Introduction
Ischemic stroke is the most common type of brain
disease in adults worldwide; it is characterized by high incidence,
disability and mortality rates (1). In China, the number of
stroke-associated deaths has notably increased over the past two
decades (1,2). Although rapid recanalization of the
relevant artery and subsequent promotion of brain revascularization
can successfully treat patients, reperfusion can increase the
production of reactive oxygen species (ROS), inflammation and
endoplasmic reticulum stress, leading to ischemia-reperfusion (I/R)
injury, such as human brain microvascular endothelial cell (hBMEC)
apoptosis (3). hBMECs, located
between the blood and the brain parenchyma, are essential for
normal neurological function and are responsible for transferring
essential nutrients and removing potentially harmful toxins
(4). I/R injury-induced hBMEC
apoptosis impairs the therapeutic effect of relevant artery
recanalization (5–7). Thus, protection of hBMECs from I/R
injury may be crucial for intervention of ischemic stroke.
MicroRNAs (miR) can bind to the 3′ untranslated
region (3′UTR) of mRNAs, and inhibit their translation and promote
their degradation, regulating a number of biological processes
(8). For example, there is
evidence to indicate that miR-15a-5p is involved in the
pathogenesis of vascular endothelial cell injury following I/R
(9). Induction of miR-15a-5p
overexpression inhibits vascularization in a mouse model of hind
limb ischemia (9). Furthermore,
increased levels of miR-15a-5p are detected in the heart following
myocardial infarction, and inhibition of miR-15a-5p decreases
infarct size and enhances cardiac function in mice that have
experienced myocardial infarction (10). Notably, miR-15a-5p can decrease
Bcl-2 expression levels and increase apoptosis of cerebral vascular
endothelial cells following oxygen-glucose deprivation and
reoxygenation (OGD-R) (11).
However, little is currently known about which factors regulate
miR-15a-5p expression levels following OGD-R in hBMECs.
Long non-coding RNAs (lncRNAs) are defined as being
>200 nucleotides in length and function as miR sponges to
regulate numerous biological processes, including apoptosis,
inflammation and I/R injury (12,13).
For example, expression levels of antisense non-coding RNA in the
INK4 locus (ANRIL) are upregulated in the cortex in a rat model of
cerebral infarction and ANRIL can activate the NF-κB signaling
pathway to promote inflammation (12). The UCA1 lncRNA expression level is
downregulated in myocardial tissues in a mouse model of I/R injury
by targeting P27 to induce apoptosis (13). On the other hand,
metastasis-associated lung adenocarcinoma transcript 1 (MALAT1) can
target the pro-apoptotic protein Bim to decrease pro-inflammatory
cytokine production in BMECs of mice with I/R-induced brain injury
(14). The small nucleolar RNA
host gene 16 (SNHG16) is a newly identified oncogenic lncRNA that
regulates the progression and metastasis of lung, breast and
bladder cancer (15,16). Previous studies have demonstrated
that lncRNAs can share miRNA response elements (MREs) and induce
gene silencing (17,18). However, it is currently unknown
whether SNHG16 can interact with miR-15a-5p to regulate the
proliferation and apoptosis of hBMECs during I/R.
The present study used hBMECs to investigate the
effect of OGD-R on proliferation, apoptosis and miR-15a-5p, Bcl-2
and SNHG16 expression levels. The potential interaction of SNHG16
with miR-15a-5p was analyzed via pull-down, luciferase and
immunoprecipitation assays.
Materials and methods
Establishment of an in vitro model of
OGD-R
The hBMEC line hCMEC/D3 was obtained from Procell
Life Science & Technology Co., Ltd. The cells were cultured in
complete DMEM containing 10% fetal bovine serum (FBS; Gibco; Thermo
Fisher Scientific, Inc.) for 24 h at 37°C in a humidified
atmosphere of 5% CO2. I/R injury of hBMECs was induced
by OGD-R (19,20). Briefly, the cells were cultured in
a medium without glucose and serum (Gibco; Thermo Fisher
Scientific, Inc.) at 37°C in a 5% CO2 and 95%
N2 hypoxic cell culture system (H35; Don Whitley
Scientific) for 4 h and then cultured in complete medium at 37°C in
5% CO2. hBMECs cultured in complete medium at 37°C in 5%
CO2 were used as the control.
Transfection with RNA
oligoribonucleotides
The human SNHG16 wild-type or its mutant at (MREs)
were cloned into the plasmid pmirGLO downstream of the Renilla
luciferase open reading frame to generate the plasmid pSNHG16-wt or
pSNHG16-mut (GenePharma), respectively. HBMECs were transfected
with pSNHG16-wt or pSNHG16-mut, hsa-miR-15a-5p mimics or scrambled
miRNA (miR-NC; ON-TARGETplus SMARTpool; GE Healthcare Dharmacon,
Inc.) at a final concentration of 100 nM using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturer's recommendation.
At 6 h post-transfection, the cells were subjected to OGD-R. The
transfection efficiency in cells was determined using reverse
transcription quantitative PCR (RT-qPCR).
Measurement of miR-15a-5p and SNHG16
using RT-qPCR
Total RNA was extracted from individual groups of
hBMECs using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.) and reverse transcribed into cDNA at 50°C for 30
min using the cDNA Reverse Transcription kit (Applied Biosystems;
Thermo Fisher Scientific, Inc.). The relative levels of target
SNHG16 to control GAPDH mRNA transcripts were determined via
RT-qPCR using SYBR Green (Thermo Fisher Scientific, Inc.) and
specific primers in an Applied Biosystems 7500 PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The sequences of
primers were: SNHG16: Forward 5′-CAGAATGCCATGGTTTCCCC-3′; reverse,
5′-TGGCAAGAGACTTCCTGAGG-3′; GAPDH: Forward,
5′-GCACCGTCAAGGCTGAGAAC-3′; reverse, 5′-ATGGTGGTGAAGACGCCAGT-3′.
Similarly, the relative levels of miR-15a-5p to control U6 small
nuclear RNA were analyzed using RT-qPCR using the TaqMan miRNA
assay kit (Applied Biosystems; Thermo Fisher Scientific, Inc.). The
thermocycling conditions were as follows: 20 sec at 95°C, followed
by 40 cycles of 15 sec at 95°C and 60 sec at 60°C, and 60°C for 1
min. The data were normalized to the control and analyzed using the
2−ΔΔCq method (21).
Cell apoptosis assay
The frequency of apoptotic hBMECs was determined by
flow cytometry. Briefly, hBMECs (1×105 cells/well) were
cultured in 10% FBS/DMEM (Gibco; Thermo Fisher Scientific, Inc.)
for 24 h at 37°C in 5% CO2 and subjected to OGD-R. Cells
were collected at the indicated time points post-reoxygenation. The
cells were stained with phycoerythrin-conjugated propidium iodide
and FITC-conjugated Annexin V (both 1:10; both BD Biosciences) for
20 min at room temperature. The percentages of apoptotic cells were
analyzed using a BD FACSCalibur flow cytometer (BD
Biosciences).
Measurement of cell proliferation
Cell proliferation was measured using MTT assay.
Briefly, hBMECs (1×104 cells/well) were cultured in 10%
FBS/DMEM (Gibco; Thermo Fisher Scientific, Inc.) for 24 h at 37°C
in 5% CO2 and subjected to OGD-R. The cells were exposed
to 10 µl of 5 mg/ml MTT for 4 h. The generated purple formazan in
individual wells was dissolved in 50 µl DMSO. Absorbance was
measured at 540 nm in a microplate reader (SpectraMax M5; Molecular
Devices, LLC).
RNA-pull-down assay
hBMECs (6×106 cells) were cultured at 37°C for 24 h
and were transfected with biotinylated miR-15a-5p, and scrambled
miRNA (Guangzhou RiboBio Co., Ltd). After hBMECs were subjected to
OGD-R, total RNAs were extracted at 48 h post-reoxygenation using
the RNeasy Plus Mini kit (Qiagen GmbH) according to the
manufacturer's instructions.. The biotinylated RNAs were absorbed
with M-280 streptaviden magnetic beads (Invitrogen; Thermo Fisher
Scientific, Inc.) for 4 h at 4°C. Next, the beads were washed with
high salt buffer (1% Triton X-100; 0.1% SDS; 20 mM Tris-HCl, pH
8.0; 2 mM EDTA; 500 mM NaCl). The bound RNAs were measured using
RT-qPCR as described above. Based on the starbase 2.0 analysis
software (starbase.sysu.edu.cn/mirLncRNA.php), a total of 146
lncRNA that potentially formed complementary base pairs with
miR-15a-5p (22). Among them, 25
lncRNAs were further analyzed according to their bioComplex and
clipReadNum (Table I).
 | Table I.Predicted miR-lncRNA
interactions. |
Table I.
Predicted miR-lncRNA
interactions.
| Gene | Target sites | bioComplex | clipReadNum | lncRNA relative
expression level |
|---|
| SNHG16 | 1 | 9 | 185 | 22.6±2.1 |
| CTBP1-AS1 | 1 | 1 | 27 | 7.3±1.6 |
| RP11-869B15.1 | 1 | 2 | 19 | 1.1±0.3 |
| RP11-169K16.9 | 1 | 1 | 12 | 3.2±0.6 |
| RP11-384K6.6 | 1 | 1 | 19 | 2.6±1.4 |
| AC074093.1 | 1 | 2 | 12 | 0.2±0.1 |
| XIST | 5 | 18 | 2,862 | 15.4±3.2 |
| RP11-549J18.1 | 1 | 7 | 128 | 4.1±0.8 |
| SENP3-EIF4A1 | 1 | 5 | 27 | 2.2±1.3 |
| CTC-228N24.3 | 1 | 6 | 72 | 3.2±0.5 |
| AC084219.4 | 2 | 10 | 193 | 14.6±2.9 |
| LINC00094 | 1 | 8 | 143 | 3.7±2.8 |
| RP11-403I13.4 | 2 | 4 | 46 | 7.9±1.4 |
| ZNRD1-AS1 | 1 | 4 | 80 | 1.1±0.2 |
| RP11-91G21.2 | 1 | 2 | 15 | 0.5±0.1 |
| RP11-379K17.11 | 1 | 32 | 4,692 | 12.5±3.3 |
| AC108142.1 | 1 | 2 | 11 | 0.3±0.1 |
| MIR497HG | 1 | 1 | 14 | 0.5±0.2 |
| HCG17 | 1 | 2 | 25 | 2.3±0.3 |
| RP13-507I23.1 | 1 | 1 | 72 | 3.2±0.4 |
| C1RL-AS1 | 1 | 2 | 87 | 4.2±0.9 |
| AP000721.4 | 1 | 9 | 233 | 7.3±2.3 |
| RP11-96D1.10 | 1 | 1 | 51 | 6.6±1.7 |
| ERVK13-1 | 1 | 1 | 25 | 5.3±1.1 |
| RP3-341D10.4 | 1 | 2 | 17 | 1.5±0.2 |
Isolation of cell cytoplasm/nucleus
fractions
The cytoplasmic and nuclear components of individual
groups of hBMECs were extracted using nuclear and cytoplasmic
extraction reagents (Thermo Fisher Scientific, Inc.) according to
the manufacturer's instructions. Briefly, 1×107 cells
re-suspended in 500 µl ice-cold cell fractionation buffer were
incubated on ice for 10 min and centrifuged at 500 × g for 5 min at
4°C. The cytoplasmic fraction in the supernatants was transferred
into a new tube. The nuclear pellet was washed with ice-cold cell
fractionation buffer and lysed in cell disruption buffer for RNA
isolation. The extracted RNAs were analyzed using RT-qPCR as
described above. The U6 and GAPDH were used as nuclear and
cytoplasmic control transcripts, respectively.
RNA immunoprecipitation (RIP)
RIP was performed using the Ago2 immunoprecipitation
assay kit (Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. Briefly, hBMECs were lyzed in RIPA
lysis buffer and centrifuged at 12,000 × g for 15 min at 4°C. The
supernatants were reacted with 10 µl beads and 2 µl argonaute 2
(Ago2) antibody or control IgG (1:100; cat. no. 17–700; EMD
Millipore) rotating overnight at 4°C. After being centrifuged at
12,000 × g for 15 min at 4°C and washed with lysis buffer, RNAs
were extracted using TRIzol® reagent and the presence of
the binding targets was characterized by RT-qPCR as described
above.
Dual luciferase reporter assay
In order to assess the SNHG16/miR-15a-5p binding
specificity, luciferase activity was measured (Promega Corporation)
according to the manufacturer's protocol. hBMECs were transfected
in triplicate with 10 ug pSNHG16-wt or pSNHG16-mut, together with
miR-15a-5p mimics or scrambled miRNA using
Lipofectamine® 2000. After transfection for 48 h, the
relative luciferase activity in individual groups of cells was
determined using a Lumat LB 9507 luminometer (Berthold
Technologies; GmbH). The data were expressed as the fold change
relative to the control groups defined as 1.0.
Western blotting
Individual groups of hBMECs were lyzed in RIPA lysis
buffer (50 mM Tris-HCl, 1% Nonidet P-40, 150 mM NaCl, 0.5% sodium
deoxycholate and 0.1% SDS) and centrifuged at 12,000 × g for 10 min
at 4°C. After the concentrations of total proteins were measured
using a BCA protein assay kit (Pierce; Thermo Fisher Scientific,
Inc.), individual samples (50 µg/lane) were analyzed using 10%
SDS-PAGE and electrophoretically transferred onto PVDF membranes
(EMD Millipore). After being blocked with 5% non-fat dry milk in
Tris buffer saline with 0.1% Tween-20 (TBST) for 2 h at room
temperature, the membranes were probed with antibodies against
Bcl-2 (1:100; cat. no. 15071) or β-actin (1:100; cat. no. 3700; all
Cell Signaling Technology, Inc.) at 4°C overnight. The bound
antibodies were detected using horseradish peroxidase-conjugated
anti-mouse IgG (1:200; cat. no. 7076; Cell Signaling Technology,
Inc.) and visualized using the enhanced chemiluminescent reagents.
The relative Bcl-2 to β-actin expression levels were determined
using densitometric analysis using the Image Lab™ Software (version
3.0; Bio-Rad Laboratories, Inc.). A total of three independent
experiments were performed.
Statistical analysis
Statistical analyses were performed using Stata 7.0
statistical software (StataCorp LP). The normal distribution of
values was expressed as mean ± standard deviation and the
difference among groups was analyzed by one-way analysis of
variance followed by Tukey's multiple comparisons as a post-hoc
test. The paired Student's t-tests was used to compare percentages
of apoptotic hBMECs and miR-15a-5p expression levels at the
indicated time points. P<0.05 was considered to indicate a
statistically significant difference.
Results
miR-15a-5p induces apoptosis of
hBMECs
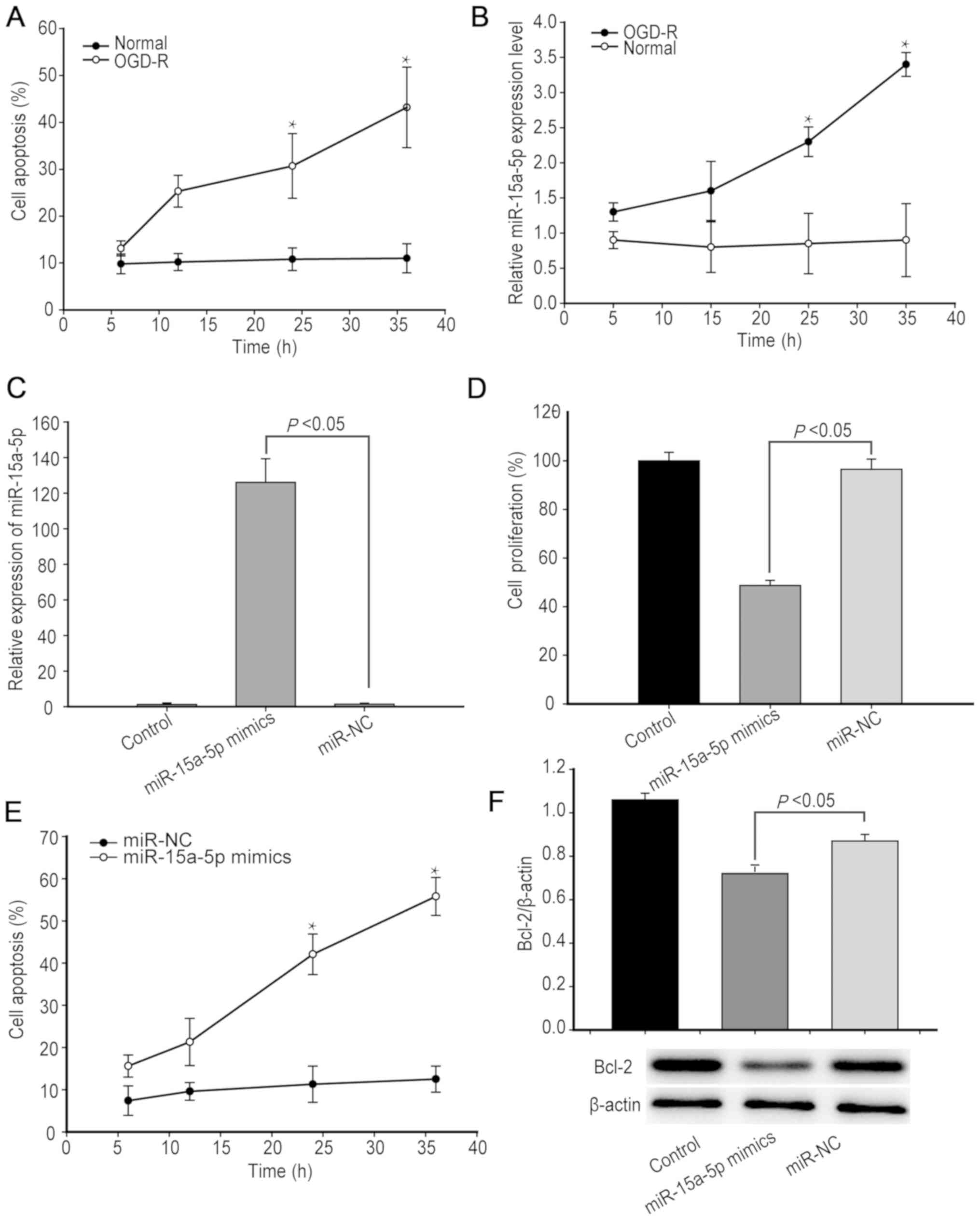
A previous study demonstrated that OGD-R can induce
apoptosis of endothelial cells (23). In order to determine the effect of
OGD-R on hBMECs in the present study, hBMECs were subjected to
OGD-R or cultured in normal conditions, and the frequency of
apoptotic hBMECs was determined using flow cytometry. The
percentage of apoptotic hBMECs increased over the measured time
points following OGD-R, whereas the percentage of control hBMECs
undergoing apoptosis remained at ~10% throughout the experiment
(Fig. 1A).
Analysis of miR-15a-5p expression levels indicated
that the relative miR-15a-5p expression levels in hBMECs increased
over time following OGD-R (Fig.
1B). In order to determine the role of miR-15a-5p, hBMECs were
transfected with, or without, miR-15a-5p mimics or miR-NC for 24 h,
and the relative levels of miR-15a-5p were determined using
RT-qPCR. Transfection with miR-15a-5p mimics increased the levels
of miR-15a-5p more than 120-fold (P<0.05; n=3; Fig. 1C). MTT and apoptosis assays
demonstrated that increased miR-15a-5p expression levels
significantly decreased the proliferation of hBMECs by triggering
hBMEC apoptosis (P<0.05, Fig.
1D-E). As Bcl-2 is an important survival factor and a target of
miR-15a-5p (11), the present
study analyzed the relative Bcl-2 expression levels using western
blotting. The results indicated that increased miR-15a-5p
expression levels decreased Bcl-2 expression levels in hBMECs
cultured in normal conditions (P<0.05; Fig. 1F). The results of the present study
indicated that OGD-R triggered hBMEC apoptosis by upregulating
miR-15a-5p expression levels to decrease Bcl-2 expression
levels.
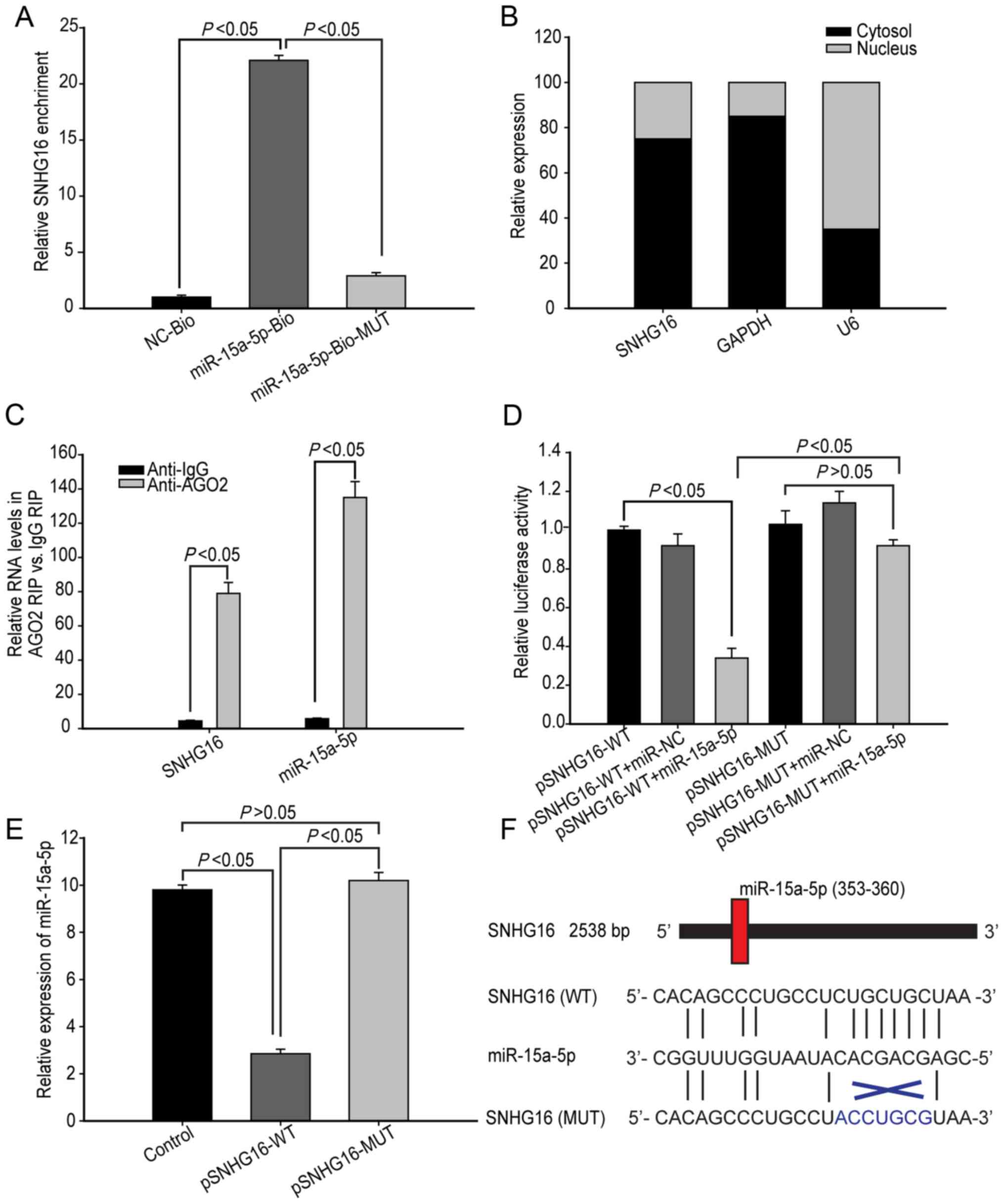
SNHG16 regulates miR-15a-5p
lncRNAs can regulate miRNA expression levels. In
order to screen potential lncRNAs that regulate miR-15a-5p
expression levels, hBMECs were cultured under OGD-R for 48 h and
potential RNAs were pulled-down. A total of 25 lncRNAs were
identified, of which SNHG16 was the most abundant, as indicated by
the results of RT-qPCR (Table I).
SNHG16 was pulled-down by biotinylated miR-15a-5p, but not by the
biotinylated miR-15a-5p mutant with a mutation at the binding site
of SNHG16, indicating that SNHG16 interacted with miR-15a-5p
(Fig. 2A). Therefore, SNHG16 was
selected for subsequent experiments.
It was hypothesized that SNHG16 may act as a
competitive endogenous RNA (ceRNA) in hBMECs by targeting
miR-15a-5p. In order to test this, the cellular localization of
SNHG16 was analyzed. As shown in Fig.
2B, SNHG16 was primarily distributed in the cytoplasm,
suggesting that SNHG16 may play a role at a post-transcriptional
level. As miRs are present in miR ribonucleoprotein complexes,
which contain Ago2 protein, RIP was used to characterize SNHG16
distribution using anti-Ago2. The present study demonstrated that
anti-Ago2 precipitated not only SNHG16, but also miR-15a-5p,
suggesting that SNHG16 may be present in the miR-15a-5p RNA-induced
silencing complex (Fig. 2C).
Furthermore, luciferase reporter assays indicated that
co-transfection with miR-15a-5p mimics, but not with control
miR-NC, significantly decreased the luciferase activity of binding
seed region in wild-type SNHG16 in hBMECs (P<0.05; n=3; Fig. 2D). However, miR-15a-5p mimics did
not significantly affect the luciferase activity in mutant SNHG16
(P>0.05; n=3; Fig. 2D).
Furthermore, overexpression of SNHG16, but not its mutant,
significantly decreased the relative miR-15a-5p expression levels
in hBMECs (P<0.05; n=3; Fig.
2E). Collectively, such data indicate that SNHG16 acts as a
ceRNA to downregulate miR-15a-5p expression levels in hBMECs. The
predicted binding sites of miR-15a-5p to SNHG16 and the coding
sequence of SNHG16 at MREs are presented in Fig. 2F.
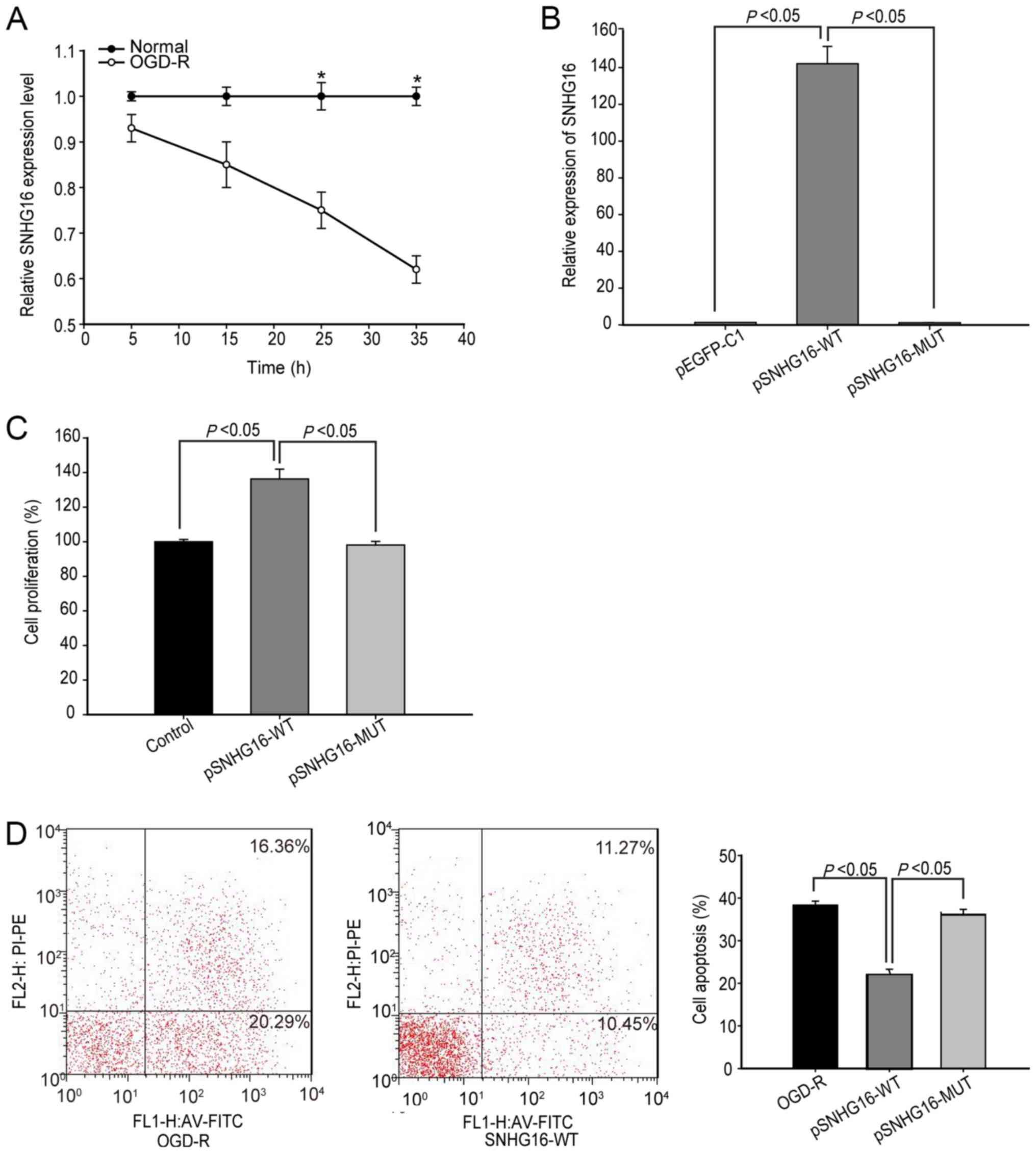
SNHG16 inhibits OGD-R induced
apoptosis in hBMECs
The present study investigated the role of SNHG16 in
regulating OGD-R induced apoptosis of hBMECs. Following
reoxygenation, the relative level of SNHG16 transcripts in hBMECs
gradually decreased compared with that in control cells cultured
under normal conditions and reached 0.58-fold at 36 h
post-reoxygenation (P<0.05; Fig.
3A). Furthermore, transfection with pSNHG16, but not its
mutant, significantly increased SNHG16 expression levels and
enhanced proliferation of hBMECs at 48 h post-transfection
(P<0.05; n=3; Fig. 3B and C).
In accordance with these results, flow cytometric analysis
indicated that transfection with pSNHG16, but not its mutant,
significantly decreased the percentages of hBMECs undergoing
OGD-R-induced apoptosis (36.3±1.5 vs. 21.1±1.9%; P<0.05; n=3;
Fig. 3D). These results indicated
that SNHG16 enhanced the proliferation of hBMECs and inhibited
OGD-R-induced apoptosis.
SNHG16 inhibits miR-15a-5p-induced
apoptosis in hBMECs
In order to further demonstrate the association
between miR-15a-5p and SNHG16 in hBMECs, the present study
determined whether SNHG16 could modulate the miR-15a-5p-regulated
proliferation and apoptosis of hBMECs following OGD-R. HBMECs were
co-transfected with pSNHG16-wt or pSNHG16-mut together with
miR-15a-5p mimics or miR-NC and subjected to OGD-R. At 48 h
post-reoxygenation, the proliferation of hBMECs was determined
using MTT assay. Transfection with pSNHG16 and miR-NC significantly
increased the proliferation of hBMECs, supporting the hypothesis
that SNHG16 promotes hBMEC proliferation (P<0.05; n=3; Fig. 4A). In contrast, transfection with
pSNHG16 and miR-15a-5p (both P<0.05, n=3) significantly
decreased the proliferation of hBMECs (Fig. 4A). Similarly, transfection with
miR-15a-5p significantly decreased the proliferation of SNHG16
mutant-overexpressing hBMECs. Flow cytometry indicated that
transfection with miR-15a-5p significantly increased the
percentages of apoptotic hBMECs that had been transfected with
either pSNHG16 or pSNHG16 mutant (P<0.05; n=3; Fig. 4B). These results indicated that
SNHG16 protects hBMECs from OGD-R-induced apoptosis by targeting
miR-15a-5p.
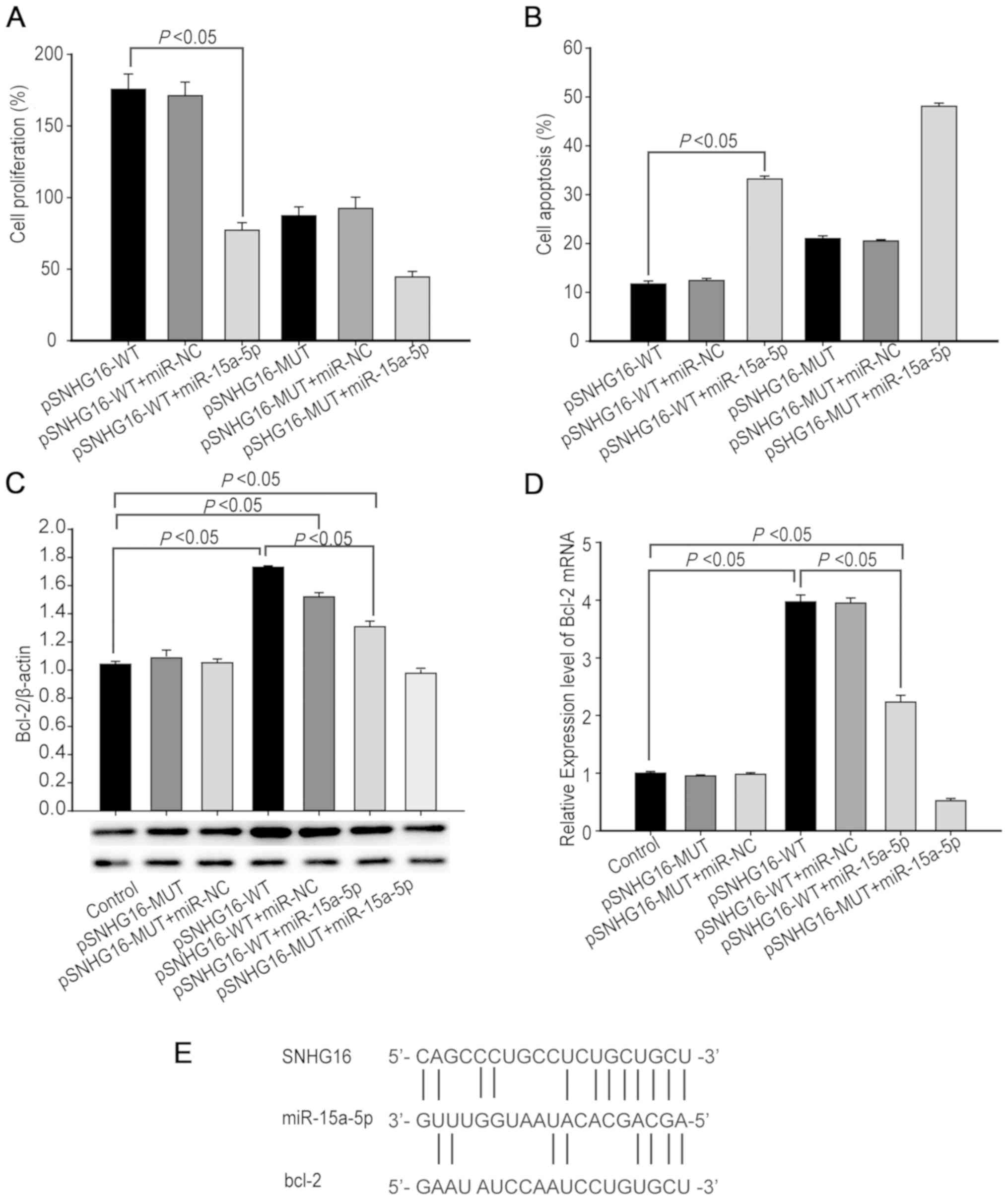 | Figure 4.SNHG16 antagonizes miR-15a-5p to
increase Bcl-2 expression levels and protect hBMECs from
OGD-R-induced apoptosis. hBMECs were transfected with pSNHG16-wt or
pSNHG16-mut, together with miR-15a-5p mimics or miR-NC and were
subjected to OGD-R. At 48 h post-reoxygenation, the proliferation,
apoptosis and Bcl-2 expression levels in hBMECs were determined.
(A) MTT analysis of cell proliferation. (B) Flow cytometric
analysis of cell apoptosis. (C) Western blotting analysis of Bcl-2
protein expression levels. (D) RT-qPCR analysis of Bcl-2 mRNA
transcripts. (E) Potential binding sites among SNHG16, miR-15a-5p
and Bcl-2 mRNA were predicted using bioinformatics using the online
software starBase v2.0. SNHG16 may act as a molecular sponge for
miR-15a-5p and regulate its target Bcl-2 expression levels.
Statistical significance was analyzed by one-way analysis of
variance followed by Tukey's multiple comparisons. Data are
presented as the mean ± standard deviation of each group of cells
from three separate experiments. SNHG, small nucleolar host gene;
miR, microRNA; hBMEC, human brain microsvascular endothelial cell;
OGD-R, oxygen-glucose deprivation and reoxygenation; NC, normalized
control; RT-qPCR, reverse transcription quantitative PCR; WT,
wild-type; MUT, mutant. |
SNHG16 positively regulates Bcl-2
expression levels in hBMECs
Bcl-2 is a regulator of cell survival and apoptosis
and a potential target of miR-15a-5p (24). Accordingly, it was hypothesized
that SNHG16 inhibited OGD-R-induced apoptosis by antagonizing
miR-15a-5p to upregulate Bcl-2 in hBMECs. In order to investigate
this, hBMECs were co-transfected with, or without, pSNHG16-wt or
pSNHG16-mut together with miR-15a-5p mimics or miR-NC and subjected
to OGD-R. At 48 h post-reoxygenation, the relative Bcl-2 expression
levels in the different groups of cells were determined using
western blotting and RT-qPCR. Transfection with either pSNHG16
mutant or together with miR-NC did not alter the relative levels of
Bcl-2 expression in hBMECs, whereas transfection with pSNHG16
mutant and miR-15a-5p notably decreased Bcl-2 expression levels in
hBMECs (P<0.05; n=3; Fig. 4C and
D). Furthermore, transfection with pSNHG16 and miR-NC
significantly increased the relative Bcl-2 expression levels while
transfection with pSNHG16 and miR-15a-5p significantly decreased
Bcl-2 expression levels in hBMECs, although their Bcl-2 expression
levels were significantly higher than those of the control cells
(P<0.05 for all). Bioinformatics demonstrated that SNHG16 could
bind to miR-15a-5p, which could, in turn, bind to the 3′UTR
sequence of Bcl-2 (Fig. 4E).
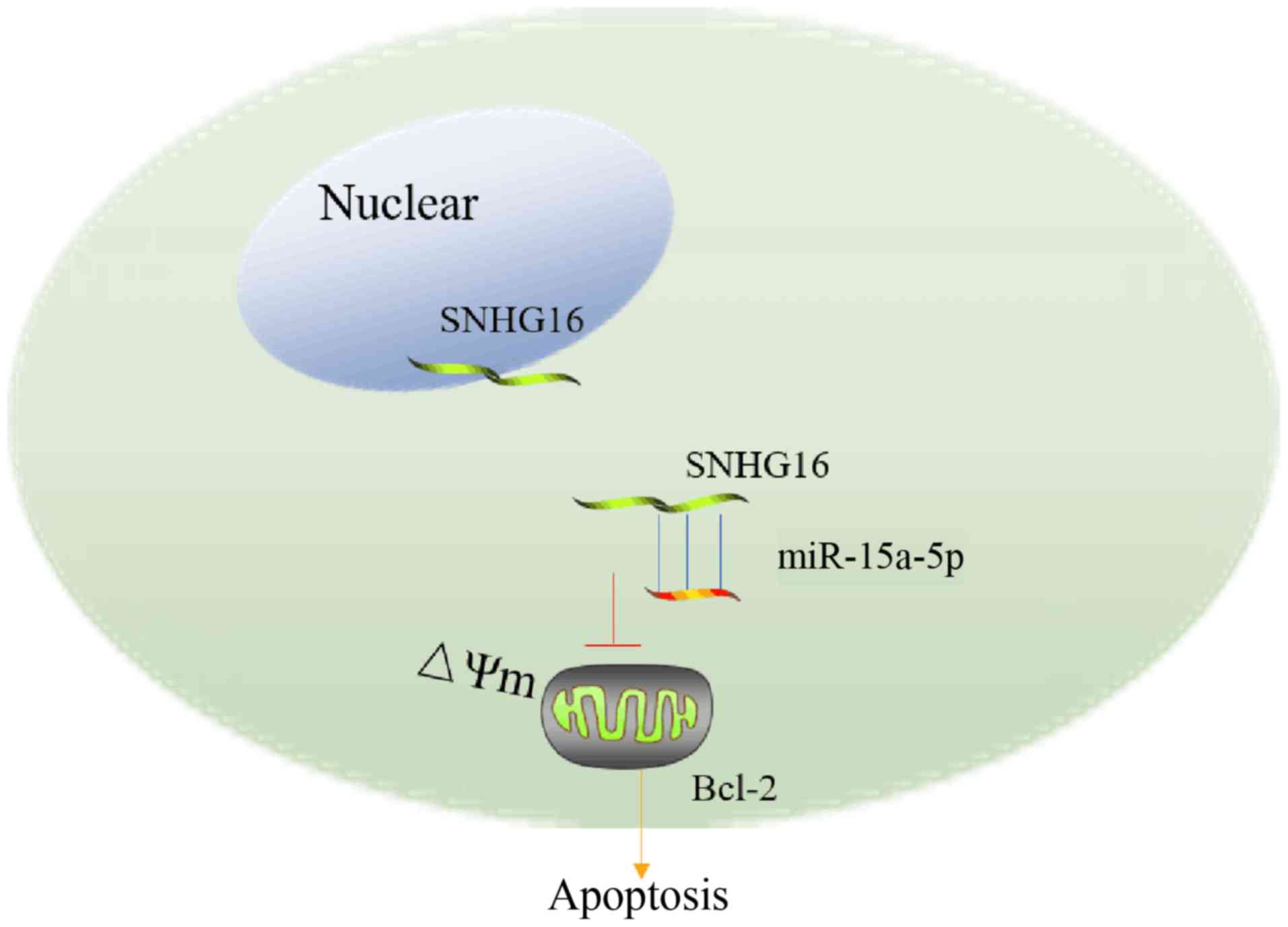
Collectively, these data indicated that there is a regulatory
signaling pathway in which SNHG16 regulates Bcl-2 via competitively
sponging miR-15a-5p (Fig. 5).
Discussion
The present study investigated the potential
function and molecular mechanisms by which SNHG16 regulates
OGD-R-induced apoptosis in hBMECs. The present study demonstrated
that SNHG16 expression levels in hBMECs decreased in a
time-dependent manner following OGD-R. Induction of SNHG16
overexpression significantly enhanced the proliferation of hBMECs
by inhibiting OGD-R-induced apoptosis. The present study further
demonstrated that SNHG16 directly interacted with miR-15a-5p to
enhance Bcl-2 expression levels in hBMECs.
In previous studies, lncRNAs have been demonstrated
to have a pivotal role in I/R injury. Microarray and RNA-seq
techniques have identified a number of abnormally expressed lncRNAs
in animal models of I/R injury and patients with ischemic stroke
(25,26). Furthermore, certain lncRNAs have
been demonstrated to regulate the pathogenesis of I/R injury. For
example, MALAT1 protects hBMECs from OGD-R-induced apoptosis by
enhancing PI3K dependent signaling (23). The present study demonstrated that
OGD-R induced apoptosis in ~40% of hBMECs. Induction of SNHG16
overexpression significantly decreased the amount of apoptotic
hBMECs following OGD-R. The results of the present study coincided
with previous observations that SNHG16 promoted proliferation,
migration and epithelial-to-mesenchymal transition in esophageal
squamous cell carcinoma cells (27).
Gene expression levels can be modulated by lncRNA at
epigenetic, transcriptional and post-transcriptional levels
(28). In addition, a number of
lncRNAs contain MREs and bind to certain miRNAs, acting as ceRNAs
to decrease the functions of miRNAs at the post-transcriptional
level (29). In order to screen
potential lncRNAs that may serve as miR-15a-5p sponges, the present
study used bioinformatics analysis and RNA-pull-down assay. The
present study demonstrated that SNHG16 was effectively pulled down
by miR-15a-5p and was primarily located in the cytoplasm of hBMECs.
Luciferase reporter assays demonstrated that miR-15a-5p can
directly bind to SNHG16 via the putative MRE. Furthermore,
induction of SNHG16 significantly decreased relative miR-15a-5p
expression levels in hBMECs. RIP and dual luciferase reporter
assays demonstrated that SNHG16 downregulated the function of
miR-15a-5p by acting as a ceRNA.
Previous studies have demonstrated that miR-15a-5p
regulates endothelial cell apoptosis during I/R injury (11,30).
I/R-induced apoptosis of cardiomyocytes can be promoted by
miR-15a-5p by targeting mothers against decapentaplegic homolog 7;
inhibition of miR-15a-5p has the opposite effect (30). The present study demonstrated that
miR-15a-5p expression levels increased in a time-dependent manner
in hBMECs following OGD-R, which was consistent with previous
reports (9,30). In addition, miR-15a-5p
overexpression significantly decreased the proliferation of hBMECs
by decreasing Bcl-2 expression levels. The results of the present
study support the hypothesis that miR-15a-5p contributes to the
pathogenesis of I/R injury by enhancing OGD-R-induced endothelial
cell apoptosis through targeting of Bcl-2 expression levels.
The present study further determined an association
between miR-15a-5p and SNHG16 in hBMECs proliferation and
apoptosis. The results of the present study indicated that SNHG16
could modulate miR-15a-5p-regulated apoptosis of hBMECs following
OGD-R and that overexpression of miR-15a-5p largely inhibited hBMEC
proliferation upregulated by SNHG16. These results indicated that
SNHG16 promotes hBMEC proliferation by inhibiting miR-15a-5p. In
contrast to the effects of miR-15a-5p, SNHG16 overexpression
significantly upregulated Bcl-2 expression levels in hBMECs
following OGD-R. Furthermore, overexpression of miR-15a-5p notably
reversed the expression levels of Bcl-2 in hBMECs upregulated by
SNHG16. Accordingly, SNHG16 may act as a ceRNA to antagonize
miR-15a-5p and decrease its inhibitory effect on Bcl-2 expression
levels in hBMECs following OGD-R. The results of the present study
support previous observations that endogenous SNHG16 competes with
miR-140-5p and miR-98 to promote the development of esophageal
squamous cell carcinoma and breast cancer (15,27).
The present study highlighted the significance of
the association between SNHG16, miR-15a-5p and transcription factor
Bcl-2 in the regulation of apoptosis in hBMECs. The results of the
present study indicate that SNHG16 may inhibit I/R-induced
apoptosis of hBMECs during cerebral I/R injury. Mechanistically,
SNHG16 may serve as a molecular sponge to target miR-15a-5p and
upregulate Bcl-2 expression levels. Thus, targeting SNHG16-based
mechanisms may provide novel therapeutic strategies and aid in drug
development for I/R injury.
Acknowledgements
Not applicable.
Funding
The present study was supported by a grant from the
Medical and Health Technology Development Program in Yancheng City,
China (grant no. YK2016067).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
MP designed the study and drafted the manuscript.
HT, ML, LQ, HY analyzed and interpreted the data. ML, LQ, HY
performed the experiments. HT critically revised the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
OGD-R
|
oxygen-glucose deprivation and
reoxygenation
|
|
RIP
|
RNA immunoprecipitation
|
|
ceRNA
|
competitive endogenous RNA
|
|
lncRNA
|
long non-coding RNA
|
References
|
1
|
Zheng L, Cheng W, Wang X, Yang Z, Zhou X
and Pan C: Overexpression of MicroRNA-145 ameliorates astrocyte
injury by targeting aquaporin 4 in cerebral ischemic stroke. BioMed
Res Int. 2017:26852017. View Article : Google Scholar
|
|
2
|
Poisson SN, Schardt TQ, Dingman A and
Bernard TJ: Etiology and treatment of arterial ischemic stroke in
children and young adults. Curr Treat Options Neurol. 16:3152014.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Emberson J, Lees KR, Lyden P, Blackwell L,
Albers G, Bluhmki E, Brott T, Cohen G, Davis S, Donnan G, et al
Stroke Thrombolysis Trialists' Collaborative Group, : Effect of
treatment delay, age, and stroke severity on the effects of
intravenous thrombolysis with alteplase for acute ischaemic stroke:
A meta-analysis of individual patient data from randomised trials.
Lancet. 384:1929–1935. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Alkabie S, Basivireddy J, Zhou L, Roskams
J, Rieckmann P and Quandt JA: SPARC expression by cerebral
microvascular endothelial cells in vitro and its influence on
blood-brain barrier properties. J Neuroinflammation. 13:2252016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Yang Q, He GW, Underwood MJ and Yu CM:
Cellular and molecular mechanisms of endothelial
ischemia/reperfusion injury: Perspectives and implications for
postischemic myocardial protection. Am J Transl Res. 8:765–777.
2016.PubMed/NCBI
|
|
6
|
Kong Q, Dai L, Wang Y, Zhang X, Li C,
Jiang S, Li Y, Ding Z and Liu L: HSPA12B attenuated acute
myocardial ischemia/reperfusion injury via maintaining
endothelialIntegrity in a PI3K/Akt/mTOR-dependent mechanism. Sci
Rep. 6:336362016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Sun ZY, Wang FJ, Guo H, Chen L, Chai LJ,
Li RL, Hu LM, Wang H and Wang SX: Shuxuetong injection protects
cerebral microvascular endothelial cells against oxygen-glucose
deprivation reperfusion. Neural Regen Res. 14:783–793. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Suárez Y and Sessa WC: MicroRNAs as novel
regulators of angiogenesis. Circ Res. 104:442–454. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hullinger TG, Montgomery RL, Seto AG,
Dickinson BA, Semus HM, Lynch JM, Dalby CM, Robinson K, Stack C,
Latimer PA, et al: Inhibition of miR-15 protects against cardiac
ischemic injury. Circ Res. 110:71–81. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yin KJ, Olsen K, Hamblin M, Zhang J,
Schwendeman SP and Chen YE: Vascular endothelial cell-specific
microRNA-15a inhibits angiogenesis in hindlimb ischemia. J Biol
Chem. 287:27055–27064. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yin KJ, Deng Z, Hamblin M, Xiang Y, Huang
H, Zhang J, Jiang X, Wang Y and Chen YE: Peroxisome
proliferator-activated receptor delta regulation of miR-15a in
ischemia-induced cerebral vascular endothelial injury. J Neurosci.
30:6398–6408. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang B, Wang D, Ji TF, Shi L and Yu JL:
Overexpression of lncRNA ANRIL up-regulates VEGF expression and
promotes angiogenesis of diabetes mellitus combined with cerebral
infarction by activating NF-κB signaling pathway in a rat model.
Oncotarget. 8:17347–17359. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Liu Y, Zhou D, Li G, Ming X, Tu YF, Tian
J, Lu H and Yu B: Long non coding RNA-UCA1 contributes to
cardiomyocyte apoptosis by suppression of p27 expression. Cell
Physiol Biochem. 35:1986–1998. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Zhang X, Tang X, Liu K, Hamblin MH and Yin
KJ: Long noncoding RNA Malat1 regulates cerebrovascular pathologies
in ischemic stroke. J Neurosci. 37:1797–1806. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cai C, Huo Q, Wang X, Chen B and Yang Q:
SNHG16 contributes to breast cancer cell migration by competitively
binding miR-98 with E2F5. Biochem Biophys Res Commun. 485:272–278.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Christensen LL, True K, Hamilton MP,
Nielsen MM, Damas ND, Damgaard CK, Ongen H, Dermitzakis E, Bramsen
JB, Pedersen JS, et al: SNHG16 is regulated by the Wnt pathway in
colorectal cancer and affects genes involved in lipid metabolism.
Mol Oncol. 10:1266–1282. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Veneziano D, Marceca GP, Di Bella S,
Nigita G, Distefano R and Croce CM: Investigating miRNA-lncRNA
Interactions: Computational Tools and Resources. Methods Mol Biol.
1970:251–277. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang W, Lou W, Ding B, Yang B, Lu H, Kong
Q and Fan W: A novel mRNA-miRNA-lncRNA competing endogenous RNA
triple sub-network associated with prognosis of pancreatic cancer.
Aging (Albany NY). 11:2610–2627. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Wang J and Chen Y, Yang Y, Xiao X, Chen S,
Zhang C, Jacobs B, Zhao B, Bihl J and Chen Y: Endothelial
progenitor cells and neural progenitor cells synergistically
protect cerebral endothelial cells from
Hypoxia/reoxygenation-induced injury via activating the PI3K/Akt
pathway. Mol Brain. 9:122016. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Alluri H, Anasooya Shaji C, Davis ML and
Tharakan B: Oxygen-glucose deprivation and reoxygenation as an in
vitro ischemia-reperfusion injury model for studying blood-brain
barrier dysfunction. J Vis Exp. 99:e526992015.
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
starBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42(D1): D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xin JW and Jiang YG: Long noncoding RNA
MALAT1 inhibits apoptosis induced by oxygen-glucose deprivation and
reoxygenation in human brain microvascular endothelial cells. Exp
Ther Med. 13:1225–1234. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hao R, Hu X, Wu C and Li N:
Hypoxia-induced miR-15a promotes mesenchymal ablation and
adaptation to hypoxia during lung development in chicken. PLoS One.
9:e988682014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Dykstra-Aiello C, Jickling GC, Ander BP,
Shroff N, Zhan X, Liu D, Hull H, Orantia M, Stamova BS and Sharp
FR: Altered expression of long noncoding RNAs in blood after
ischemic stroke and proximity to putative stroke risk loci. Stroke.
47:2896–2903. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Zhang J, Yuan L, Zhang X, Hamblin MH, Zhu
T, Meng F, Li Y, Chen YE and Yin KJ: Altered long non-coding RNA
transcriptomic profiles in brain microvascular endothelium after
cerebral ischemia. Exp Neurol. 277:162–170. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Zhang K, Chen J, Song H and Chen LB:
SNHG16/miR-140-5p axis promotes esophagus cancer cell
proliferation, migration and EMT formation through regulating ZEB1.
Oncotarget. 9:1028–1040. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Ren W and Yang X: Pathophysiology of long
non-coding RNAs in ischemic stroke. Front Mol Neurosci. 11:962018.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ebert MS, Neilson JR and Sharp PA:
MicroRNA sponges: Competitive inhibitors of small RNAs in mammalian
cells. Nat Methods. 4:721–726. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yang Y, Ding S, Xu G, Chen F and Ding F:
MicroRNA-15a inhibition protects against
hypoxia/reoxygenation-induced apoptosis of cardiomyocytes by
targeting mothers against decapentaplegic homolog 7. Mol Med Rep.
15:3699–3705. 2017. View Article : Google Scholar : PubMed/NCBI
|