Introduction
With the population aging, the incidence of diabetes
mellitus is gradually increasing (1). Diabetes may lead to various
complications, among which foot ulcer is a major complication that
may seriously affect the quality of life, lead to a prolonged
hospital stay and may even result in lower limb amputation
(2). Vascular lesions in diabetes
are difficult to heal due to impaired angiogenesis and deregulated
vascular homeostasis (3).
Long non-coding RNAs (lncRNAs), a class of
non-coding RNAs longer than 200 nucleotides regulate gene
expression at multiple levels, including epigenetic regulation,
transcriptional regulation and post-transcription regulation
(4). Emerging evidence has
indicated that lncRNAs also have crucial roles in the development
and progression of skin wound healing (5). lncRNA5322 may promote the
proliferation and differentiation of hair follicle stem cells by
targeting the microRNA (miR)-21-mediated PI3K/AKT signaling pathway
in these cells (6). Overexpression
of lncRNA AC067945.2 is able to reduce collagen expression in skin
fibroblasts through vascular endothelial growth factor (VEGF) and
Wnt signaling pathways (7). lncRNA
X-inactive specific transcript (XIST) is inversely regulated by
miR-29a and overexpression of lncRNA XIST in denatured dermis may
promote human skin fibroblast proliferation and migration, as well
as extracellular matrix synthesis (8). Previous studies have confirmed that
lncRNAs are closely linked to angiogenesis (9). However, the role of angiogenesis
pathway-associated lncRNAs in wound healing in diabetes has
remained largely elusive.
In the present study, human skin fibroblasts were
cultured under high-glucose conditions in vitro to mimic a
diabetic environment and angiogenic pathway-associated lncRNA
expression profiles were compared between the high and normal
glucose groups. Several candidate angiogenesis pathway-associated
lncRNAs and mRNAs were analyzed by reverse
transcription-quantitative PCR (RT-qPCR). In addition, the
University of California Santa Cruz (UCSC) Genome Browser was used
to identify lncRNAs and their potential target mRNAs in the
angiogenic pathway.
Materials and methods
Isolation of human fibroblasts and
cell culture
The present study was approved by the Biomedical
Ethics Committee of the Affiliated Hospital of Nanchang University
(Nanchang, China). Written informed consent was obtained from all
patients. Human skin tissues were obtained from the remaining skin
samples of three patients (one female who was 25 years old; two
males, age, 20–32 years) who underwent skin grafting at the
Department of Burns Surgery of the First Affiliated Hospital of
Nanchang University (Nanchang, China) between January 2016 and June
2018. During this time period, the skin cells were isolated from
fresh tissues, and only the skin cells were one patient were
cultured at a time.
The tissues were washed three times with PBS and a
penicillin/streptomycin solution (Beijing Solarbio Science and
Technology Co., Ltd.). The tissues were subsequently cut into 10×10
mm sections and were digested with 0.25% trypsin + EDTA (Gibco;
Thermo Fisher Scientific, Inc.) at 4°C for 8 h. Dulbecco's modified
Eagle's medium (DMEM) with glucose (5.5 mM) supplemented with 10%
fetal bovine serum (FBS; GE Healthcare) was added to terminate the
digestion. The epidermis was removed with tweezers and the dermal
tissues were rinsed with PBS and sliced into small pieces (0.5–1
mm3). The tissue pieces were then placed on a
25-cm2 Petri dish (Corning, Inc.) to which 5 ml of
culture medium containing DMEM with glucose (5.5 mM) supplemented
with 100 U/ml penicillin plus 100 mg/ml streptomycin and 10% FBS
were added. The dish was then placed in an incubator containing air
with 5% CO2 and saturated humidity at 37°C. The culture
medium was replaced every 3 days. The fibroblasts exhibited fusion
after 14 days and were then sub-cultured on a 25-cm2
Petri dish. The subsequent experiments were performed with
third-generation fibroblasts. Fibroblasts (2×105) that
were seeded on 6-well plates were subjected to different
treatments: Fibroblasts cultured in conditioned medium with 5.5 mM
glucose were used as the control group and fibroblasts cultured in
conditioned medium with 50 mM glucose were used as the experimental
group. Each group was set up in three wells. After 48 h of culture,
the total RNA was extracted.
RNA extraction and quality
control
Total RNA was isolated with TRIzol reagent
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol. A NanoDrop ND-1000 spectrophotometer
(NanoDrop Technologies; Thermo Fisher Scientific, Inc.) was used to
estimate RNA quantity. RNA integrity and genomic DNA contamination
were assessed by standard denaturing agarose gel
electrophoresis.
Microarray and data analysis
The Agilent Array platform 2100 (Agilent
Technologies, Inc.) was employed for the microarray analysis.
Sample preparation and microarray hybridization were performed
according to the manufacturer's protocols. In brief, mRNA was
purified from total RNA following the removal of ribosomal RNA with
an mRNA-ONLY™ Eukaryotic mRNA Isolation kit (Epicentre).
Subsequently, each sample was amplified and transcribed into
fluorescent complementary (c)RNA using the Arraystar Flash RNA
Labeling protocol. The labeled cRNAs were hybridized onto the
LncPath™ Human Angiogenesis Array (8×15 K; Arraystar), which
simultaneously detected the expression of 828 lncRNAs and their 251
potential coding targets associated with the angiogenic signaling
pathway. After washing the slides, the arrays were scanned using
the Agilent Scanner G2505C (Agilent Technologies, Inc.). The
Agilent Feature Extraction software (version 11.0.1.1; Agilent
Technologies, Inc.) was used to analyze the acquired array images.
Quantile normalization and subsequent data processing were
performed using GeneSpring GX software (version 12.1, Agilent
Technologies, Inc.). Differentially expressed lncRNAs and mRNAs
with statistical significance between the two groups were
identified through volcano plot filtering. The differentially
expressed lncRNAs and mRNAs between the two samples were identified
through fold change filtering. Hierarchical clustering was
performed to display the distinguishable lncRNA and mRNA expression
pattern between the samples. The microarray analysis was performed
with the assistance of KangChen Biotech. To detect the functions of
the lncRNAs, the GENCODE annotation (version 21) was used (10). For analysis of the lncRNAs and
their potential protein-coding gene targets in the angiogenic
pathway, the UCSC Genome Browser was used to identify the mRNAs
associated with angiogenic pathways near the transcriptional region
of lncRNA (http://genomeucsc.edu/).
RT-qPCR
Total RNA was extracted from each group with TRIzol
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and then
reverse-transcribed using an RT Reagent kit (Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol. lncRNA
expression levels in each group were estimated by RT-qPCR using an
ABI Q6 (Applied Biosystems; Thermo Fisher Scientific, Inc.).
Certain candidate lncRNAs were validated by PCR. The primers used
in the present study are listed in Table I. The total RNA (1 µg) was
transcribed into cDNA. PCR was performed in a total reaction volume
of 10 µl, including 5.0 µl of 2X Master Mix (Roche), 1.0 µl of cDNA
template, 0.3 µl of PCR forward primer (10 mM), 0.3 µl of PCR
reverse primer (10 mM) and 4.4 µl of double-distilled water. qPCR
was performed using an initial denaturation step of 10 min at 95°C,
followed by 40 cycles of 15 sec at 95°C and 60 sec at 60°C. Each
RT-qPCR was performed in triplicate and all of the samples were
normalized to GAPDH. The relative expression levels of the
candidate genes were calculated using the 2−∆∆Cq method
(11).
 | Table I.Primers used for reverse
transcription-quantitative PCR. |
Table I.
Primers used for reverse
transcription-quantitative PCR.
| Seqname | Gene symbol | Sequence (5′-3′) | Annealing temperature
(°C) | Product length
(bp) |
|---|
| – | GAPDH | F:
AGAAGGCTGGGGCTCATT | 60 | 158 |
|
|
| R:
TGCTAAGCAGTTGGTGGTG |
|
|
| ENST00000407916 | RP4-791C19.1 | F:
CCAAGGGCAGACAAGTTACAATG | 60 | 121 |
|
|
| R:
TGGGTTTAACAGATTCCAGTACTCC |
|
|
| ENST00000534518 | CTD-2589O24.1 | F:
GCCATCGCCGCCAGTGTATGT | 60 | 58 |
|
|
| R:
GGTAAAAGTAGATGCAGTACCTGATA |
|
|
| NM_003377 | VEGFB | F:
GATCCGGTACCCGAGCAGT | 60 | 72 |
|
|
| R:
TTAGGTCTGCATTCACACTGGC |
|
|
| NM_000899 | KITLG | F:
ATGACCTTGTGGAGTGCGTGA | 60 | 150 |
|
|
| R:
CAGATGCCACTACAAAGTCCTTGA |
|
|
| NM_000602 | SERPINE1 | F:
GGTGAAGACACACACAAAAGGTAT | 60 | 102 |
|
|
| R:
CTCCAAAACTGCTGAAACTACA |
|
|
Statistical analysis
All statistical analyses were performed using the
SPSS 17.0 software package (SPSS, Inc.). Unpaired Student's t-tests
were used to analyze the differentially expressed lncRNAs or mRNAs
obtained in the microarray and PCR. P<0.05 was considered to
indicate statistical significance. The threshold value set to
designate differentially expressed lncRNAs and mRNAs was a fold
change of ≥1.2 (P<0.05).
Results
Morphological characteristics of human
skin fibroblasts
Human skin fibroblasts were isolated from skin
tissues and cultured in DMEM with 10% FBS and 1,000 mg/l glucose.
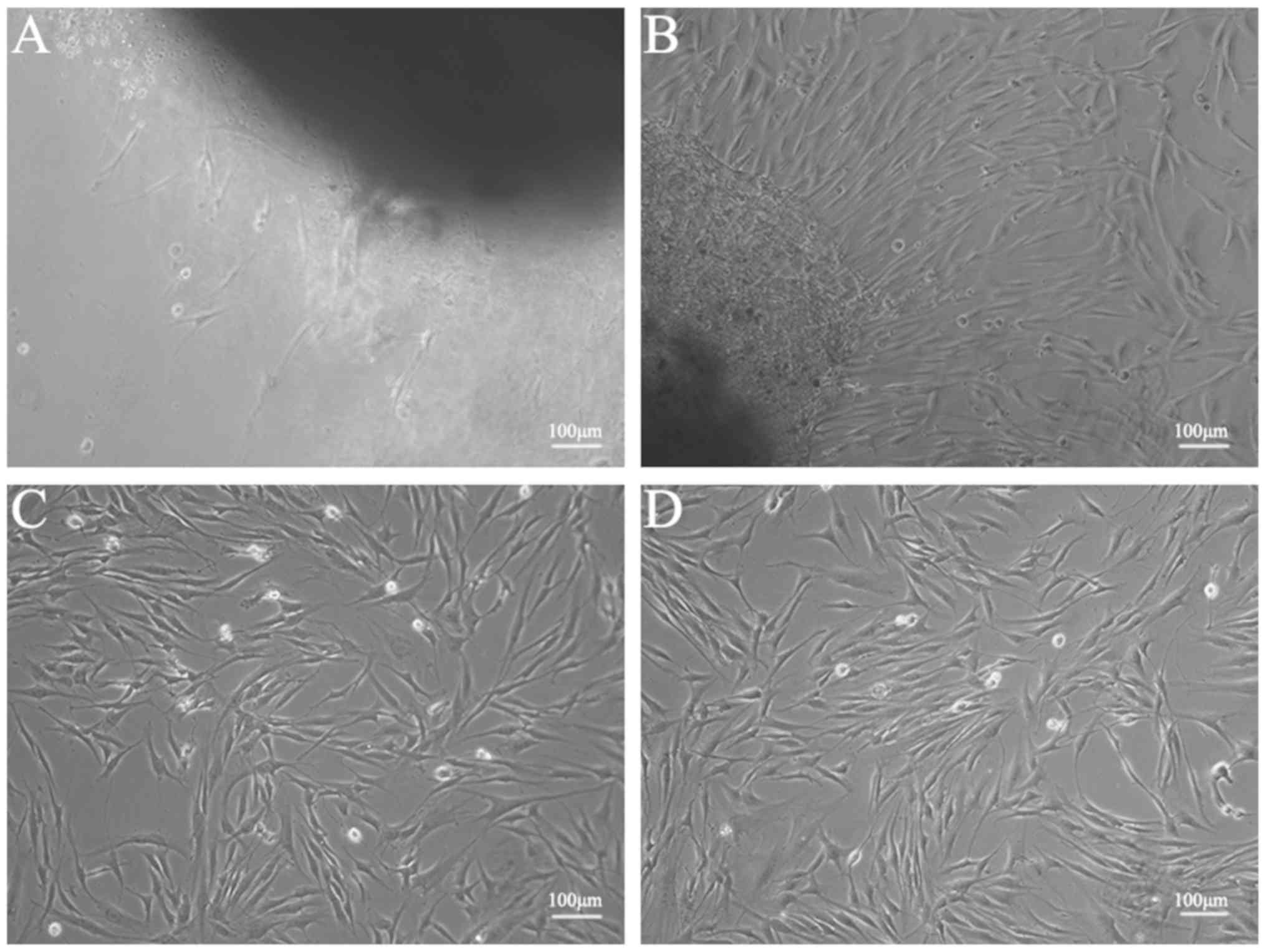
Following 7 days of culture, it was possible to see the fibroblast
cells that had migrated from the tissue pieces (Fig. 1A). The fibroblasts exhibited a long
spindle-shaped morphology with larger cell bodies. Following 14
days of culture, the fibroblasts covered the majority of the
surface area of the 25-cm2 Petri dish (Fig. 1B). The experiments were performed
with third-generation fibroblasts. No difference in cell morphology
was observed between cells cultured under high and normal glucose
conditions (Fig. 1C and D).
RNA quantification and quality
assurance
A NanoDrop ND-1000 spectrophotometer was used to
accurately determine the quantity of RNA. The optical density at
260 nm (OD260)/OD280 ratios of total RNA were
close to 2.0 for pure RNA and the OD260/OD230
ratios of the total RNA samples were >1.8. RNA has a maximum
absorption peak at a wavelength of 260 nm and proteins have a
maximum wavelength of 280 nm (12). Therefore, the
(OD260)/OD280 ratio was used to assess
protein contamination, while the OD260/OD230
ratio was used to assess organic compound contamination. The 18S
and 28S ribosomal RNA bands, which were assessed using denaturing
1% agarose gel electrophoresis, were clearly visible in the RNA
samples with an intensity ratio of ≥2:1. These results were in
accordance with the requirements of the microarray analysis.
Overview of lncRNA profiles
Using microarray analysis, the angiogenesis
pathway-associated lncRNA expression profiles of human skin
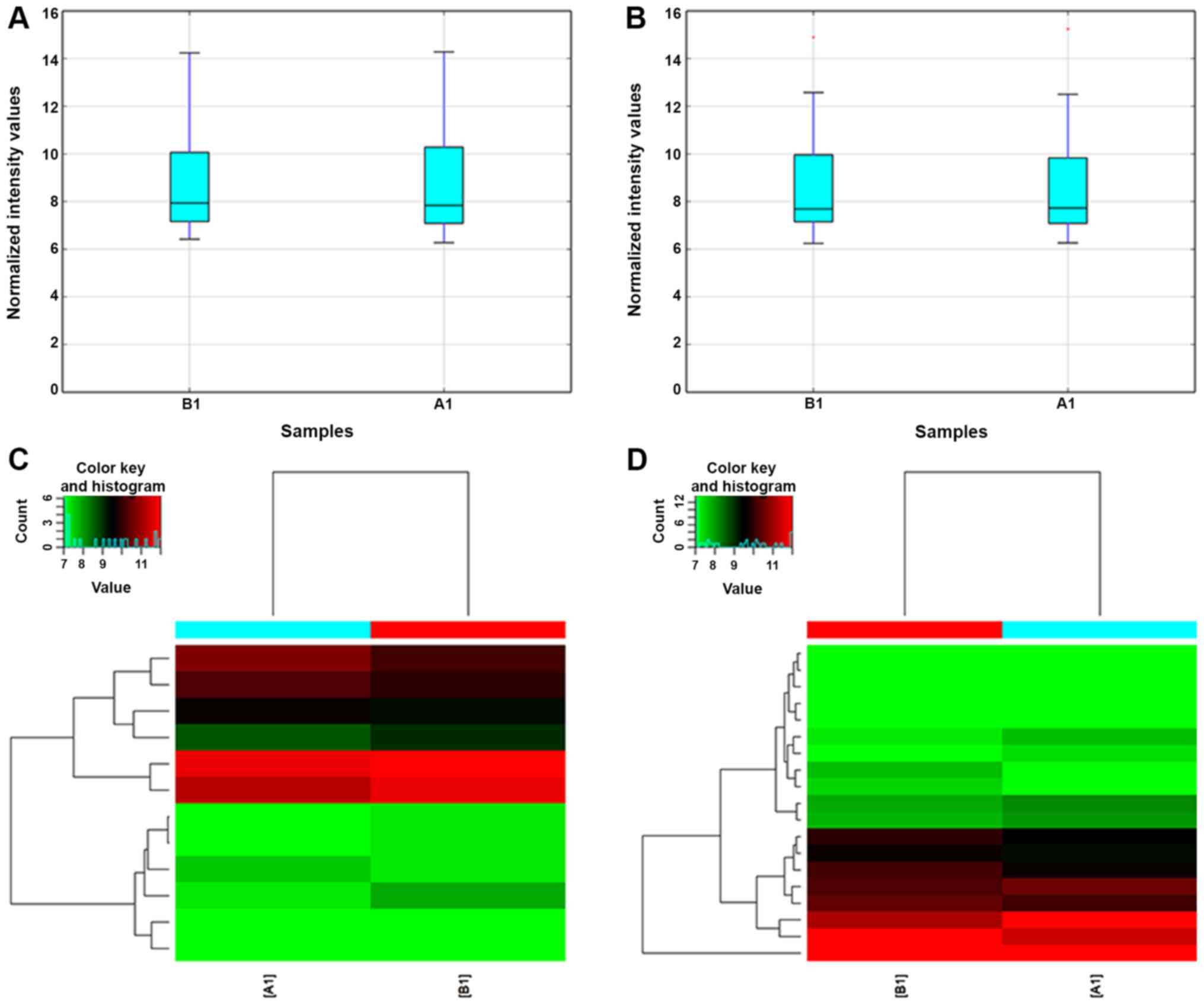
fibroblasts under high glucose conditions were obtained (Figs. 2 and 3). The box plot reflects the gene
expression data before and after standardization of the original
data. After normalization, the median of the overall data were at
the same level, indicating that the results of the data
normalization were good (Fig. 2A and
B).
Hierarchical cluster analysis is used to determine
similarities between data for classification and is more commonly
used in gene chip data analysis. It uses a series of calculations
to first identify the two groups that have the closest association
(e.g. whether the gene expression behavior is correlated), and then
identify the two groups that have similar associations and merge
them until all of the groups are combined into one group. The
expression of the selected differential genes was used to calculate
the correlation between samples. Cluster analysis of the
differentially expressed genes is able to comprehensively and
intuitively indicate the relationship and differences between
samples. Genes clustered in the same group may have similar
biological functions (Fig. 2C and
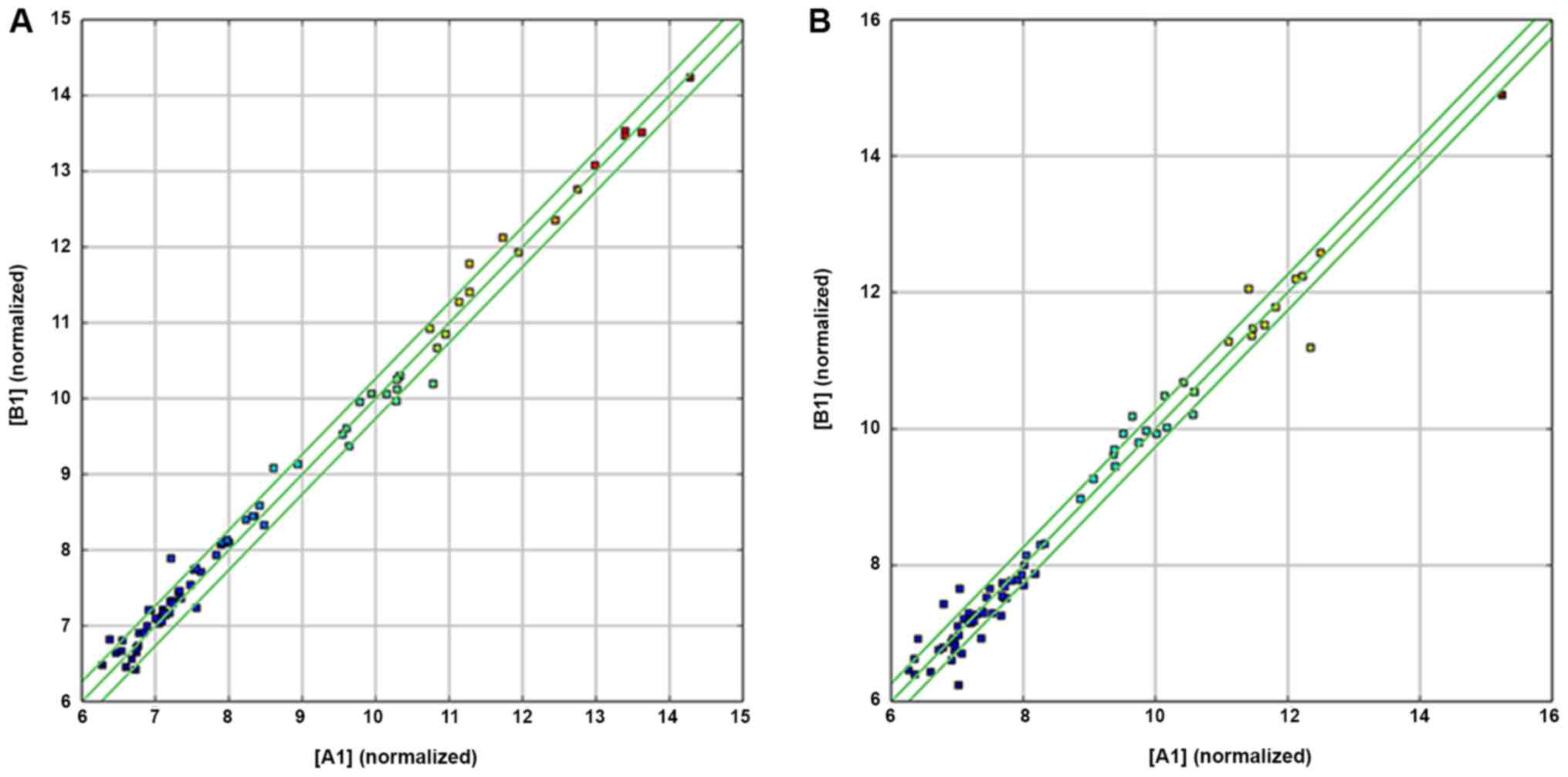
D). A scatter plot of the chip data is frequently used to
evaluate the overall distribution of the two groups of data. The
raw data analyzed by the chip is standardized and converted into
log2, which is drawn in a two-dimensional rectangular coordinate
system plane. Each point in the scatter plot represents a probe
signal. The X-axis and Y-axis values correspond to the strength of
the probe signal in different samples (Fig. 3A and B). The expression profiles
indicated that 14 lncRNAs were differentially expressed (fold
change ≥1.2, P<0.05) in the high and normal glucose groups.
Among these, 7 lncRNAs were upregulated and 7 lncRNAs were
downregulated in the high glucose group compared to the normal
glucose group (P<0.05; Table
II).
 | Table II.Differentially expressed angiogenesis
pathway-associated long non-coding RNAs in human skin fibroblasts
under high vs. normal glucose conditions. |
Table II.
Differentially expressed angiogenesis
pathway-associated long non-coding RNAs in human skin fibroblasts
under high vs. normal glucose conditions.
| Seqname | Probe ID | Fold change | Direction of
regulation |
|---|
|
ENST00000487582 | ASPWP0001058 | 1.2074108 | Up |
|
ENST00000560769 | ASPWP0002798 | 1.3577658 | Up |
|
ENST00000279573 | ASPWP0007625 | 1.5992882 | Up |
| uc009viz.2 | ASPWP0008050 | 1.2333524 | Up |
| AF080092 | ASPWP0008612 | 1.4161263 | Up |
|
ENST00000447329 | ASPWP0233858 | 1.3118266 | Up |
| TCONS_00020502 | ASPWP0242229 | 1.3868367 | Up |
|
ENST00000438325 | ASPWP0006381 | −1.5034269 | Down |
|
ENST00000503199 | ASPWP0007394 | −1.2359937 | Down |
|
ENST00000377951 | ASPWP0099869 | −1.2420934 | Down |
|
ENST00000447355 | ASPWP0131110 | −1.2034646 | Down |
|
ENST00000380194 | ASPWP0181987 | −1.2499400 | Down |
|
ENST00000407916 | ASPWP0171081 | −1.225475 | Down |
|
ENST00000534518 | ASPWP0089206 | −1.2235197 | Down |
Overview of mRNA profiles in the
angiogenic pathway
In total, 22 mRNAs were determined to be
differentially expressed in the high vs. normal glucose groups,
including 9 upregulated mRNAs and 13 downregulated mRNAs (Table III, Figs. 2D and 3B).
 | Table III.Differentially expressed angiogenesis
pathway-associated mRNAs in human skin fibroblasts under high vs.
normal glucose conditions. |
Table III.
Differentially expressed angiogenesis
pathway-associated mRNAs in human skin fibroblasts under high vs.
normal glucose conditions.
| Seqname | Probe ID | Fold change | Direction of
regulation |
|---|
| NM_002982 | ASPWP0003551 | 1.4274665 | Up |
| NM_002006 | ASPWP0005206 | 1.2722540 | Up |
| NM_170744 | ASPWP0006739 | 1.4474682 | Up |
| NM_021219 | ASPWP0008287 | 1.5533651 | Up |
| NM_002982 | ASPWP0009471 | 1.5335374 | Up |
| NM_003256 | ASPWP0010787 | 1.2091137 | Up |
| NM_002006 | ASPWP0010964 | 1.3310729 | Up |
|
ENST00000367976 | ASPWP0011091 | 1.5619674 | Up |
| NM_006153 | ASPWP0011949 | 1.2470495 | Up |
| NM_000899 | ASPWP0000190 | −1.2406115 | Down |
| NM_002422 | ASPWP0001242 | −2.2271753 | Down |
| NM_001511 | ASPWP0005163 | −1.2857458 | Down |
| NM_001305 | ASPWP0008354 | −1.7152201 | Down |
| NM_005228 | ASPWP0009560 | −1.2898507 | Down |
| NM_000321 | ASPWP0010047 | −1.3559353 | Down |
| NM_002659 | ASPWP0010442 | −1.2347152 | Down |
| NM_001278 | ASPWP0011456 | −1.2372123 | Down |
| NM_002422 | ASPWP0011539 | −1.2705653 | Down |
| NM_001143818 | ASPWP0011670 | −1.3228819 | Down |
| NM_002576 | ASPWP0009278 | −1.23502 | Down |
| NM_003377 | ASPWP0012308 | −1.2222489 | Down |
| NM_000602 | ASPWP0000680 | −1.2365507 | Down |
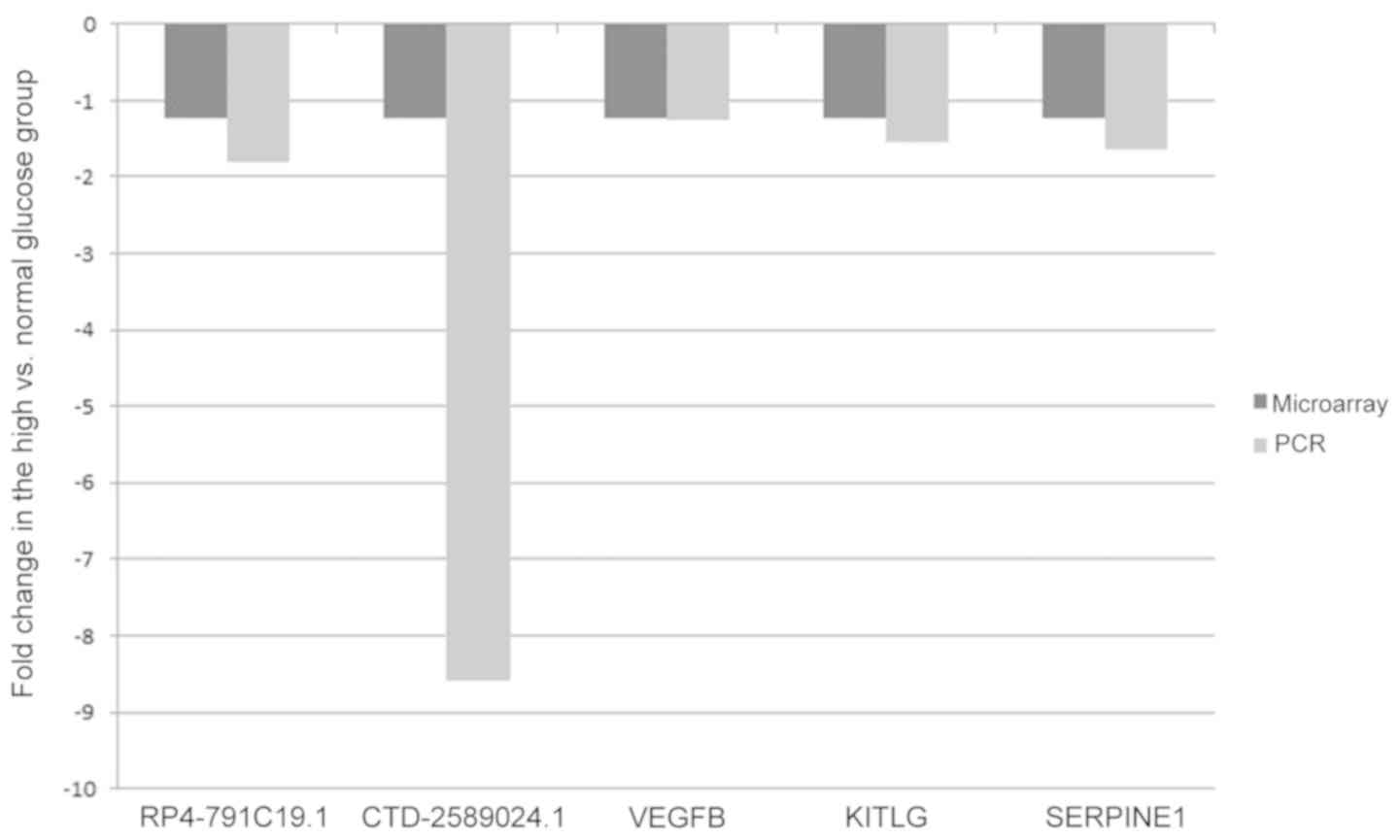
RT-qPCR validation
Certain differentially expressed lncRNAs
(RP4-791C19.1 and CTD-2589O24.1) and mRNAs [vascular endothelial
growth factor B (VEGFB), KIT ligand (KITLG) and serpin family E
member 1 (SERPINE1)] were selected to validate the microarray
results by RT-qPCR using the 2−∆∆Cq method with GAPDH as
the reference gene. The results suggested that the PCR results were
consistent with the microarray data (Fig. 4).
Analysis of lncRNAs and their
protein-coding gene targets in the angiogenic pathway
Various angiogenic pathway-associated lncRNAs were
predicted to regulate the RNA expression in the high-glucose group
(Table IV). It was indicated that
the downregulated lncRNAs (RP4-791C19.1 and CTD-2589O24.1) may act
on their target gene epidermal growth factor receptor (EGFR) and
p21 (RAC1) activated kinase 1 (PAK1), respectively, as enhancers
and cis-regulate their expression under high-glucose
conditions.
 | Table IV.Genes whose mRNA expression was
predicted to be regulated by angiogenesis pathway-associated long
non-coding RNAs under high-glucose conditions. |
Table IV.
Genes whose mRNA expression was
predicted to be regulated by angiogenesis pathway-associated long
non-coding RNAs under high-glucose conditions.
| Gene symbol | Direction of
regulation | Genomic
relationship | mRNA symbol |
|---|
| XLOC_009823 | Up | Downstream | KITLG |
| CTD-2589O24.1 | Down | Downstream | PAK1 |
| RP11-350N15.4 | Up | Overlapping | FGFR1 |
| RP11-395B7.7 | Up | Downstream | SERPINE1 |
| RP4-791C19.1 | Down | Downstream | EGFR |
Discussion
The present study provides novel insight into the
molecular mechanisms of wound healing in diabetes mellitus.
Angiogenesis is the process of new capillary formation (13), which involves multiple stages,
including endothelial cell proliferation, migration, tube
formation, micro-vessel development and branching, as well as
tissue remodeling (14).
Angiogenesis is a highly regulated process that supplies cells with
the nutrients and gases through blood vessels required for wound
healing (15). Impaired wound
healing is one of the major complications of diabetes that
increases morbidity, mortality and health expenditure (16). Therefore, it is necessary to gain a
more in-depth understanding of the molecular mechanisms of wound
healing in diabetes to identify appropriate treatment modalities
and promote wound healing.
Angiogenic imbalance contributes to difficulty in
wound healing in patients with diabetes (17). It has been indicated that lncRNAs,
which are a class of non-coding RNAs longer than 200 nucleotides
and localized in the nucleus or cytoplasm, are involved in
different regulatory mechanisms, including chromatin remodeling,
protein scaffolding and translational control (18). Long non-coding RNAs have emerged as
the critical regulators of angiogenesis in various diseases. For
instance, a knockout strategy suggested a functional role of the
lncRNA Fendrr to interfere with chromatin modifications and thereby
developmental signaling in the heart (19). The lncRNA associated with
microvascular invasion in hepatocellular carcinoma promotes
angiogenesis, and lncRNA MVIH serves as a predictor of poor
recurrence-free survival following hepatectomy (20). However, the functions of
angiogenesis pathway-associated lncRNAs in the mechanisms of wound
healing in patients with diabetes have remained largely
elusive.
Effects of high glucose on the biological behavior
of fibroblasts have been previously reported in the literature.
Buranasin et al (21)
indicated that fibroblast migration was significantly inhibited in
cells cultured at higher glucose levels (50 mM), resulting in
prolonged wound closure; cell proliferation at 50 mM glucose was
significantly reduced compared with that in the control group (5.5
mM). Similarly, in the present study, higher glucose levels (50 mM)
were used as the experimental group to mimic a diabetic environment
in vitro and was used together with a control group with
normal glucose levels (5.5 mM) to examine the angiogenesis
pathway-associated lncRNA expression profiles in the high and
normal glucose groups. Analysis of the microarray data suggested
that 14 angiogenesis pathway-associated lncRNAs and 22 mRNAs were
differentially expressed between the two groups. Among these, 7
lncRNAs and 9 mRNAs were indicated to be upregulated and 7 lncRNAs
and 13 mRNAs were downregulated in the high glucose group compared
to the normal glucose group. Several candidate angiogenesis
pathway-associated lncRNAs (RP4-791C19.1 and CTD-2589O24.1) and
mRNAs (VEGFB, KITLG and SERPINE1) were verified by RT-qPCR and the
results were consistent with the microarray data, implying that the
results of the microarray were reliable.
In the present study, the angiogenesis
pathway-associated lncRNAs RP4-791C19.1 and CTD-2589O24.1 were
identified to be downregulated in the high-glucose group, and the
also associated mRNAs EGFR and PAK1 were also downregulated in the
high-glucose group. Their differential expression changed in the
same direction. Furthermore, as respective potential target genes
of lncRNAs RP4-791C19.1 and CTD-2589O24.1, EGFR and PAK1 were
identified. To detect the functions of the lncRNAs, the GENCODE
annotation was used. In addition, the UCSC Genome Browser was used
to predict the associations of and mechanisms regulating the
lncRNAs and the angiogenesis pathway-associated target genes.
Of note, lncRNAs RP4-791C19.1 and CTD-2589O24.1
transcribed from chromosome 7 and 11, respectively, were indicated
to act as enhancers to regulate their neighboring protein-coding
genes. Enhancers are frequently defined as cis-acting DNA sequences
that increase gene transcription. They generally function
independently of the direction and distance from their target
promoters (22). Enhancer-like
lncRNAs may activate proximal promoters and stimulate the
transcription of their nearby coding genes (23). Accumulating evidence indicates that
depletion of enhancer-like lncRNAs may lead to decreased expression
of their nearby coding genes (24). It may therefore be hypothesized
that downregulation of the angiogenesis pathway-associated lncRNAs
RP4-791C19 and CTD-2589O24.1 may lead to decreased expression of
their associated genes, EGFR and PAK1, respectively. Accumulating
evidence has also demonstrated that EGFR and PAK1 participate in
new capillary blood vessel formation and have a critical role in
angiogenesis. For instance, upregulation of EGFR leads to poor
survival through the activation of angiogenesis in glioblastoma
(25). Effective inhibition of
EGFR activation was indicated to ameliorate gastric tumor
development through a delay in growth, induction of apoptosis, as
well as inhibition of metastasis and angiogenesis (26). PAK1 is the best-characterized
member of an evolutionarily conserved family of serine/threonine
kinases, which has a key role in the regulation of cell
morphogenesis, motility, mitosis and angiogenesis (27).
There are several limitations to this study. First,
the functions of the differentially expressed angiogenesis
pathway-associated lncRNAs under high-glucose conditions were
predicted but the exact functional roles of these lncRNAs were not
verified and illustrated. Analysis of the potential gene targets of
the lncRNAs implicated in human angiogenesis pathways demonstrated
that lncRNAs RP4-791C19.1 and CTD-2589O24.1 may act on their target
genes, EFGR and PAK1, respectively, as enhancers and cis-regulate
their expression. As the next step, the detailed functional roles
and underlying regulatory mechanisms of these lncRNAs in the
angiogenesis signaling pathway should be illustrated. In future
research, the functions of the identified lncRNAs may be
investigated by overexpression and RNA interference methods in
vivo or in vitro. As another limitation, cell motility,
the cell cycle and proliferation were not assessed at higher
concentrations of glucose in the present study. Dermal fibroblasts
have essential roles in wound healing. However, they lose their
normal regenerative functions under certain pathologic conditions,
e.g. in chronic diabetic wounds (28). In the present study, high-glucose
conditions mimicking the diabetic environment, as well as a low
glucose concentration resembling the normal physiological
environment were used. Culture under high-glucose conditions may
reduce the angiogenic potential of dermal fibroblasts, which may
explain for the mechanism of diabetic wound healing from the
perspective of vascularization. However, skin-derived fibroblasts
represent a part of angiogenetic processes and endothelial cells
are also involved. In future studies, differentially expressed
angiogenesis pathway-associated lncRNAs in endothelial cells under
high-glucose conditions should also be investigated. In recent
years, high-glucose conditions were used to mimic the diabetic
environment [e.g., Duru et al (29)]; however, wound-healing in a
diabetic patient is far more complex than just a hyperglycemic
micro-environment. It involves continuous inflammation and chronic
capillary damage (30). In further
research, factors including a hyperglycemic microenvironment,
inflammatory agents and chronic capillary damage will be combined
to mimic a diabetic environment. As another limitation, there may
be differences between in vivo and in vitro
experiments. Therefore, further studies in animal models, e.g. a
streptozotocin-induced rat model of diabetes, may be required to
confirm the roles of these differentially expressed angiogenesis
pathway-associated lncRNAs in wound healing in vivo. In the
future, these lncRNAs may serve as novel therapeutic targets for
the treatment of wounds in patients with diabetes in routine
clinical practice.
In conclusion, in the present study, 14 angiogenesis
pathway-associated lncRNAs and 22 mRNAs were identified to be
differentially expressed under high-glucose conditions. Among them,
the downregulated lncRNAs RP4-791C19.1 and CTD-2589O24.1 may act on
their respective target genes EGFR and PAK1 as enhancers and
cis-regulate their expression. In the future, it is necessary to
confirm and illustrate the detailed functional roles and underlying
regulatory mechanisms of these lncRNAs in the angiogenesis
signaling pathway to provide new directions to promote wound
healing in diabetes.
Acknowledgements
Not applicable.
Funding
The current study was supported by the Chinese
National Natural Science Foundation (grant nos. 81860340 and
81460293), the Special Fund for Graduate Innovation Project of
Nanchang University (grant no. CX2017249) and the Special Fund for
Graduate Innovation Project of Jiangxi province (grant no.
YC2019-B019).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LT performed the experiments and wrote the
manuscript. QH and YH analyzed data and edited the manuscript. DL
designed the experiment and revised the manuscript. All authors
have read and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Biomedical
Ethics Committee of the Affiliated Hospital of Nanchang University.
All patients provided informed written consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Hu C and Jia W: Diabetes in China:
Epidemiology and genetic risk factors and their clinical utility in
personalized medication. Diabetes. 67:3–11. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tellechea A, Leal EC, Kafanas A, Auster
ME, Kuchibhotla S, Ostrovsky Y, Tecilazich F, Baltzis D, Zheng Y,
Carvalho E, et al: Mast cells regulate wound healing in diabetes.
Diabetes. 65:2006–2019. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Davidson EP, Coppey LJ, Shevalye H,
Obrosov A and Yorek MA: Vascular and neural complications in type 2
diabetic rats: Improvement by sacubitril/valsartan greater than
valsartan alone. Diabetes. 67:1616–1626. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Herter EK and Xu LN: Non-coding RNAs: New
players in skin wound healing. Adv Wound Care (New Rochelle).
6:93–107. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Cai B, Zheng Y, Ma S, Ma S, Xing Q, Wang
X, Yang B, Yin G and Guan F: Long noncoding RNA regulates hair
follicle stem cell proliferation and differentiation through
PI3K/AKT signal pathway. Mol Med Rep. 17:5477–5483. 2018.PubMed/NCBI
|
|
7
|
Chen L and Li J, Li Q, Li X, Gao Y, Hua X,
Zhou B and Li J: Overexpression of LncRNA AC067945.2 down-regulates
collagen expression in skin fibroblasts and possibly correlates
with the VEGF and Wnt signalling pathways. Cell Physiol Biochem.
45:761–771. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Guo L and Huang X, Liang P, Zhang P, Zhang
M, Ren L, Zeng J, Cui X and Huang X: Role of XIST/miR-29a/LIN28A
pathway in denatured dermis and human skin fibroblasts (HSFs) after
thermal injury. J Cell Biochem. 119:1463–1474. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Kumar MM and Goyal R: LncRNA as a
therapeutic target for angiogenesis. Curr Top Med Chem.
17:1750–1757. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Harrow J, Denoeud F, Frankish A, Reymond
A, Chen CK, Chrast J, Lagarde J, Gilbert JGR, Storey R, Swarbreck
D, et al: GENCODE: Producing a reference annotation for ENCODE.
Genome Biol. 7 (Suppl 1):S4.1–S9. 2006. View Article : Google Scholar
|
|
11
|
Schmittgen TD and Livak KJ: Analyzing
real-time PCR data by the comparative C(T) method. Nat Protoc.
3:1101–1108. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wilfinger WW, Mackey K and Chomczynski P:
Effect of pH and ionic strength on the spectrophotometric
assessment of nucleic acid purity. Biotechniques. 22:474–476,
478–481. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Nambiar DK, Kujur PK and Singh RP:
Angiogenesis assays. Methods Mol Biol. 1379:107–115. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Potente M, Gerhardt H and Carmeliet P:
Basic and therapeutic aspects of angiogenesis. Cell. 146:873–887.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Martin P and Nunan R: Cellular and
molecular mechanisms of repair in acute and chronic wound healing.
Br J Dermatol. 173:370–378. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong Y, Rodrigues M, Kwon SH, Li X, Sigen
A, Brett EA, Elvassore N, Wang W and Gurtner GC: Acceleration of
diabetic wound regeneration using an in situ-formed stem-cell-based
skin substitute. Adv Healthc Mater. 7:e18004322018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wang W, Yan X, Lin Y, Ge H and Tan Q:
Wnt7a promotes wound healing by regulation of angiogenesis and
inflammation: Issues on diabetes and obesity. J Dermatol Sci.
2018.(Online ahead of print). View Article : Google Scholar
|
|
18
|
Mercer TR and Mattick JS: Structure and
function of long noncoding RNAs in epigenetic regulation. Nat
Struct Mol Biol. 20:300–307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Klattenhoff CA, Scheuermann JC, Surface
LE, Bradley RK, Fields PA, Steinhauser ML, Ding H, Butty VL, Torrey
L, Haas S, et al: Braveheart, a long noncoding RNA required for
cardiovascular lineage commitment. Cell. 152:570–583. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yuan SX, Yang F, Yang Y, Tao QF, Zhang J,
Huang G, Yang Y, Wang RY, Yang S, Huo XS, et al: Long noncoding RNA
associated with microvascular invasion in hepatocellular carcinoma
promotes angiogenesis and serves as a predictor for hepatocellular
carcinoma patients' poor recurrence-free survival after
hepatectomy. Hepatology. 56:2231–2241. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Buranasin P, Mizutani K, Iwasaki K,
Mahasarakham CPN, Kido D, Takeda K and Izumi Y: High
glucose-induced oxidative stress impairs proliferation and
migration of human gingival fibroblasts. PLoS One. 13:e2018552018.
View Article : Google Scholar
|
|
22
|
Pennacchio LA, Bickmore W, Dean A, Nobrega
MA and Bejerano G: Enhancers: Five essential questions. Nat Rev
Genet. 14:288–295. 2013. View
Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ørom UA, Derrien T, Beringer M, Gumireddy
K, Gardini A, Bussotti G, Lai F, Zytnicki M, Notredame C, Huang Q,
et al: Long noncoding RNAs with enhancer-like function in human
cells. Cell. 143:46–58. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lai F, Orom UA, Cesaroni M, Beringer M,
Taatjes DJ, Blobel GA and Shiekhattar R: Activating RNAs associate
with Mediator to enhance chromatin architecture and transcription.
Nature. 494:497–501. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ngo MT and Harley B: Perivascular signals
alter global gene expression profile of glioblastoma and response
to temozolomide in a gelatin hydrogel. Biomaterials. 198:122–134.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wu Y, Yuan M, Su W, Zhu M, Yao X, Wang Y,
Qian H, Jiang L, Tao Y, Wu M, et al: The constitutively active PKG
II mutant effectively inhibits gastric cancer development via a
blockade of EGF/EGFR-associated signalling cascades. Ther Adv Med
Oncol. 10:19603666752018. View Article : Google Scholar
|
|
27
|
Li LH, Wu GY, Lu YZ, Chen XH, Liu BY,
Zheng MH and Cai JC: p21-activated protein kinase 1 induces the
invasion of gastric cancer cells through c-Jun NH2-terminal
kinase-mediated activation of matrix metalloproteinase-2. Oncol
Rep. 38:193–200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Jung N, Yu J, Um J, Dubon MJ and Park KS:
Substance P modulates properties of normal and diabetic dermal
fibroblasts. Tissue Eng Regen Med. 13:155–161. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Duru EA, Fu Y and Davies MG: Role of S-1-P
receptors and human vascular smooth muscle cell migration in
diabetes and metabolic syndrome. J Surg Res. 177:e75–e82. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao R, Liang H, Clarke E, Jackson C and
Xue M: Inflammation in chronic wounds. Int J Mol Sci. 17:20852016.
View Article : Google Scholar
|