
manuscript. All authors read and approved the manuscript and agree
to be accountable for all aspects of the research in ensuring that
the accuracy or integrity of any part of the work are appropriately
investigated and resolved.
Ethics approval and consent to
participate
All experiments involving the use of animals in this
study were approved by the Ethics Committee of Shanxi Medical
University (Taiyuan, Shanxi, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bagga S, Bracht J, Hunter S, Massirer K,
Holtz J, Eachus R and Pasquinelli AE: Regulation by let-7 and lin-4
miRNAs results in target mRNA degradation. Cell. 122:553–563. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Eulalio A, Rehwinkel J, Stricker M,
Huntzinger E, Yang SF, Doerks T, Dorner S, Bork P, Boutros M and
Izaurralde E: Target-specific requirements for enhancers of
decapping in miRNA-mediated gene silencing. Genes Dev.
21:2558–2570. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Valencia-Sanchez MA, Liu J, Hannon GJ and
Parker R: Control of translation and mRNA degradation by miRNAs and
siRNAs. Genes Dev. 20:515–524. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Liu X, Xiao J, Zhu H, Wei X, Platt C,
Damilano F, Xiao C, Bezzerides V, Boström P, Che L, et al: miR-222
is necessary for exercise-induced cardiac growth and protects
against pathological cardiac remodeling. Cell Metab. 21:584–595.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Mehta A, Mann M, Zhao JL, Marinov GK,
Majumdar D, Garcia-Flores Y, Du X, Erikci E, Chowdhury K and
Baltimore D: The microRNA-212/132 cluster regulates B cell
development by targeting Sox4. J Exp Med. 212:1679–1692. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Guay C, Kruit JK, Rome S, Menoud V, Mulder
NL, Jurdzinski A, Mancarella F, Sebastiani G, Donda A, Gonzalez BJ,
et al: Lymphocyte-derived exosomal microRNAs promote pancreatic β
cell death and may contribute to Type 1 diabetes development. Cell
Metab. 29:348–361.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Singh PB, Pua HH, Happ HC, Schneider C,
von Moltke J, Locksley RM, Baumjohann D and Ansel KM: MicroRNA
regulation of type 2 innate lymphoid cell homeostasis and function
in allergic inflammation. J Exp Med. 214:3627–3643. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Valenti MT, Dalle Carbonare L and Mottes
M: Role of microRNAs in progenitor cell commitment and osteogenic
differentiation in health and disease (Review). Int J Mol Med.
41:2441–2449. 2018.PubMed/NCBI
|
|
10
|
Wang Y, Liang Y and Lu Q: MicroRNA
epigenetic alterations: Predicting biomarkers and therapeutic
targets in human diseases. Clin Genet. 74:307–315. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goldring MB and Marcu KB: Epigenomic and
microRNA-mediated regulation in cartilage development, homeostasis,
and osteoarthritis. Trends Mol Med. 18:109–118. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang Y, Huang X and Yuan Y: MicroRNA-410
promotes chondrogenic differentiation of human bone marrow
mesenchymal stem cells through down-regulating Wnt3a. Am J Transl
Res. 9:136–145. 2017.PubMed/NCBI
|
|
13
|
Zhao C, Miao Y, Cao Z, Shi J, Li J, Kang
F, Dou C, Xie Z, Xiang Q and Dong S: MicroRNA-29b regulates
hypertrophy of murine mesenchymal stem cells induced toward
chondrogenesis. J Cell Biochem. 120:8742–8753. 2019. View Article : Google Scholar
|
|
14
|
Chen S, Xu Z, Shao J, Fu P and Wu H:
MicroRNA-218 promotes early chondrogenesis of mesenchymal stem
cells and inhibits later chondrocyte maturation. BMC Biotechnol.
19:62019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wüst S, Dröse S, Heidler J, Wittig I,
Klockner I, Franko A, Bonke E, Günther S, Gärtner U, Boettger T, et
al: Metabolic maturation during muscle stem cell differentiation is
achieved by miR-1/133a-mediated inhibition of the Dlk1-Dio3 Mega
gene cluster. Cell Metab. 27:1026–1039.e6. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen JF, Mandel EM, Thomson JM, Wu Q,
Callis TE, Hammond SM, Conlon FL and Wang DZ: The role of
microRNA-1 and microRNA-133 in skeletal muscle proliferation and
differentiation. Nat Genet. 38:228–233. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Zhao Y, Ransom JF, Li A, Vedantham V, von
Drehle M, Muth AN, Tsuchihashi T, McManus MT, Schwartz RJ and
Srivastava D: Dysregulation of cardiogenesis, cardiac conduction,
and cell cycle in mice lacking miRNA-1-2. Cell. 129:303–317. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Li P, Wei X, Guan Y, Chen Q, Zhao T, Sun C
and Wei L: MicroRNA-1 regulates chondrocyte phenotype by repressing
histone deacetylase 4 during growth plate development. FASEB J.
28:3930–3941. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
J Mohler: Requirements for Hedgehog, a
segmental polarity gene, in patterning larval and adult cuticle of
drosophila. Genetics. 120:1061–1072. 1988.PubMed/NCBI
|
|
20
|
Jahan E, Matsumoto A, Rafiq AM, Hashimoto
R, Inoue T, Udagawa J, Sekine J and Otani H: Fetal jaw movement
affects Ihh signaling in mandibular condylar cartilage development:
The possible role of Ihh as mechanotransduction mediator. Arch Oral
Biol. 59:1108–1118. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou J, Wei X and Wei L: Indian Hedgehog,
a critical modulator in osteoarthritis, could be a potential
therapeutic target for attenuating cartilage degeneration disease.
Connect Tissue Res. 55:257–261. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wei F, Zhou J, Wei X, Zhang J, Fleming BC,
Terek R, Pei M, Chen Q, Liu T and Wei L: Activation of Indian
hedgehog promotes chondrocyte hypertrophy and upregulation of
MMP-13 in human osteoarthritic cartilage. Osteoarthritis Cartilage.
20:755–763. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ma RS, Zhou ZL, Luo JW, Zhang H and Hou
JF: The Ihh signal is essential for regulating proliferation and
hypertrophy of cultured chicken chondrocytes. Comp Biochem Physiol
B Biochem Mol Biol. 166:117–122. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen X, Macica CM, Nasiri A and Broadus
AE: Regulation of articular chondrocyte proliferation and
differentiation by indian hedgehog and parathyroid hormone-related
protein in mice. Arthritis Rheum. 58:3788–3797. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Deng A, Zhang H, Hu M, Liu S, Wang Y, Gao
Q and Guo C: The inhibitory roles of Ihh downregulation on
chondrocyte growth and differentiation. Exp Ther Med. 15:789–794.
2018.PubMed/NCBI
|
|
26
|
Kurio N, Saunders C, Bechtold TE, Salhab
I, Nah HD, Sinha S, Billings PC, Pacifici M and Koyama E: Roles of
Ihh signaling in chondroprogenitor function in postnatal condylar
cartilage. Matrix Biol. 67:15–31. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Mirando AJ, Dong Y, Kim J and Hilton MJ:
Isolation and culture of murine primary chondrocytes. Methods Mol
Biol. 1130:267–277. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wei L, Fleming BC, Sun X, Teeple E, Wu W,
Jay GD, Elsaid KA, Luo J, Machan JT and Chen Q: Comparison of
differential biomarkers of osteoarthritis with and without
posttraumatic injury in the Hartley guinea pig model. J Orthop Res.
28:900–906. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Guan Y, Chen Q, Yang X, Haines P, Pei M,
Terek R, Wei X, Zhao T and Wei L: Subcellular relocation of histone
deacetylase 4 regulates growth plate chondrocyte differentiation
through Ca2+/calmodulin-dependent kinase IV. Am J
Physiol Cell Physiol. 303:C33–C40. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Wei L, Kanbe K, Lee M, Wei X, Pei M, Sun
X, Terek R and Chen Q: Stimulation of chondrocyte hypertrophy by
chemokine stromal cell-derived factor 1 in the chondro-osseous
junction during endochondral bone formation. Dev Biol. 341:236–245.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhou J, Chen Q, Lanske B, Fleming BC,
Terek R, Wei X, Zhang G, Wang S, Li K and Wei L: Disrupting the
Indian hedgehog signaling pathway in vivo attenuates surgically
induced osteoarthritis progression in Col2a1-CreERT2; Ihhfl/fl
mice. Arthritis Res Ther. 16:R112014. View
Article : Google Scholar : PubMed/NCBI
|
|
33
|
Sumiyoshi K, Kubota S, Ohgawara T, Kawata
K, Nishida T, Shimo T, Yamashiro T and Takigawa M: Identification
of miR-1 as a micro RNA that supports late-stage differentiation of
growth cartilage cells. Biochem Biophys Res Commun. 402:286–290.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Leung VY, Gao B, Leung KK, Melhado IG,
Wynn SL, Au TY, Dung NW, Lau JY, Mak AC, Chan D, et al: SOX9
governs differentiation stage-specific gene expression in growth
plate chondrocytes via direct concomitant transactivation and
repression. PLoS Genet. 7:e10023562011. View Article : Google Scholar : PubMed/NCBI
|