Introduction
Globally, glaucoma is the second leading cause of
vision loss after cataracts (1);
it is characterized by optic neuropathy and loss of visual field.
By 2020, it is estimated that 11.1 million individuals will suffer
from bilateral blindness due to primary glaucoma (2). Primary open-angle glaucoma (POAG) is
more common in individuals of African and European descent
(3), and in China its incidence is
less than that of primary closed-angle glaucoma (4). However, in recent years, its
incidence rate has increased in parallel with the increased
incidence of myopia (incidence of POAG: 23% higher) (5) and metabolic diseases (incidence of
POAG: 49% higher) (6). The
pathogenesis of POAG is usually slow, and the majority of patients
will not exhibit obvious symptoms. Since patients may not be aware
of the loss of vision until the advanced stages of the disease,
genetic screening is important for early diagnosis, prevention and
treatment (1). The primary risk
factors for POAG include age, myopia, family history, high mental
stress, diabetes, smoking, drinking and increased intraocular
pressure (IOP) (7). Owing to
familial predispositions, the prevalence of POAG in first-degree
relatives is 7–10 times higher compared with the general population
(8). POAG-associated gene
mutations have been reported in myocilin (MYOC), optineurin
(OPTN), tANK binding kinase 1 (TBK1), WD repeat
domain 36 (WDR36) and ankyrin repeat and SOCS box containing
10 (ASB10) (9). MYOC
was the first gene found to be associated with POAG. Johnson et
al (10) and Sheffield et
al (11) mapped the chromosome
region 1q21-q31 of the POAG locus, GLC1A, which is
associated with both juvenile- and adult-onset POAG. At present,
771 nucleotide substitutions have been reported in the MYOC
gene. Among these, 331 substitutions are disease-causing mutations
(DCM) (12). MYOC has been
investigated for >20 years and is the most common mutated gene
in patients with glaucoma (13,14).
Although there are several studies on the function of WT
MYOC, its function remains incompletely understood, and the
mechanisms by which mutations in MYOC result in POAG,
remains to be investigated in several cases. It has been reported
that a possible cause of POAG in patients with a mutated
MYOC gene is upregulated expression of the mutated troponin
subtype in the endoplasmic reticulum of trabecular reticulum (TM)
cells, and its retention in the endoplasmic reticulum, which leads
to trabecular reticulum stress, dysfunction and apoptosis (15–18).
TM cell loss is associated with the disturbance of aqueous outflow
regulation and increased IOP, and ultimately leads to loss of
vision (19). In this study, we
characterized the clinical results of a Chinese POAG family and
studied its molecular basis, to expand the MYOC mutation spectrum
in the Chinese population.
Materials and methods
Clinical observations and
diagnosis
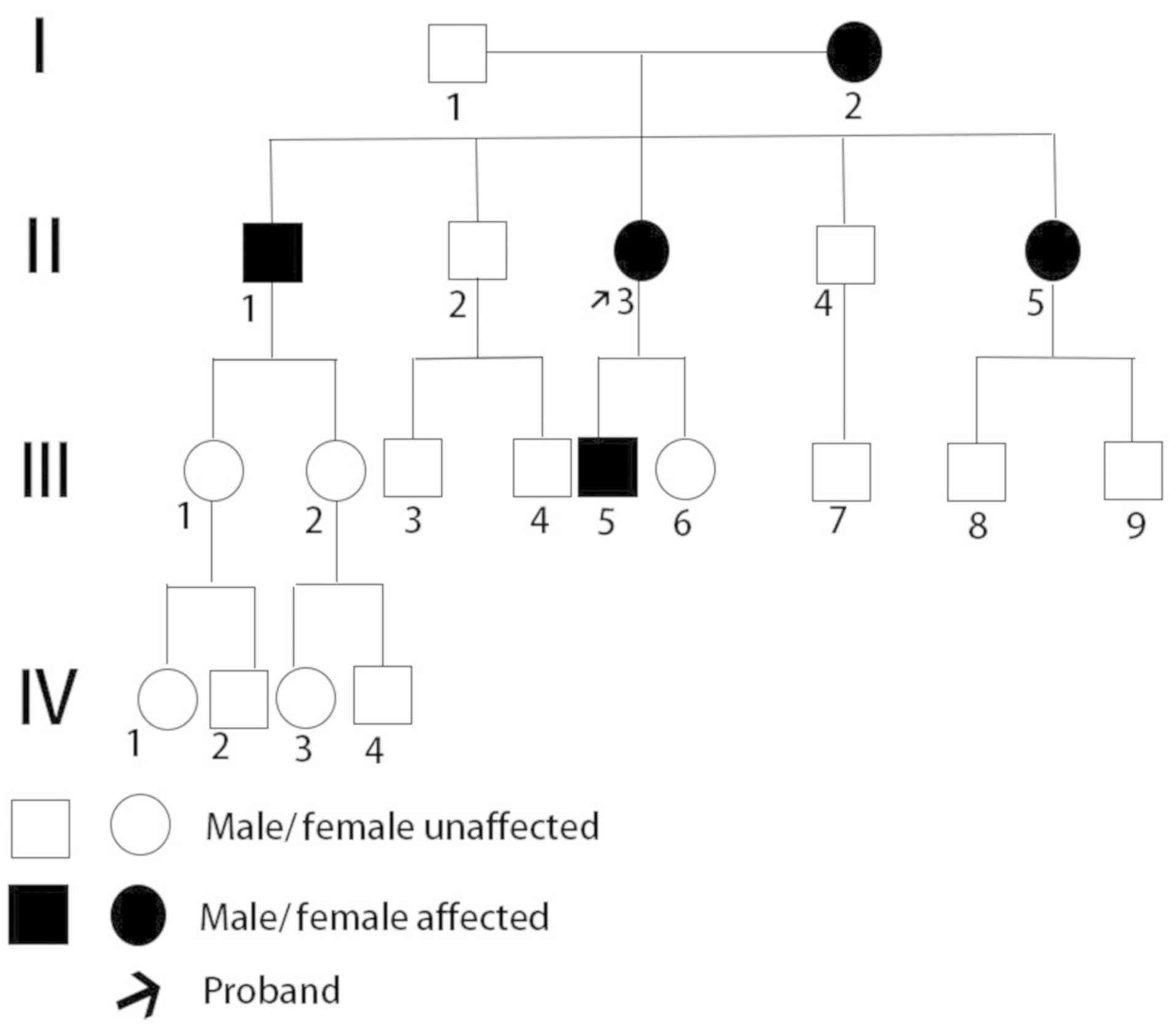
A large 20-member family spanning four generations
(Fig. 1) was enrolled in the
present study at the Department of Ophthalmology, Wuhan Union
Hospital (Wuhan, China), between January and February 2018. The
study adhered to the principles of the Declaration of Helsinki and
was approved by the Ethics Committee of Wuhan Union Hospital. After
obtaining written informed consent from all participants or their
legal guardians, where required, their medical history was
collected. Of the participants, 10 members of the family underwent
comprehensive ophthalmologic examination, including visual acuity,
IOP, slit-lamp bio-microscopy, direct fundus examination,
gonioscopy, standard automated perimetry and retinal nerve fiber
layer (RNFL) by optical coherence tomography. Additionally, two
patients (I2, II1), with low vision and five unaffected children
(III9, IV1, IV2, IV3, IV4), underwent slit-lamp bio-microscopy, IOP
measurements and direct fundus examination. The three unaffected
members were unable to undergo any examination. The diagnostic
criteria for POAG were based on at least two of the following
glaucoma characteristics, with the opening of the anterior chamber
angle, excluding any secondary glaucoma: Characteristic
glaucomatous changes of the optic disc, visual field defects and
high IOP (>21 mmHg) (20).
Ocular hypertension (OHT) was defined as IOP >22 mmHg and
long-term follow-up without optic disc damage or visual field
impairment (21). Unaffected
individuals exhibit IOP values in the normal range (≤21 mmHg) and
lack of optic nerve damage. Based on the World Health Organizations
standards stated in 1992 (22),
visual acuity ≤3/60 and/or visual field <10° in the eye with
comparatively better vision was diagnosed as blindness. The
University of São Paulo Glaucoma Visual Field Staging System
(USP-GVFSS): Early visual field defect, visual field index (VFI)
>91%; moderate visual field defect, 91%≥ VFI >78%; severe
visual field defect, VFI ≤78% (23).
Genetic detection using whole-exome
sequencing (WES)
Peripheral blood (2 ml) was collected from each
individual into EDTA-tubes (Becton, Dickinson and Company). Genomic
DNA was extracted from leucocytes in the blood samples using a
Blood Genome Extraction kit (Tiangen Biotech Co., Ltd.) according
to the manufacturer's protocol. Genomic DNA was quantitated by
Qubit 3.0 Fluorometer (Thermo Fisher Scientific, Inc.). An NEBNext
Ultra II DNA Library Prep kit (New England BioLabs, Inc.) was used
for library preparation. Sample DNA (e.g., concentration of DNA
were 96.2 (III8), 116 (III9) and 106 ng/ul (IV4) restpectively) was
the subjected to NextSeq 500 Sequencing system (Illumina, Inc.) to
perform 150 bp pair-end sequencing by Genokon Medical Laboratory
(Xiamen, China).
Following sequencing, quality controls were
performed to remove low-quality data by Trimmomatic (http://www.usadellab.org/cms/uploads/supplementary/Trimmomatic/Trimmomatic-0.36.zip)
(24). Clean reads were aligned to
the reference human genome (GRCh37/hg19) using the Burrows-Wheeler
Alignment tool (25). GATK
(software.broadinstitute.org/gatk) was used to identify
single-nucleotide polymorphisms and insertions or deletions
(indels). Subsequently, ANNOVAR (https://annovar.openbioinformatics.org/en/latest/user-guide/download/,
version number 20191024) (26) was
used to annotate genetic variants with functional information.
Common variants were filtered out, such as variants of intergenic,
intronic, upstream, downstream, or synonymous variants and variants
with minor allele frequency (MAF) >1% in the 1000 Genomes
Project (27), the ExAC database
(http://exac.broadinstitute.org/) and
gnomAD (https://gnomad.broadinstitute.org/). SIFT (http://sift.jcvi.org), MutationAssessor (http://mutationassessor.org/r3/) PROVEAN
(28), Mutation Taster2
(http://www.mutationtaster.org) and CADD
(https://annovar.openbioinformatics.org/en/latest/user-guide/download/;
version no. hg19_dbnsfp33a_20170221) (29) were used for pathogenicity
prediction of each variant. Exomiser (30) and Phenolyzer (31) were used to perform
genotype-phenotype analyses. Finally, the interpretation of
variants was performed to identify the potential mutations, in
accordance with the American College of Medical Genetics and
Genomics Standards and Guidelines (32).
Sanger sequencing
The potential mutation of the proband and the
remaining family members were validated using Sanger sequencing.
DNA was extracted from peripheral blood samples from the proband
and other family members. DNA sequence including the candidate
mutation was amplified using the following primers: MYOC forward,
5′-TGTAGTCTCGGCTCACAG-3′ and reverse, 5′-TGATAGGATAGAGGGCTTT-3′.
The PCR cycle consisted of an initial denaturation step of 5 min at
98°C followed by 35 cycles of 30 sec at 98°C, 30 sec at 53°C, and
45 sec at 72°C, and a final step at 72°C for 5 min. All PCR
products were separated and then directly sequenced using BigDye
Terminator v.3.1 Mix (Applied Biosystems; Thermo Fisher Scientific,
Inc.) and analyzed by capillary electrophoresis using an ABI Prism
3500 Genetic Analyzer (Applied Biosystems; Thermo Fisher
Scientific, Inc.).
Multiple sequence alignment analysis
and protein structural & functional analysis
The amino acid sequence of MYOC across different
species (Xenopus tropicalis: XP_002934195.3; Homo sapiens:
NP_000252.1; Macaca mulatta: XP_001099905.2; Rattus norvegicus:
NP_110492.1; Mus musculus: NP_034995.3) were aligned by Clustal
Omega (https://www.ebi.ac.uk/Tools/msa/clustalo/). The
structural impact of missense variants on protein was predicted and
analyzed by Phyre2 (http://www.sbg.bio.ic.ac.uk/~phyre2/html/page.cgi?id=index).
Results
Clinical findings
The pedigree of the family investigated in the
present study is shown in Fig. 1.
The II3 proband experienced symptoms of eye swelling, headache and
nausea for the first time at 28 years of age. The proband was
diagnosed with POAG by her local GP. The results of her ophthalmic
examination are described in Table
I. The highest IOP was 50 and 20 mmHg [oculus dexter (OD) and
oculus sinister (OS)], respectively), and the cup-to-disc ratio was
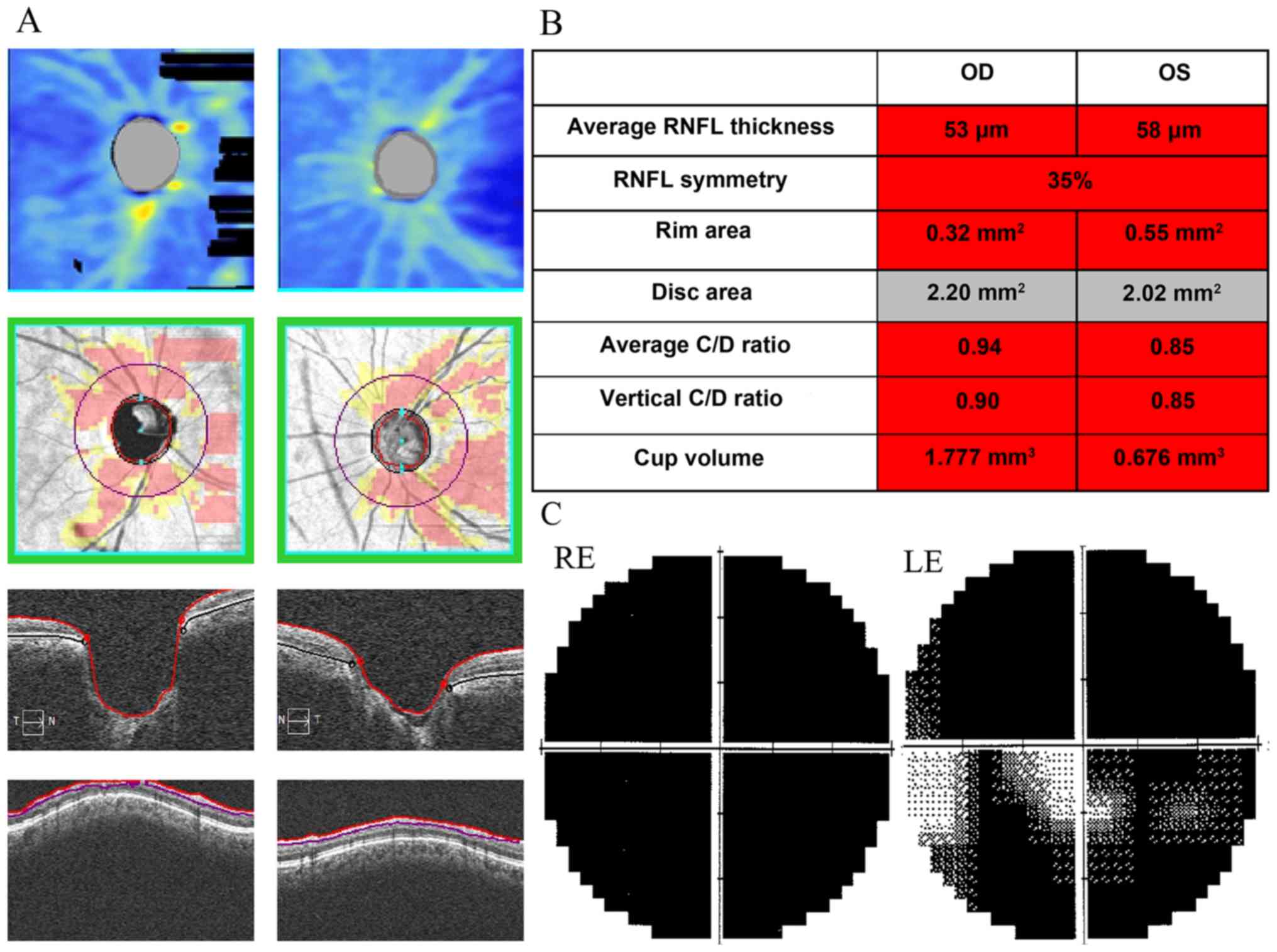
0.9 and 0.8 (OD and OS, respectively). Optical coherence tomography
revealed a significant thinning of the patient's RNFL thickness
(Fig. 2A and B). The patient
exhibited severe visual field damage (Fig. 2C) and low vision (OD, light
perception; OS: 1.0). Of the remaining 19 family members examined,
four were diagnosed with POAG, one exhibited OHT, whereas the
remaining 14 were unaffected and exhibited normal clinical
features, without obvious signs of glaucoma; the ophthalmic
examination results of five patients with POAG and one individual
with OHT are listed in Table I.
All five patients with POAG had a wide anterior chamber angle and
normal iris. Among these five patients, three were female. The mean
age of all POAG patients at diagnosis was 26.2±4.12 years (range,
22–33 years). All patients showed symptoms of eye distention,
headache and vision loss before diagnosis. The mean maximum IOP
values were 40.2±6.53 mmHg (range, 8–55 mmHg) and 39.2±15.51 mmHg
(range, 20–60 mmHg) (OD and OS, respectively); two patients were
blind. Severe visual field defects were observed in three eyes of
two patients (II3, III5). A total of nine eyes in five patients
(I2, II1, II3, II5, III5) underwent surgery (80%), at an average
age of 27.2±3.54 years (range, 22–33 years). No optic disc or
visual field defects were observed for the OHT individuals
(III9).
 | Table I.Demographic data and clinical
characteristics of patients with the D208Y mutation. |
Table I.
Demographic data and clinical
characteristics of patients with the D208Y mutation.
| A, Patients with
POAG |
|---|
|
|---|
| Pedigree number
(n=5) | Sex | Age at study,
years | IOP at study, nCT
OD/OS, mm hg | Age at diagnosis,
years | BCVA, OD/OS | C/D ratio,
OD/OS | Visual field
damage, OD/OS | Highest IOP, nCT
OD/OS, mm hg | Operation eye/age,
years |
|---|
| I2 | Female | 75 | 18/17 | 33 | NLP/NLP | 1.0/1.0 | NA | 55/60 | OU/33 |
| II1 | Male | 54 | 10/18 | 22 | FC/50 cm; HM/30
cm | 0.9/0.9 | NA | 53/55 | OU/22 |
| II3 | Female | 50 | 17/15 | 28 | LP/1.0 | 0.9/0.8 | S/S | 50/20 | OU/28 |
| II5 | Female | 43 | 18/15 | 22 | 1.0/1.0 | 0.7/0.6 | E/E | 35/30 | OU/27 |
| III5 | Male | 26 | 8/31 | 26 | 1.0/0.08 | 0.5/0.9 | E/S | 8/31 | OS/26 |
| Mean | – | 49.6±15.91 | – | 26.2±4.12 | – | – | – |
40.2±17.57/39.2±15.51 | – |
|
| B, Unaffected
family members |
|
| Pedigree number
(n=3) | Sex | Age at study,
years | IOP at study,
nCT OD/OS, mm hg | Age at
diagnosis, years | BCVA,
OD/OS | C/D ratio,
OD/OS | Visual field
damage, OD/OS | Highest IOP, nCT
OD/OS, mm hg | Operation
eye/age, years |
|
| III2 | Male | 29 | 14/17 | – | 1.0/1.0 | 0.3/0.4 | Normal | 14/17 | – |
| III6 | Male | 11 | 13/16 | – | 1.0/1.0 | 0.3/0.3 | Normal | 13/16 | – |
| III8 | Male | 10 | 18/19 | – | 1.0/1.0 | 0.3/0.3 | Normal | 18/19 | – |
|
| C, Patients with
ocular hypertension (n=1) |
|
| Pedigree
number | Sex | Age at study,
years | IOP at study,
nCT OD/OS, mm hg | Age at
diagnosis, years | BCVA,
OD/OS | C/D ratio,
OD/OS | Visual field
damage, OD/OS | Highest IOP, nCT
OD/OS, mm hg | Operation
eye/age, years |
|
| III9 | Male | 17 | 28/29 | – | 0.2/0.12 | 0.5/0.4 | Normal | 28/29 | – |
Mutation screening of MYOC in
POAG
WES was performed for the human genome, covering
>20,000 genes and 85% of the human heritage diseases. The
detection range included mutation types such as single nucleotide
variants and indels. MYOC is the most common POAG-related
pathogenic gene (14); the
reference sequence of the MYOC gene can be found in the NCBI gene
databank (ID: 4653). After comparing with the reference sequence, a
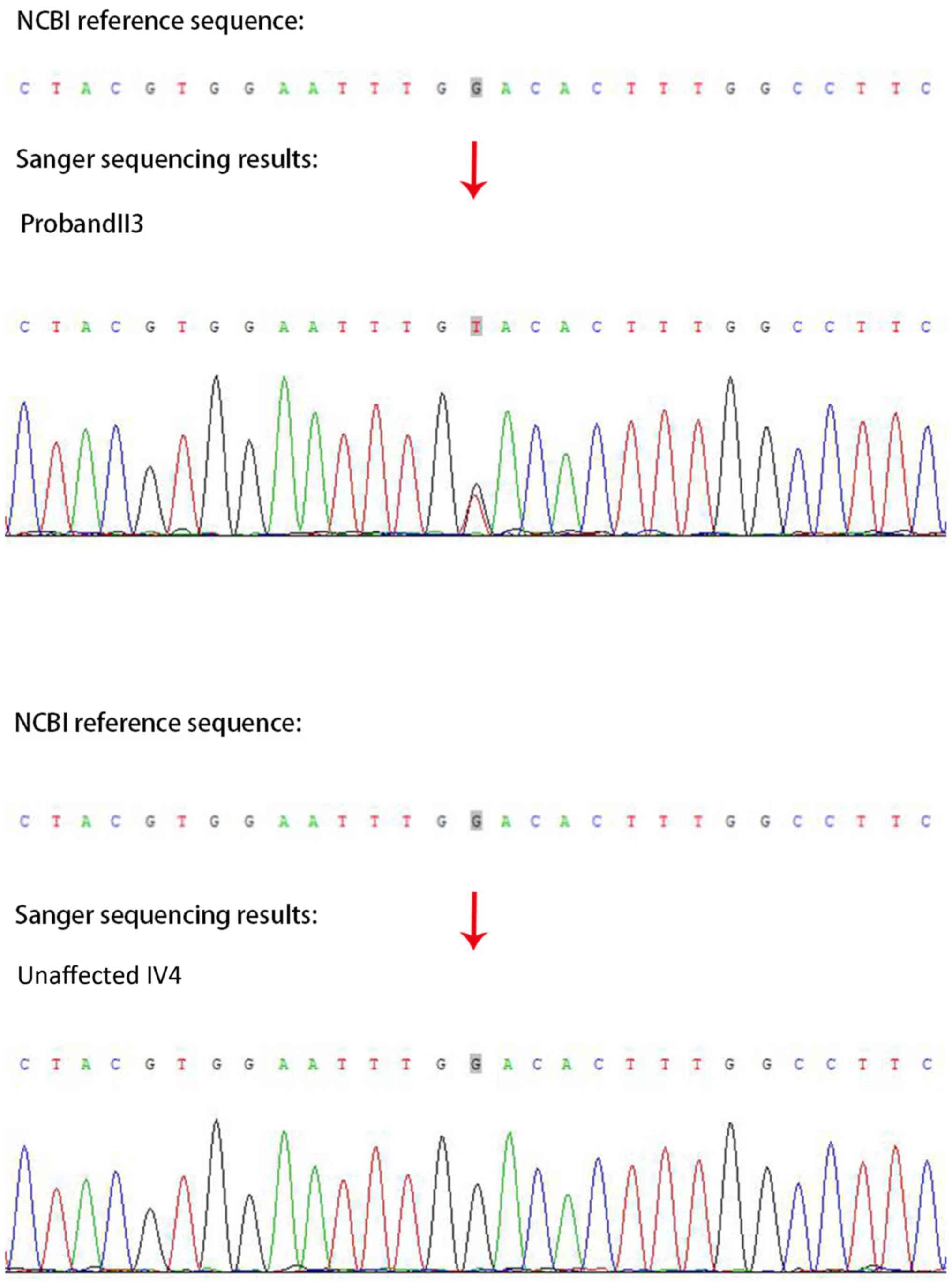
heterozygous MYOC missense mutation (c.G622T: p.D208Y) was detected
in the proband with POAG (II3; Fig.
3). The disease associated with the MYOC gene variation in the
OMIM database is the primary open-angle glaucoma 1A type and is an
autosomal dominant genetic (https://www.omim.org/entry/137750). The mutation (G to
T) was located in the coding region of exon 2 at base 622, leading
to the mutation of amino acid 208 of the encoded protein from
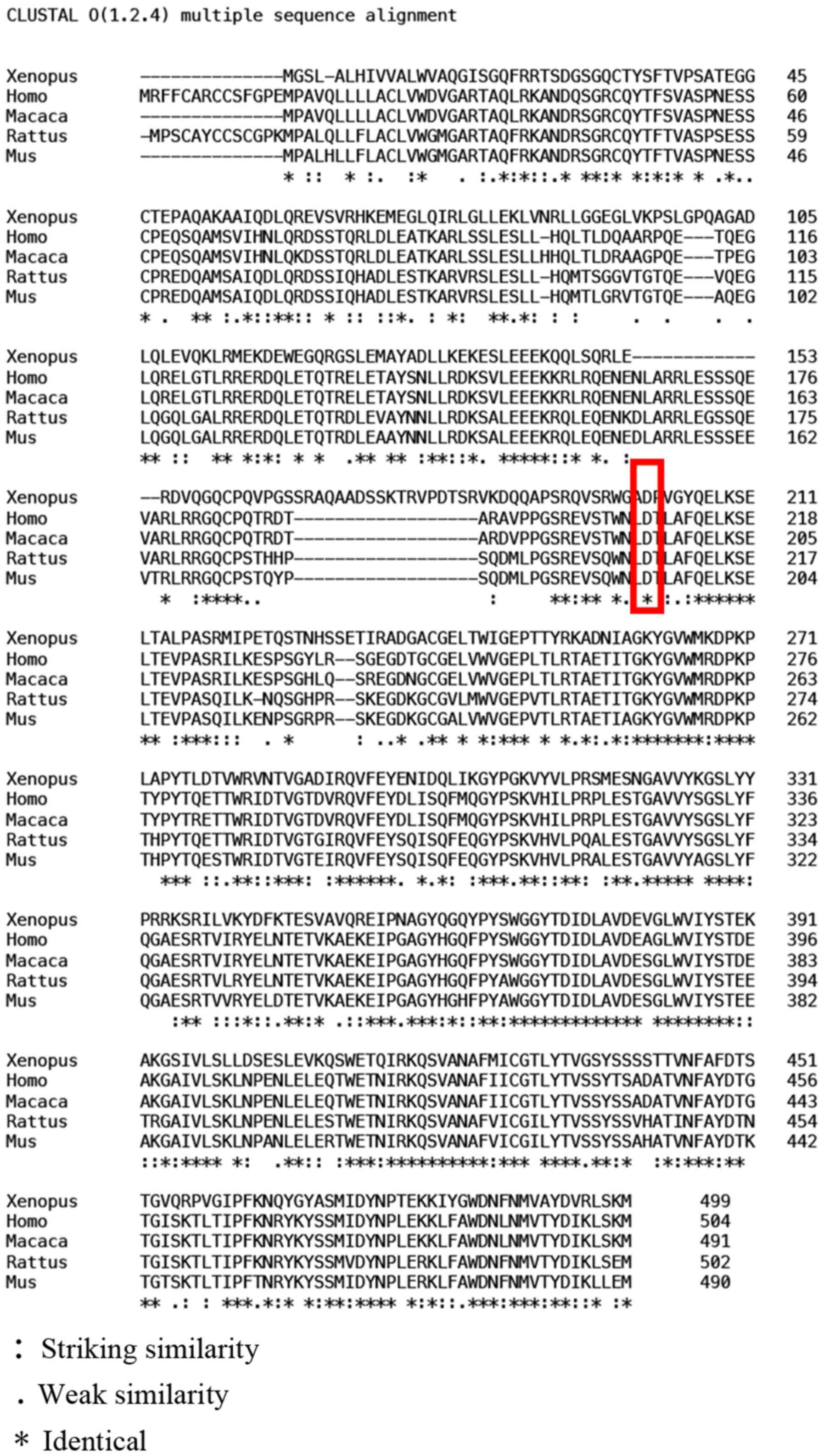
aspartic acid to tyrosine. Multiple sequence alignment analyses
indicated that this gene shows a high degree of conservation among
different species (Fig. 4). This
detected mutation locus was not found in several databases
(ClinVar, 1000 genomes, ExAC and gnomAD). Protein function
prediction software SIFT and MutationAssessor revealed that the
mutation was deleterious. Based on protein structural and
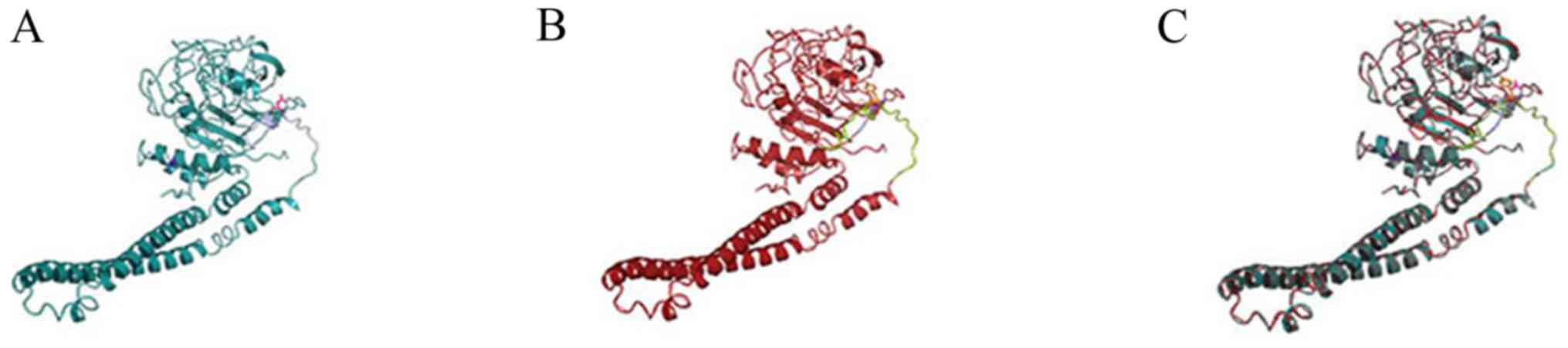
functional analysis with Phyre2, the aspartic acid was replaced by
tyrosine due to the base mutation, but the secondary structure of
the mutant protein did not change significantly compared with that
of the wild type (Fig. 5). sA
publicly available version of the database query HGMD, included in
the reported MYOC gene variants, missense mutation rare benign
variation (hgmd.cf.ac.uk/ac/index.php). Of the 20 family members,
five individuals with POAG (I2, II1, II3, II5 and III5), one
individual with OHT (III9) and three unaffected family members
(III2, III6 and III8) were found to carry the mutation, whereas the
remaining unaffected members did not harbor the mutation.
Therefore, the mutation prevalence in this family was 45%. Three
mutation carriers (III2, III6 and III8) did not exhibit elevated
IOP values or glaucomatous defects (Table I).
Discussion
In the present study, a novel c.G622T: p.D208Y
heterozygous mutation of the MYOC gene was identified in a
Chinese family with a high incidence of POAG. In one previous
study, a heterozygous variation of p.D208E in MYOC was found
in one patient with OHT and two patients with POAG, although its
relationship with POAG could not be clarified as it was also found
in a 50-year-old individual without POAG (33). In the current pedigree, five
patients with POAG and four unaffected individuals were found to
harbor the p.D208Y mutation in MYOC. The incomplete
penetrance suggests that, depending on the mode of inheritance, a
DCM may cause related diseases in some individuals, but not in all
DCM carriers. Age-related penetrance of POAG was defined as the
ratio of the total number of POAG patients carrying the mutated
gene to the total number of mutation carriers in a specific age
group. The penetrance of the D208Y mutation was calculated as 0% in
individuals <20 years old, 55.6% in patients 20–35 years old,
11.1% in patients 31–35 years old and 0% >45 years old. The
penetrance of the majority of MYOC mutations is incomplete,
which may be associated with age-related gene expression,
environmental exposure time and gene-gene or gene-environment
interactions (34). Compared with
other evaluated MYOC mutations, the penetrance of the
p.D208Y mutation in this family was low. In previous studies, the
penetrance of MYOC p.Q368X was low, with 56.4% (35) of such patients developing glaucoma
at 40 years of age and 78% developing glaucoma at 70 years of age
(36). By contrast, MYOC
p.P370L reached full penetrance by 27 years of age (37). It is possible therefore that the
three non-affected carriers and one carrier with OHT of the mutated
gene in the present study were too young to show signs of glaucoma.
As aspartic acid is negatively charged and hydrophilic, whereas
tyrosine is not charged and hydrophilic, the mutation of p.D208Y
may change the local charge density of this protein. However, the
exact function of MYOC and the physiological and pathological
effects of MYOC in POAG remain unclear. For the cases in the
present study, complete ophthalmological surveillance with optic
disc photography, tonometry and automated perimetry every 6 months
is recommended. This would facilitate further determination of
whether the MYOC mutated gene is a pathogenic gene of
POAG.
POAG is asymptomatic in its early stages and is
often detected in the advanced stage with severe visual field
damage and high IOP. Owing to the genetic characteristics of POAG,
the association between its genotype and phenotype is of great
significance for predicting the phenotypic variation range of
specific mutations and for better diagnosis and treatment. Further
studies on a larger number of families from different ethnic
backgrounds are required to establish the genotypic-phenotypic
associations for this blindness-causing disease. Although no
pathogenic characteristics of p.D208Y were identified in the
present study, the results expand the mutational spectrum of
MYOC-induced POAG, which may be of clinical significance for
disease prediction.
Acknowledgements
The authors would like to thank Mr. Wenlong Xie and
Mrs. Fengfeng Zhang (Genokon Institute of Medical Science and
Laboratory, Xiamen, China) for their help performing whole exome
sequencing and sanger sequencing.
Funding
This study was supported by a financial grant from
The Wuhan Science and Technology Bureau (grant no.
02.07.17040008.05).
Availability of data and materials
The data that support the findings of the present
study are available from GenBank (accession no. MN335319), but
restrictions apply to the availability of these data, which were
used under license for the current study, and thus are not publicly
available. The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request and with permission from GenBank.
Authors' contributions
WF and FC conceived and designed the study. WF, FC
and WZ conducted clinical examinations. WF, WL, CD and YG analyzed
and interpreted the data. WF wrote the manuscript. WF, FC, CD and
YG reviewed and revised the manuscript. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
The study adhered to the principles of the
Declaration of Helsinki and was approved by the Ethics Committee of
Union Hospital (Tongji Medical College, Huazhong University of
Science and Technology, Wuhan, China). Written informed consent was
obtained from all participants or their legal guardians, where
required.
Patient consent for publication
Written informed consent for publication was
obtained from all participants or their legal guardians, where
required.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Quigley HA: Glaucoma. Lancet.
377:3263–1377. 2011. View Article : Google Scholar
|
|
2
|
Quigley HA and Broman AT: The number of
people with glaucoma worldwide in 2010 and 2020. Br J Ophthalmol.
90:262–267. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kwon YH, Fingert JH, Kuehn MH and Alward
WL: Primary open-angle glaucoma. N Engl J Med. 360:1113–1124. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Cheng JW, Cheng SW, Ma XY, Cai JP, Li Y
and Wei RL: The prevalence of primary glaucoma in mainland China: A
systematic review and meta-analysis. J Glaucoma. 22:301–306. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Marcus MW, de Vries MM, Junoy Montolio FG
and Jansonius NM: Myopia as a risk factor for open-angle glaucoma:
A systematic review and meta-analysis. Ophthalmology.
118:1989–1994.e2. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhou M, Wang W, Huang W and Zhang X:
Diabetes mellitus as a risk factor for open-angle glaucoma: A
systematic review and meta-analysis. PLoS One. 9:e1029722014.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stewart WC: The effect of lifestyle on the
relative risk to develop open-angle glaucoma. Curr Opin Ophthalmol.
6:3–9. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Budde WM: Heredity in primary open-angle
glaucoma. Curr Opin Ophthalmol. 11:101–106. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Raymond V: Molecular genetics of the
glaucomas: Mapping of the first five ‘GLC’ loci. Am J Hum Genet.
60:272–277. 1997.PubMed/NCBI
|
|
10
|
Johnson AT, Drack AV, Kwitek AE, Cannon
RL, Stone EM and Alward WL: Clinical features and linkage analysis
of a family with autosomal dominant juvenile glaucoma.
Ophthalmology. 100:524–529. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sheffield VC, Stone EM, Alward WL, Drack
AV, Johnson AT, Streb LM and Nichols BE: Genetic linkage of
familial open angle glaucoma to chromosome 1q21-q31. Nat Genet.
4:47–50. 1993. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Rangachari K, Bankoti N, Shyamala N,
Michael D, Sameer Ahmed Z, Chandrasekaran P and Sekar K: Glaucoma
Pred: Glaucoma prediction based on Myocilin genotype and phenotype
information. Genomics. 111:696–699. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Stone EM, Fingert JH, Alward WL, Nguyen
TD, Polansky JR, Sunden SL, Nishimura D, Clark AF, Nystuen A,
Nichols BE, et al: Identification of a gene that causes primary
open angle glaucoma. Science. 275:668–670. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang K, Gaitsch H, Poon H, Cox NJ and
Rzhetsky A: Classification of common human diseases derived from
shared genetic and environmental determinants. Nat Genet.
49:1319–1325. 2017. View
Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu Y and Vollrath D: Reversal of mutant
myocilin non-secretion and cell killing: Implications for glaucoma.
Hum Mol Genet. 13:1193–1204. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Zode GS, Kuehn MH, Nishimura DY, Searby
CC, Mohan K, Grozdanic SD, Bugge K, Anderson MG, Clark AF, Stone EM
and Sheffield VC: Reduction of ER stress via a chemical chaperone
prevents disease phenotypes in a mouse model of primary open angle
glaucoma. J Clin Invest. 121:3542–3553. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jacobson N, Andrews M, Shepard AR,
Nishimura D, Searby C, Fingert JH, Hageman G, Mullins R, Davidson
BL, Kwon YH, et al: Non-secretion of mutant proteins of the
glaucoma gene myocilin in cultured trabecular meshwork cells and in
aqueous humor. Hum Mol Genet. 10:117–125. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Joe MK, Sohn S, Hur W, Moon Y, Choi YR and
Kee C: Accumulation of mutant myocilins in ER leads to ER stress
and potential cytotoxicity in human trabecular meshwork cells.
Biochem Biophys Res Commun. 312:592–600. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tamm ER: Myocilin and glaucoma: Facts and
ideas. Prog Retin Eye Res. 21:395–428. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yao YH, Wang YQ, Fang WF, Zhang L, Yang JH
and Zhu YH: A recurrent G367R mutation in MYOC associated with
juvenile open angle glaucoma in a large Chinese family. Int J
Ophthalmol. 11:369–374. 2018.PubMed/NCBI
|
|
21
|
Pasutto F, Keller KE, Weisschuh N, Sticht
H, Samples JR, Yang YF, Zenkel M, Schlötzer-Schrehardt U, Mardin
CY, Frezzotti P, et al: Variants in ASB10 are associated with
open-angle glaucoma. Hum Mol Genet. 21:1336–1349. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
World Health Organization (WHO), .
International statistical classification of diseases and related
health problems - 10th revision (ICD-10). WHO; Geneva: 2010,
https://www.who.int/classifications/icd/ICD10Volume2_en_2010.pdf
|
|
23
|
Susanna R Jr and Vessani RM: Staging
glaucoma patient: Why and how? Open Ophthalmol J. 3:59–64. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bolger AM, Lohse M and Usadel B:
Trimmomatic: A flexible trimmer for Illumina sequence data.
Bioinformatics. 30:2114–2120. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li H and Durbin R: Fast and accurate short
read alignment with Burrows-Wheeler transform. Bioinformatics.
25:1754–1760. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang K, Li M and Hakonarson H: ANNOVAR:
Functional annotation of genetic variants from high-throughput
sequencing data. Nucleic Acids Res. 38:e1642010. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
1000 Genomes Project Consortium, ; Auton
A, Brooks LD, Durbin RM, Garrison EP, Kang HM, Korbel JO, Marchini
JL, McCarthy S, McVean GA and Abecasis GR: A global reference for
human genetic variation. Nature. 526:68–74. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Choi Y and Chan AP: PROVEAN web server: A
tool to predict the functional effect of amino acid substitutions
and indels. Bioinformatics. 31:2745–2747. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kircher M, Witten DM, Jain P, O'Roak BJ,
Cooper GM and Shendure J: A general framework for estimating the
relative pathogenicity of human genetic variants. Nat Genet.
46:310–315. 2014. View
Article : Google Scholar : PubMed/NCBI
|
|
30
|
Robinson PN, Köhler S, Oellrich A; Sanger
Mouse Genetics Project, ; Wang K, Mungall CJ, Lewis SE, Washington
N, Bauer S, Seelow D, et al: Improved exome prioritization of
disease genes through cross-species phenotype comparison. Genome
Res. 24:340–348. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yang H, Robinson PN and Wang K:
Phenolyzer: Phenotype-based prioritization of candidate genes for
human diseases. Nat Methods. 12:841–843. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Richards S, Aziz N, Bale S, Bick D, Das S,
Gastier-Foster J, Grody WW, Hegde M, Lyon E, Spector E, et al:
Standards and guidelines for the interpretation of sequence
variants: A joint consensus recommendation of the american college
of medical genetics and genomics and the association for molecular
pathology. Genet Med. 17:405–424. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lam DS, Leung YF, Chua JK, Baum L, Fan DS,
Choy KW and Pang CP: Truncations in the TIGR gene in individuals
with and without primary open-angle glaucoma. Invest Ophthalmol Vis
Sci. 41:1386–1391. 2000.PubMed/NCBI
|
|
34
|
Lei L, Li S, Liu X and Zhang C: The
clinical feature of myocilin Y437H mutation in a Chinese family
with primary open-angle glaucoma. Br J Ophthalmol. 103:1524–1529.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Craig JE, Baird PN, Healey DL, McNaught
AI, McCartney PJ, Rait JL, Dickinson JL, Roe L, Fingert JH, Stone
EM and Mackey DA: Evidence for genetic heterogeneity within eight
glaucoma families, with the GLC1A Gln368STOP mutation being an
important phenotypic modifier. Ophthalmology. 108:1607–1620. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Allingham RR, Wiggs JL, De La Paz MA,
Vollrath D, Tallett DA, Broomer B, Jones KH, Del Bono EA, Kern J,
Patterson K, et al: Gln368STOP myocilin mutation in families with
late-onset primary open-angle glaucoma. Invest Ophthalmol Vis Sci.
39:2288–2295. 1998.PubMed/NCBI
|
|
37
|
Shimizu S, Lichter PR, Johnson AT, Zhou Z,
Higashi M, Gottfredsdottir M, Othman M, Moroi SE, Rozsa FW,
Schertzer RM, et al: Age-dependent prevalence of mutations at the
GLC1A locus in primary open-angle glaucoma. Am J Ophthalmol.
130:165–177. 2000. View Article : Google Scholar : PubMed/NCBI
|