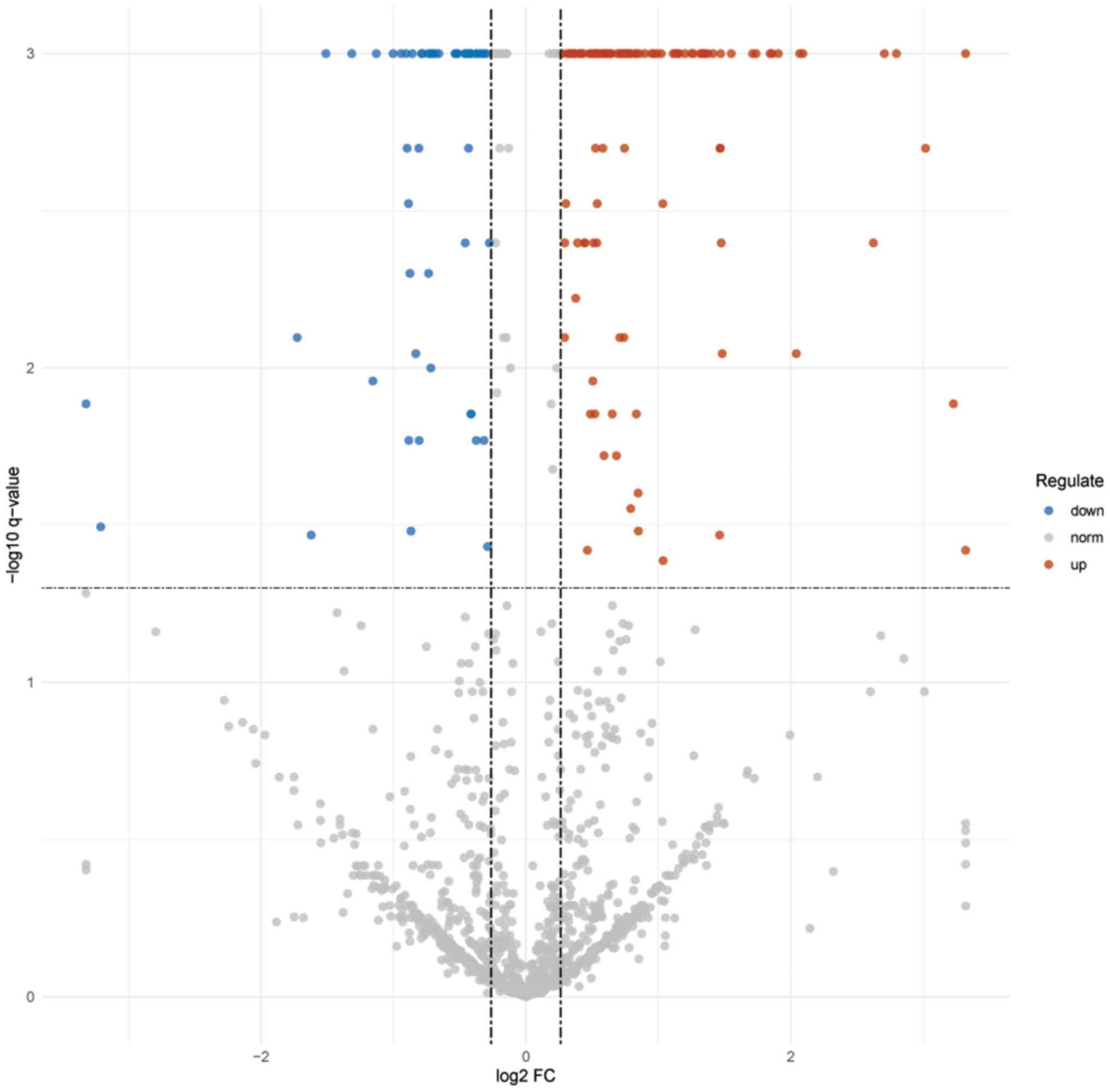
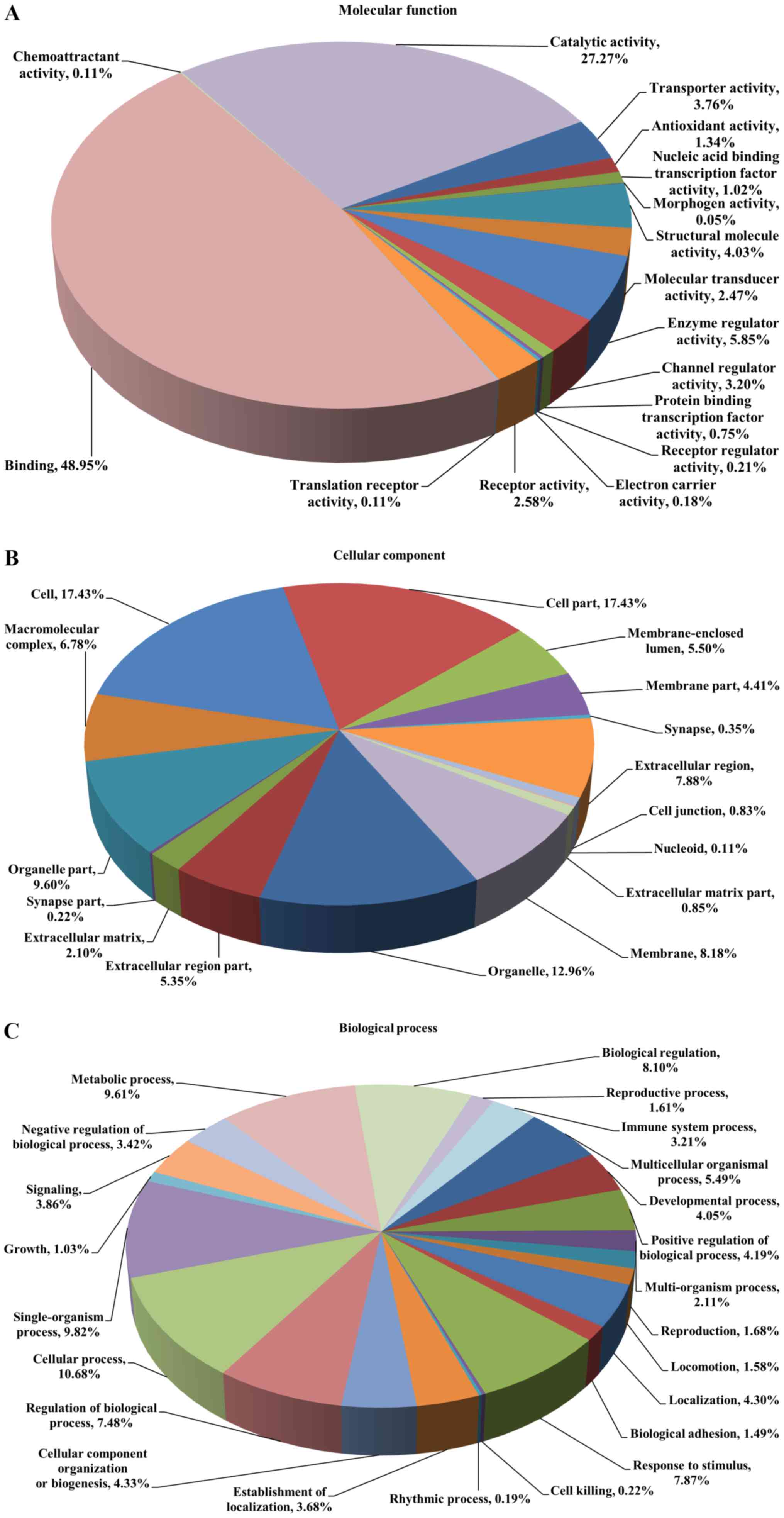
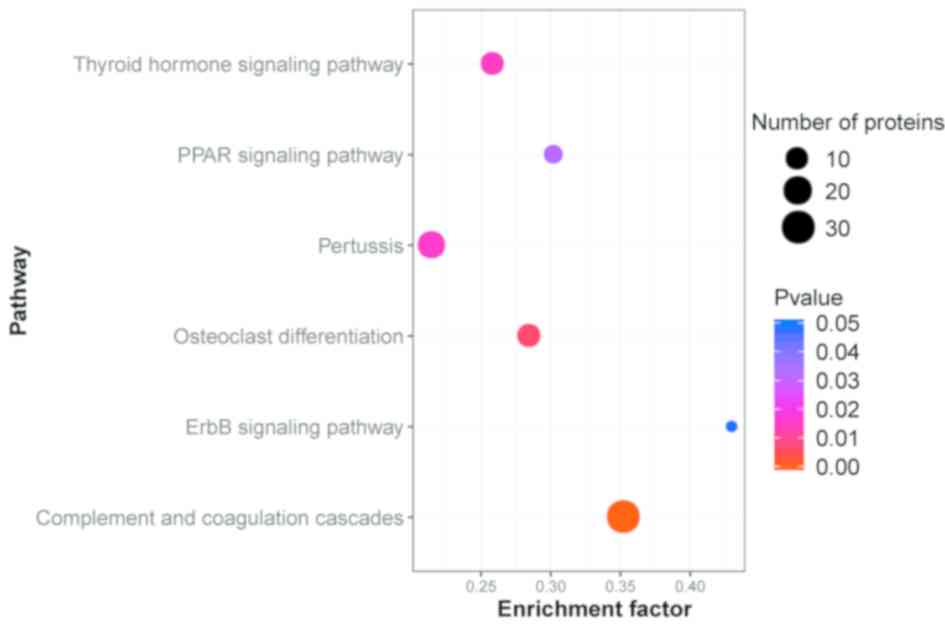
|
1
|
Wolfe RA, Ashby VB, Milford EL, Ojo AO,
Ettenger RE, Agodoa LY, Held PJ and Port FK: Comparison of
mortality in all patients on dialysis, patients on dialysis
awaiting transplantation, and recipients of a first cadaveric
transplant. N Engl J Med. 341:1725–1730. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Hariharan S, Kasiske B, Matas A, Cohen A,
Harmon W and Rabb H: Surrogate markers for long-term renal
allograft survival. Am J Transplant. 4:1179–1183. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Josephson MA: Monitoring and managing
graft health in the kidney transplant recipient. Clin J Am Soc
Nephrol. 6:1774–1780. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Saundh BK, Baker R, Harris M, Welberry
Smith MP, Cherukuri A and Hale A: Early BK polyomavirus (BKV)
reactivation in donor kidney is a risk factor for development of
BKV-associated nephropathy. J Infect Dis. 207:137–141. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sui W, Zhang R, Chen J, He H, Cui Z, Ou M,
Li W, Qi S, Wen J, Lin X and Dai Y: Quantitative proteomic analysis
of Down syndrome in the umbilical cord blood using iTRAQ. Mol Med
Rep. 11:1391–1399. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Tremlett H, Dai DL, Hollander Z, Kapanen
A, Aziz T, Wilson-McManus JE, Tebbutt SJ, Borchers CH, Oger J and
Cohen Freue GV: Serum proteomics in multiple sclerosis disease
progression. J Proteomics. 118:2–11. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Lin XC, Sui WG, Qi SW, Tang DE, Cong S,
Zou GM, Zhang Y, Li H, Chen WB, Cheng ZQ and Dai Y: Quantitative
proteomic profiling of renal tissue in human chronic rejection
biopsy samples after renal transplantation. Transplant Proc.
47:323–331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ross PL, Huang YN, Marchese JN, Williamson
B, Parker K, Hattan S, Khainovski N, Pillai S, Dey S, Daniels S, et
al: Multiplexed protein quantitation in Saccharomyces cerevisiae
using amine-reactive isobaric tagging reagents. Mol Cell
Proteomics. 3:1154–1169. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Choe L, D'Ascenzo M, Relkin NR, Pappin D,
Ross P, Williamson B, Guertin S, Pribil P and Lee KH: 8-plex
quantitation of changes in cerebrospinal fluid protein expression
in subjects undergoing intravenous immunoglobulin treatment for
Alzheimer's disease. Proteomics. 7:3651–3660. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Spanos C and Moore JB: Sample preparation
approaches for iTRAQ labeling and quantitative proteomic analyses
in systems biology. Methods Mol Biol. 1394:15–24. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Desouza LV, Voisin SN and Siu KW:
iTRAQ-labeling for biomarker discovery. Methods Mol Biol.
1002:105–114. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Applied Biosystems iTRAQ™ Reagents
Amine-modifying labeling reagents for multiplexed relative and
absolute protein quantification chemistry reference guide. Applied
Biosystems. Part Number 4351918 Rev. A. 2004.
|
|
13
|
Zhang L, Jiang H, Xu G, Chu N, Xu N, Wen
H, Gu B, Liu J, Mao S, Na R, et al: iTRAQ-based quantitative
proteomic analysis reveals potential early diagnostic markers of
clear-cell Renal cell carcinoma. Biosci Trends. 10:210–219. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wu D, Zhu D, Xu M, Rong R, Tang Q, Wang X
and Zhu T: Analysis of transcriptional factors and regulation
networks in patients with acute renal allograft rejection. J
Proteome Res. 10:175–181. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Freue GV, Sasaki M, Meredith A, Günther
OP, Bergman A, Takhar M, Mui A, Balshaw RF, Ng RT, Opushneva N, et
al: Proteomic signatures in plasma during early acute renal
allograft rejection. Mol Cell Proteomics. 9:1954–1967. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wen B, Zhou R, Feng Q, Wang Q, Wang J and
Liu S: IQuant: An automated pipeline for quantitative proteomics
based upon isobaric tags. Proteomics. 14:2280–2285. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Brosch M, Yu L, Hubbard T and Choudhary J:
Accurate and sensitive peptide identification with mascot
percolator. J Proteome Res. 8:3176–3181. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shen L, Liao L, Chen C, Guo Y, Song D,
Wang Y, Chen Y, Zhang K, Ying M, Li S, et al: Proteomics Analysis
of Blood Serums from Alzheimer's Disease Patients Using iTRAQ
Labeling Technology. J Alzheimers Dis. 56:361–378. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J, Yu L, Huang X, Wang Y and Zhao J:
Comparative proteome analysis of saccular intracranial aneurysms
with iTRAQ quantitative proteomics. J Proteomics. 130:120–128.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Wang X, Zhi Q, Liu S, Xue SL, Shen C, Li
Y, Wu C, Tang Z, Chen W, Song JL, et al: Identifcation of specifc
biomarkers for gastric adenocarcinoma by ITRAQ proteomic approach.
Sci Rep. 6:388712016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Chen J, Ge L, Liu A, Yuan Y, Ye J, Zhong
J, Liu L and Chen X: Identification of pathways related to FAF1/H.
pylori-associated gastric carcinogenesis through an integrated
approach based on iTRAQ quantification and literature review. J
Proteomics. 131:163–176. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Perez JD, Sakata MM, Colucci JA, Spinelli
GA, Felipe CR, Carvalho VM, Cardozo KH, Medina-Pestana JO,
Tedesco-Silva H Jr, Schor N and Casarini DE: Plasma proteomics for
the assessment of acute renal transplant rejection. Life Sci.
158:111–120. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Williams WW, Taheri D, Tolkoff-Rubin N and
Colvin RB: Clinical role of the renal transplant biopsy. Nat Rev
Nephrol. 8:110–121. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Nankivell BJ and Alexander SI: Rejection
of the kidney allograft. N Engl J Med. 363:1451–1462. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Schwarz A, Gwinner W, Hiss M, Radermacher
J, Mengel M and Haller H: Safety and adequacy of renal transplant
protocol biopsies. Am J Transplant. 5:1992–1996. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Muenchhoff J, Poljak A, Song F, Raftery M,
Brodaty H, Duncan M, McEvoy M, Attia J, Schofield PW and Sachdev
PS: Plasma protein profiling of mild cognitive impairment and
Alzheimer's disease across two independent cohorts. J Alzheimers
Dis. 43:1355–1373. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Feng X, Zhang J, Chen WN and Ching CB:
Proteome profiling of Epstein-Barr virus infected nasopharyngeal
carcinoma cell line: Identification of potential biomarkers by
comparative iTRAQ-coupled 2D LC/MS-MS analysis. J Proteomics.
74:567–576. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jing L, Parker CE, Seo D, Hines MW,
Dicheva N, Yu Y, Schwinn D, Ginsburg GS and Chen X: Discovery of
biomarker candidates for coronary artery disease from an APOE-knock
out mouse model using iTRAQ-based multiplex quantitative
proteomics. Proteomics. 11:2763–2776. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ye H, Sun L, Huang X, Zhang P and Zhao X:
A proteomic approach for plasma biomarker discovery with 8-plex
iTRAQ labeling and SCX-LC-MS/MS. Mol Cell Biochem. 343:91–99. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Couttas TA, Raftery MJ, Erce MA and
Wilkins MR: Monitoring cytoplasmic protein complexes with blue
native gel electrophoresis and stable isotope labelling with amino
acids in cell culture: Analysis of changes in the 20S proteasome.
Electrophoresis. 32:1819–1823. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Albaum SP, Hahne H, Otto A, Haußmann U,
Becher D, Poetsch A, Goesmann A and Nattkemper TW: A guide through
the computational analysis of isotope-labeled mass
spectrometry-based quantitative proteomics data: An application
study. Proteome Sci. 9:302011. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jin J, Ku YH and Kim Y and Kim Y, Kim K,
Lee JY, Cho YM, Lee HK, Park KS and Kim Y: Differential proteome
profiling using iTRAQ in microalbuminuric and normoalbuminuric type
2 diabetic patients. Exp Diabetes Res. 2012:1686022012. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Leong S, Nunez AC, Lin MZ, Crossett B,
Christopherson RI and Baxter RC: iTRAQ-based proteomic profiling of
breast cancer cell response to doxorubicin and TRAIL. J Proteome
Res. 11:3561–3572. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Ou M, Zhang X, Dai Y, Gao J, Zhu M, Yang
X, Li Y, Yang T and Ding M: Identification of potential
microRNA-target pairs associated with osteopetrosis by deep
sequencing, iTRAQ proteomics and bioinformatics. Eur J Hum Genet.
22:625–632. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Cernoch M and Viklicky O: Complement in
kidney transplantation. Front Med (Lausanne). 4:662017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Thadhani R, Pascual M and Bonventre JV:
Acute renal failure. N Engl J Med. 334:1448–1460. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Hourcade DE: The role of properdin in the
assembly of the alternative pathway C3 convertases of complement. J
Biol Chem. 281:2128–2132. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lesher AM, Nilsson B and Song WC:
Properdin in complement activation and tissue injury. Mol Immunol.
56:191–198. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Miwa T, Sato S, Gullipalli D, Nangaku M
and Song WC: Blocking properdin, the alternative pathway, and
anaphylatoxin receptors ameliorates renal ischemia-reperfusion
injury in decay-accelerating factor and CD59 doubleknockout mice. J
Immunol. 190:3552–3559. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Han M, Fan L, Qin Z, Lavingia B and
Stastny P: Alleles of keratin 1 in families and populations. Hum
Immunol. 74:1453–1458. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Remotti F, Fetsch JF and Miettinen M:
Keratin 1 expression in endothelia and mesenchymal tumors: An
immunohistochemical analysis of healthy and neoplastic tissues. Hum
Pathol. 32:873–879. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Collard CD, Montalto MC, Reenstra WR,
Buras JA and Stahl GL: Endothelial oxidative stress activates the
lectin complement pathway: Role of cytokeratin 1. Am J Pathol.
159:1045–1054. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Sun Q, Liu Z, Chen J, Chen H, Wen J, Cheng
D and Li L: Circulating anti-endothelial cell antibodies are
associated with poor outcome in renal allograft recipients with
acute rejection. Clin J Am Soc Nephrol. 3:1479–1486. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Sun Q, Cheng Z, Cheng D, Chen J, Ji S, Wen
J, Zheng C and Liu Z: De novo development of circulating
anti-endothelial cell antibodies rather than pre-existing
antibodies is associated with post-transplant allograft rejection.
Kidney Int. 79:655–662. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Sun Q, Liu Z, Yin G, Chen H, Chen J and Li
L: Detectable circulating antiendothelial cell antibodies in renal
allograft recipients with C4d-positive acute rejection: A report of
three cases. Transplantation. 79:1759–1762. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Glotz D, Lucchiari N, Pegaz-Fiornet B and
Suberbielle-Boissel C: Endothelial cells as targets of allograft
rejection. Transplantation. 82 (Suppl 1):S19–S21. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Qin Z, Zou Y, Lavingia B and Stastny P:
Identification of endothelial cell surface antigens encoded by
genes other than HLA. A combined immunoprecipitation and proteomic
approach for the identification of antigens recognized by
antibodies against endothelial cells in transplant recipients. Hum
Immunol. 74:1445–1452. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Guo X, Hu J, Luo W, Luo Q, Guo J, Tian F,
Ming Y and Zou Y: Analysis of sera of recipients with allograft
rejection indicates that keratin 1 is the target of
anti-endothelial antibodies. J Immunol Res. 2017:86798412017.
View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Nordestgaard BG, Chapman MJ, Ray K, Borén
J, Andreotti F, Watts GF, Ginsberg H, Amarenco P, Catapano A,
Descamps OS, et al: Lipoprotein (a) as a cardiovascular risk
factor: Current status. Eur Heart J. 31:2844–2853. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Smolders B, Lemmens R and Thijs V:
Lipoprotein (a) and stroke: A meta-analysis of observational
studies. Stroke. 38:1959–1966. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Shimoyamada H, Fan J, Watanabe T and
Nagata M: Accelerated atherosclerosis with apoLipoprotein (a) and
oxidized low-density lipoprotein deposition in acute rejection of
transplanted kidney analogous to atherosclerosis. Clin Transplant.
16 (Suppl 8):S35–S39. 2002. View Article : Google Scholar
|
|
52
|
White P and Cooke N: The multifunctional
properties and characteristics of vitamin D-binding protein. Trends
Endocrinol Metab. 11:320–327. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Speeckaert M, Huang G, Delanghe JR and
Taes YE: Biological and clinical aspects of the vitamin D binding
protein (Gc-globulin) and its polymorphism. Clin Chim Acta.
372:33–42. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Daiger SP, Schanfield MS and
Cavalli-Sforza LL: Group-specific component (Gc) proteins bind
vitamin D and 25-hydroxyvitamin D. Proc Natl Acad Sci USA.
72:2076–2080. 1975. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Mathieu C and Jafari M: Immunomodulation
by 1,25-dihydroxyvitamin D3: Therapeutic implications in
hemodialysis and renal transplantation. Clin Nephrol. 66:275–283.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Gao Y, Wu K, Xu Y, Zhou H, He W, Zhang W,
Cai L, Lin X, Fang Z, Luo Z, et al: Characterization of acute renal
allograft rejection by human serum proteomic analysis. J Huazhong
Univ Sci Technolog Med Sci. 29:585–591. 2009. View Article : Google Scholar : PubMed/NCBI
|