Introduction
Breast cancer (BC) is currently the most common
cancer type worldwide. For instance, according to the 2015 World
Health Organization World Cancer Report, there are ~14,000,000 new
cases and 8,200,000 BC-related mortalities worldwide (1). Over the last few decades in China,
the incidence of BC has increased and account for 15% of all new
cancer cases in women. The age of onset has decreased and breast
cancer is the most diagnosed cancer between the ages of 30 and 59
years old (2,3). Despite the significant achievements
in BC therapy, as a result of developments in modern technology,
the precise molecular mechanisms underlying the progression of BC
remain unknown (4,5). Therefore, the identification of
biomarkers is required for a more effective diagnosis and treatment
of BC.
A previous study (6) developed a humanized mouse model of
BC, in which healthy human breast tissues were implanted into the
flank of the mice. Human BC cells were then injected into the
implanted human breast tissues, and the results of this study
revealed that human BC cells proliferate well in human breast
tissues and metastasize into distantly implanted human tissues
(6). Moreover, human BC cells
obtained via primary culture from the primary human BC lesion in
the mouse model, demonstrate a more aggressive behavior in
vitro (7). In another previous
study, microarray expression analysis was performed and multiple
differentially expressed genes were screened, including ribosomal
protein (RP) L32 (8). RPL32
encodes an RP that is a component of the 60S subunit. Furthermore,
this protein belongs to the L32E family of RPs and is located in
the cytoplasm.
RPs are components of ribosomes involved in protein
translation and ribosome assembly (9). According to the size of the subunits
they are derive from, RPs are termed large or small subunit RPs.
However, it has been revealed that certain RPs are expressed in
tissue-specific patterns and can differentially contribute to
ribosome composition, affect ribosomal RNA processing and regulate
translation (10). Previous
studies have reported that perturbations of several individual RPs
occur in numerous types of human cancer, including cancer of the
brain, pancreas, bladder and other tissues (11–17).
These studies have rapidly established mutations in RPs as a novel,
underexplored class of oncogenic factors. For example, it has been
shown that the expression of RPL22 is significantly downregulated
at the mRNA and protein level in non-small cell lung cancer
(18). Furthermore, RPL31
modulates prostate cancer cell proliferation via the p53 pathway
(19). RPS15A also promotes
malignant transformation and predicts the outcome of colorectal
cancer via the misregulation of the p53 signaling pathway (20).
In the present study, the expression of RPL32, whose
biological and clinical significance is yet to be elucidated, was
evaluated in human breast tumor tissues, the SUM 1315 human BC line
and an in vivo mouse model.
Materials and methods
Cell culture
MCF-10A human breast epithelial and BT474,
MDA-MB-231 and HCC-1937 human BC cell lines were obtained from the
American Type Culture Collection. The human BC SUM 1315 cell line,
an estrogen receptor-, progestogen- and human epidermal growth
factor receptor 2-negative BC cell line, was provided by Dr Stephen
Ethier (University of Michigan). The cells were cultured in a
humidified atmosphere of 5% CO2 at 37°C and with
complete high glucose DMEM (Wisent Biotechnology), supplemented
with 10% FBS (Wisent Biotechnology), 100 U/ml penicillin and 100
µg/ml streptomycin (Beyotime Institute of Biotechnology).
Transfection of green fluorescent
protein (GFP)
SUM 1315 cells were grown in a 6-well cell culture
cluster. When cells reached ~80% confluence, the culture medium was
removed, and 2 ml plenti-GFP lentivirus (provided by Professor
Beicheng Sun, Nanjing Drum Tower Hospital, the Affiliated Hospital
of Nanjing University Medical School, Jiangsu, China) combined with
12 µl polybrene (Sigma-Aldrich; Merck KGaA) were added. After
incubation for 4 h, 2 ml culture medium was added. After 24 h, the
mixed medium was replaced by the fresh culture medium for further
culturing and passaging.
Tissue microarray and
immunohistochemical staining
The microarray of BC tissue (all from female
patients with BC), which contained 128 samples of infiltrating
ductal carcinoma and six samples of infiltrating ductal carcinoma
with infiltrating lobular carcinoma (HBre-Duc150-Sur-01), was
purchased from Outdo Biotech Co., Ltd. The mean age of patients was
53.3±13.2 years old and the mean tumor size was 3.4±1.8 cm. A total
of 11 patients had tumor Grade I, 47 tumor Grade II and 76 tumor
Grade III (according to 7th Tumor, Node, Metastasis staging system;
American Joint Committee on Cancer). A total of eight patients were
clinical Stage I BC, 80 clinical Stage II and 46 clinical Stage
III. The patients whose samples were included in the microarray
underwent surgery between January 2001 and August 2004 in local
hospitals.
Tissue microarray sections (thickness, 5 µm) were
dewaxed and the endogenous peroxidase was quenched with 3%
H2O2 in methanol for 30 min at room
temperature. Prior to staining, non-specific binding was blocked by
incubation with 10% BSA (Beijing Solarbio Science & Technology
Co., Ltd.) in PBS at 37°C for 1 h. Tissue sections were incubated
with a primary antibody against RPL32 (1:1,000, cat. no. ab229758;
Abcam) in PBS containing 1% BSA at 4°C overnight, followed by
incubation with horseradish peroxidase (HRP)-conjugated anti-rabbit
(1:500) or anti-mouse (1:500) antibodies (cat. nos. BS13278 or
BS12478; Bioworld Technology, Inc.) for 1 h at room temperature.
The color was then developed by incubation with an ImmunoPure Metal
Enhanced Diaminobenzidine Substrate kit (Thermo Fisher Scientific,
Inc.). Following incubation, the tissue sections were washed three
times with PBS for 10 min. The tissue sections were finally
counterstained with 1% hematoxylin for 5 min at room
temperature.
To determine the expression of RPL32, cytosolic
staining of yellowish or brownish granules was graded as follows:
i) 0, background staining; ii) 1, faint staining; iii) 2, moderate
staining; and iv) 3, strong staining. In addition, positive
staining areas in entire tissue sections were graded as follows: i)
0, <5%; ii) 1, 5–25%; iii) 2, 26–50%; iv) 3, 51–75%; and v) 4,
76–100%. Combining these two parameters, 0–2 and ≥3 were defined as
negative and positive staining, respectively. The stained tissue
microarray samples were assessed using an ECLIPSE microscope (Nikon
Corporation) and scored with a semi-quantitative scale by two
independent blinded investigators.
Cell transfection
The commercial lentiviral construct LV3-RPL32-siRNA
vector (RPL32 siRNA; GeneChem, Inc.) was modified to knockdown
RPL32 (target sequence, 5′-CTCACAATGTTTCCTCCAA-3′) in the SUM 1315
human BC lines. The LV3 empty construct (control siRNA) served as
the negative control. SUM 1315 cells stably transfected with the
negative control were termed ‘1315-CON’ and those subjected to
RPL32 knockdown were ‘1315-KD’. Cells were plated in 6-well plates
at 30–40% confluence and infected with the lentivirus, according to
the manufacturer's instructions. Polybrene (5 µg/ml; Sigma-Aldrich;
Merck KGaA) was added to the lentivirus to enhance the infection
efficiency. Stable pooled populations of BC cells were generated
using puromycin (3 µg/ml) for 2 weeks. Then, RPL32 expression was
analyzed using reverse transcription-quantitative (RT-q)PCR and
western blotting.
RT-qPCR
Total RNA was isolated from tissues using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and cDNA was synthesized using PrimeScript RT reagent (Takara
Bio, Inc.), following the manufacturer's instructions. The PCR
amplification conditions used were 94°C for 30 sec, 35 cycles of
94°C for 30 sec, 55°C for 30 sec and 72°C for 1 min, followed by
72°C for 10 min. β-actin (ATCB) was used as the endogenous
reference gene. The following PCR primers were used: ATCB forward,
5′-GCTGTGCTATCCCTGTACGC-3′ and reverse, 5′-TGCCTCAGGGCAGCGGAACC-3′;
and RPL32 forward, 5′-CTGGTCCACAACGTCAAGGAG-3′ and reverse,
5′-CACGATGGCTTTGCGGTTC-3′.
All PCR reactions were performed using the
fluorescent SYBR-Green I methodology. RT-qPCR was performed using a
StepOnePlus RT PCR system (Thermo Fisher Scientific, Inc.) using
FastStart Universal SYBR Green Master (Roche Diagnostics),
according to the manufacturer's instructions. The RT-qPCR
conditions consisted of an initial denaturation step at 95°C for 10
min, followed by 40 cycles of 15 sec at 95°C and 1 min at 60°C. A
melting curve was set at 95°C for 15 sec, 60°C for 15 sec and 95°C
for 15 sec at the end of each run to verify the specificity.
Relative quantification was calculated using the 2−ΔΔCq
method (21) and normalized based
on the expression of ACTB.
Western blotting
Cells were scraped from the plates (cell density
80–90%), rinsed with ice-cold PBS and lysed in cell lysis buffer
containing protease and phosphatase inhibitors to obtain total
protein extracts (cat. no. P0013C; Beyotime Institute of
Biotechnology). The protein concentrations were determined using a
bicinchoninic acid protein assay (Thermo Fisher Scientific, Inc.)
and equal amounts of protein (60 µg) were separated using SDS-PAGE
and transferred to polyvinylidene fluoride membranes. The
percentage of SDS-PAGE for RPL32, matrix metalloproteinase (MMP)-2
and MMP-9 was 12, 10 and 10%, respectively. The membranes were then
blocked with 5% non-fat milk at room temperature for 2 h. Following
which, membranes were washed three times with TBS buffer with 0.1%
Tween-20 and then incubated with the following primary antibodies
overnight at 4°C: Anti-RPL32 (cat. no. ab229758; Abcam), anti-MMP-2
(cat. no. ab92536; Abcam), anti-MMP-9 (cat. no. ab38898; Abcam),
anti-β-actin (cat. no. BS1002; Bioworld Technology, Inc.) and
anti-GAPDH (cat. no. AP0063; Bioworld Technology, Inc.). Then,
membranes were incubated with HRP-conjugated secondary anti-rabbit
(1:2,000, cat. no. BS13278; Bioworld Technology, Inc.) and
anti-mouse antibodies (1:2,000, cat. no. BS12478; Bioworld
Technology, Inc.) for 1 h at room temperature. The antibodies were
diluted according to the manufacturer's instructions.
Immunoreactive proteins were visualized using the SuperSignal West
Femto Maximum Sensitivity Substrate kit (Thermo Fisher Scientific,
Inc.). The signals were detected using FluorChem E System
(ProteinSimple). The density of each band was quantified using
Photoshop CC 2017 release (Adobe Systems, Inc.) and plotted using
GraphPad Prism 5 (GraphPad Software, Inc.). The levels of the
proteins were normalized to those of the corresponding total
protein; the levels of all the other targeted proteins were
normalized to those of GAPDH and β-actin.
Cell Counting Kit (CCK)-8 cell
viability assay
Cell viability was assessed using a CCK-8 (Dojindo
Molecular Technologies, Inc.), according to the manufacturer's
instructions. Cells (3×103/well) were seeded into
96-well plates and cultured overnight. At 0, 24, 48 and 72 h, the
CCK-8 reagent (10 µl) was added to each well and incubated at 37°C
for 1 h. The absorbance was then measured at 450 nm (optical
density value) using an automated microplate reader (Tecan Group,
Ltd.) Each experiment was performed ≥3 times independently.
Cell invasion and migration
assays
Cells that were to be tested for invasion were
collected by trypsinization, washed with PBS, resuspended in
conditioned medium and then added to the upper chamber of
Matrigel-coated (precoating at 37°C for 30 min) invasion filter
inserts (8-µm pore size) at 5×104 cells/well.
Conditioned medium without serum (600 µl) was plated into the lower
compartment of the invasion chamber. The chambers were incubated at
37°C for 24 h in 5% CO2. Following incubation, the
filter inserts were removed from the wells and the cells on the
upper side of the filter were removed mechanically by wiping, using
a cotton swab. The filters were fixed for 10 min with 4%
paraformaldehyde and stained with 1% hematoxylin and eosin for 10
min at room temperature. The cells invading through the Matrigel
were located on the underside of the filter. Images of three random
fields were captured with a fluorescent microscope (LSM710) at
magnification ×100, and the number of cells was counted to
calculate the mean number of cells per field that had
transmigrated.
Cells (5×105) were seeded in 6-well
plates and grown to 90% confluence in 2 ml growth medium. The cells
were carefully wounded using a 5-mm-wide tip and cellular debris
was removed by washing with DMEM. The cells were cultured in DMEM
serum-free medium. Cell migration into the wound area was imaged at
the indicated time points (0 and 48 h) with a fluorescent
microscope (LSM710) at magnification ×100, and analyzed by ImageJ
software (v1.8.0; National Institutes of Health).
Animal model construction
All procedures involving mice and the corresponding
experimental protocols were approved by the Animal Care and Use
Committee of Nanjing Medical University (approval no.
IACUC-1706010). A total of 18 female nude mice (BALB/c Nude; age,
4–5-weeks; body weight ~14–16 g) were purchased from the Model
Animal Research Center of Nanjing University and randomly divided
into two groups (1315-CON and 1315-KD, n=9 mice/group).
Randomization was created using the standard = RAND() function in
Microsoft Excel (Office 2016 edition). Mice were housed in groups
of 4–5. Environmental conditions in the animal lab (Nanjing Medical
University) were a temperature of 21°C, humidity of 55% and under
specific pathogen free conditions. Cages (330×205×130
mm3), bedding and drinking water were autoclaved and
changed regularly. Food was sterilized by irradiation. Food and
water were available ad libitum. The mice were maintained in
a 12-h light/dark cycle. During housing, animals were monitored
daily for health status.
Mice were anesthetized by intraperitoneal injection
with pentobarbital sodium (40 mg/kg body weight; Sigma
Fenstertechnik GmbH & Co.). Each group received a subcutaneous
injection of 5×105 1315-CON or 1315-KD cells into the
mammary fat pad. Tumor size and animal weight were measured every 2
days. Prior to sacrifice, the orthotopic tumor mass of mice was
measured in two dimensions with calipers and the tumor volume was
calculated according to the equation (length × width × width)/2.
After 28 days, all the mice were sacrificed by intraperitoneal
injection with pentobarbital sodium (100 mg/kg body weight). The
mouse death was verified 10 min after injection, for ≥5 min using
the following criteria: i) No breath; ii) no heartbeat; iii) no
nerve reflexes; and iv) muscles relaxed. The primary endpoint were
the incidence of tumor formation and metastases. The criteria for
determining the time of mouse euthanized included: i) Body weight
loss >20%; ii) loss of appetite for 24 h; iii) the food intake
decreased to half; iv) too weak to stand; v) infection; and vi) low
body temperature.
Statistical analysis
All the data for the different experimental groups
are presented as the mean ± SD, and were obtained from ≥3
independent experiments. The statistical analysis was performed
using SPSS version 19.0 (IBM Corp.). The gray values of protein
bands on the western blots (mean ± SD) were normalized to those of
GADPH or b-actin, and compared using an unpaired Student's t-test.
All in vitro experiments were performed in triplicate.
Differences between each group in vitro were analyzed using
Student's t-tests and one-way ANOVA followed by Dunnett's post hoc
test. P<0.05 was considered to indicate a statistically
significant difference.
Results
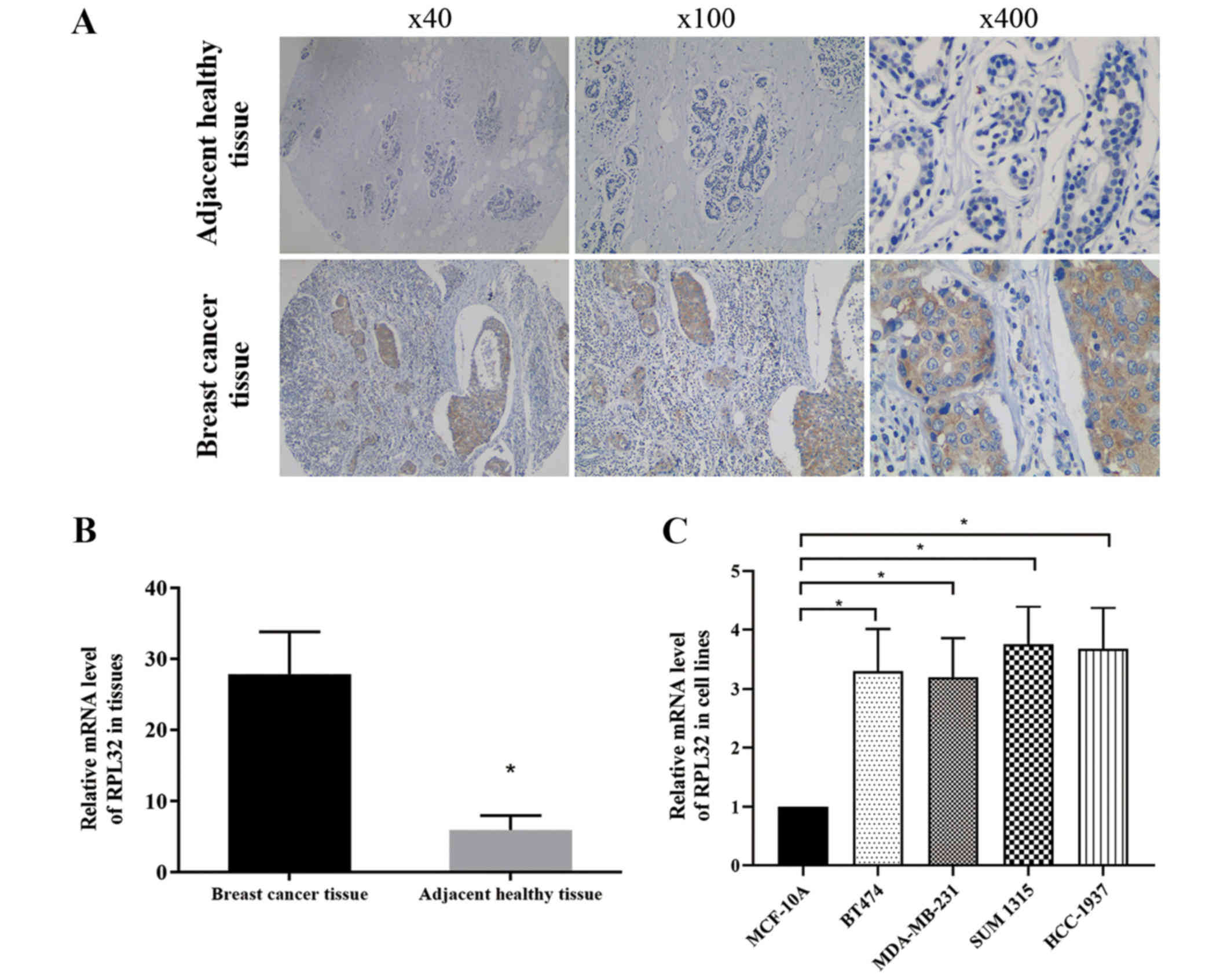
RPL32 is upregulated in BC
Immunohistochemical staining was used to assess the
RPL32 expression in 134 pairs of BC and matched adjacent healthy
tissues. The results demonstrated strong RPL32 staining in most BC
specimens and a weak staining in matched healthy tissues (Fig. 1A). RT-qPCR was then used to detect
the relative mRNA expression of RPL32 in BC and matched adjacent
healthy tissues, and the results were consistent with those of the
immunohistochemical assay (Fig.
1B).
RT-qPCR was also used to compare the RPL32 gene
expression among breast epithelial and BC cell lines. It was
identified that RPL32 expression significantly higher in BC cell
lines compared with the normal breast epithelial cell line
(Fig. 1C).
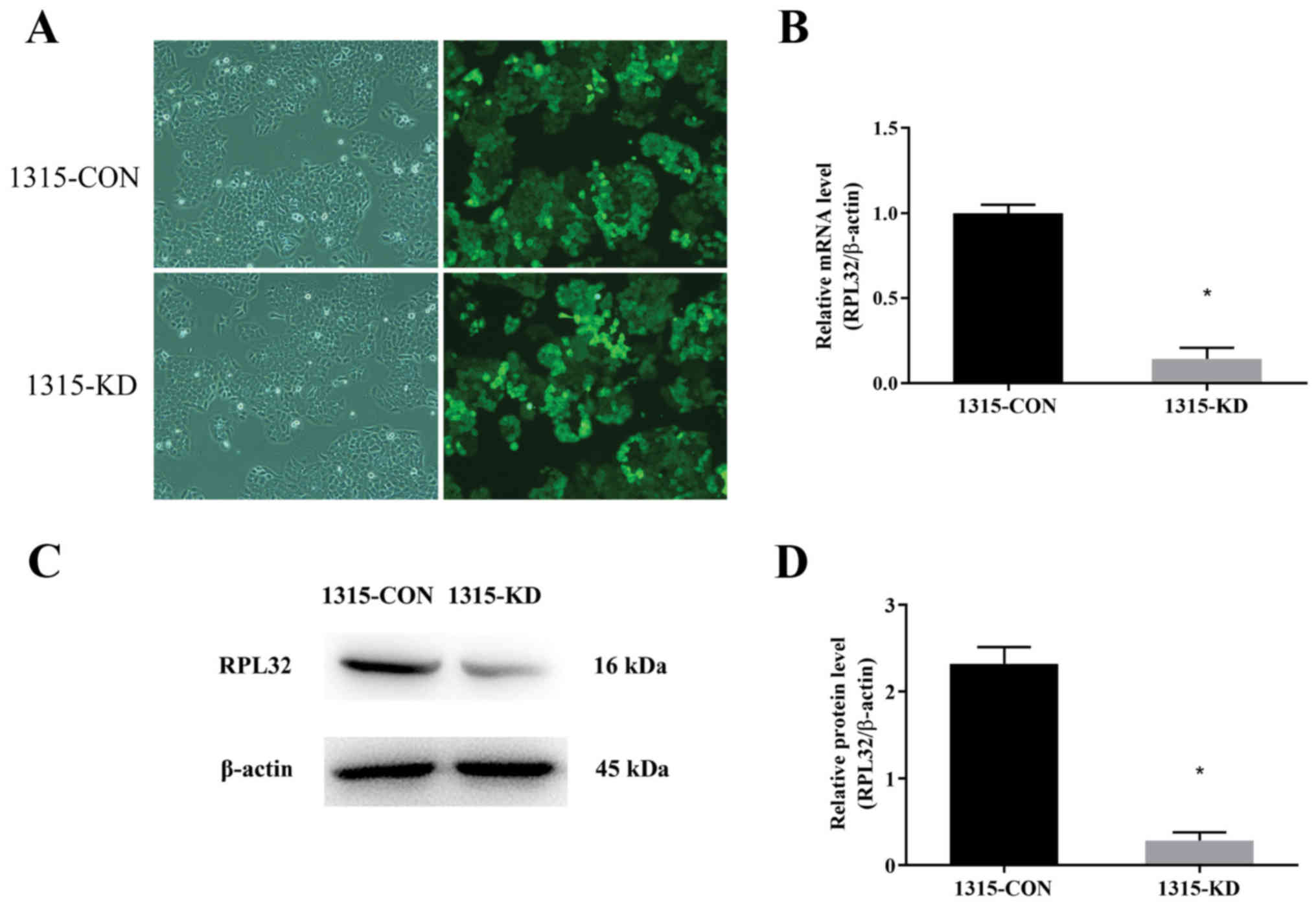
Lentivirus-delivered siRNA silencing
specifically inhibits the expression of RPL32 at the mRNA and
protein levels
To study the role of RPL32 in BC,
lentivirus-delivered siRNA targeting RPL32 was used in SUM 1315
cells. RPL32 and control siRNAs were constructed. The infection
efficiency of the lentivirus was assessed by detecting the GFP
expression 3 days after transduction. As presented in Fig. 2A, >90% of the cells were
infected. In addition, RT-qPCR and western blotting were used to
analyze the knockdown efficiency of RPL32 in SUM 1315 cells. It was
demonstrated that RPL32 mRNA expression was reduced by ~90%
following RPL32 silencing compared with the 1315-CON group
(Fig. 2B). The RPL32 protein
expression was also significantly decreased by siRNA treatment
(Fig. 2C and D); the expression of
RPL32 in the 1315-KD group was ~45% of that in the 1315-CON group.
These results indicated that lentivirus-delivered siRNA silencing
specifically inhibited the expression of RPL32 at the mRNA and
protein levels.
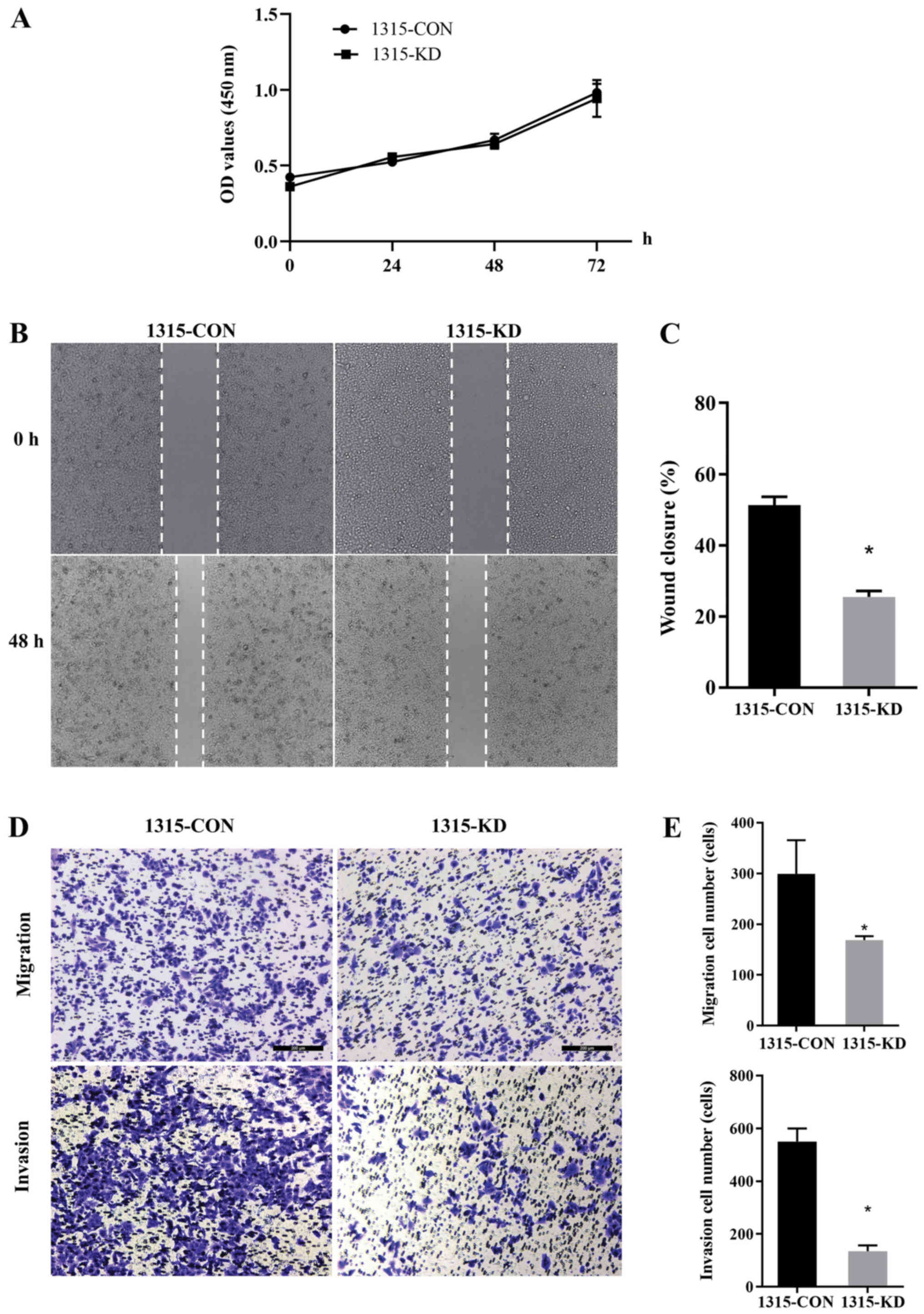
Knockdown of RPL32 inhibits migration
and invasion of SUM 1315 cells
A CCK-8 assay was performed and 1315-KD cells did
not show any difference in proliferation compared with 1315-CON
cells. This indicated that RPL32 had no impact on in vitro
cell proliferation (Fig. 3A). To
assess whether RPL32 silencing influenced cell migration and
invasion, wound-healing and Transwell assays were performed using
SUM 1315 cells. 1315-KD cells exhibited a delayed wound closure of
scratch gaps after 24 h (Fig. 3B and
C). Transwell assays demonstrated a reduced invasive ability of
RPL32-silenced SUM 1315 cells. Moreover, fewer 1315-KD cells
invaded the filter, with or without Matrigel, compared with
1315-CON cells (Fig. 3D and E).
These data suggested that RPL32 knockdown inhibited the migration
and invasion of SUM 1315 cells.
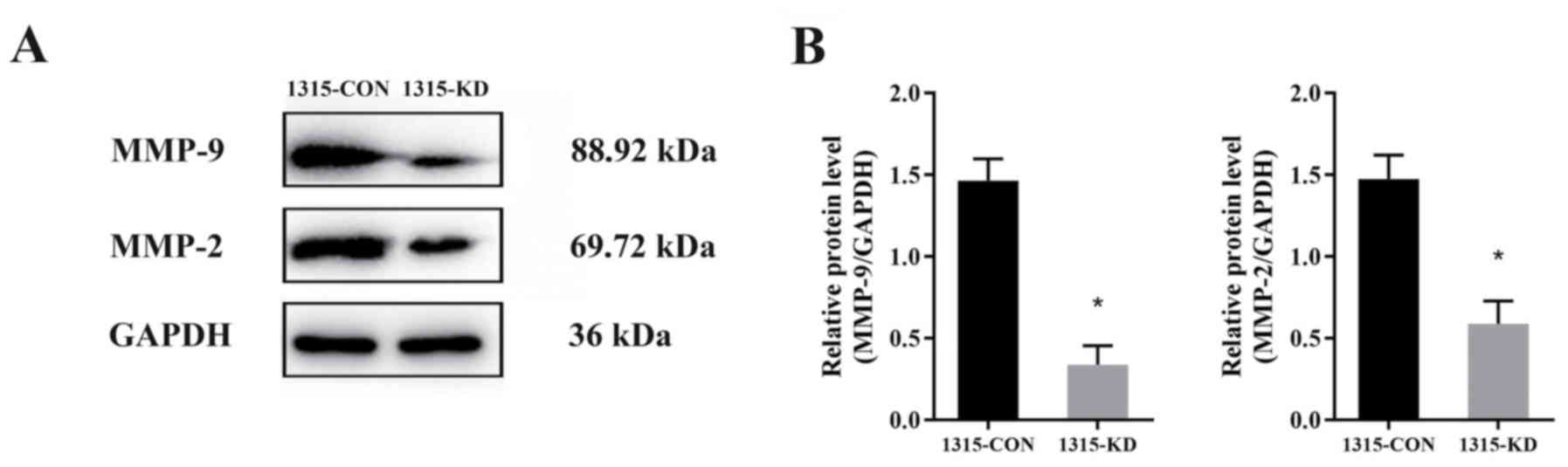
Knockdown of RPL32 decreases the
expression levels of MMP-2 and MMP-9
To investigate the molecular mechanisms via which
RPL32 silencing inhibits SUM 1315 cell migration and invasion,
western blot analysis was performed to detect the expression levels
of MMP-2 and MMP-9 (Fig. 4A). The
data identified that the expression levels of MMP-9 and MMP-2 was
decreased by ~30 and 40%, respectively (Fig. 4B). Thus, it was indicated that
RPL32 may promote SUM 1315 cell migration and invasion via the MMP
signaling pathway.
RPL32 knockdown suppresses SUM 1315
cell metastasis in vivo
To examine the in vitro observations in
vivo, BC mouse xenografts were generated using 1315-CON and
1315-KD cells. All mice were healthy prior to tumor cell injection.
Then, 28 days after cell injection, all xenograft tumors were
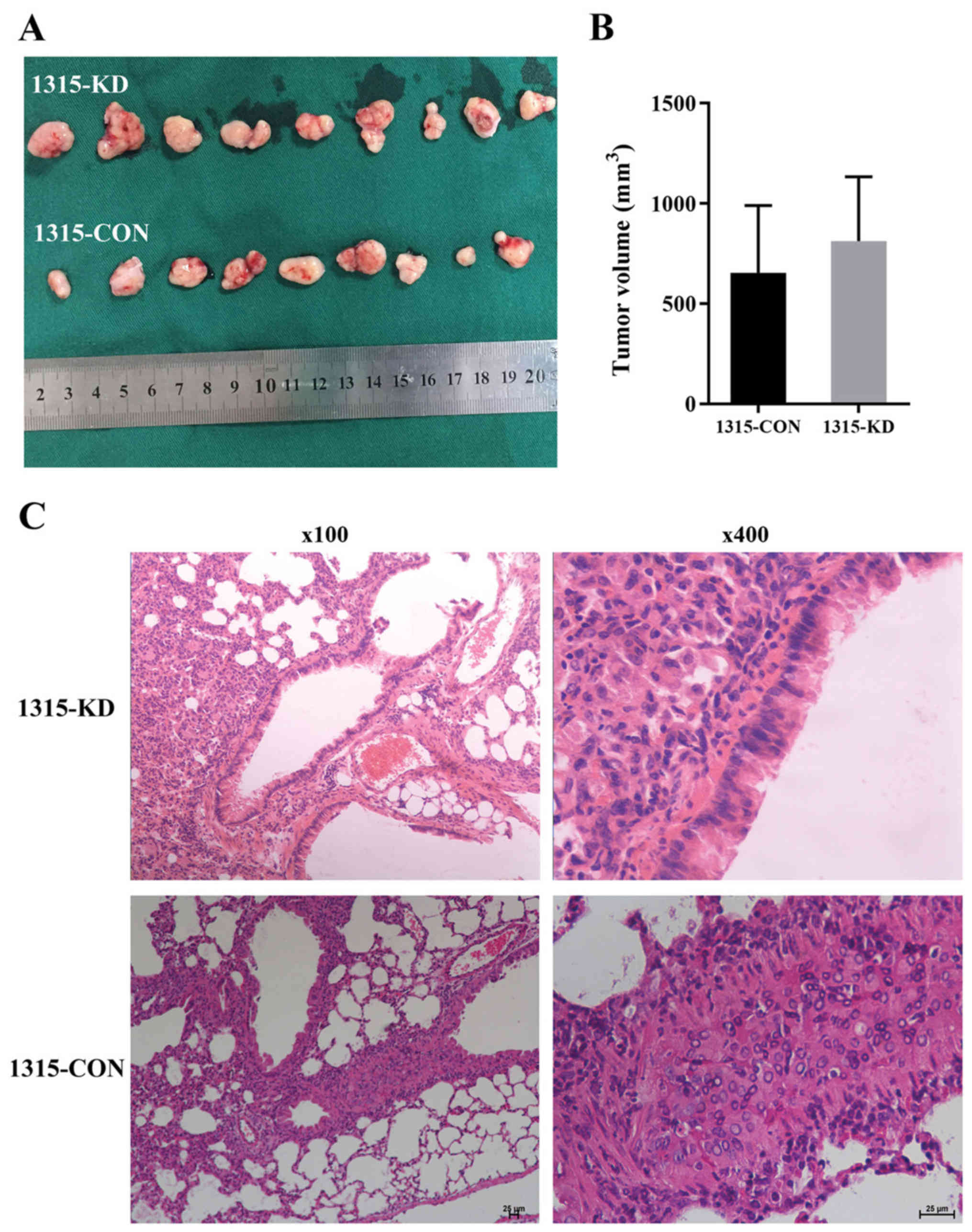
measured and resected (Fig. 5A).
The results demonstrated that the maximum tumor diameter was 14.9
mm and the weight was 1.7 g (9.1% body weight). The maximum
percentage of weight loss in the study was 18.5%. There was no
significant difference in tumor volume between the 1315-CON and
1315-KD groups (Fig. 5B).
To evaluate the effect of RPL32 on BC tumor
metastasis in vivo, lung and liver tissues were removed from
all xenograft mice. Histological analysis of the lungs identified
less metastases in the 1315-KD compared with the 1315-CON group. It
was found that 5/9 (55.56%) mice developed lung metastasis in the
1315-CON group, while no (0%) mice developed lung metastasis in the
1315-KD group (Fig. 5C).
Furthermore, no liver metastasis was detected in either group (data
not shown). Collectively, these data suggested that RPL32 silencing
could effectively suppress BC metastasis in vivo.
Discussion
Thus far, several RPs have been reported to be
associated with certain types of cancer, including cancer of the
brain, pancreas and bladder (11–17).
However, to the best of our knowledge, few studies have focused on
RPs in BC. Previous studies have revealed the oncogenic activity of
RPs in BC. For instance, it has been shown that the oncogenic role
of RPL39 may be mediated via inducible nitric oxide synthase
(22). Another study reported that
the overexpression of RPL19 could sensitize BC cells to endoplasmic
reticulum stress-induced cell death by activating the unfolded
protein response (23). Moreover,
a DNA microarray experiment demonstrated that the expression of
RPL32, a 60S RP, was correlated with non-small cell lung and
prostate cancer types (24,25).
However, the role of RPL32 in BC remains to be elucidated.
In the present study, it was found that the RPL32
expression was increased in BC tissues compared with healthy
tissues. In addition, RPL32 expression was higher in BC cell lines
compared with normal breast epithelial cell line. These results
indicated that RPL32 may be associated with BC. Thus, it was
hypothesized that RPL32 may serve an important role in BC
tumorigenesis.
On the basis of the present findings, a lentivirus
carrying an RPL32 siRNA was used to silence the expression of
RPL32. Wound-healing assays demonstrated that RPL32-silenced cells
exhibited a delayed wound closure of scratch gaps compared with the
control cells. In addition, RPL32 silencing significantly reduced
the invasion ability of the cells. These results suggested an
oncogenic effect of RPL32 on cell migration and invasion in BC.
However, no difference in cell proliferation was identified between
the groups.
To further examine the underlying mechanism of the
RPL32-mediated promotion of cell migration and invasion, western
blot analysis was performed to detect MMPs that are associated with
cancer cell migration and invasion. Moreover, compared with control
cells, the expression levels of MMP-2 and MMP-9 in RPL32-silenced
cells were significantly decreased. Thus, these results indicated
that RPL32 may decrease BC cell migration and invasion by
downregulating MMP expression.
To assess the results in vivo, a mouse BC
model was constructed. The estimated effect size and level of
variability were used to determine the required sample size for
future studies. Considering this fact, as well as with the desire
to reduce the use of animals, 18 mice were used. 1315-CON and
1315-KD cells were injected into the mammary fat pad of
4–5-week-old nude mice. Then, 28 days later, the tumors were
resected and lung and liver tissues were collected. The primary
breast tumor volumes were compared between mouse models. No
difference was observed between the 1315-CON and 1315-KD mouse
groups. This in vivo result was consistent with the previous
in vitro result, which indicated that RPL32 was not
associated with tumor proliferation. However, histological analysis
demonstrated that cancer cells could be found in lung tissues from
the 1315-CON group. In contrast, no cancer cells were identified in
lung tissues from the 1315-KD group and no cancer cells were found
in liver tissues from both groups. These results indicated that
RPL32 silencing could significantly reduce cancer cell metastasis,
thus suggesting that RPL32 may be a novel molecular diagnostic
target for BC. However, the key molecular mechanism via which RPL32
affects the occurrence and development of BC remains to be
elucidated. Moreover, it should be noted that the present study has
some limitations. First, only one breast cancer cell line was
tested and data of RPL32 upregulation was lacking. Second, the
underlying mechanism still needs to be further investigated. In
addition, the clinical application of RPL32 marker requires a
clinical study.
To the best of our knowledge, the present study was
the first to demonstrate the oncogenic function of RPL32 in BC. The
molecular mechanism underlying the effect of RPL32 on cell
migration and invasion in BC was also preliminarily evaluated.
Collectively, it was concluded that PRL32 was a potential target
for molecular targeted treatment of BC.
Acknowledgements
Not applicable.
Funding
This research was supported in part by the National
Natural Science Foundation of China (grant no. 81302305), Natural
Science Foundation of Jiangsu Province (grant no. BK20131027),
Jiangsu Province Six Talents Summit Project (grant no. WSW-001),
Youth Talent Project (grant no. FRC201308) and a project funded by
the Priority Academic Program Development of Jiangsu Higher
Education Institutions (grant no. PAPD 0271).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JW, SW and LW participated in the study design. LX,
CJ, QZ and RC contributed to data collection and analysis. All
authors were involved in the writing of the article. JW critically
reviewed the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All participants provided written informed consent
prior to participating in the study. The Institutional Animal Care
and Use Committee of Nanjing Medical University approved all the
tests conducted in this paper (approval no. IACUC-1706010) and
animal procedures were conducted in accordance with the ethical
standards in the 1964 Helsinki Declaration. This study was approved
by the Ethics and Research Committee of The First Affiliated
Hospital of Nanjing Medical University (approval no.
2013-SRFA-108).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
McGuire S: World cancer report 2014.
Geneva, Switzerland: World Health Organization, International
Agency for Research on cancer, WHO Press, 2015. Adv Nutr.
7:418–419. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Song QK, Li J, Huang R, Fan JH, Zheng RS,
Zhang BN, Zhang B, Tang ZH, Xie XM, Yang HJ, et al: Age of
diagnosis of breast cancer in China: Almost 10 years earlier than
in the United States and the European Union. Asian Pac J Cancer
Prev. 15:10021–10025. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Song QK, Wang XL, Zhou XN, Yang HB, Li YC,
Wu JP, Ren J and Lyerly HK: Breast cancer challenges and screening
in China: Lessons from current registry data and population
screening studies. Oncologist. 20:773–779. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Gray J and Druker B: Genomics: The breast
cancer landscape. Nature. 486:328–329. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ban KA and Godellas CV: Epidemiology of
breast cancer. Surg Oncol Clin N Am. 23:409–422. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Wang J, Xia TS, Liu XA, Ding Q, Du Q, Yin
H and Wang S: A novel orthotopic and metastatic mouse model of
breast cancer in human mammary microenvironment. Breast Cancer Res
Treat. 120:337–344. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zheng MJ, Wang J, Xu L, Zha XM, Zhao Y,
Ling LJ and Wang S: Human mammary microenvironment better regulates
the biology of human breast cancer in humanized mouse model. Med
Oncol. 32:4272015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zheng M, Wang J, Ling L, Xue D, Wang S and
Zhao Y: Screening and analysis of breast cancer genes regulated by
the human mammary microenvironment in a humanized mouse model.
Oncol Lett. 12:5261–5268. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Fatica A and Tollervey D: Making
ribosomes. Curr Opin Cell Biol. 14:313–318. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Xue S and Barna M: Specialized ribosomes:
A new frontier in gene regulation and organismal biology. Nat Rev
Mol Cell Biol. 13:355–369. 2012. View
Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yong WH, Shabihkhani M, Telesca D, Yang S,
Tso JL, Menjivar JC, Wei B, Lucey GM, Mareninov S, Chen Z, et al:
Ribosomal proteins RPS11 and RPS20, two stress-response markers of
glioblastoma stem cells, are novel predictors of poor prognosis in
glioblastoma patients. PLoS One. 10:e01413342015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Shi C, Wang Y, Guo Y, Chen Y and Liu N:
Cooperative down-regulation of ribosomal protein L10 and NF-κB
signaling pathway is responsible for the anti-proliferative effects
by DMAPT in pancreatic cancer cells. Oncotarget. 8:35009–35018.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Paquet ÉR, Hovington H, Brisson H, Lacombe
C, Larue H, Têtu B, Lacombe L, Fradet Y and Lebel M: Low level of
the X-linked ribosomal protein S4 in human urothelial carcinomas is
associated with a poor prognosis. Biomark Med. 9:187–197. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Jung Y, Lee S, Choi HS, Kim SN, Lee E,
Shin Y, Seo J, Kim B, Jung Y, Kim WK, et al: Clinical validation of
colorectal cancer biomarkers identified from bioinformatics
analysis of public expression data. Clin Cancer Res. 17:700–709.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Russo A, Saide A, Smaldone S, Faraonio R
and Russo G: Role of uL3 in multidrug resistance in p53-mutated
lung cancer cells. Int J Mol Sci. 18:5472017. View Article : Google Scholar
|
|
16
|
Fan H, Li J, Jia Y, Wu J, Yuan L, Li M,
Wei J and Xu B: Silencing of ribosomal protein L34 (RPL34) inhibits
the proliferation and invasion of esophageal cancer cells. Oncol
Res. 25:1061–1068. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sim EU, Chan SL, Ng KL, Lee CW and
Narayanan K: Human ribosomal proteins RPeL27, RPeL43, and RPeL41
are upregulated in nasopharyngeal carcinoma cell lines. Dis
Markers. 2016:51795942016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yang M, Sun H, Wang H, Zhang S, Yu X and
Zhang L: Down-regulation of ribosomal protein L22 in non-small cell
lung cancer. Med Oncol. 30:6462013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Maruyama Y, Miyazaki T, Ikeda K, Okumura
T, Sato W, Horie-Inoue K, Okamoto K, Takeda S and Inoue S: Short
hairpin RNA library-based functional screening identified ribosomal
protein L31 that modulates prostate cancer cell growth via p53
pathway. PLoS One. 9:e1087432014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen J, Wei Y, Feng Q, Ren L, He G, Chang
W, Zhu D, Yi T, Lin Q, Tang W, et al: Ribosomal protein S15A
promotes malignant transformation and predicts poor outcome in
colorectal cancer through misregulation of p53 signaling pathway.
Int J Oncol. 48:1628–1638. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Bisaccia F, De Palma A and Palmieri F:
Identification and purification of the tricarboxylate carrier from
rat liver mitochondria. Biochim Biophys Acta. 977:171–176. 1989.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hong M, Kim H and Kim I: Ribosomal protein
L19 overexpression activates the unfolded protein response and
sensitizes MCF7 breast cancer cells to endoplasmic reticulum
stress-induced cell death. Biochem Biophys Res Commun. 450:673–678.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Dmitriev AA, Kashuba VI, Haraldson K,
Senchenko VN, Pavlova TV, Kudryavtseva AV, Anedchenko EA, Krasnov
GS, Pronina IV, Loginov VI, et al: Genetic and epigenetic analysis
of non-small cell lung cancer with NotI-microarrays. Epigenetics.
7:502–513. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Karan D, Kelly DL, Rizzino A, Lin MF and
Batra SK: Expression profile of differentially-regulated genes
during progression of androgen-independent growth in human prostate
cancer cells. Carcinogenesis. 23:967–975. 2002. View Article : Google Scholar : PubMed/NCBI
|