Introduction
Apolipoprotein M (apoM) was discovered by Xu and
Dahlback in 1999 (1). It is
predominantly expressed in the liver and kidney, but also weakly
expressed in colorectal tissues, and is a constituent of plasma
high-density lipoproteins (HDLs) (2). As a component of HDLs, apoM is of
vital importance for preβ-HDL formation in reverse cholesterol
transport (3), and is suspected to
be involved in anti-inflammation-associated atherogenic inhibition
(4). A reduction in plasma apoM
has been demonstrated in obese patients with sepsis and systemic
inflammatory response syndrome (5). Additionally, apoM mRNA expressions
levels have been revealed to be regulated by a number of
inflammatory mediators (including platelet-activating factors)
in vivo and/or in vitro (6,7).
Other studies have revealed that the level of apoM is markedly
correlated with that of inflammatory factors and adhesion molecules
in vitro (5,8). Notably, the human apoM gene is
located at the major histocompatibility complex class III region of
chromosome 6 (chromosome 17 in mice); this region also contains a
number of other genes associated with immune and inflammatory
responses (9). Previous studies
have demonstrated that apoM attenuates lipopolysaccharide
(LPS)-associated acute lung injury and that the number of splenic
CD4+ T-lymphocytes is decreased in
apoM−/− mice (10,11).
These studies indicated that apoM may be involved in immune and
inflammatory responses in vivo.
Although the relationship between apoM and cytokines
associated with the inflammatory response has previously been
identified (5,8,12,13),
the majority of these investigations were carried out in
vitro or are clinical case studies, thus the regulatory
mechanism of apoM remains to be elucidated. Nuclear factor-κB
(NF-κB) is a classic member of the Rel family of transcription
factors that regulates a variety of cellular functions, including
inflammatory, innate and adaptive immune responses, cell
proliferation and development (14,15).
In fact, NF-κB can regulate the downstream expression of
pro-inflammatory genes and protein adhesion molecules, including
interleukin (IL)-1β, IL-6, intercellular adhesion molecule-1
(ICAM-1) and vascular cell adhesion molecule-1 (VCAM-1) (8,16,17).
Conversely, the primary function of IL-10 is to inhibit the
macrophage-associated generation of cytokines and chemokines, by
inhibiting the activation of NF-κB and other related molecules
(18).
The correlation between the expression levels of
apoM and IL-10 has rarely been directly studied and the upstream
and downstream effects of the association between NF-κB and IL-10
were previously unclear. To clarify the underlying mechanisms of
apoM in inflammation, an acute inflammatory model of wild-type
(apoM+/+) and apoM deficient
(apoM−/−) mice was previously established
(10,11). Preliminary experiments in the
present study confirmed the establishment of the inflammatory model
following LPS administration and that the expression levels of
iNOS, NF-κB, ICAM-1, VCAM-1, IL-1β, IL-6 and the multifunctional
cytokine IL-10 were increased in apoM−/− and
apoM+/+ mice. Furthermore, the expression levels
of pro-and anti-inflammatory factors, as well as adhesion molecules
were compared in the presence and absence of apoM. The interaction
between apoM and LPS, and the subsequent effects on cytokine
release, were also assessed.
Materials and methods
Generation of apoM-deficient mice
C57BL/6 apoM −/− mice were
generated by homologous recombination at the Model Animal Research
Center of Nanjing University (China) as previously described
(10). A total of 36 male C57BL/6
apoM +/+ and apoM−/− mice, aged
6–8 weeks and weighing ~25 g were housed in a specific
pathogen-free animal experiment center at Soochow University. Food
and water was provided ad libitum, and the conditions for a
regular and stress-free life period were provided, including
moderate temperature (20–25°C), standard 12-h light/dark cycle and
suitable relative humidity (55–60%).
Animals and experimental design
In the present study, experimental procedures
involving animals conformed to the Jiangsu Laboratory Animal
Management Ordinance and were approved by the Ethics Committee of
the Third Affiliated Hospital of Soochow University. A total of 18
male apoM+/+ and 18 male
apoM−/− mice were divided into the following
three groups: i) Saline group (control)-receiving intraperitoneal
(i.p.) injections of 0.9% saline (100 µl, pyrogen-free; n=6); ii) 5
mg/kg LPS group-receiving i.p. injections of LPS (100 µl, 5 mg/kg;
Escherichia coli serotype O111:B4; Sigma-Aldrich; Merck
KGaA; MO; n=6); and iii) 10 mg/kg LPS group-receiving i.p.
injections of LPS (100 µl, 10 mg/kg; n=6) (10,11,19).
The mice were randomly assigned to receive three i.p. injections,
once every 24 h and were sacrificed by cervical dislocation 24 h
after the third injection. Blood and liver tissue samples were
collected for further, immediate experimentation. To separate the
serum, whole blood was left for ≥30 min at room temperature to
allow coagulation, centrifuged at 1,000 × g for 10 min at room
temperature and subsequently cryopreserved at −80°C. The sera were
used to detect serum iNOS, NF-κB, IL-1β, IL-6 and IL-10 levels and
the liver tissue samples were used to detect iNOS, NF-κB, IL-1β,
ICAM-1 and VCAM-1 expression levels.
Genotype identification of apoM
knockout mice
Mouse DNA extraction
The tail tip of each mouse (~0.5 cm) was processed
using the Ezup Column Animal Genomic DNA Extraction kit (Sangon
Biotech Co., Ltd.) according to the manufacturer's protocol.
Specimens with absorbance values (A260/280 nm) of
1.8-1.9 were stored at −20°C and used for subsequent
experimentation.
Primer and probe design
The base sequences of apoM (NC_000083.6, http://www.ncbi.nlm.nih.gov/nuccore/NC_000083.6)
and Lox-Neo-LoxP (NC_000086.7, http://www.ncbi.nlm.nih.gov/nuccore/NC_000086.7)
were obtained from GenBank (NCBI). The PCR primers and probe sets
(Table I) were designed using
Primer Premier 5.00 (Premier Biosoft International) and synthesized
by Sangon Biotech Co., Ltd. The mouse apoM+/+
probe was labeled with a fluorescent 6-carboxyfluorescein (6-FAM)
group and the apoM−/− probe was labeled with a
3H-II phthalocyanine dye (CY5) at the 5′-end, as previously
described (20).
 | Table I.Mouse genotype identification primers
and probes for dual probe PCR. |
Table I.
Mouse genotype identification primers
and probes for dual probe PCR.
| Name | Primer/probe | Sequence
(5′-3′) | Product length
(base pairs) |
|---|
|
apoM+/+ mouse | Forward primer |
CGGAAGTGGACATACCGATTG | 105 |
|
| Reverse primer |
AAACGGCTAAGGGAGACGAGA |
|
|
| Probe |
FAM-CTGAAGGTTGGTTCCCACCAGCCC-BHQ1 |
|
|
apoM−/− mouse | Forward primer |
GCTCCTGCCGAGAAAGTATCC | 98 |
|
| Reverse primer |
CGATGTTTCGCTTGGTGGTC |
|
|
| Probe |
CY5-TGCAATGCGGCGGCTGCATACGCT-BHQ2 |
|
Duplex fluorescence real-time PCR for mouse
genotype identification
Duplex fluorescence real-time PCR was performed
using the LightCycler 480 II PCR system (Roche Diagnostics GmbH) to
enable dual-channel fluorescence detection. The amplification
reactions contained 2.5 µl 10X PCR Buffer (including
Mg++), 2 µl 2.5 mM dNTPs, 0.1 µl of each primer and
probe (100 µM; Table I), 0.125 µl
Taq polymerase (5 U/µl), 2 µl template (replaced by water in the no
template controls) and nuclease-free water up to a final volume of
25 µl. The thermocycling conditions were: 95°C for 3 min, 40 cycles
at 95°C for 10 sec and 60°C for 15 sec and 40°C for 30 sec.
Fluorescence signals were acquired at 60°C and detected using the
FAM (465–510 nm, apoM+/+ mice) and CY5 (618–660
nm, apoM−/− mice) channels. When the Ct value was
≤35, PCR amplification was considered to be successful; when the Ct
value was >40, or there was no value, the detection result was
considered to be negative; when a Ct value >35 and <40 was
obtained, a retest was recommended. A single
apoM+/+ and apoM−/− PCR product
were selected to verify the methodology (cloning and sequencing,
performed by Shanghai Biotech Co., Ltd.), as previously described
(20).
Hematoxylin and eosin (H&E) Staining of
hepatic tissues
Hepatic samples were fixed in 10% formalin for 24 h
at room temperature and then embedded in paraffin. Hepatic sections
were cut into 5-µm-thick slices and used for H&E staining
according to the established method (21). All slides were examined under an
inverted microscope (Olympus IX73, magnification, ×200) by an
experienced pathologist.
Reverse transcription-quantitative (RT-q) PCR
quantification of mRNA expression levels of iNOS, NF-κB, IL-1β,
ICAM-1 and VCAM-1 in liver tissues
Total RNA was extracted from liver tissues using an
RNA purification kit (Shanghai Shenergy Biocolor Bioscience and
Technology Co., Ltd.) according to the manufacturer's instructions.
The quality and quantity of the RNA were determined by
spectrophotometry (Eppendorf) at 260/280 nm. A total of 2 µg RNA
was reverse-transcribed using the RevertAid First-strand cDNA
Synthesis kit per the manufacturer's protocol (Thermo Fisher
Scientific, Inc.); the PCR primer and probe sets (listed in
Table II) were designed using
Primer Premier 5.00 (Premier Biosoft International) and synthesized
by Sangon Biotech Co., Ltd. GAPDH was used as the reference
control. The amplification reactions contained (SNBC Bioon, Inc.)
2.5 µl 10X PCR buffer (including Mg++), 2 µl 2.5 mM
dNTPs, 0.1 µl of each primer (100 µM), 0.05 µl probe (100 µM),
0.125 µl 5 U/µl Taq polymerase, 2 µl template (replaced by water in
the no template controls) and nuclease-free water to a final volume
of 25 µl. The thermocycling conditions were: 95°C for 3 min,
followed by 40 cycles at 95°C for 5 sec and 60°C for 15 sec. The
relative mRNA expression level was determined using the
2−ΔΔCq method (22).
 | Table II.Primers and fluorescent probes for
reverse transcription-quantitative PCR. |
Table II.
Primers and fluorescent probes for
reverse transcription-quantitative PCR.
| Name | Primer/probe | Sequence
(5′-3′) |
|---|
| iNOS | Forward primer |
TGCCACGGACGAGACGG |
|
| Reverse primer |
TGTTGCTGAACTTCCAGTCATTGT |
|
| Probe |
FAM-AGAGATTGGAGGCCTTGTGTCAGCC-TAMRA |
| NF-κB | Forward primer |
GAGAAGAACAAGAAATCCTACCCAC |
|
| Reverse primer |
TCCATTTGTGACCAACTGAACG |
|
| Probe |
FAM-CAACTATGTGGGGCCTGCAAAGGTT-TAMRA |
| IL-1β | Forward primer |
GCAGGCAGTATCACTCATTGTGG |
|
| Reverse primer |
GAGTCACAGAGGATGGGCTCTTC |
|
| Probe |
FAM-TGGAGAAGCTGTGGCAGCTACCTGTGT-TAMRA |
| ICAM-1 | Forward primer |
ATCACCGTGTATTCGTTTCCG |
|
| Reverse primer |
GTGAGGTCCTTGCCTACTTGCT |
|
| Probe |
FAM-AGTGTGGAGCTGAGACCTCTGCCAGCCT-TAMRA |
| VCAM-1 | Forward primer |
CTGAACCCAAACAGAGGCAGA |
|
| Reverse primer |
GGTATCCCATCACTTGAGCAGG |
|
| Probe |
FAM-CATCTGGGTCAGCCCCTCTCCTATACTAGA-TAMRA |
| GAPDH | Forward primer |
TCTTGTGCAGTGCCAGCCT |
|
| Reverse primer |
TGAGGTCAATGAAGGGGTCG |
|
| Probe |
FAM-AGGTCGGTGTGAACGGATTTGGC-TAMRA |
Determination of the protein expression levels of
iNOS, NF-κB, IL-1β, ICAM-1 and VCAM-1 using western blotting
Hepatic protein samples were extracted using a total
protein extraction kit (BestBio Biotechnology) and protein
quantified using the BCA method (BestBio Biotechnology). Serum
samples were diluted (1:25) and loaded onto 8% SDS-page gels and
subsequently transferred onto 0.45 µm PVDF membranes (EMD
Millipore). The membranes were blocked in 1X PBST containing 3%
bovine serum albumin (Multi Sciences, Inc.) for 2 h at room
temperature. The membranes were incubated with primary antibodies
against iNOS (1:300; cat. no. 18985-1-AP; ProteinTech Group, Inc.),
NF-κB (1:1,000; cat. no. 8242; Cell Signaling Technology, Inc.),
IL-1β (1:1,000; cat. no. 12242; Cell Signaling Technology, Inc.),
ICAM-1 (1:300; cat. no. sc-18853; Santa Cruz Biotechnology, Inc.),
VCAM-1 (1:300; cat. no. sc-13160; Santa Cruz Biotechnology, Inc.),
GAPDH (1:3,000; cat. no. ARG10112, Arigo Biolaboratories Corp.) and
Transferrin (1:500; cat. no. 17435-1-AP; ProteinTech Group, Inc.)
overnight at 4°C. Then, the membranes were incubated with secondary
antibodies of HRP-conjugated Affinipure Goat Anti-Mouse IgG (H+L)
(1:3,000; cat. no. SA00001-1, ProteinTech Group, Inc.) and
HRP-conjugated Affinipure Goat Anti-Rabbit IgG (H+L) (1:3,000; cat.
no. SA00001-2, ProteinTech Group, Inc.) for 1 h at room
temperature. The protein blots were observed using an ECL
chemiluminescent kit (ECL plus; Thermo Fisher Scientific, Inc.) and
the gray values of the bands were measured using Clinx ChemiCapture
3300 (Shanghai Clinx Science Instruments Co., Ltd.) and Quantity
One software v4.6.2 (Bio-Rad Laboratories, Inc.).
Detection of serum iNOS
Serum iNOS was detected using an NOS typing assay
kit (cat. no. A014-1; Nanjing Jiancheng Bioengineering Institute)
according to the manufacturer's protocol. The absorbance was
measured at a wavelength of 530 nm and NOS activity was calculated
based on the absorbance value.
Detection of serum ICAM-1 and VCAM-1
Serum ICAM-1 and VCAM-1 were detected using
enzyme-linked immunosorbent assay kits (cat. nos. H065 and H066;
Nanjing Jiancheng Bioengineering Institute) according to the
manufacturer's protocols. The absorbance was measured at a
wavelength of 450 nm.
Determination of IL-6 and IL-10 expression levels
by Luminex assay
Prior to the bioassay, samples were thawed on ice
and homogenized by vortexing for 10 sec, followed by centrifugation
at 1,000 × g for 10 min at room temperature. The concentrations of
serum IL-6 and IL-10 were detected using a Luminex 200 system
(Luminex Corporation) with the MILLIPLEX® Map kit and
the Human Magnetic Bead MAPmate™ Buffer kit (EMD Millipore). The
wells of the standard, control and background samples were assayed
in duplicate and standard curves were extended down to <1.0
pg/ml. The assays were performed using a Luminex 200 multiplexing
instrument (Luminex Corporation) according to the manufacturer's
instructions, at 4°C with shaking 1,000 rpm for 16 h on a plate
shaker (Eppendorf) and using a hand-held magnetic block for each
wash step. In summary, each well of the 96-well microtiter plates
was pre-wetted with wash buffer, prior to the addition of 25 µl
sample and 25 µl of the appropriate buffer (Assay Buffer, Standard,
or Control 1 or 2) to the background, standard and control wells
(according to the standard curve). The plates were then incubated
with 25 µl pre-mixed antibody-immobilized beads overnight on an
orbital shaker at 4°C. The plates were washed twice with wash
buffer and 25 µl biotinylated detector antibody was added to each
well and incubated for 1 h at room temperature. Without further
washing, 25 µl streptavidin-phycoerythrin solution was added to
each well and the plates were incubated for 30 min at room
temperature on a plate shaker, protected from direct light. Prior
to analysis, the beads were washed twice in wash buffer and
resuspended in 150 µl/well Luminex sheath fluid.
Statistical analysis
The data are expressed as the means ± standard error
or the mean. Statistical analysis was conducted using Prism
software, version 6.0 (GraphPad Software, Inc.). Comparisons
between two groups were statistically evaluated using the
Mann-Whitney test and those between three groups were analyzed
using Ordinary one-way ANOVA followed by Tukey's post hoc test.
Cross interaction was analyzed by two-way ANOVA. P<0.05 was
considered to indicate a statistically significant difference.
Results
Analysis of PCR amplification curves
for mouse genotype identification
The PCR amplification curves were consistent with
the effective amplification results (Ct value ≤35). Mice with a
significant ‘s’ type amplification curve in the FAM channel and no
amplification in the CY5 channel were identified as apoM
+/+. Mice with a significant ‘s’ type amplification
curve in the CY5 channel and no amplification in the FAM channel
were identified as apoM−/−. Mice with a prominent
‘S’ type amplification curve in both channels were apoM
heterozygous (apoM+/−; Fig. S1).
PCR product cloning and sequencing
alignment
An apoM+/+ and an
apoM−/− PCR product were selected for cloning and
sequencing (performed by Shanghai Biotech Co., Ltd.). Sequence
alignment indicated that the sequences of the
apoM+/+ and apoM−/− PCR
products in the subsequent recombinant plasmids completely matched
those of their respective sequences (Fig. S2).
Confirmation of the successful
establishment of the inflammatory model following LPS
administration
Following the administration of 10 mg/kg LPS to the
apoM−/− and apoM+/+ mice,
H&E staining revealed significant inflammatory pathological
changes in the liver, including the infiltration of inflammatory
cells into the liver portal area. The reaction to LPS treatment was
more pronounced in apoM−/− mice than in
apoM+/+ mice; the degree of inflammatory cell
exudate was greater and the number of Kupffer cells was increased
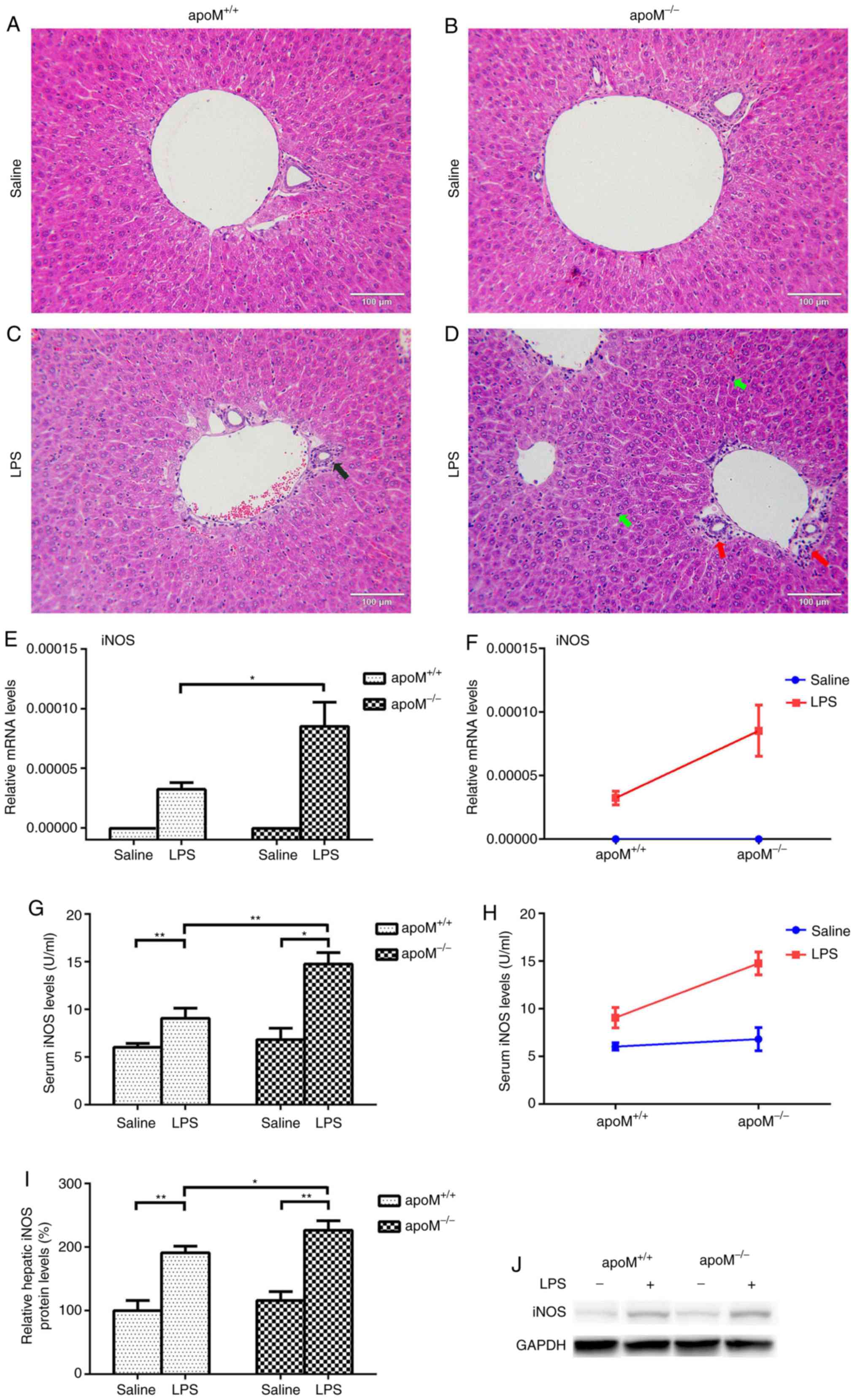
(Fig. 1A-D). As a clear indicator
of inflammation, the mRNA and protein levels of iNOS were
significantly increased in the liver and serum of LPS-treated mice,
especially in the apoM−/− group (Fig. 1E-J; P<0.05).
Effects of LPS and/or apoM on NF-κB
and IL-1β expression levels in apoM−/− and
apoM+/+ mice
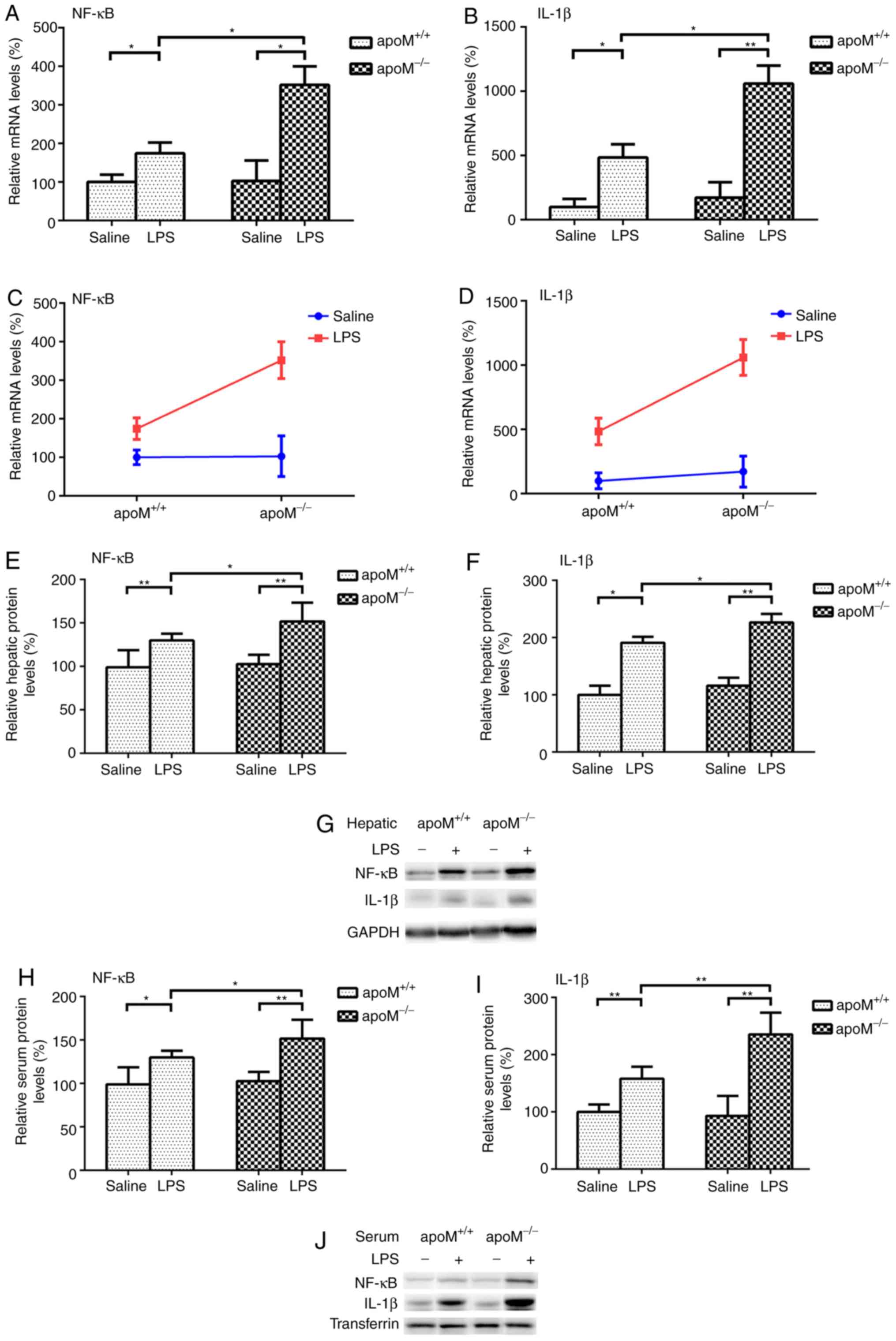
There were no significant differences in the
expression levels of NF-κB and IL-1β between the
apoM−/− and apoM+/+ mice of the
saline control group. Following the administration of 10 mg/kg LPS,
the expression levels of NF-κB mRNA increased 3.42- (P<0.05) and
1.74-fold (P<0.05) and those of IL-1β mRNA increased 6.18-
(P<0.01) and 4.84-fold (P<0.05), in the
apoM−/− and apoM+/+ mice,
respectively, when compared with the saline group (Fig. 2A and B). Following LPS treatment,
the mRNA expression levels of hepatic NF-κB and IL-1β in the
apoM−/− mice were 2.02 and 2.19 times that of
those in the apoM+/+ mice (Fig. 2A and B; P<0.05). Two-way ANOVA
revealed that the effects of apoM and LPS interaction on the
expression levels of hepatic NF-κB and IL-1β mRNA were
statistically significant (Fig. 2C and
D; P<0.05). NF-κB and IL-1β protein expression levels were
also significantly increased in the liver and serum of LPS-treated
mice, particularly in the apoM−/− group (Fig. 2E-J; P<0.05).
Effects of LPS and/or apoM on ICAM-1
and VCAM-1 expression levels in apoM−/− and
apoM+/+ mice
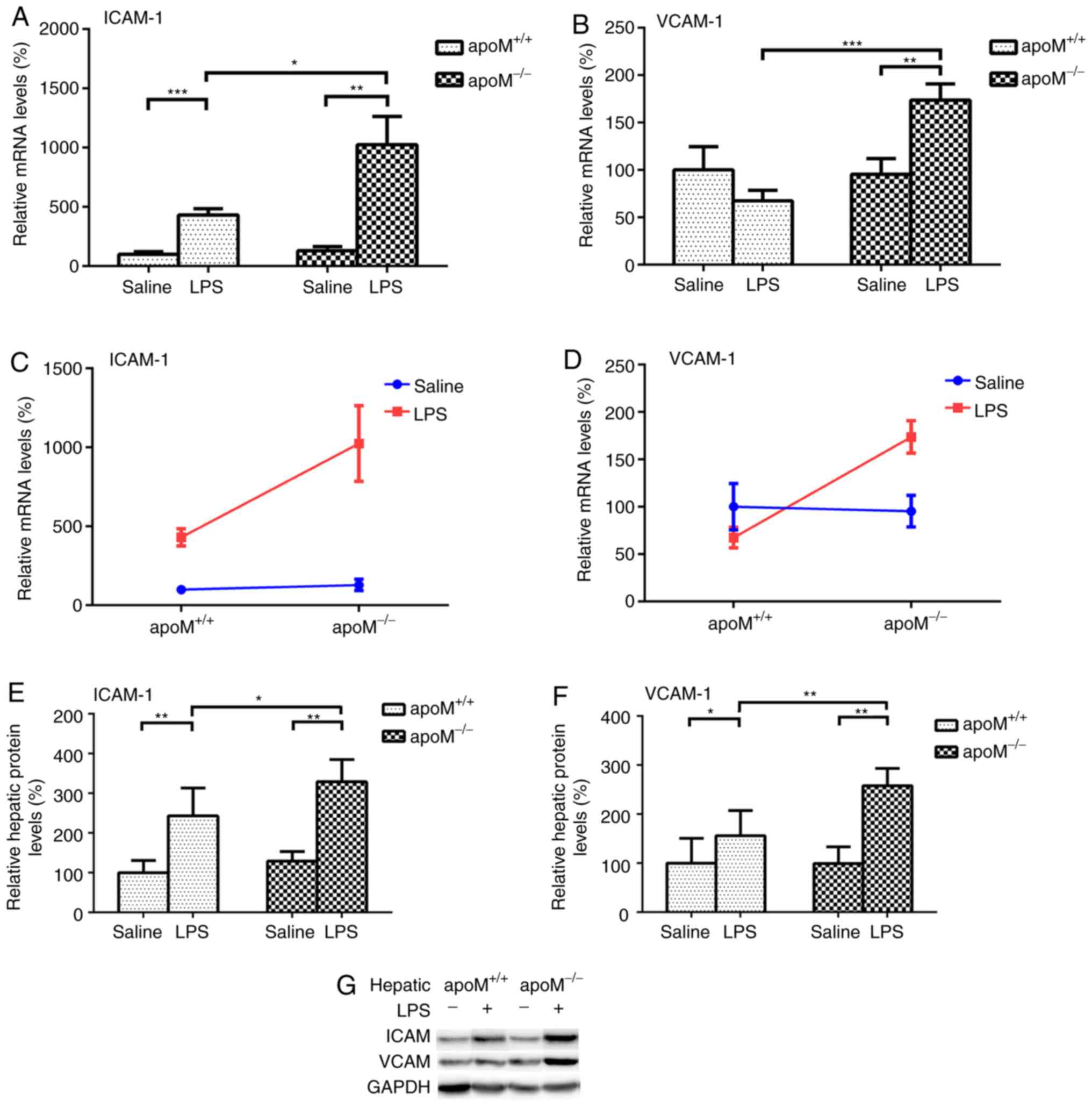
There were no significant differences in the
expression levels of ICAM-1 and VCAM-1 between
apoM−/− and apoM+/+ mice in the
saline control group. Compared with this group, the injection of 10
mg/kg LPS resulted in a 7.95- (P<0.01) and 4.30-fold
(P<0.001) increase in the expression levels of ICAM-1 mRNA in
apoM−/− and apoM+/+ mice,
respectively (Fig. 3A); the
expression level of VCAM-1 increased 1.82-fold (Fig. 3B; P<0.01) in the
apoM−/− mice, though there was no significant
difference in VCAM-1 expression in the apoM+/+
mice. Additionally, LPS administration increased the mRNA
expression levels of hepatic ICAM-1 and VCAM-1 in the
apoM−/− mice to 2.38 and 2.57 times that of the
apoM+/+ mice (Fig.
3A and B; P<0.05 and P<0.001, respectively). Two-way
ANOVA revealed that the effects of the apoM and LPS interaction on
hepatic ICAM-1 and VCAM-1 mRNA expression levels was statistically
significant (Fig. 3C and D;
P<0.05). ICAM-1 and VCAM-1 protein levels were also
significantly increased in the livers of LPS-treated mice,
especially those in the apoM−/− group (Fig. 3E-G; P<0.05). The protein mass of
ICAM-1 and VCAM-1 in serum were not detected using western blotting
and enzyme-linked immunosorbent assay.
Effects of LPS and/or apoM on serum
cytokine levels of IL-6 and IL-10 in apoM−/− and
apoM+/+ mice
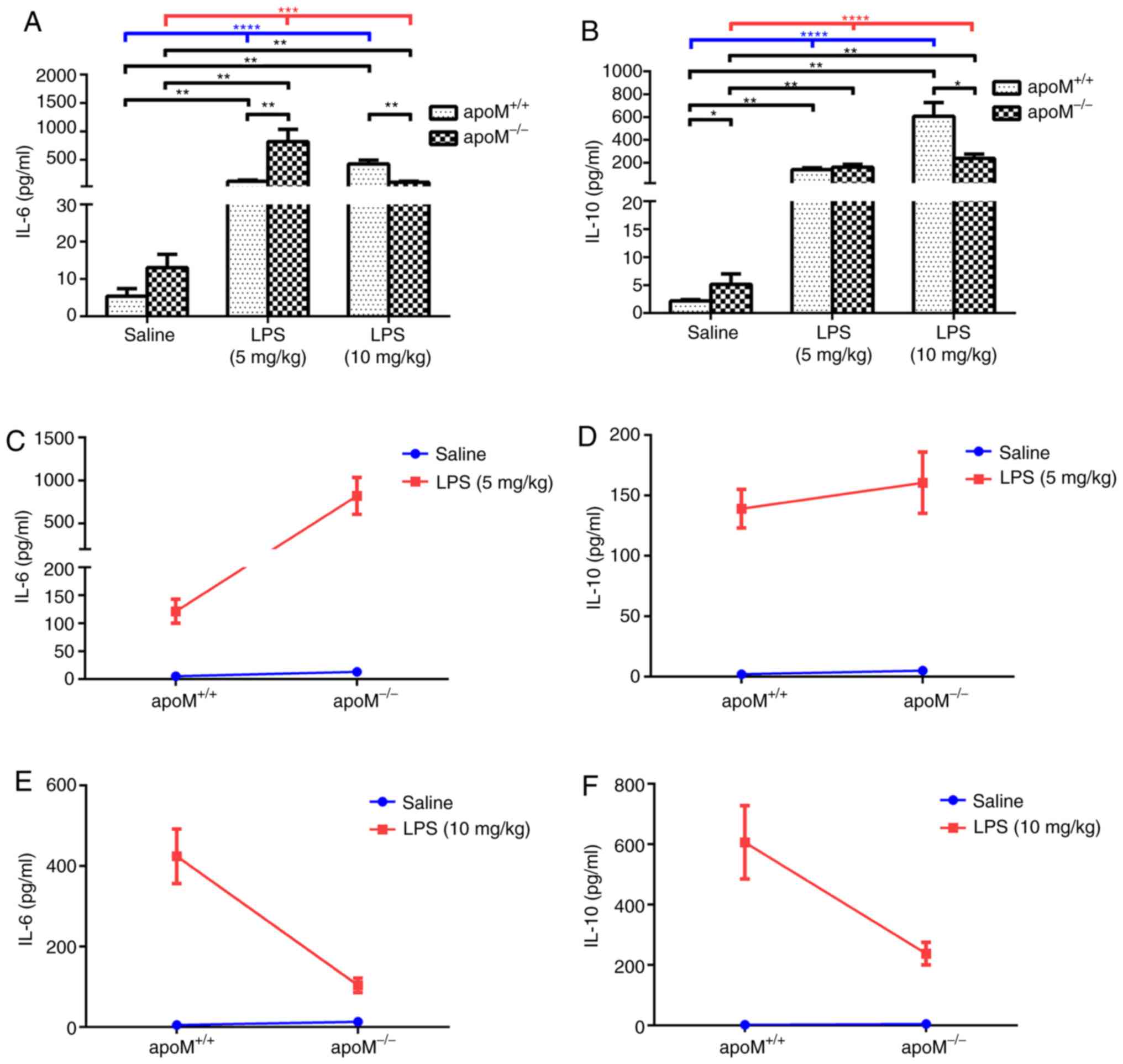
Following treatment with LPS, the serum IL-6 and
IL-10 levels were significantly increased in both
apoM+/+ and apoM−/− mice
(Fig. 4A and B; P<0.001).
Furthermore, serum IL-6 and IL-10 levels were increased in the LPS
group, compared with the saline group (Fig. 4A and B; P<0.01), although there
was no significant difference between the serum IL-6 levels of the
apoM+/+ and apoM−/− mice in the
saline group. After a 5 mg/kg injection of LPS, the serum levels of
IL-6 in the apoM−/− mice were considerably higher
than those in the apoM+/+ mice (P<0.01).
However, after 10 mg/kg LPS, the serum levels of IL-6 in the
apoM−/− mice were lower than those in the
apoM+/+ mice (Fig. 4A;
P<0.01).
The levels of serum IL-10 in the
apoM−/− mice were higher than those in the
apoM+/+ mice of the saline group (P<0.05), but
no significant difference was observed between the
apoM+/+ and apoM−/− mice in the
5 mg/kg LPS-treatment group. However, the serum IL-10 levels in
apoM−/− mice were notably lower than those in the
apoM+/+ mice following treatment with 10 mg/kg
LPS (Fig. 4B; P<0.05).
Furthermore, two-way ANOVA revealed that the effects of apoM and
LPS interaction on serum IL-6 levels in the 5 mg/kg LPS group were
statistically significant (Fig.
4C; P<0.01), whilst there were no significant differences in
the levels of IL-10 (Fig. 4D). The
effects of the apoM and LPS interaction on serum IL-6 and IL-10
levels in the 10 mg/kg LPS group were also statistically
significant (Fig. 4E and F;
P<0.001 and P<0.01, respectively). Interaction analyses
between apoM and LPS in the two different dose groups indicated
that an increase in LPS resulted in a decrease in IL-6 and IL-10
serum expression levels in apoM−/− mice, compared
with apoM+/+ mice.
Discussion
Inflammation is predominantly involved in the host
defense against infection and cellular damage (23,24).
It can widely occur in vascularized tissues and is closely
associated with the occurrence and development of tumors and
diseases such as atherosclerosis and diabetes (25). Various inflammatory factors and
adhesion molecules serve important roles in the development of
inflammation (23,26). In recent years, the
anti-inflammatory properties of apoM have received increasing
attention, with a number of studies revealing its ability to
inhibit inflammation in atherogenesis, chronic infection, pneumonia
and bowel cancer (2,11,27,28).
However, the effects of apoM on the expression of adhesion
molecules during inflammation, particularly the multifunctional
cytokine IL-10, are less well studied and to date, the underlying
mechanisms remained unknown. In the present study, apoM was
revealed to significantly inhibit the expression of iNOS, NF-κB,
IL-1β, ICAM-1 and VCAM-1 in an LPS-induced mouse model.
Furthermore, the expression level of serum IL-6 and IL-10 was
increased in mice induced with different concentrations of LPS.
Notably, the effects of apoM on IL-10 and IL-6 were reversed with
increasing concentrations of LPS.
Our previous study (11) demonstrated that the expression
levels of IL-1β and IL-6 were significantly increased in the lung
tissues of 10 mg/kg LPS-induced apoM−/−mice,
compared with the apoM+/+ group, suggesting that
apoM exerts anti-inflammatory effects in LPS-induced acute lung
injury. These results also revealed that LPS treatment increased
the expression levels of IL-1β, IL-6 and IL-10, and that NF-κB
expression in the 10-mg/kg LPS group also increased significantly
in apoM−/−, compared with
apoM+/+ mice. However, further analysis showed
that when treated with 5 mg/kg LPS, apoM inhibited the effects of
LPS on IL-6, resulting in an anti-inflammatory effect. By contrast,
in the 10 mg/kg LPS group, apoM promoted the effects of LPS on IL-6
and IL-10. Previous studies have revealed that IL-6 and IL-10
possess both pro- and anti-inflammatory properties (29,30).
The reason may be that in the early stages, apoM retards
inflammation by inhibiting the production of IL-6. In more severe
cases, apoM may exert its anti-inflammatory effects by increasing
the expression levels of both IL-6 and IL-10, although the specific
mechanism of this inference requires further investigation.
The transcription factor NF-κB exerts protective and
anti-apoptotic effects in the liver, and is considered to be a key
inducible, rather than constitutively expressed regulator, of
inflammation that is responsive to pro-inflammatory cytokines
(31,32). Previous studies have revealed that
NF-κB is a key factor regulating the expression of various genes,
including IL-1β, IL-6, ICAM-1 and VCAM-1; it can be regulated by
IL-10 (8,16,18)
and as such, is integral to inflammatory reactions such as the
immune response, smooth muscle cell proliferation and angiogenesis.
A number of studies have suggested that apoM is able to inhibit the
activity of NF-κB and the expression of inflammatory factors
through various biological pathways, dependent or independent of
sphingosine 1-phosphate (5,8,12,33–35).
Gao et al (8) revealed that
apoM reduced the expression levels of ICAM-1 and VCAM-1 by
inhibiting NF-κB activity. Therefore, apoM was speculated to
inhibit the expression of pro-inflammatory factors and adhesion
molecules by preventing NF-κB activation, thereby exerting a
protective effect against inflammation. As aforementioned, studies
(18,30) have shown that the primary function
of IL-10 is to inhibit the production of cytokines and chemokines
in macrophages. This is achieved by inhibiting a number of
inflammatory molecules, including NF-κB (18), which can also regulate the
expression of IL-10 (30). To the
best of the authors' knowledge, the direct upstream and downstream
relationship between apoM and IL-10 has yet to be elucidated, thus,
in the present study, the expression levels of IL-10 in
apoM−/− mice were determined and further analyses
were performed to confirm the interaction between IL-10 and apoM.
Serum ICAM-1 and VCAM-1 are generally detected by enzyme-linked
immunosorbent assay (36,37), but in the present study, they were
not detected using western blotting and enzyme-linked immunosorbent
assay, probably because their expression levels were too low.
In summary, the inflammatory response is a complex
process with mutual, network-like regulation between cytokines and
its regulatory balance may be associated with the development and
severity of the inflammatory response. The aforementioned results
indicated that apoM is involved in the regulation of inflammatory
factors and adhesion molecules in acute LPS-induced inflammation.
Furthermore, apoM may act by retarding the expression of
inflammatory factors and adhesion molecules through the inhibition
of NF-κB. These findings have expanded the network of research on
inflammatory development, providing greater opportunities to treat
inflammatory pathologies and suggesting that apoM may be a
potential target for the clinical treatment of inflammation
(particularly vascular inflammation).
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
This study was supported by the National Natural
Science Foundation of China (grant no. 81370372), the Natural
Science Foundation of Jiangsu province (grant nos. BK20191158 and
BK20151179) and the Jiangsu Provincial Youth Medicine Key Talent
Project (grant no. QNRC2016282).
Availability of data and materials
The datasets used during the present study are
available from the corresponding author upon reasonable
request.
Authors' contributions
XZ, GL and NX made substantial contributions to the
conception and design of the study. YS performed the experiments,
analyzed the data and contributed to writing the manuscript. YY
performed the genotype identification experiments. HongyL, JZ and
HongL detected the inflammatory cytokines and adhesion molecule
expression levels. YL analyzed the data. All authors read and
approved the final version of the manuscript.
Ethics approval and consent to
participate
In the present study, experimental procedures
involving animals conformed to the Jiangsu Laboratory Animal
Management Ordinance and were approved by the Ethics Committee of
the Third Affiliated Hospital of Soochow University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ApoM
|
apolipoprotein M
|
|
HDL
|
high-density lipoprotein
|
|
ICAM-1
|
intercellular adhesion molecule-1
|
|
IL
|
interleukin
|
|
iNOS
|
inducible nitric oxide synthase
|
|
LPS
|
lipopolysaccharide
|
|
NF-κB
|
nuclear factor-κB
|
|
RT-qPCR
|
reverse transcription-quantitative
PCR
|
|
VCAM-1
|
vascular cell adhesion molecule-1
|
References
|
1
|
Xu N and Dahlback B: A novel human
apolipoprotein (apoM). J Biol Chem. 274:3117–31290. 1999.
View Article : Google Scholar
|
|
2
|
Luo G, Zhang X, Mu Q, Chen L, Zheng L, Wei
J, Berggren-Soderlund M, Nilsson-Ehle P and Xu N: Expression and
localization of apolipoprotein M in human colorectal tissues.
Lipids Health Dis. 9:1022010. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Feng YH, Zheng L, Wei J, Yu MM, Zhang J,
Luo GH and Xu N: Increased apolipoprotein M induced by lack of
scavenger receptor BI is not activated via HDL-mediated cholesterol
uptake in hepatocytes. Lipids Health Dis. 17:2002018. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Wei J, Yu Y, Feng Y, Zhang J, Jiang Q,
Zheng L, Zhang X, Xu N and Luo G: Negative correlation between
serum levels of homocysteine and apolipoprotein M. Curr Mol Med.
19:120–126. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li T, Yang L, Zhao S and Zhang S:
Correlation between apolipoprotein M and inflammatory factors in
obese patients. Med Sci Monit. 24:5698–5703. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Luo G, Shi Y, Zhang J, Mu Q, Qin L, Zheng
L, Feng Y, Berggren-Söderlund M, Nilsson-Ehle P, Zhang X and Xu N:
Palmitic acid suppresses apolipoprotein M gene expression via the
pathway of PPARβ/δ in HepG2 cells. Biochem Biophys Res Commun.
445:203–207. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ren K, Tang ZL, Jiang Y, Tan YM and Yi GH:
Apolipoprotein M. Clin Chim Acta. 446:21–29. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Gao JJ, Hu YW, Wang YC, Sha YH, Ma X, Li
SF, Zhao JY, Lu JB, Huang C, Zhao JJ, et al: ApoM suppresses
TNF-α-induced expression of ICAM-1 and VCAM-1 through inhibiting
the activity of NF-κB. DNA Cell Biol. 34:550–556. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Huang XS, Zhao SP, Hu M and Luo YP:
Apolipoprotein M likely extends its anti-atherogenesis via
anti-inflammation. Med Hypotheses. 69:136–140. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang Z, Luo G, Feng Y, Zheng L, Liu H,
Liang Y, Liu Z, Shao P, Berggren-Söderlund M, Zhang X and Xu N:
Decreased splenic CD4(+) T-lymphocytes in apolipoprotein M gene
deficient mice. Biomed Res Int. 2015:2935122015. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhu B, Luo GH, Feng YH, Yu MM, Zhang J,
Wei J, Yang C, Xu N and Zhang XY: Apolipoprotein M protects against
lipopolysaccharide-induced acute lung injury via
sphingosine-1-phosphate signaling. Inflammation. 41:643–653. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Frej C, Mendez AJ, Ruiz M, Castillo M,
Hughes TA, Dahlbäck B and Goldberg RB: A shift in ApoM/S1P between
HDL-particles in women with type 1 diabetes mellitus is associated
with impaired anti-inflammatory effects of the ApoM/S1P complex.
Arterioscler Thromb Vasc Biol. 37:1194–1205. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ruiz M, Frej C, Holmer A, Guo LJ, Tran S
and Dahlbäck B: High-density lipoprotein-associated apolipoprotein
M limits endothelial inflammation by delivering
sphingosine-1-phosphate to the sphingosine-1-phosphate receptor 1.
Arterioscler Thromb Vasc Biol. 37:118–129. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sun SC: The non-canonical NF-κB pathway in
immunity and inflammation. Nat Rev Immunol. 17:545–558. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
King KE, George AL, Sakakibara N, Mahmood
K, Moses MA and Weinberg WC: Intersection of the p63 and NF-κB
pathways in epithelial homeostasis and disease. Mol Carcinog.
58:1571–1580. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen KY and Wang LC: Stimulation of IL-1β
and IL-6 through NF-κB and sonic hedgehog-dependent pathways in
mouse astrocytes by excretory/secretory products of fifth-stage
larval angiostrongylus cantonensis. Parasit Vectors. 10:4452017.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Nonaka K, Kajiura Y, Bando M, Sakamoto E,
Inagaki Y, Lew JH, Naruishi K, Ikuta T, Yoshida K, Kobayashi T, et
al: Advanced glycation end-products increase IL-6 and ICAM-1
expression via RAGE, MAPK and NF-κB pathways in human gingival
fibroblasts. J Periodontal Res. 53:334–344. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Conti P, Kempuraj D, Kandere K, Di
Gioacchino M, Barbacane RC, Castellani ML, Felaco M, Boucher W,
Letourneau R and Theoharides TC: IL-10, an inflammatory/inhibitory
cytokine, but not always. Immunol Lett. 86:123–129. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song X, Ren Z and Zhang C: Antioxidant,
anti-inflammatory and renoprotective effects of acidic-hydrolytic
polysaccharides by spent mushroom compost (Lentinula edodes)
on LPS-induced kindey injury. Int J Biol Macromol. 151:1267–1276.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang Y, Lu Z, Yun L, Li P, Jun Z, Jiang W,
Miao M and Guang L: Establishment of real-time quantitative PCR
method for identification of apolipoprotein M knockout mice. Chin J
Clin Lab Sci. 33:32015.
|
|
21
|
Wu BM, Liu JD, Li YH and Li J: Margatoxin
mitigates CCl4induced hepatic fibrosis in mice via macrophage
polarization, cytokine secretion and STAT signaling. Int J Mol Med.
45:103–114. 2020.PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kuprash DV and Nedospasov SA: Molecular
and cellular mechanisms of inflammation. Biochemistry (Mosc).
81:1237–1239. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen B, Ma Y, Xue X, Wei J, Hu G and Lin
Y: Tetramethylpyrazine reduces inflammation in the livers of mice
fed a high fat diet. Mol Med Rep. 19:2561–2568. 2019.PubMed/NCBI
|
|
25
|
Sayers SR, Beavil RL, Fine NHF, Huang GC,
Choudhary P, Pacholarz KJ, Barran PE, Butterworth S, Mills CE,
Cruickshank JK, et al: Structure-functional changes in eNAMPT at
high concentrations mediate mouse and human beta cell dysfunction
in type 2 diabetes. Diabetologia. 63:313–323. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Lassailly G, Bou Saleh M, Leleu-Chavain N,
Ningarhari M, Gantier E, Carpentier R, Artru F, Gnemmi V, Bertin B,
Maboudou P, et al: Nucleotide-binding oligomerization domain 1
(NOD1) modulates liver ischemia reperfusion through the expression
adhesion molecules. J Hepatol. 70:1159–1169. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Wolfrum C, Poy MN and Stoffel M:
Apolipoprotein M is required for prebeta-HDL formation and
cholesterol efflux to HDL and protects against atherosclerosis. Nat
Med. 11:418–422. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
28
|
Adib-Conquy M and Cavaillon JM: Host
inflammatory and anti-inflammatory response during sepsis. Pathol
Biol (Paris). 60:306–313. 2012.(In French). View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Scheller J, Chalaris A, Schmidt-Arras D
and Rose-John S: The pro- and anti-inflammatory properties of the
cytokine interleukin-6. Biochim Biophys Acta. 1813:878–888. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Iyer SS and Cheng G: Role of interleukin
10 transcriptional regulation in inflammation and autoimmune
disease. Crit Rev Immunol. 32:23–63. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baeck C and Tacke F: Balance of
inflammatory pathways and interplay of immune cells in the liver
during homeostasis and injury. EXCLI J. 13:67–81. 2014.PubMed/NCBI
|
|
32
|
Ben-Neriah Y and Karin M: Inflammation
meets cancer, with NF-κB as the matchmaker. Nat Immunol.
12:715–723. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ma X, Hu YW, Zhao ZL, Zheng L, Qiu YR,
Huang JL, Wu XJ, Mao XR, Yang J, Zhao JY, et al: Anti-inflammatory
effects of propofol are mediated by apolipoprotein M in a
hepatocyte nuclear factor-1α-dependent manner. Arch Biochem
Biophys. 533:1–10. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Galvani S, Sanson M, Blaho VA, Swendeman
SL, Obinata H, Conger H, Dahlbäck B, Kono M, Proia RL, Smith JD and
Hla T: HDL-bound sphingosine 1-phosphate acts as a biased agonist
for the endothelial cell receptor S1P1 to limit vascular
inflammation. Sci Signal. 8:ra792015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zheng Z, Zeng Y, Zhu X, Tan Y, Li Y, Li Q
and Yi G: ApoM-S1P modulates Ox-LDL-induced inflammation through
the PI3K/Akt signaling pathway in HUVECs. Inflammation. 42:606–617.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Mierzejewska P, Zabielska MA, Kutryb-Zajac
B, Tomczyk M, Koszalka P, Smolenski RT and Slominska EM: Impaired
L-arginine metabolism marks endothelial dysfunction in
CD73-deficient mice. Mol Cell Biochem. 458:133–142. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Li F, Zhang T, He Y, Gu W, Yang X, Zhao R
and Yu J: Inflammation inhibition and gut microbiota regulation by
TSG to combat atherosclerosis in ApoE(−/-) mice. J Ethnopharmacol.
247:1122322020. View Article : Google Scholar : PubMed/NCBI
|