Introduction
Carotid restenosis has been associated with an
increased risk of ischaemic stroke and with increasing morbidity
and mortality (1,2). Studies investigating the pathogenesis
of carotid restenosis are vital for the development of novel
prevention and treatment strategies. Abnormal proliferation and
migration of vascular smooth muscle cells (VSMCs) is a common
pathogenetic factor in carotid artery stenosis. Inhibition of the
hypertrophy of VSMCs might be an effective method for treatment of
carotid artery stenosis.
Docosahexaenoic acid (DHA) is an important component
of fish oil, which is generally used in the prevention of
cardiovascular disease (3). A
number of previous studies demonstrated the anti-inflammatory and
lipid-regulating effects of DHA in cardiovascular disease (4,5). The
mechanism underlying the DHA protective effect in cardiovascular
diseases requires additional investigation. DHA has been
demonstrated to regulate the VSMC cycle by stimulating apoptosis,
and by inhibiting proliferation and migration (6,7). In
another previous study, the beneficial effect of DHA on the
migration of VSMCs was partially dependent on the regulation of
matrix metalloproteinase (MMP)-2 and MMP-9 (8). Moreover, DHA could increase nitric
oxide (NO) production through activation of the p44/p42/MAPK
signalling in VSMCs (6). Another
mechanism of the protective effect of DHA involves regulation of
VSMC proliferation through the inhibition of phosphorylation of the
cyclin- dependent kinase 2/cyclin E complex (9). However, the mechanism underlying the
inhibitory effect of DHA on the proliferation and migration of
VSMCs requires further investigation.
The protective mechanism of DHA in vascular
pathology has been partially elucidated, which suggested that
biological changes were predominantly caused by an abnormal
expression of genes involved in proliferation, migration, cell
cycle and apoptosis (10–13). Moreover, several previous studies
have reported that non-coding RNAs, including microRNAs, regulate
these biological processes. For instance, microRNA-155 is a
multifunctional non-coding RNA which regulates the pathological
processes involved in cardiovascular disease (14). Indeed, microRNA-155 participates in
the regulation of haematopoietic lineage differentiation, vascular
remodelling, viral infection and inflammation (15–18).
Genetic deficiency of microRNA-155 in cardiomyocytes prevents
cardiac hypertrophy and failure (19). Furthermore, mircroRNA-155 has been
shown to act as a tumour suppressor and an oncogene in a various
cancer types (20,21). MicroRNA-155 can regulate glucose
usage through the PIK3R1-PDK/AKT-FOXO3a-cMYC axis, according to a
previous study on 50 triple-negative breast cancer specimens
(22). However, it is unclear
whether microRNA-155 can modulate the VSMC phenotype, or
participate in the protective effect of DHA in carotid
restenosis.
In the present study, DHA had an inhibitory effect
on the proliferation and migration of VSMCs, which was mediated by
regulation of miR-155 expression. The present results could be
applied in the clinical prevention of carotid restenosis. In
addition, the present study identified a positive effect of
microRNA-155 on the proliferation and migration of VSMCs, in
agreement with previous studies (23,24).
The phenotype changes of the DHA-stimulated VSMCs were reversed by
inhibiting the expression of microRNA-155. Overall, these effects
may involve regulation of the miR-155 target gene suppressor of
cytokine signalling 1 (Socs1).
Materials and methods
Cell culture
VSMCs were isolated from the aorta of 8-week-old
male Wistar rats (180–270 g) (Vital River), according to a
previously published protocol (8).
The Ethics Committee of Hwa Mei Hospital, University of Chinese
Academy of Sciences (Ningbo No. 2 Hospital) approved the present
study. Rats were housed at 20–25°C with a 12 h light/dark cycle and
free access to food and water. Briefly, adventitia was quickly
removed from a sacrificed rat for enzymatic digestion after
removing the fat tissue and mechanically extruding the media. Cells
were cultured in DMEM, supplemented with 10% FBS (both Gibco;
Thermo Fisher Scientific, Inc.) and 100 U/ml penicillin and 0.1
mg/ml streptomycin antibiotics. All cultured cells were maintained
at 37°C in a humidified 5% CO2 incubator. The cells were
passaged 3–7 times before performing the experiments. The cell type
and the maintenance of the differentiated state were confirmed by
the presence of a well-structured network of actin stress fibres,
according to the immunochemistry data (data not shown).
MicroRNA transfection
The microRNA-155 mimics, mimics negative control,
microRNA-155 inhibitors and inhibitor negative control were
obtained from Guangzhou RiboBio Co., Ltd. (sequences not
available). VSMCs were seeded into 6-well cell culture plates at
density of 5×106 cells per well prior to transfection.
Following a 20-h incubation, cells were transfected with miRNA at
final concentration of 50 nM using Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. After a 6-h incubation with miRNAs,
the medium was changed to fresh complete medium. Transfected cells
were grown for 24 h prior to subsequent experiments.
RNA isolation and reverse
transcription-quantitative PCR (RT-qPCR)
VSMCs were treated with 50 µM DHA (Sigma-Aldrich;
Merck KGaA) for 2 h, then cells were transfected with microRNA-155
mimics and inhibitors before harvesting for RNA isolation. VSMCs
were centrifuged at 4°C with 700 × g for 5 min for harvesting after
cells were transfected for 24 h. RNA was extracted using
TRIzol® (Invitrogen; Thermo Fisher Scientific, Inc.),
according to the manufacturer's protocol. The concentration, purity
and integrity of the RNA were measured using an ultraviolet
spectrophotometer, as well as 0.8% agarose gel electrophoresis.
Reverse transcription was performed using an RT kit (MicroRNA
Reverse Transcription kit; Applied Biosystems; Thermo Fisher
Scientific, Inc.) with 1 µg total RNA. The RT conditions were as
follows: 25°C for 10 min, 37°C for 120 min, and 85°C for 5 min. The
PCR primer sequences for miR-155 were: Forward,
5′-CTGTTAATGCTAATCGTGATAG-3′ and reverse, 5′-GCAGGGTCCGAGGT-3′. The
PCR primer sequences for Socs1 were 5′-CTGCGGCTTCTATTGGGGAC-3′ and
5′-AAAAGGCAGTCGAAGGTCTCG-3′. The PCR primers of snoRNA U6 (internal
control) were: Forward, 5′-GCGCGTCGTGAAGCGTTC-3′ and reverse,
5′-GTGCAGGGTCCGAGGT-3′. The PCR primers of GAPDH (internal control)
were: Forward, 5′-CGGAGTCAACGGATTTGGTCGTAT-3′ and reverse,
5′-AGCCTTCTCCATGGTGGTGAAGAC-3′. The fluorophore was SYBR Green
(Roche Applied Science). The real-time PCR conditions were as
follows: 95°C for 10 min, 60°C for 10 sec, 72°C for 30 sec (40
cycles) and 4°C for 5 min. The real-time PCR amplification scheme
was performed according to the manufacturer's instructions (Roche
Applied Science). The concentration of microRNAs were calculated by
the 2−ΔΔCq method (25).
Cell viability
After the VSMCs were transfected with the miR-155
mimics and inhibitors, the VSCMs were grown in 96-well cell culture
plates at 5×105 cells per well. The VSMCs were then
harvested and kept in the same medium for the detection of
proliferation, following standard culture for 24, 48 or 72 h. Cell
proliferation was assessed using a Cell Counting Kit-8 (CCK-8)
assay (Abcam). All procedures were performed according to the
manufacturer's protocol. Absorbance was detected using a microplate
reader at 450 nm.
Wound healing assay
VSMCs were treated with 50 µM DHA and transfected
with microRNA-155 mimics and inhibitors before evaluating their
migration in the wound heal assay. After transfection for 6 h, a
sterile 10-µl white tip was used to draw a straight line across the
3.5 cm dish, with the tip on the bottom of the plate, and the
growth medium was replaced with serum-free medium. At this time
point, 0 h, images were recorded using a light microscope
(magnification, ×40). Subsequently, microscopic images were
recorded at 24 and 48 h after wounding.
Luciferase reporter assay
Socs1, the target gene of microRNA-155, was
predicted by inputting the microRNA sequence in TargetScan web
(targetscan.org/). The Socs1-3′ untranslated
region (UTR) DNA fragment, including the putative microRNA-155
binding sequence, was cloned into the PGL3-basic (Promega
Corporation) (hereafter referred to as PGL3/Socs1-3′UTR).
Additionally, a mutant Socs1-3′UTR plasmid was constructed by
mutating the miR-155 binding sequence (referred to as
PGL3/Socs1-3′UTR mutant thereafter). VSMCs were plated in a 24-well
plate at 1×105 cells per well before transfection using
Lipofectamine 3000) (Thermo Fisher Scientific, Inc.). Each
transfection contained the vector DNA and pRL-SV40 plasmid. The
pGL3-basic vector was used as a negative control. PRL-TK (Promega
Corporation) was used as the internal control. Luciferase assays
were performed 48 h after transfection, and independent experiments
were repeated three times for each plasmid construct according to
the manufacturer's protocol (Promega Corporation). The luciferase
activity was measured via fluorescence spectrophotometer (Luminex
Corporation). The ratio between luciferase activity and
Renilla luciferase activity represented the value of
measured sample.
Western blotting
VSMCs were treated with 50 µM DHA and transfected
with the microRNA-155 mimics and inhibitors before harvesting for
protein isolation. Proteins were isolated in the protein extraction
buffer (Beyotime Institute of Biotechnology). The protein
concentration was measured by BCA method. Protein samples (20
µg/lane) were separated by SDS-PAGE on 10% gels and transferred to
a nitrocellulose membrane. The membranes were incubated with 5%
fat-free milk in TBS with Tween® 20 for 1 h at room
temperature and with primary antibodies overnight at 4°C. The PVDF
membranes were washed with TBST buffer and incubated with
peroxidase-conjugated specific secondary antibodies from Abcam
[Ki67 (1:5,000; cat. no. ab15580), Socs1 (1:4,000; cat. no.
ab62584) and actin (1:3,000; cat. no. ab6276)] with shaking for 1
h. The enhanced chemiluminescence solution (Sigma-Aldrich; Merck
KGaA) was the prepared in a dark room, and the exposure time of the
film was determined according to the chemiluminescence intensity.
Densitometric analysis was performed using Image Lab software
(version 3.0; Bio-Rad Laboratories, Inc.).
Statistical analysis
Results are presented as the mean ± standard
deviation of three replicate experiments. For multiple comparisons,
statistical analysis was conducted using ANOVA, followed by Sidak's
post hoc test. A Student's t-test was used for statistical analysis
of two groups. All the experiments were repeated at least for three
times. P<0.05 was considered to indicate a statistically
significant difference. IBM SPSS Statistics software (version 21.0;
IBM Corp.) was used to perform all the statistical analyses.
Results
DHA inhibits the proliferation and
migration of VSMCs
VSMCs were stimulated with 50 µM DHA to evaluate its
effects on proliferation and migration. VSMC proliferation
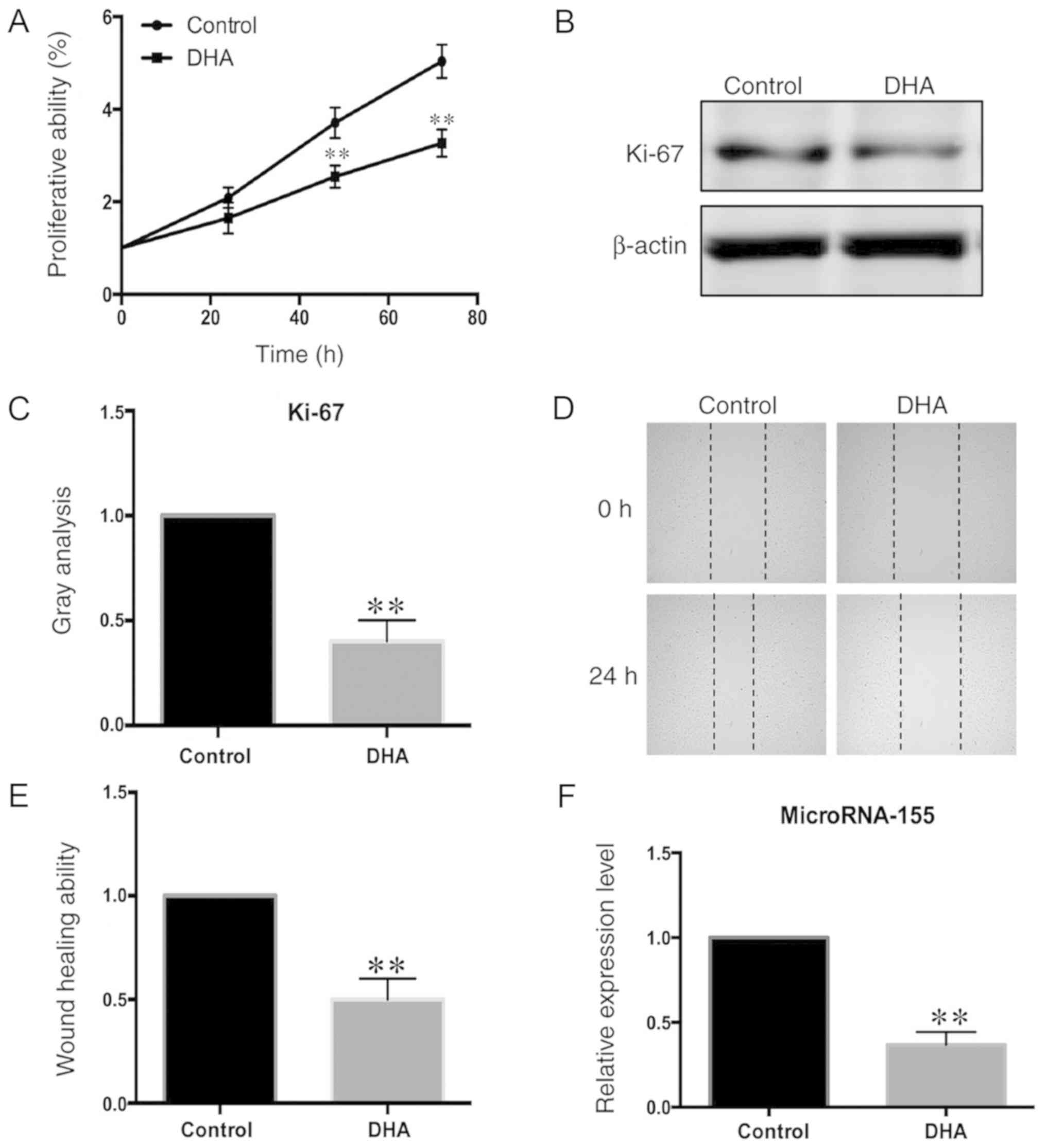
following DHA treatment was evaluated using the CCK-8 assay and by
measuring the Ki-67 protein levels (Fig. 1A and B). The proliferation of VSMCs
treated with DHA for 48 h was significantly decreased, compared
with untreated cells (Fig. 1A).
The effect of DHA on VSMC migration was then assessed, using a
wound healing assay. The migration of VSMCs treated with DHA was
also decreased, compared with untreated cells (Fig. 1C-E).
Expression levels of microRNA-155 are
decreased in DHA-stimulated VSMCs
MicroRNA-155 can regulate the immune response and
development of T cells (26,27).
In order to determine whether microRNA-155 might participate in the
regulation of the biological activities of VSMCs, the expression
levels of microRNA-155 in DHA-stimulated VSMCs were measured using
RT-qPCR. The expression levels of microRNA-155 were significantly
decreased in DHA-treated cells, compared with untreated cells
(Fig. 1F).
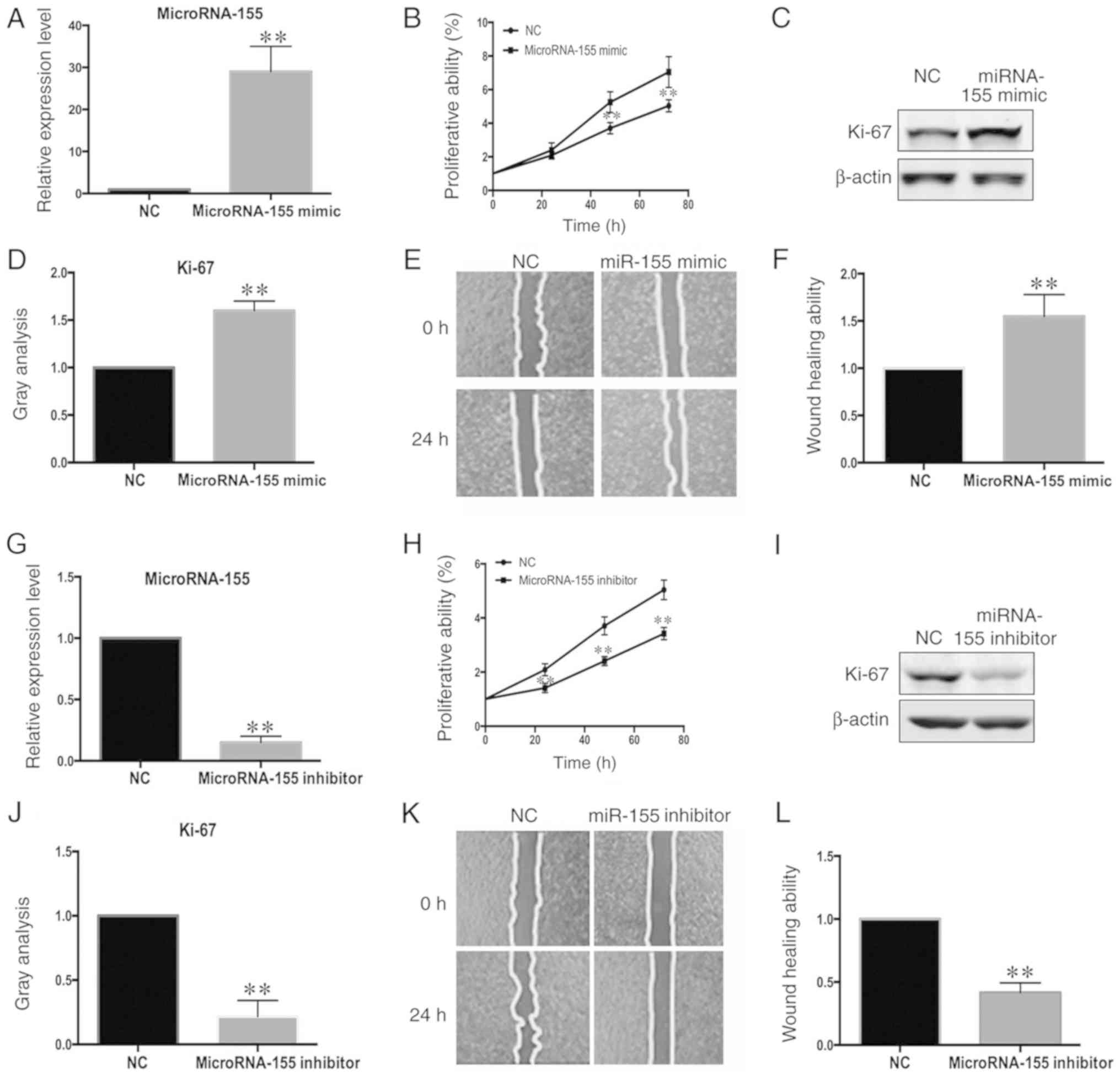
MicroRNA-155 regulates the
proliferation and migration of VSMCs
The role of miR-155 in the regulation of VSMC
proliferation and migration was also investigated. The
proliferation of VSMCs transfected with miR-155 mimic (Fig. 2A) and microRNA-155-inhibitor
(Fig. 2G) was evaluated using a
CCK-8 assay and by measuring the protein levels of Ki-67. The
proliferation of VSMCs was increased by the microRNA-155 mimic
(Fig. 2B-D). Conversely, the
proliferation of VSMCs transfected with the microRNA-155 inhibitor
was significantly decreased (Fig.
2H-J). These results demonstrated that microRNA-155 can
regulate the proliferation of VSMCs. The potential effect of
microRNA-155 on VSMC migration was then assessed. After confirming
the efficiency of overexpression and interference by RT-qPCR
(Fig. 2A and G), the migration of
VSMCs was evaluated using a wound healing assay. The migration of
VSMCs was increased following microRNA-155 overexpression (Fig. 2E and F) and was decreased following
transfection of microRNA-155-inhibitor (Fig. 2K and L). These results suggested
that microRNA-155 can regulate the migration potential of
VSMCs.
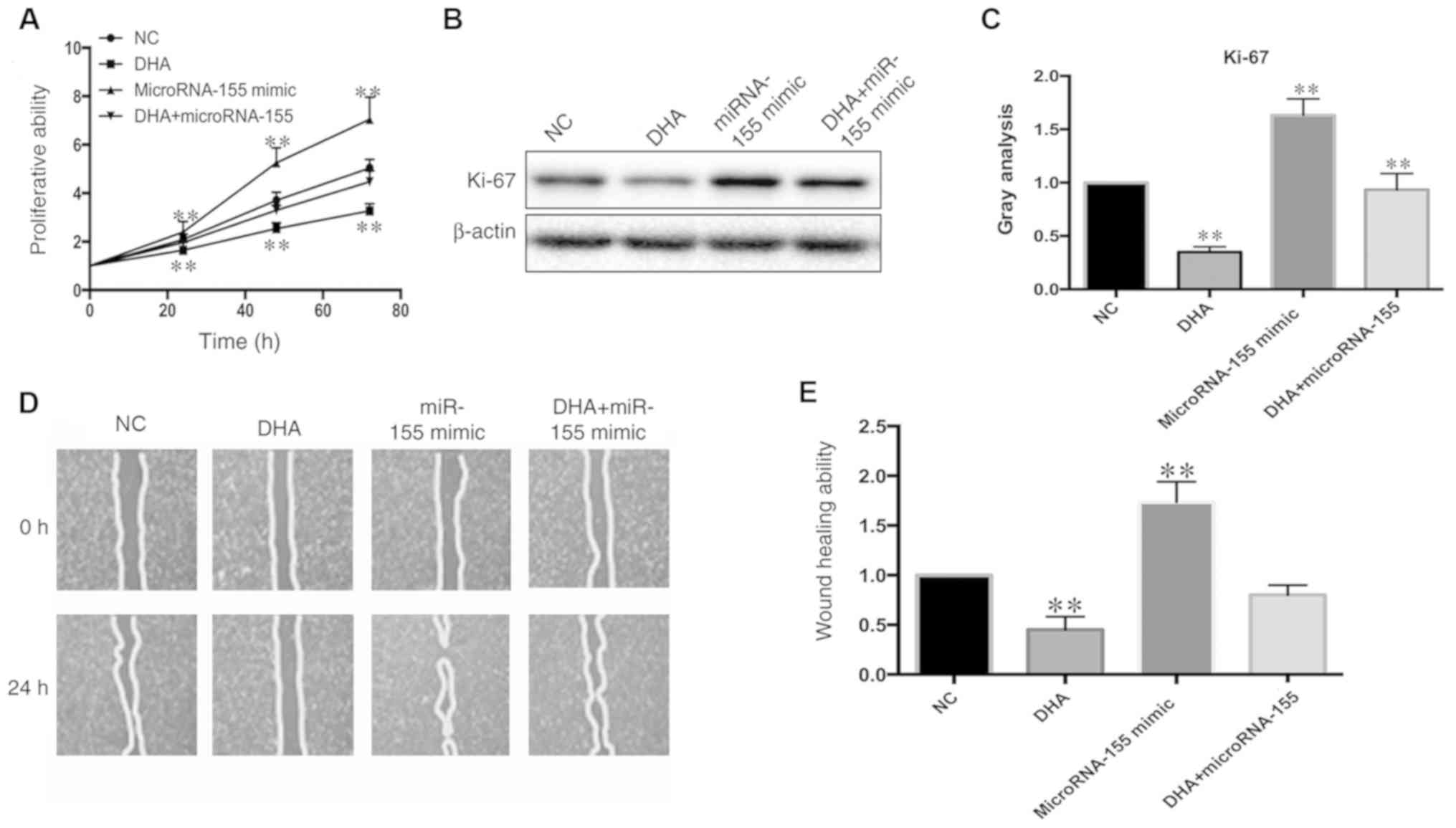
Restoration of the expression level of
microRNA-155 partially reverses the inhibition of VSMC
proliferation and migration
MicroRNA-155 plays an essential role in the
regulation of VSMC proliferation and migration (7). In order to investigate whether a
decrease in proliferation and migration in the DHA-stimulated VSMCs
was caused by microRNA-155 downregulation, VSMCs were transfected
with microRNA-155 mimic, then treated with DHA (50 µM). The
proliferation of VSMCs was analysed using a CCK-8 assay and
measurement of Ki-67 protein levels. The inhibition of VSMC
proliferation was partially reversed by microRNA-155 overexpression
(Fig. 3A-C). In agreement with
this, the migration of DHA-stimulated VSMCs was also reversed
according to the wound healing assay (Fig. 3D and E). Thus, overexpression of
microRNA-155 restored the proliferation and migration of
DHA-stimulated VSMCs.
Socs1 is the target gene of
microRNA-155 in VSMCs
Identification of the target gene of microRNA-155 is
important for understanding the biological functions of
microRNA-155, and is required to investigate a possible mechanism
of the regulatory effects of microRNA-155 on VSMC proliferation and
migration. According to the target gene prediction data generated
by TargetScan (targetscan.org/), Socs1 was a
candidate target based on a study in macrophages (28). Socs1 is a suppressor of cytokine
signalling, which is required for cell growth and survival
(28). The mRNA levels of Socs1
were significantly increased following transfection with the
microRNA-155-mimics, compared with the control (Fig. 4A). By contrast, the expression of
Socs1 was increased in the absence of microRNA-155 (Fig. 4B). In order to find direct evidence
for the interaction between Socs1 and microRNA-155, a luciferase
plasmid containing the Socs1 gene 3′UTR (PGL3/Socs1-3′UTR) was
constructed. Transfection of the microRNA-155-mimic significantly
suppressed the luciferase activity of PGL3/Socs1-3′UTR (Fig. 4C) but had no effect on the
PGL3/Socs1-3′UTR mutant plasmid (Fig.
4D), suggesting that Socs1 is the direct target gene of
microRNA-155. In order to demonstrate that Socs1 is regulated by
DHA through microRNA-155, the expression levels of Socs1 were
measured in DHA-stimulated VSMCs. Both the protein and mRNA levels
of Socs1 were increased following DHA stimulation (Fig. 4E-G).
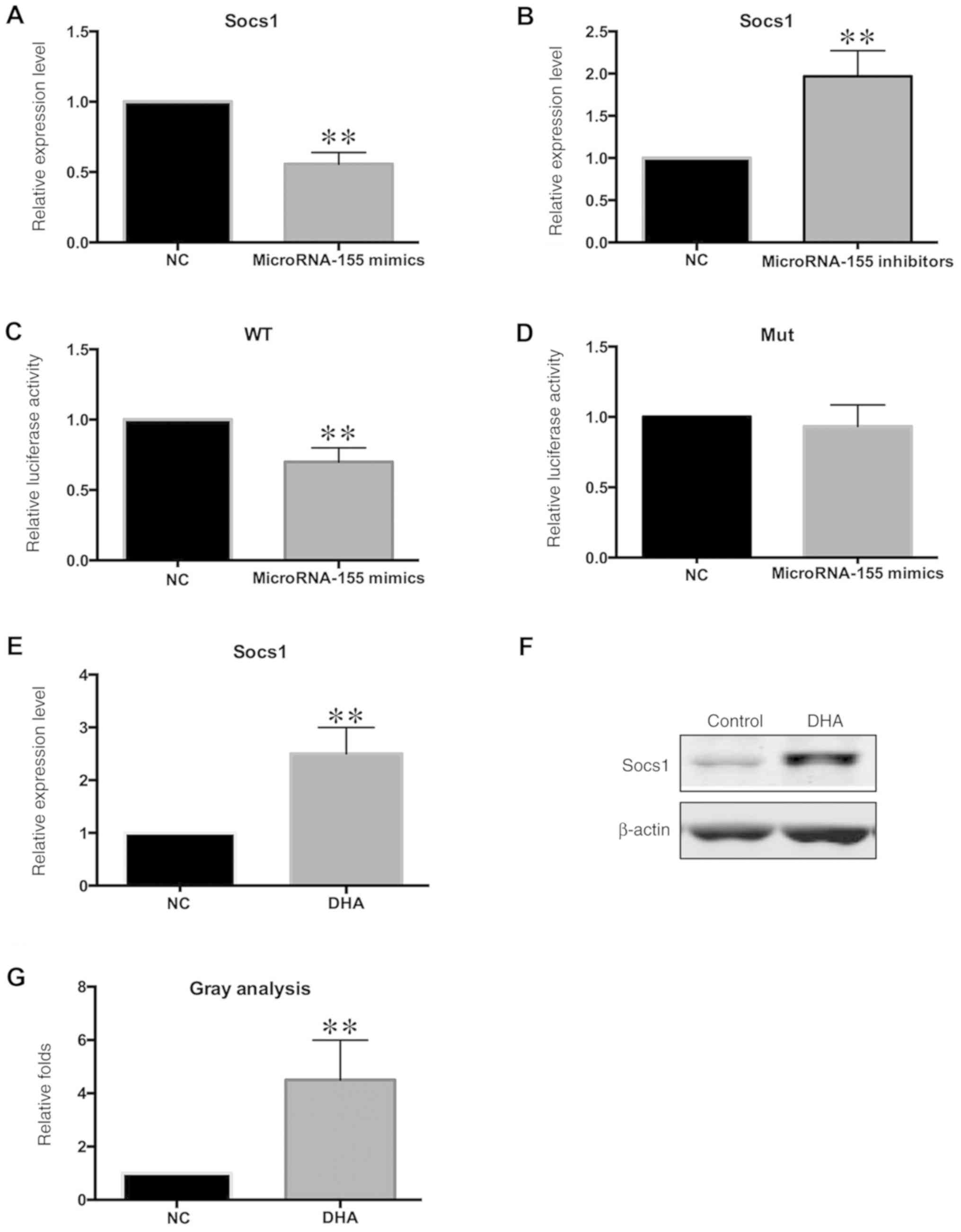 | Figure 4.Socs1 is the target gene of
microRNA-155. VSMCs were transfected with miR-155-mimic and
inhibitor. Expression levels of Socs1 was analysed by RT-qPCR. (A)
Expression level of Socs1 was decreased in
miR-155-mimic-transfected cells, vs. NC. (B) Expression level of
Socs1 was increased in miR-155-inhibitor-transfected cells,
compared with NC. (C) Luciferase plasmid WT PGL3/Socs1-3′UTR was
transfected, together with miR-155-mimic. Luciferase activity of WT
PGL3/Socs1-3′UTR was significantly decreased. (D) Luciferase
plasmid Mut PGL3/Socs1-3′UTR was transfected, together with
miR-155-mimic. Luciferase activity of Mut PGL3/Socs1-3′UTR remained
unchanged. VSMCs were treated with 50 µM DHA, and Socs1 expression
levels were analysed by RT-qPCR and western blotting. (E)
Expression levels of Socs1 were increased in the DHA-stimulated
VSMCs, compared with NC. (F) Protein levels of Socs1 were increased
in the DHA-stimulated VSMCs, compared with NC. (G) Gray analysis of
the relative protein levels of Socs1. **P<0.01 compared with NC.
VSMCs, vascular smooth muscle cells; DHA, docosahexaenoic acid; NC,
negative control; RT-qPCR, reverse transcription-quantitative PCR;
WT, wild-type; Mut, mutant; miR, microRNA; Socs1, suppressor of
cytokine signalling. |
Discussion
In the present study, the protective mechanism of
DHA in the inhibition of VSMC proliferation and migration was
investigated. Previous studies on the protective effect of DHA in
cardiovascular diseases focused on changes in gene expression
(29,30). The present study demonstrated that
the inhibitory effect of DHA on the proliferation and migration of
VSMCs is mediated by microRNA-155 downregulation. MicroRNA-155 is a
multifunctional non-coding RNA that plays an important role in the
pathogenesis of carotid restenosis (31). However, the Socs1 target gene of
microRNA-155 can regulate various signalling pathways, which may
contribute to the function of microRNA-155 in the regulation of
VSMC proliferation and migration.
A recent study demonstrated that low DHA levels were
significantly associated with lipid-rich coronary and carotid
plaques, yet the mechanism of DHA in the stabilization of carotid
plaque and its beneficial effects in patients with carotid
restenosis require additional investigation (32). DHA is an important component of ω-3
fatty acids, which can enhance Gefitinib sensitivity and induce
apoptosis in PC-9/GR cells (33).
According to nutritional statistical data, low levels of ω-3 fatty
acids are associated with cerebral small vessel diseases,
hypertension, cardiovascular dysfunction and acute ischaemic stroke
(34,35). The present study focused on the
protective function of DHA in carotid restenosis by evaluating its
effect on VSMC hyperplasia through the regulation of proliferation
and migration.
MicroRNA-155 is one of the most multifunctional
non-coding RNAs, and it participates in the regulation of numerous
biological functions in various cell types, including cells of the
immune system (36). In the
present study, microRNA-155 regulated the proliferation and
migration of VSMCs, possibly through its target gene, Socs1. As a
multifunctional microRNA, microRNA-155 could also participate in
other biological processes. Numerous previous studies have been
published on the function of microRNA-155 and the mechanisms
underlying its effects (36–38).
MicroRNA-155 could inhibit apoptosis by regulating PTEN signalling
in psoriasis (37). The function
of microRNA-155 in cardiac hypertrophy was demonstrated in a mouse
model, suggesting that loss of microRNA-155 expression might
prevent the progression of heart failure (38). In cancer cells, microRNA-155
deficiency can prevent tumour growth by enhancing the function and
recruitment of tumour suppressor cells in the tumour
micro-environment (39). The
function of microRNA-155 is mediated by regulation of a series of
signalling pathways. In fibrosis, microRNA-155 was strongly
associated with the activation of Wnt/β-catenin signalling
(40). Moreover, a previous study
demonstrated that the immunoregulatory function of microRNA-155 was
mediated by the negative regulation of Akt and Stat5 signalling
(41). Socs1 is not the only known
target gene of microRNA-155 (42);
various signalling pathways regulated by microRNA-155 may
contribute to the phenotype of DHA-stimulated VSMCs. The present
study predominantly focused on the mechanism of microRNA-155
regulation of cell proliferation and migration.
Socs1 is an important target gene of microRNA-155
according to numerous previous studies (43,44).
However, its functions in microvascular endothelial cells and VSMCs
remain poorly studied. Several previous studies focused on the
regulatory role of Socs1 in the immune system; for instance, Socs1
was demonstrated to be involved in the inflammatory response in
atherogenesis (45,46). In cancer cells, the Socs1 gene acts
a tumour suppressor and is frequently silenced by hypermethylation
of the promoter (47). Socs1
tumour suppressor activity is mediated by enhancing p53 tumour
suppressor activity and by inhibiting the Met receptor (47). In the present study, Socs1
expression levels were increased in DHA-stimulated VSMCs,
suggesting that it may serve a role in the proliferation and
migration of VSMCs. However, the underlying mechanism requires
further investigation, and the function of Socs1 in VSMCs still
requires extensive evaluation.
In conclusion, the present study demonstrated that
DHA could inhibit the proliferation and migration of VSMCs, which
may be a possible mechanism for the protective effect of DHA in
carotid restenosis. To the best of our knowledge, the present study
is the first to indicate that changes in microRNA expression may
influence the activity of VSMCs. The function of microRNA-155 might
be varied in different cell types. Further studies and experiments
are required, and should focus on functional studies of
microRNA-155 in vascular disease.
Acknowledgements
Not applicable.
Funding
The present study was supported by Ningbo Health
Branding Subject Fund (grant no. PPXK2018-01), Ningbo Natural
Science Fund (grant no. 2019A610345) and Hwamei Scientific Research
Fund (grant no. 2018HMKY56).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
DL and XY designed and initiated the present study.
XY and CX carried out the cell culture experiments. QX performed
RT-qPCR analysis in cultured cells. DL and XY wrote the manuscript.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by The Ethics
Committee of Hwa Mei Hospital, University of Chinese Academy of
Sciences (Ningbo No. 2 Hospital).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Bonati LH, Ederle J, Dobson J, Engelter S,
Featherstone RL, Gaines PA, Beard JD, Venables GS, Markus HS,
Clifton A, et al: Length of carotid stenosis predicts
peri-procedural stroke or death and restenosis in patients
randomized to endovascular treatment or endarterectomy. Int J
Stroke. 9:3396–305. 2014. View Article : Google Scholar
|
|
2
|
Plummer C, Henderson RD, O'Sullivan JD and
Read SJ: Ischemic stroke and transient ischemic attack after head
and neck radiotherapy: A review. Stroke. 42:2410–2418. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schunck WH, Konkel A, Fischer R and
Weylandt KH: Therapeutic potential of omega-3 fatty acid-derived
epoxyeicosanoids in cardiovascular and inflammatory diseases.
Pharmacol Ther. 183:177–204. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Iverson C, Bacong A, Liu S, Baumgartner S,
Lundstrom T, Oscarsson J and Miner JN: Omega-3-carboxylic acids
provide efficacious anti-inflammatory activity in models of
crystal-mediated inflammation. Sci Rep. 8:12172018. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Trattner S, Ruyter B, Ostbye TK,
Kamal-Eldin A, Moazzami A, Pan J, Gjoen T, Brännäs E, Zlabek V and
Pickova J: Influence of dietary sesamin, a bioactive compound on
fatty acids and expression of some lipid regulating genes in Baltic
Atlantic salmon (Salmo salar L.) juveniles. Physiol Res.
60:125–137. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Hirafuji M, Machida T, Tsunoda M, Miyamoto
A and Minami M: Docosahexaenoic acid potentiates interleukin-1beta
induction of nitric oxide synthase through mechanism involving
p44/42 MAPK activation in rat vascular smooth muscle cells. Br J
Pharmacol. 136:613–619. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhang J, Zhao F, Yu X, Lu X and Zheng G:
MicroRNA-155 modulates the proliferation of vascular smooth muscle
cells by targeting endothelial nitric oxide synthase. Int J Mol
Med. 35:1708–1714. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Delbosc S, Glorian M, Le Port AS, Bereziat
G, Andreani M and Limon I: The benefit of docosahexanoic acid on
the migration of vascular smooth muscle cells is partially
dependent on Notch regulation of MMP-2/-9. Am J Pathol.
172:1430–1440. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Terano T, Tanaka T, Tamura Y, Kitagawa M,
Higashi H, Saito Y and Hirai A: Eicosapentaenoic acid and
docosahexaenoic acid inhibit vascular smooth muscle cell
proliferation by inhibiting phosphorylation of Cdk2-cyclinE
complex. Biochem Biophys Res Commun. 254:502–506. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Newell M, Baker K, Postovit LM and Field
CJ: A critical review on the effect of docosahexaenoic acid (DHA)
on cancer cell cycle progression. Int J Mol Sci. 18:17842017.
View Article : Google Scholar
|
|
11
|
Rani K and Aung NY: Docosahexaenoic acid
inhibits vascular smooth muscle cell proliferation induced by
glucose variability. Open Biochem J. 11:56–65. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Oono K, Takahashi K, Sukehara S, Kurosawa
H, Matsumura T, Taniguchi S and Ohta S: Inhibition of PC3 human
prostate cancer cell proliferation, invasion and migration by
eicosapentaenoic acid and docosahexaenoic acid. Mol Clin Oncol.
7:217–220. 2017.PubMed/NCBI
|
|
13
|
Geng L, Zhou W, Liu B, Wang X and Chen B:
DHA induces apoptosis of human malignant breast cancer tissues by
the TLR-4/PPAR-α pathways. Oncol Lett. 15:2967–2977.
2018.PubMed/NCBI
|
|
14
|
Sun Y, Wang K, Ye P, Wu J, Ren L, Zhang A,
Huang X, Deng P, Wu C, Yue Z, et al: MicroRNA-155 promotes the
directional migration of resident smooth muscle progenitor cells by
regulating monocyte chemoattractant protein 1 in transplant
arteriosclerosis. Arterioscler Thromb Vasc Biol. 36:1230–1239.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Li X, Kong D, Chen H, Liu S, Hu H, Wu T,
Wang J, Chen W, Ning Y, Li Y and Lu Z: miR-155 acts as an
anti-inflammatory factor in atherosclerosis-associated foam cell
formation by repressing calcium-regulated heat stable protein 1.
Sci Rep. 6:217892016. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Jin C, Cheng L, Lu X, Xie T, Wu H and Wu
N: Elevated expression of miR-155 is associated with the
differentiation of CD8+ T cells in patients with HIV-1. Mol Med
Rep. 16:1584–1589. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Pacurari M and Tchounwou PB: Role of
MicroRNAs in renin-angiotensin-aldosterone system-mediated
cardiovascular inflammation and remodeling. Int J Inflam.
2015:1015272015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Izzard L, Dlugolenski D, Xia Y, McMahon M,
Middleton D, Tripp RA and Stambas J: Enhanced immunogenicity
following miR-155 incorporation into the influenza a virus genome.
Virus Res. 235:115–120. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Heymans S, Corsten MF, Verhesen W, Carai
P, van Leeuwen RE, Custers K, Peters T, Hazebroek M, Stöger L,
Wijnands E, et al: Macrophage microRNA-155 promotes cardiac
hypertrophy and failure. Circulation. 128:1420–1432. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Bhattacharya S, Chalk AM, Ng AJ, Martin
TJ, Zannettino AC, Purton LE, Lu J, Baker EK and Walkley CR:
Increased miR-155-5p and reduced miR-148a-3p contribute to the
suppression of osteosarcoma cell death. Oncogene. 35:5282–5294.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ji Y, Wrzesinski C, Yu Z, Hu J, Gautam S,
Hawk NV, Telford WG, Palmer DC, Franco Z, Sukumar M, et al: miR-155
augments CD8+ T-cell antitumor activity in lymphoreplete hosts by
enhancing responsiveness to homeostatic gammac cytokines. Proc Natl
Acad Sci USA. 112:476–481. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Kim S, Lee E, Jung J, Lee JW, Kim HJ, Kim
J, Yoo HJ, Lee HJ, Chae SY, Jeon SM, et al: microRNA-155 positively
regulates glucose metabolism via PIK3R1-FOXO3a-cMYC axis in breast
cancer. Oncogene. 37:2982–2991. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yang LX, Liu G, Zhu GF, Liu H, Guo RW, Qi
F and Zou JH: MicroRNA-155 inhibits angiotensin II-induced vascular
smooth muscle cell proliferation. J Renin Angiotensin Aldosterone
Syst. 15:109–116. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang G, Zhong L, Luo H and Wang S:
MicroRNA-155-3p promotes breast cancer progression through
down-regulating CADM1. Onco Targets Ther. 12:7993–8002. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang CR, Zhu HF and Zhu Y: Knockout of
microRNA-155 ameliorates the Th17/Th9 immune response and promotes
wound healing. Curr Med Sci. 39:954–964. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Knolle MD, Chin SB, Rana BMJ, Englezakis
A, Nakagawa R, Fallon PG, Git A and McKenzie ANJ: MicroRNA-155
protects group 2 innate lymphoid cells from apoptosis to promote
type-2 immunity. Front Immunol. 9:22322018. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Saleh M, Friedl A, Srivastava M, Soliman
H, Secombes CJ and El-Matbouli M: STAT3/SOCS3 axis contributes to
the outcome of salmonid whirling disease. PLoS One.
15:e02344792020. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Oono K, Ohtake K, Watanabe C, Shiba S,
Sekiya T and Kasono K: Contribution of Pyk2 pathway and reactive
oxygen species (ROS) to the anti-cancer effects of eicosapentaenoic
acid (EPA) in PC3 prostate cancer cells. Lipids Health Dis.
19:152020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Watanabe Y and Tatsuno I: Prevention of
cardiovascular events with omega-3 polyunsaturated fatty acids and
the mechanism involved. J Atheroscler Thromb. 27:183–198. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Farina FM, Hall IF, Serio S, Zani S,
Climent M, Salvarani N, Carullo P, Civilini E, Condorelli G, Elia L
and Quintavalle M: miR-128-3p is a novel regulator of vascular
smooth muscle cell phenotypic switch and vascular diseases. Circ
Res. 126:e120–e135. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Nakagawa I, Park HS, Yokoyama S, Wada T,
Yamada S, Motoyama Y, Kichikawa K and Nakase H: Pretreatment with
and ongoing use of omega-3 fatty acid ethyl esters reduce the
slow-flow phenomenon and prevent in-stent restenosis in patients
undergoing carotid artery stenting. J Vasc Surg. 66:122–129. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ding X, Ge L, Yan A, Ding Y, Tao J, Liu Q
and Qiao C: Docosahexaenoic acid serving as sensitizing agents and
gefitinib resistance revertants in EGFR targeting treatment. Onco
Targets Ther. 12:10547–10558. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Song TJ, Chang Y, Shin MJ, Heo JH and Kim
YJ: Low levels of plasma omega 3-polyunsaturated fatty acids are
associated with cerebral small vessel diseases in acute ischemic
stroke patients. Nutr Res. 35:368–374. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Morin C, Rousseau E, Blier PU and Fortin
S: Effect of docosahexaenoic acid monoacylglyceride on systemic
hypertension and cardiovascular dysfunction. Am J Physiol Heart
Circ Physiol. 309:H93–H102. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Yee D, Shah KM, Coles MC, Sharp TV and
Lagos D: MicroRNA-155 induction via TNF-α and IFN-γ suppresses
expression of programmed death ligand-1 (PD-L1) in human primary
cells. J Biol Chem. 292:20683–20693. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Xu L and Leng H, Shi X, Ji J, Fu J and
Leng H: miR-155 promotes cell proliferation and inhibits apoptosis
by PTEN signaling pathway in the psoriasis. Biomed Pharmacother.
90:524–530. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Seok HY, Chen J, Kataoka M, Huang ZP, Ding
J, Yan J, Hu X and Wang DZ: Loss of MicroRNA-155 protects the heart
from pathological cardiac hypertrophy. Circ Res. 114:1585–1595.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang J, Yu F, Jia X, Iwanowycz S, Wang Y,
Huang S, Ai W and Fan D: MicroRNA-155 deficiency enhances the
recruitment and functions of myeloid-derived suppressor cells in
tumor microenvironment and promotes solid tumor growth. Int J
Cancer. 136:E602–E613. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wan YC, Li T, Han YD, Zhang HY, Lin H and
Zhang B: MicroRNA-155 enhances the activation of Wnt/β-catenin
signaling in colorectal carcinoma by suppressing HMG-box
transcription factor 1. Mol Med Rep. 13:2221–2228. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Tu YX, Wang SB, Fu LQ, Li SS, Guo QP, Wu
Y, Mou XZ and Tong XM: Ovatodiolide targets chronic myeloid
leukemia stem cells by epigenetically upregulating hsa-miR-155,
suppressing the BCR-ABL fusion gene and dysregulating the
PI3K/AKT/mTOR pathway. Oncotarget. 9:3267–3277. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang L, Wang W, Li X, He S, Yao J, Wang
X, Zhang D and Sun X: MicroRNA-155 promotes tumor growth of human
hepatocellular carcinoma by targeting ARID2. Int J Oncol.
48:2425–2434. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Liu D, Han P, Gao C, Gao W, Yao X and Liu
S: microRNA-155 modulates hepatic stellate cell proliferation,
apoptosis, and cell cycle progression in rats with alcoholic
hepatitis via the MAPK signaling pathway through targeting SOCS1.
Front Pharmacol. 11:2702020. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Chen L, Ming X, Li W, Bi M, Yan B, Wang X,
Yang P and Yang B: The microRNA-155 mediates hepatitis B virus
replication by reinforcing SOCS1 signalling-induced autophagy. Cell
Biochem Funct. 38:436–442. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ye J, Guo R, Shi Y, Qi F, Guo C and Yang
L: miR-155 regulated inflammation response by the SOCS1-STAT3-PDCD4
axis in Atherogenesis. Mediators Inflamm. 2016:80601822016.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Yang Y, Yang L, Liang X and Zhu G:
MicroRNA-155 promotes atherosclerosis inflammation via targeting
SOCS1. Cell Physiol Biochem. 36:1371–1381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Bouamar H, Jiang D, Wang L, Lin AP, Ortega
M and Aguiar RC: MicroRNA 155 control of p53 activity is context
dependent and mediated by Aicda and Socs1. Mol Cell Biol.
35:1329–1340. 2015. View Article : Google Scholar : PubMed/NCBI
|