Introduction
Chondrocyte differentiation is a pivotal event
during bone formation in most mammals, and it comprises of two
processes (1). Intramembranous
ossification is an essential process during which mesenchymal stem
cells directly differentiate into osteoblasts and is responsible
for the development of irregular bones, such as the skull bone,
clavicle and parts of the jaw (1).
Endochondral ossification is the second important process of
chondrocyte differentiation, responsible for the formation of long
bones and vertebrate skeleton (2),
which is controlled by a series of events (3). These events are initiated by the
establishment of a chondrogenic template and culminated by its
replacement with the coordinated activity of osteoblasts,
osteoclasts and endothelial cells (3,4).
Chondrocyte maturation ultimately leads to hypertrophy, which is an
essential contributor to longitudinal bone growth (5,6).
However, abnormal chondrocyte hypertrophy is involved in the
pathogenesis of osteoarthritis and skeletal dysplasia (7). Meanwhile, histone deacetylase 4
(HDAC4) is known as a member of a large family of HDAC that alter
gene expression by catalyzing the removal of acetyl groups from
core histones, which are responsible for chondrocyte hypertrophy
and bone formation (8,9). However, the regulatory mechanism
underpinning HDAC4 function remains largely unclear. Therefore, a
better understanding of the mechanism underlying HDAC4-mediated
regulation of endochondral ossification in chondrocyte hypertrophy
is necessary for the improved management of skeletal growth
diseases.
A previous study demonstrated that HADC4 plays an
important role in the formation of the skeletal system (10). Knockout of HDAC4, a repressor of
matrix metalloproteinase-13 (MMP-13) transcription, results in
increased MMP-13 expression in vitro and in vivo
(11). MMP-13 plays a critical
role in endochondral ossification and bone remodeling (12). Chen et al (13) reported that HDAC4 inhibits the
expression of MMP-13 and prevents chondrocyte hypertrophy by
inhibiting the activity of runt-related transcription factor-2
(Runx2), a transcription factor essential during endochondral bone
formation. Wnt5a is a secreted glycoprotein belonging to the
Wingless/integrase 1 family of secreted glycoproteins; it is highly
conserved among species and plays key roles in processes regulating
embryonic development and postnatal tissue homeostasis (14). Wnt5a-knockout mice
reportedly demonstrate perinatal lethality primarily due to
respiratory failure and present extensive developmental
abnormalities and the Wnt5a signaling pathway can regulate
fundamental cellular processes, including cell proliferation and
survival (15). Therefore, HDAC4
is considered as a central regulator of chondrocyte hypertrophy and
growth plate development, and the complete deletion of HDAC4 is
lethal (9,10,16–18).
To investigate the roles of HDAC4 in postnatal skeletal
development, the present study constructed a mouse strain with
conditional HDAC4-knockout in collagen type 2α1
(Col2α1)-expressing cells.
Materials and methods
Generation of floxed HDAC4
animals
The animal experiments and protocol were approved by
the Institutional Animal Care and Use Committee of Rhode Island
Hospital (Providence, United States of America). To investigate the
physiological role of HDAC4 in hypertrophic chondrocytes, the
HDAC4 gene was deleted in Col2α1-Cre mice using the Cre/loxP
gene targeting technique (six female mice, mean weight 20 g, from
University of Texas Southwestern Medical Center, mice transgenic
for Cre in Col2α1-expressing chondrocytes). Col2α1-Cre mice were
mated with HDAC4fl/fl (six male mice, mean weight 20 g,
provided by Dr Olson, University of Texas Southwestern Medical
Center) animals to obtain HDAC4fl/−, Col2α1-Cre mice
(9). Mice transgenic for Cre in
Col2α1-expressing chondrocytes (Col2α1-Cre) have been previously
reported (19). These mice were
subsequently interbred with HDAC4fl/fl animals, and
their offspring (HDAC4d/d, Col2α1-Cre) and
HDAC4fl/fl animals were analyzed at postnatal (P) days
2, 4, 6, 8, 10 and 21 after birth. All mice were housed in a cage
with free access to tap water and food. The housing environment was
set at 22±2°C and 60±10% humidity in a 12-h light/dark cycle. The
health and behavior of mice were monitored and recorded on a daily
basis. A total of 40 mice were divided into two groups
(HDAC4fl/fl group and HDAC4d/d; Col2α1-Cre
group; n=20 for each group). The mice were euthanized by an
overdose of carbon dioxide according to the American Veterinary
Medical Association (AVMA) guidelines (20). The experiment endpoint was set when
mice were postnatal 21 days old. The mice are first put into the
euthanasia chamber and CO2 was directed into the top of
the euthanasia chamber. Weep holes were drilled into the euthanasia
chamber lid to allow room air to escape as CO2 filled
the chamber. One-hundred percent compressed CO2 gas was
used to euthanize mice. When the flow rate displaced no ≤30% of the
chamber volume/minute for apparent death, death was confirmed by
absence of breathing or heartbeat. In this way, the mice had no
pain, no nerve reaction and no signs of bleeding. The growth
progress of mice was recorded at postnatal (P) days 2, 4, 6 and 8,
10 and 21.
Genotyping and PCR
Genomic DNA was isolated from skin tissue from the
tail of the mice using the QIAamp DNA Micro kit (Qiagen GmbH) and
used for PCR genotype identification of HDAC4d/d,
Col2α1-Cre transgenic mice. Routine mouse genotyping was performed
using PCR, using the DNA polymerase Hotstar from Qiagen GmbH. The
following primer pairs were used for the Cre allele: Forward,
5′-ATCCGAAAAGAAAACGTTGA-3′ and reverse, 5′-ATCCAGGTTACGGATATAGT-3′.
PCR was initiated for 15 min at 95°C, then 30 cycles of
denaturation at 95°C for 30 sec, primer annealing for 40 sec at
55°C and a final extension step at 72°C for 1 min. The HDAC4
allele and target gene deletion were identified using the following
primers: Forward, 5′-ATCTGCCCACCAGAGTATGTG-3′ and reverse,
5′-CTTGTTGAGAACAAACTCCTGCAGCT-3′. PCR was initiated for 15 min at
95°C, then 35 cycles of denaturation at 94°C for 40 sec, primer
annealing for 40 sec at 58.5°C and a final extension step at 72°C
for 1 min. The expected product sizes for the wild-type
HDAC4, floxed and Cre alleles were 480, 620 and 328 bp,
respectively.
Cell proliferation assay
Cell proliferation of chondrocytes was assessed
using the in vivo bromodeoxyuridine (BrdU) assay as
previously described (21).
Briefly, mice were intraperitoneally injected with 25 µl/25 g body
weight BrdU at P3 and P5. The animals were then euthanized at P8,
P14, and P21 for specimen collection followed by
immunohistochemistry (22).
Histology
After the animals were euthanized at P8, P14, and
P21, their right knee joints (n=3) were harvested and immersed in
10% formalin for 72 h at room temperature. The specimens were
processed without decalcification and embedded in a single block of
Paraplast X-tra (Thermo Fisher Scientific, Inc.). Thereafter, 6-µm
coronal sections were mounted on slides. Safranin O/Fast green
staining for 2 h at room temperature was performed to evaluate the
developmental growth plate and hypertrophic differentiation at P8
and P21, whereas Von Kossa staining for 2 h at room temperature was
used to evaluate tissue mineralization at P8 and P14.
Photomicrographs were obtained using a Nikon Ri1 microscope (Nikon
Corporation) (23,24).
Whole-body staining and skeletal
analysis
The mice were euthanized before the experiment
began. The mineralization pattern of the skeleton was analyzed at
P10 using whole-body staining as previously described (25). Briefly, mice were skinned under
anesthesia, eviscerated and fixed in 95% ethanol for 1–2 days.
Subsequently, acetone was used to remove fat. Skeletons were
stained using Alizarin red S/Alcian blue for 3 days at 37°C and
sequentially immersed in 1% aqueous KOH at room temperature until
the skeletons were clearly visible. Mineralized bones were
visualized through the staining (23).
Microcomputed tomography (µCT)
Three tibial tissues isolated from each group (8-day
old mice) were fixed in 70% ethanol for 24 h at room temperature
and prepared for high resolution µCT (SkyScan 1172). Images were
obtained using the following parameters: 40 kV, 80 µA, Pixel
size=8.5 µm, 2,000×1,332 matrix, six averages and a 0.5-mm aluminum
filter. Images were reconstructed using a threshold of 0–0.09, beam
hardening correction of 35, ring artifact correction of 7 and
Gaussian smoothing (factor=1). From the reconstructed images of the
tibia, the 637-µm region immediately proximal to the growth plate
was examined for the trabecular bone. The volume images of each
group were analyzed using CTAn software (v1.7.1, SkyScan, Burker)
to determinate the trabecular bone microstructure and trabecular
thickness (Tb.Th), trabecular separation (Tb.Sp).
Immunohistochemistry
BrdU and proliferating cell nuclear antigen (PCNA)
are typical proliferation markers that can indicate the
proliferative activity of chondrocytes. BrdU is a synthetic
nucleoside used to detect cellular proliferation by incorporating
the place of thymine and pairs with guanine (26). PCNA is a multifunctional protein
present in the nuclei of eukaryotic cells and has a crucial role in
DNA replication and DNA repair systems (27). Matrix metalloproteinase (MMP)-13 is
marker associated with endochondral ossification and bone repair
(28). Runx2 and osteoprotegerin
(OPG) are typical mineralization markers. Runx2 is a transcription
factor essential for osteoblast differentiation and chondrocyte
maturation (29). OPG is a key
protective factor of bony tissue and is responsible for osteoclast
formation and postnatal bone growth (30). To determine the expressions of
these markers, 6-µm coronal sections of right knee joints were
placed on positively charged glass slides (Thermo Fisher
Scientific, Inc.). The sections were dried on a hotplate to
increase tissue adherence. Immunohistochemistry was performed using
the 3,3′diaminobenzidine (DAB), streptavidin-peroxidase (SP) and
the DAB Histostain-SP immunohistochemistry kit (Zymed; Thermo
Fisher Scientific, Inc.). Sections were deparaffinized and
rehydrated using xylene and an alcohol gradient. Endogenous
peroxidase was blocked by treating the sections with 3% hydrogen
peroxide in methanol (Sigma-Aldrich; Merck KGaA) for 30 min at room
temperature. The sections were then digested with 5 mg/ml
hyaluronidase in phosphate-buffered saline (PBS) (Sigma-Aldrich;
Merck KGaA) for 20 min and incubated with specific antibodies
against MMP-13 (1:500, cat. no. ab219620, Abcam), BrdU (1:100, cat.
no. ab8955, Abcam), PCNA (1:10,000, cat. no. ab29, Abcam), Runx2
(1:1,000, cat. no. 192256, Abcam), OPG (1:100, cat. no. 203061,
Abcam) and CD34 (1:2,500, cat. no. 81289, Abcam) at 4°C overnight.
After treatment with biotinylated secondary antibody and SP
conjugate (500 µl, Histostain®-Plus 3rd Gen IHC
Detection kit, cat. no. D859673, Zymed; Thermo Fisher Scientific,
Inc.) at 37°C for 30 min, the sections were developed in DAB
chromogen (Zymed; Thermo Fisher Scientific, Inc.) and then
counterstained with hematoxylin (Zymed; Thermo Fisher Scientific,
Inc.) for 2 min at room temperature. Images were captured using a
Nikon Ri1 light microscope (Nikon Corporation) at ×4 magnification.
Percentages of positive staining for BrdU, PCNA, MMP-13, Runx2, OPG
and CD34 in both the groups were semi quantified using the
Image-Pro Plus 7.0 software (Media Cybernetics, Inc.) as described
previously (31). Mean values from
three independent measurements were used for statistical analysis
(n=3).
In situ hybridization (ISH)
ISH was used to determine type X collagen (ColX) and
Wnt5a expression levels as an indicator of HDAC4-downregualtion.
ColX is a critical marker that represents terminal differentiation
during chondrogenesis (32). Wnt5a
a member of Wnt family that plays a vital role in regulating
skeletal development (33). ISH
was performed on paraffin sections prepared from tibial tissues at
P8 as described previously (34).
Accordingly, 10-µm sections were placed on positively charged glass
slides (Thermo Fisher Scientific, Inc.) and subsequently dried on a
hotplate to increase tissue adherence. These sections were then
deparaffinized and rehydrated using conventional methods. Briefly,
the tibial sections were incubated with proteinase K (cat. no.
P8107S; New England BioLabs, Inc.) at 60°C for 15 min and then
rinsed with diethyl pyrocarbonate (DEPC)-PBS for 5 min. Thereafter,
the tibial sections were incubated with 5 mg/ml hyaluronidase at
37°C for 15 min and then rinsed with DEPC-PBS for 5 min. The
sections were prehybridized for 1 h and hybridized for 16 h with a
0.5-µl probe (Col X and Wnt5a, from Department of Orthopedics, The
Warren Alpert Medical School of Brown University) at 50°C. The
sections were washed with 5X saline sodium citrate (SSC) for 5 min
at room temperature and 4X SSC for 30 min at 50°C. Thereafter, the
tibial sections were incubated with Tris-HCL-NaCl-EDTA buffer at
37°C for 10 min; washed with 2XSSC for 10 min, 0.2XSSC for 10 min
and 0.1XSSC buffer for 10 min. Following this, the sections were
incubated with a blocking buffer for 1 h at room temperature.
Digoxin antibody (1:200, ca. no. ab420, Abcam) was added to the
sections for 2 h at room temperature. The reaction was stopped with
tris-EDTA (TE) buffer (pH 8.0) for 10 min. The sections were then
washed with distilled water and counterstained with hematoxylin
(Zymed; Thermo Fisher Scientific, Inc.) for 2 min at room
temperature. Images were captured using a Nikon Ri1 light
microscope (Nikon Corporation) at ×4 magnification.
Reverse transcription-quantitative
(RT-q)PCR
The mRNA levels of HDAC4, PCNA, MMP-13, Runx2,
OPG and osteocalcin were quantified using RT-qPCR. Total RNA
was isolated using TRIzol (Life Technologies; Thermo Fisher
Scientific, Inc.) from the sternum cartilage using the RNeasy
isolation kit (Qiagen, Inc.). Accordingly, 1 µg total RNA was
reverse transcribed into cDNA using the iScripTM cDNA synthesis kit
(Bio-Rad Laboratories, Inc.), with reaction conditions: 25°C 5 min,
42°C for 30 min, 85°C for 5 min and kept at 4°C. Thereafter, 50
ng/µl of the resulting cDNA was used as the template to quantify
the relative mRNA content using the QuantiTect SYBR Green PCR kit
(Qiagen, Inc.) with the DNA Engine Opticon 2 Continuous
Fluorescence Detection system (MJ Research; Bio-Rad Laboratories,
Inc.). qPCR was initiated for 30 sec at 94°C, then 40 cycles of
denaturation at 9°C for 30 sec, primer annealing for 3 sec at 60°C
and a final extension step at 72°C for 30 sec. Primer pairs that
were used for quantitative detection of gene expression are listed
in Table I, and 18S rRNA was used
as the internal control. The cycle threshold values for target
genes were measured and calculated using a computer software (MJ
Research; Bio-Rad Laboratories, Inc.). Relative transcript levels
were calculated using the 2−ΔΔCq method where ΔΔCq=ΔCq
E-ΔCq C, ΔCq E=Cqexp-Cq18S, and ΔCq C=CqCCq 18S (35).
 | Table I.Primer sequences for reverse
transcription-polymerase chain reaction. |
Table I.
Primer sequences for reverse
transcription-polymerase chain reaction.
| Mouse gene | Primer sequence,
5′-3′ |
|---|
| HDAC4 |
|
|
Forward |
ATCTGCCCACCAGAGTATGTG |
|
Reverse |
CTTGTTGAGAACAAACTCCTGCAGCT |
| MMP-13 |
|
|
Forward |
GGACCTTCTGGTCTTCTGGC |
|
Reverse |
GGATGCTTAGGGTTGGGGTC |
| Runx2 |
|
|
Forward |
CCGCACGCAAACCGCACCAT |
|
Reverse |
CGCTCCGGCCCACAAATCTC |
| PCNA |
|
|
Forward |
GCCGAGATCTCAGCCATATT |
|
Reverse |
ATGTACTTAGAGGTACAAAT |
| OPG |
|
|
Forward |
ATTGGCTGAGTGTTTTGGTGG |
|
Reverse |
CGCTGCTTTCACAGAGGTCA |
| Osteocalcin |
|
|
Forward |
ATGAGAGCCCTCAGACTCCTC |
|
Reverse |
CGGGCCGTAGAAGCGCCGATA |
| 18s rRNA |
|
|
Forward |
CGGCTACCACATCCAAGGAA |
|
Reverse |
CGCTCCGGCCCACAAATCTC |
Statistical analysis
Data from at least three separate experiments are
presented as mean ± standard deviation. Differences between groups
were analyzed using unpaired t-tests and P<0.05 was considered
to indicate a statistically significant difference. Statistical
analyses were performed using SPSS software v18.0 (SPSS Inc.).
Results
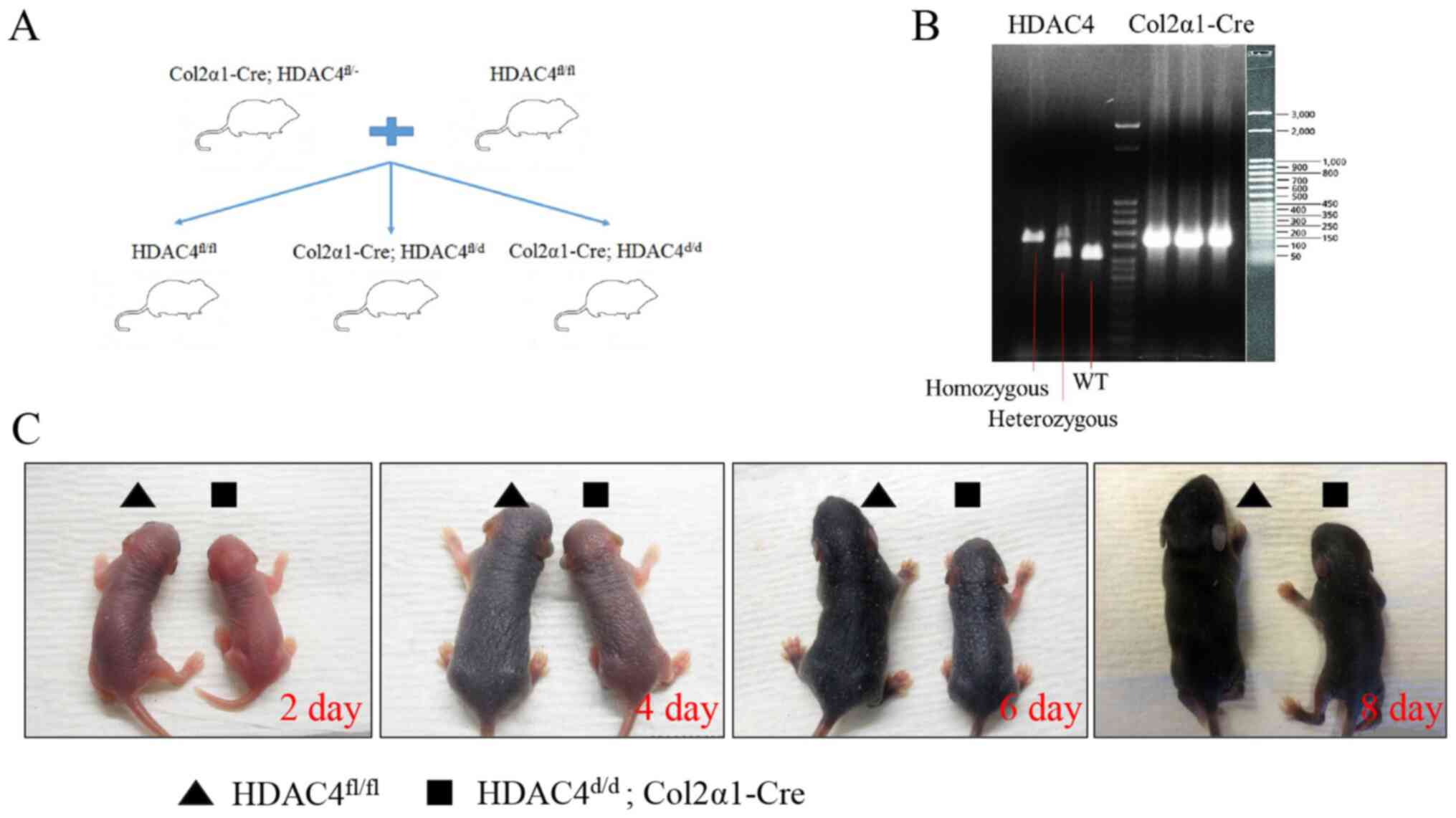
Generation of HDAC4, Col2α1-Cre
mice
To investigate the function of HDAC4 in vivo,
the mouse HDAC4 gene was deleted using homologous
combination. The targeting strategy is presented in Fig. 1A. Wild-type, homologous and
heterozygous mice were confirmed using qPCR. Product sizes for
wild-type HDAC4, floxed and Cre alleles were 480, 620 and
328 bp, respectively (Fig. 1B).
The HDAC4d/d, Col2α1-Cre mice survived until P21, were
severely runted and smaller compared with HDAC4fl/fl
mice at P2, P4, P6 and P8 (Fig.
1C). These results indicated that the development of
HDAC4d/d, Col2α1-Cre mice was slower compared with that
of HDAC4fl/fl mice.
Shortened growth plates and disordered
hypertrophic chondrocyte zones in HDAC4d/d, Col2α1-Cre
mice
Chondrocyte hypertrophy is widely observed in
endochondral ossification (5).
During bone formation, chondrocytes were enlarged, and they
secreted more ColX compared with the early stage of the
ossification (5). Here, Safranin
O/Fast green staining was performed on paraffin sections from
HDAC4fl/fl and HDAC4d/d, Col2α1-Cre mice at
P8 (femur and tibia) (Fig. 2A-C)
and P21 (tibia; Fig. 2D). As shown
in Fig. 2A2-C2 and Fig. 2A4-C4, HDAC4d/d,
Col2α1-Cre mice had fewer prehypertrophic and hypertrophic
chondrocyte zones compared with the HDAC4fl/fl mice
(Fig. 2A1-C1 and A3-C3) at P8. In
addition, the hypertrophic chondrocyte zone appeared smaller, and
chondrocytes underwent degradation in the HDAC4d/d,
Col2α1-Cre mice group (Fig. 2A2-C2 and
A4-C4) compared with the HDAC4fl/fl mice (Fig. 2A1-C1 and A3-C3) at P8. As presented
in Fig. 2D-2, HDAC4d/d,
Col2α1-Cre mice showed decreased proteoglycans zones (marked by
yellow arrows), premature ossification (marked by black arrows)
compared with the HDAC4fl/fl mice (Fig. 2D-1) at P21. These results indicated
that the shortened growth plates and disordered hypertrophic
chondrocyte zones in HDAC4d/d, Col2α1-Cre mice compared
with that of HDAC4fl/fl mice.
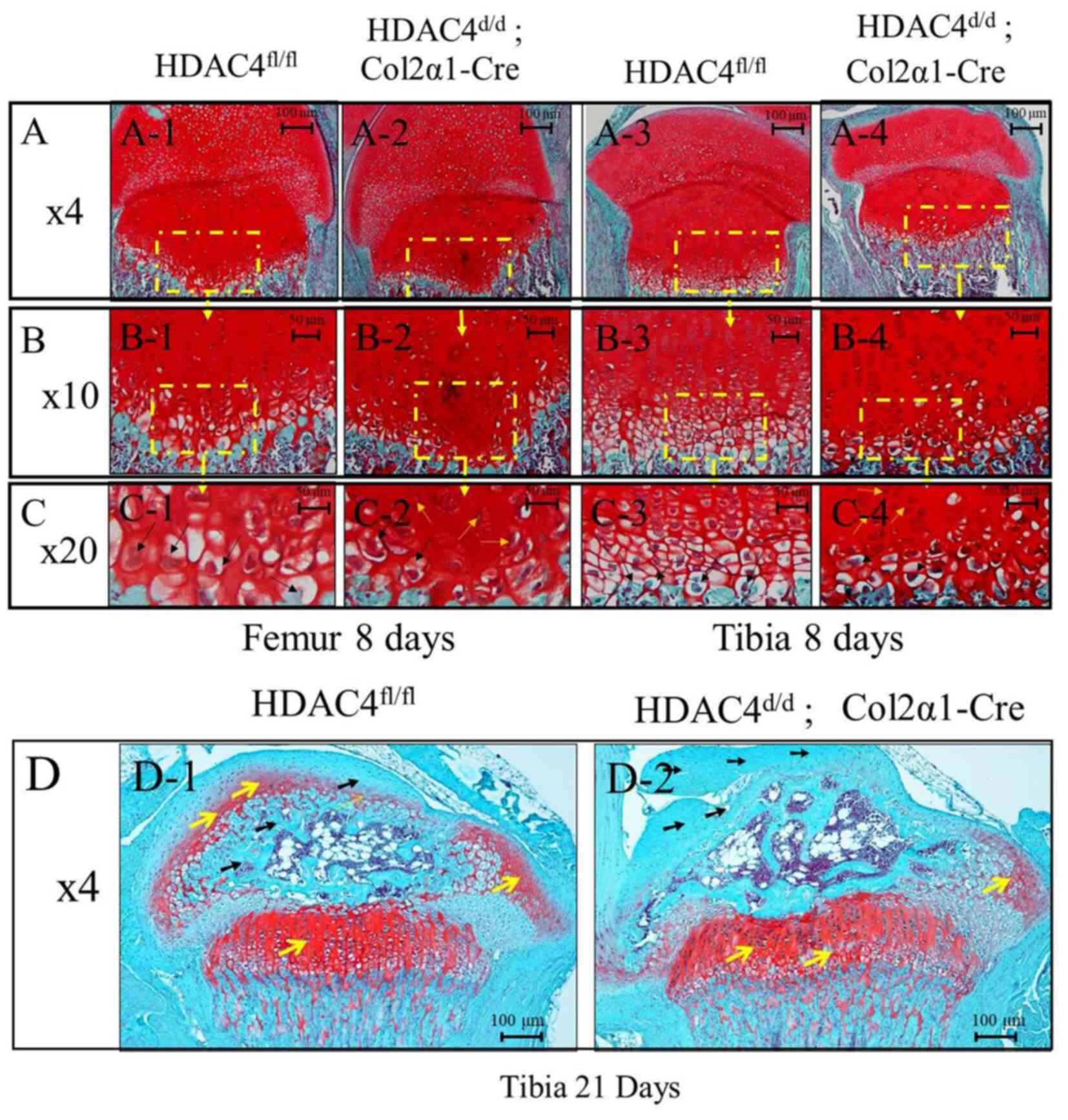 | Figure 2.Safranin O/Fast green staining on
paraffin sections of the femur and tibia tissues at P8 and P21.
(A-C) Safranin O/Fast green staining showing femur and tibia tissue
sections (magnification, ×4, ×10 and ×20 respectively) at P8. The
HDAC4d/d, Col2α1-Cre mice had fewer cartilage tissues in
the hypertrophy zone at P8 compared with those in the
HDAC4fl/fl mice. Black arrows represent hypertrophic
chondrocytes, and orange arrows represent chondrocytes degradation.
Scar bars, 100 or 50 µm. (D) Safranin O/Fast green staining showing
tibia tissue sections (magnification, ×4) at P21. The
HDAC4d/d, Col2α1-Cre mice had fewer proteoglycans but
more ossification at P21 compared with the HDAC4fl/fl
mice. The yellow arrow represents zone of proteoglycans, the black
arrow represents zone of ossification, Scale bars, 100 µm. P,
postnatal day; HDAC4, histone deacetylase 4; Col2α1, collagen type
2α1; fl, floxed; d, down. |
Accelerated vascular invasion and
mineralization due to HDAC4-knockout
During hypertrophic chondrocyte mineralization,
blood vessels are induced by hypertrophic chondrocytes that
secreted vascular endothelial growth factor (36). Von Kossa staining was performed to
assess mineralization and vascular invasion of the tissue in
HDAC4fl/fl mice and HDAC4d/d, Col2α1-Cre mice
at P8 (Fig. 3A and B) and P14
(Fig. 3C and D). As encircled in
Fig. 3B1, vascular invasion of the
growth plates in HDAC4d/d, Col2α1-Cre mice began to
appear at P8, whereas HDAC4fl/fl mice had no vascular
formation at P8 (Fig. 3A1). More
importantly, HDAC4d/d, Col2α1-Cre mice developed an
earlier secondary center of ossification (red circled area) and
growth plates (yellow circled area; Fig. 3D and D1) compared with
HDAC4fl/fl mice (Fig. 3C
and C1) at P14. Moreover, the HDAC4d/d, Col2α1-Cre
mice had thicker cortical (Fig.
3D2) and cancellous bones (Fig.
3D3) compared with HDAC4fl/fl mice (Fig. 3C2 and C3). These results indicated
that the vascular invasion and mineralization were accelerated in
HDAC4d/d, Col2α1-Cre mice compared with
HDAC4fl/fl mice.
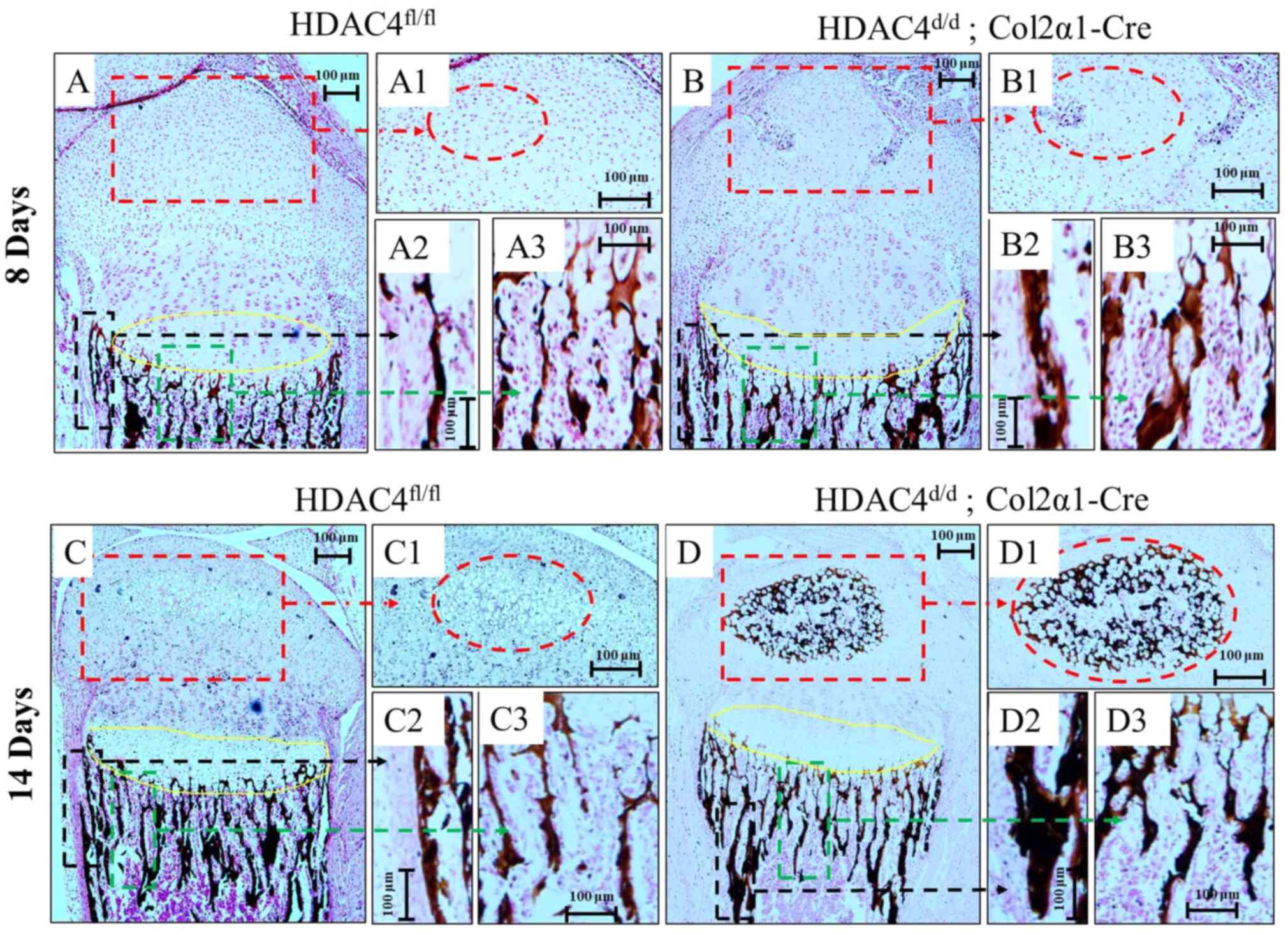 | Figure 3.Mineralization and vascular invasion
of tissues at (A and B) P8 and (C and D) P14 using Von Kossa
staining. (B1, the red circled regions) vascular invasion and (D1,
the red circled regions) secondary center of ossification were
observed earlier in the HDAC4d/d, Col2α1-Cre mice
compared with in HDAC4fl/fl mice (A1 and C1, the red
circled regions). The HDAC4d/d, Col2α1-Cre mice had (B2
and D2) thicker cortical and (B3 and D3) cancellous bones compared
with the (A2, C2, A3 and C3) HDAC4fl/fl mice. Yellow
circled areas represent growth plates. Black circled areas
represent cortical bones. Green circled areas represent cancellous
bones. Boxes indicate area magnified in adjacent figures. Scale
bars, 100 µm. P, postnatal day; HDAC4, histone deacetylase 4;
Col2α1, collagen type 2α1; fl, floxed; d, down. |
Abnormal and premature mineralization
due to HDAC4 deletion
The detailed skeletal phenotypes of the
HDAC4fl/fl and HDAC4d/d, Col2α1-Cre groups
were analyzed using Alizarin red and Alcian blue staining at P10
(Fig. 4A and B). Alizarin red and
Alcian blue staining of the whole skeleton of HDAC4d/d,
Col2α1-Cre mice revealed obvious malformed and premature
mineralization of the endochondral skeleton, including the sternum
(Fig. 4D), xiphisternum (Fig. 4D), tibia (Fig. 4F) and intervertebral disk (Fig. 4H) compared with
HDAC4fl/fl mice(Fig. 4C, E
and G). Ectopic ossification of the endochondral cartilage
prevented normal expansion of the rib cage (Fig. 4D) µCT analysis showing the
HDAC4d/d, Col2α1-Cre mice (Fig. 4J) have more cancellous bone than
HDAC4fl/fl mice (Fig.
4I) at P8. The Tb.Th was significantly greater in the
HDAC4d/d, Col2α1-Cre mice compared with
HDAC4fl/fl mice, as measured by µCT (P<0.05; Fig. 4K). Simultaneously, Tb.Sp was
significantly reduced in the HDAC4d/d, Col2α1-Cre mice
compared with that in HDAC4fl/fl mice (P<0.05;
Fig. 4L). These results indicated
that abnormal and premature mineralization occurs in the knockout
mouse.
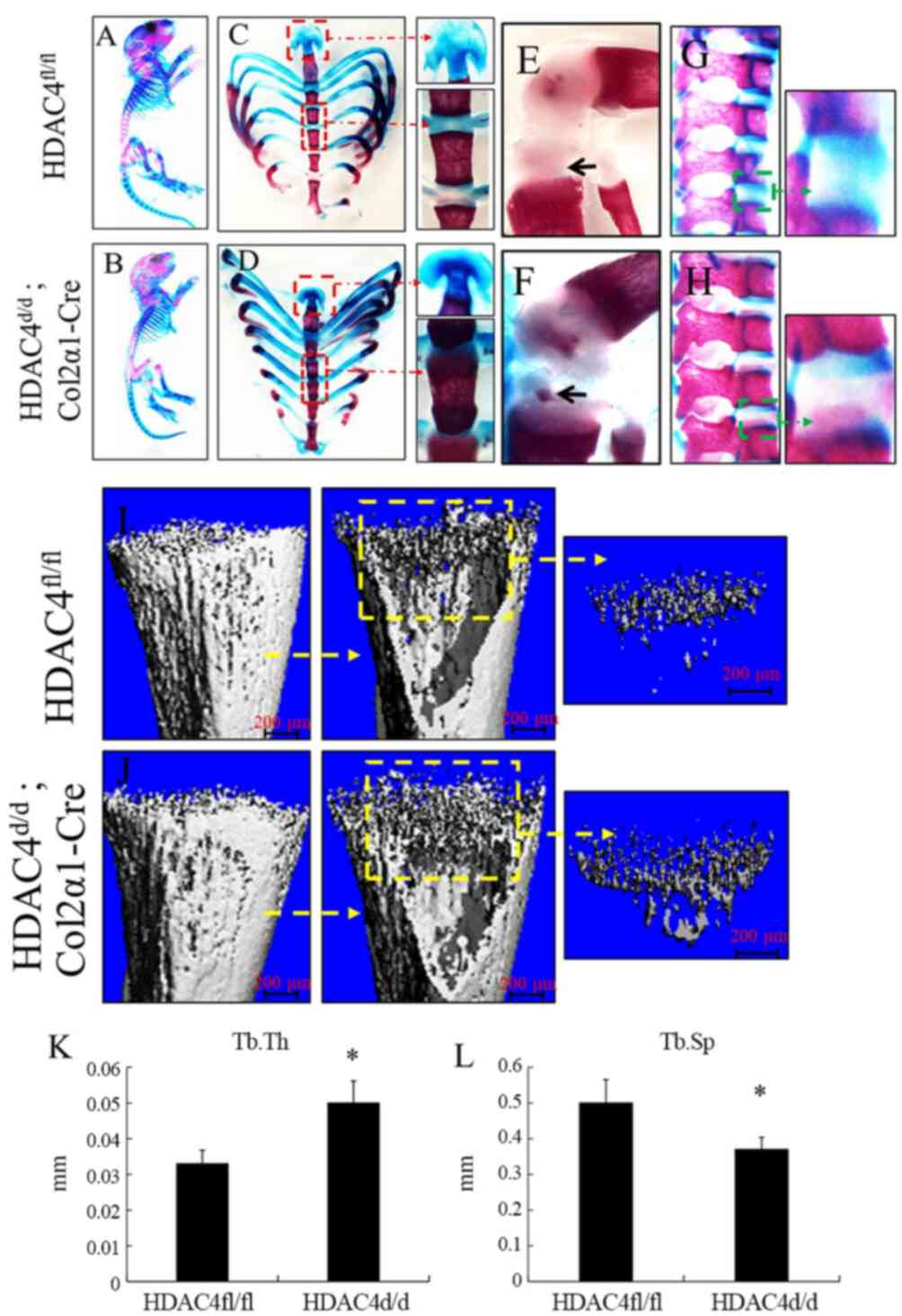 | Figure 4.Premature ossification in
HDAC4d/d, Col2α1-Cre mice. (A-H) Alizarin red and Alcian
blue stained skeletons of the HDAC4fl/fl and
HDAC4d/d, Col2α1-Cre mice at P10, the red boxes and
arrow represent xiphoid process and sternum, the green boxes and
arrow represent lumbar vertebrae, black arrows represent tibia
cartilage. (I and J) µCT analysis showing the bone image of tibial
tissues from the HDAC4fl/fl and the HDAC4d/d,
Col2α1-Cre mice at P8, yellow boxes and arrow represent cancellous
bone. (K and L) Tb.Th, trabecular thickness; Tb.Sp, trabecular
separation. in the HDAC4fl/fl and HDAC4d/d,
Col2α1-Cre mice were evaluated using µCT analysis. Each assay was
performed in triplicates for each group. Scale bars, 200 µm.
*P<0.05 vs. HDAC4fl/fl mice. P, postnatal day; HDAC4,
histone deacetylase 4; Col2α1, collagen type 2α1; fl, floxed; d,
down; Tb.Th, trabecular thickness; Tb.Sp, trabecular
separation. |
Decreased chondrocyte proliferation
due to HDAC4 deletion via Wnt5a signaling pathway inhibition and
increased chondrocyte hypertrophy
Immunohistochemistry was performed to determine the
presence of BrdU, PCNA, MMP-13, Runx2, OPG and CD34 in
HDAC4fl/fl and HDAC4d/d, Col2α1-Cre mice
(Fig. 5A-L). The percentages areas
positively expressing MMP-13, Runx2, OPG and CD34, were
significantly higher in the HDAC4d/d, Col2α1-Cre mice
compared with the HDAC4fl/fl mice (all P<0.05;
Fig. 5M-R). Conversely, the
expressions of typical proliferative markers, including BrdU and
PCNA, were significantly lower in the HDAC4d/d,
Col2α1-Cre group compared with the HDAC4fl/fl group
(P<0.05; Fig. 5M-R). ISH
confirmed increased ColX and decreased Wnt5a expression (red boxes
and arrows) in the HDAC4d/d, Col2α1-Cre mice (Fig. 5S-Z). These results indicated that
HDAC4 deletion reduce chondrocyte proliferation via Wnt5a signaling
pathway inhibition and increased chondrocyte hypertrophy.
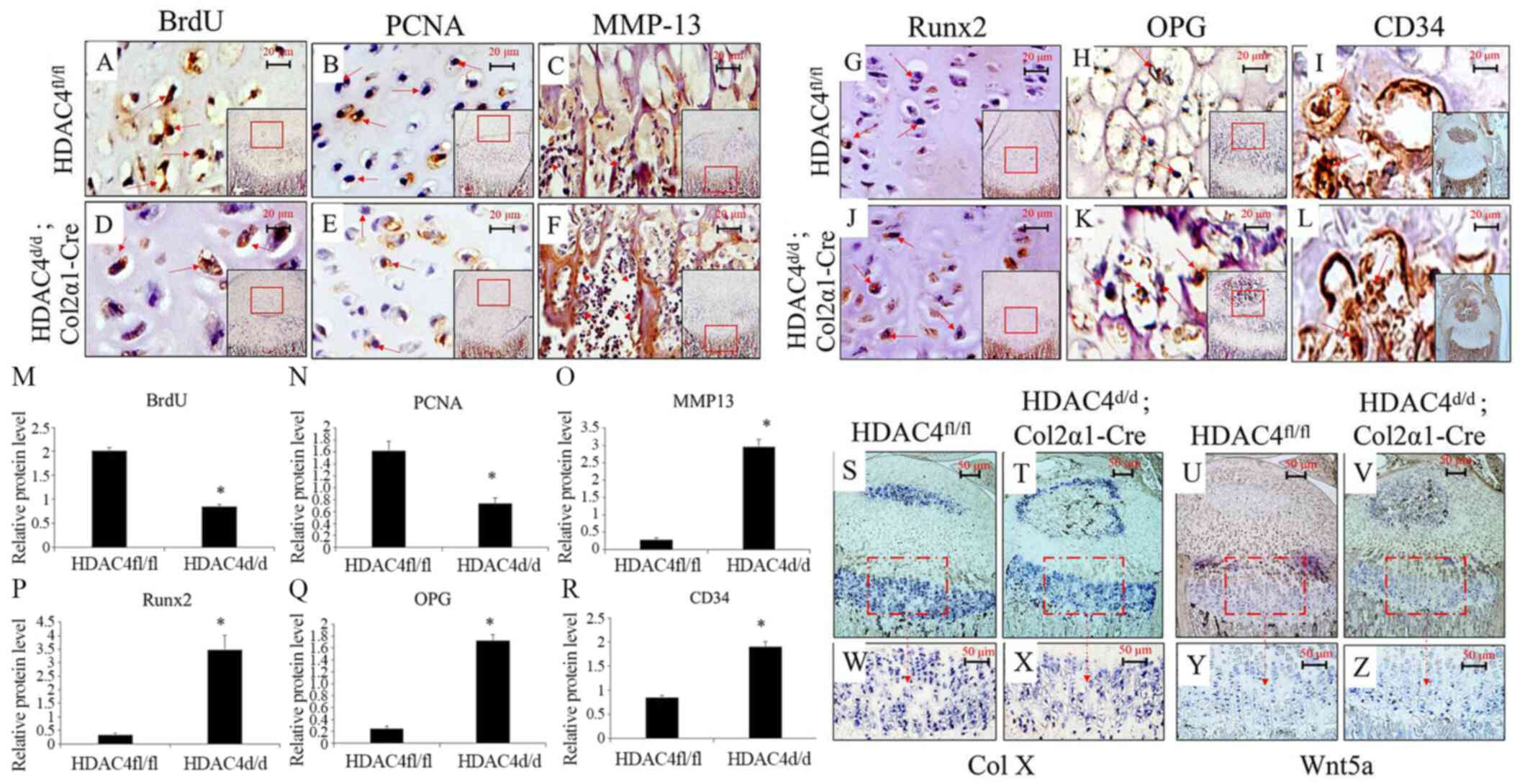 | Figure 5.Decreased chondrocyte proliferation
due to HDAC4 deletion. (A-L) Immunohistochemical staining of
the tibial articular cartilage of HDAC4fl/fl and
HDAC4d/d, Col2α1-Cre mice at P8 for determining the
expressions of BrdU, PCNA, MMP-13, Runx2, OPG. and CD34 Positive
signals (brown staining) are indicated by red arrows. Scale bars,
20 µm. (M-R) Semiquantitative analysis of positively expressed
areas for BrdU, PCNA, MMP-13, Runx2, OPG and CD34. (S-Z) In
situ hybridization on paraffin sections prepared for
determining the expressions of Col X and Wnt5a in tibial tissues at
P14. The HDAC4d/d, Col2α1-Cre mice had decreased
expressions of BrdU, PCNA and Wnt5a but increased expressions of
MMP-13, Runx2, OPG, Col X and CD34. Boxed areas in S-V indicate the
regions shown in W-Z, respectively. Scale bars, 50 µm. *P<0.05
vs. HDAC4fl/fl mice. P, postnatal day; HDAC4, histone
deacetylase 4; Col2α1, collagen type 2α1; fl, floxed; d, down;
BrdU, bromodeoxyuridine; PCNA, proliferating cell nuclear antigen;
MMP-13, matrix metalloproteinase-13; Runx2, runt-related
transcription factor 2; Col X, type X collagen; OPG,
osteoprotegerin. |
Genetic changes associated with
increased hypertrophy and mineralization and decreased
proliferation in HDAC4d/d, Col2α1-Cre mice
qPCR analyses indicated that the mRNA levels of
HDAC4 and PCNA were lower and those of MMP-13,
Runx2, OPN and osteocalcin were higher in the
HDAC4d/d, Col2α1-Cre mice compared with those of
HDAC4fl/fl mice (all P<0.05; s. 6A-F). These results
indicated that there are genetic changes associated with increased
hypertrophy and mineralization and decreased proliferation in
HDAC4d/d, Col2α1-Cre mice.
Discussion
The deletion of HDAC4 results in precocious
lethality in mice (9); however,
the mechanism underlying HDAC4-mediated postnatal regulation of
growth plate development remains unclear. A previous study
demonstrated that HDAC4 is a critical negative regulator of
chondrocyte hypertrophy due its binding to and inhibition of Runx2
(37). To further investigate the
role of chondrocyte-derived HDAC4 in postnatal skeletal
development, the present study selectively ablated the HDAC4
gene in collagen type 2-expressing cells using the Cre/loxP
gene targeting technique.
The findings of the current study demonstrated that
HDAC4-deletion in Col2α1-expressing cells results in
severely runted and slow-growing mice postdelivery, which is
consistent with the result reported by a previous study (9). In addition, the expressions of
typical proliferative markers BrdU and PCNA were significantly
lower in the HDAC4d/d, Col2α1-Cre mice compared with in
their HDAC4fl/fl control littermates, which suggested
HDAC4-deletion is associated with decreased chondrocyte growth. ISH
demonstrated that the HDAC4d/d, Col2α1-Cre mice had a
downregulated expression of Wnt5a compared with that in the
HDAC4fl/fl mice. Recent studies have also reported that
Wnt signaling is one of the numerous signaling pathways involved in
the regulation of diverse processes underpinning fundamental and
normal development, such as apoptosis, proliferation,
differentiation and polarity (38,39).
Wnt5a-knockout mice demonstrate perinatal
lethality primarily due to respiratory failure and presented
extensive developmental abnormalities (15). Wnt5a and its signaling pathway can
regulate fundamental cellular processes, including proliferation
and survival. These results suggest that Wnt5a is a potential
target for treating proliferation-related diseases (40). The present findings suggested that
HDAC4-deletion results in decreased chondrocyte
proliferation partially due to the inhibition of Wnt5a
expression.
The HDAC4d/d, Col2α1-Cre mice exhibited
premature and disordered hypertrophic chondrocytes as well as
increased levels of MMP-13 and type X collagen. MMP-13 and collagen
type X are essential for maintaining chondrocyte hypertrophy and
allowing ossification (41,42).
It is universally accepted that hypertrophic chondrocytes implicate
the final step in the maturation process of chondrocytes, which
eventually results in apoptosis of chondrocytes and replacement of
hypertrophic chondrocytes by bone tissues (43). Ossification begins when
hypertrophic chondrocytes undergo programmed cell death, and the
calcified cartilage is invaded by blood vessels, osteoblasts,
osteoclasts and mesenchymal precursors, thereby forming primary and
secondary ossification centers (3,4). The
degradation and remodeling of the cartilage matrix is necessary for
vascular invasion, whereas angiogenesis has been implicated as a
crucial step in endochondral ossification (5). The present study identified that
HDAC4 gene ablation can accelerate the mineralization and
ectopic ossification of endochondral cartilages. The expressions of
typical mineralization markers, including Runx2, OPN and
osteocalcin, were increased in the HDAC4d/d, Col2α1-Cre
mice. Therefore, it was hypothesized that premature and disordered
chondrocyte hypertrophy in HDAC4-knockout mice accelerates
the malformation of bones and results in runted mice.
Notably, the vascular invasion and secondary center
of ossification of the growth plates began to appear at P8 in the
HDAC4d/d, Col2α1-Cre mice, whereas that in the control
group began to appear at P14. Vascular invasion of the primarily
avascular hypertrophic chondrocyte zone of the growth plate is a
prerequisite for endochondral bone formation and occurs in a
sequence of events (44). Blood
vessels supplying bones orchestrate the process of bone development
and remodeling as well as regulate skeleton regeneration by
delivering nutrients, oxygen, hormones or growth factors to bone
cells (45). This process requires
a dynamic and close interaction between hypertrophic chondrocytes
and vascular invasion. The expressions of typical vascular
formation markers, such as CD34 (a widely used marker of
hematopoietic and endothelial progenitor cells), were reportedly
increased in the tibia of the HDAC4d/d, Col2α1-Cre mice
(46). The findings of the present
study demonstrated that knockout of the HDAC4 gene
accelerates endochondral bone formation by promoting vascular
invasion.
There are some limitations in this experiment. The
number of animals was small and no cell experiment was conducted.
More research is needed on the HDAC4 pathway and in the future
Wnt5a loss and gain of function studies in vitro and in
vivo should be performed to confirm the regulating effect of
the HDAC4-Wnt5a pathway. HDAC4-Wnt5a-β-catenin signaling activity
should be analyzed as it is speculated that HDAC4-Wnt5a-β-catenin
may be an indispensable role in the homeostasis of articular
cartilage (47). In the present
study, the HDAC4d/d, Col2α1-Cre mice displayed a marked
phenotype characterized by decreased chondrocyte hypertrophy, early
blood invasion and premature bone formation, leading to abnormal
growth and runted mice. These results demonstrated that
chondrocyte-derived HDAC4 is primarily responsible for the
regulation of chondrocyte differentiation and endochondral bone
formation.
Acknowledgements
The authors would like to thank Dr Olson (University
of Texas Southwestern Medical Center) who provided the
HDAC4fl/fl mice and technical support.
Funding
The current study was supported by The National
Institute of General Medical Sciences, National Institutes of
Health (grant no. P30GM122732), RIH Orthopedic Foundation and The
National Natural Science Foundation of China (grant no. 81704103,
mainly used for the author's study expenses in the United
States).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
LW and HZ designed the study. GD, CX and XS
performed the experiments. YS, PL and XWe collected the
experimental data. MZ, SW, LG, XWa and NW analyzed the experimental
data. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Animal Care and Use Committee of Rhode Island Hospital.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
ISH
|
in situ hybridization
|
|
PBS
|
phosphate-buffered saline
|
|
HDAC4
|
histone deacetylase 4
|
References
|
1
|
Li J and Dong S: The signaling pathways
involved in chondrocyte differentiation and hypertrophic
differentiation. Stem Cells Int. 2016:24703512016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
James CG, Stanton LA, Agoston H, Ulici V,
Underhill TM and Beier F: Genome-wide analyses of gene expression
during mouse endochondral ossification. PLoS One. 5:e86932010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Mackie E, Ahmed YA, Tatarczuch L, Chen KS
and Mirams M: Endochondral ossification: How cartilage is converted
into bone in the developing skeleton. Int J Biochem Cell Biol.
40:46–62. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Mackie EJ, Tatarczuch L and Mirams M: The
skeleton: A multi-functional complex organ: The growth plate
chondrocyte and endochondral ossification. J Endocrinol.
211:109–121. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun MM and Beier F: Chondrocyte
hypertrophy in skeletal development, growth, and disease. Birth
Defects Res C Embryo Today. 102:74–82. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang W, Chen J, Zhang S and Ouyang HW:
Inhibitory function of parathyroid hormone-related protein on
chondrocyte hypertrophy: The implication for articular cartilage
repair. Arthritis Res Ther. 14:2212012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Rim YA, Nam Y and Ju JH: The role of
chondrocyte hypertrophy and senescence in osteoarthritis initiation
and progression. Int J Mol Sci. 21:23582020. View Article : Google Scholar
|
|
8
|
De Ruijter AJ, Van Gennip AH, Caron HN,
Stephan KE and Van Kuilenburg AB: Histone deacetylases (HDACs):
Characterization of the classical HDAC family. Biochem J.
370:737–749. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Vega RB, Matsuda K, Oh J, Barbosa AC, Yang
X, Meadows E, McAnally J, Pomajzl C, Shelton JM, Richardson JA, et
al: Histone deacetylase 4 controls chondrocyte hypertrophy during
skeletogenesis. Cell. 119:555–566. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nakatani T, Chen T, Johnson J, Westendorf
JJ and Partridge NC: The deletion of hdac4 in mouse osteoblasts
influences both catabolic and anabolic effects in bone. J Bone
Miner Res. 33:1362–1375. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nakatani T, Chen T and Partridge NC:
MMP-13 is one of the critical mediators of the effect of HDAC4
deletion on the skeleton. Bone. 90:142–151. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Fu J, Li S, Feng R, Ma H, Sabeh F, Roodman
GD, Wang J, Robinson S, Guo XE, Lund T, et al: Multiple
myeloma-derived MMP-13 mediates osteoclast fusogenesis and
osteolytic disease. J Clin Invest. 126:1759–1772. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Chen W, Sheng P, Huang Z, Meng F, Kang Y,
Huang G and Zhang Z, Liao W and Zhang Z: MicroRNA-381 regulates
chondrocyte hypertrophy by inhibiting histone deacetylase 4
expression. Int J Mol Sci. 17:13772016. View Article : Google Scholar
|
|
14
|
Lories RJ, Corr M and Lane NE: To wnt or
not to wnt: The bone and joint health dilemma. Nat Rev Rheumatol.
9:328–339. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Kikuchi A, Yamamoto H, Sato A and
Matsumoto S: Wnt5a: Its signalling, functions and implication in
diseases. Acta Physiol (Oxf). 204:17–33. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang Z, Qin G and Zhao TC: HDAC4:
Mechanism of regulation and biological functions. Epigenomics.
6:139–150. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Huang HM, Li XL, Tu SQ, Chen XF, Lu CC and
Jiang LH: Effects of roughly focused extracorporeal shock waves
therapy on the expressions of bone morphogenetic protein-2 and
osteoprotegerin in osteoporotic fracture in rats. Chin Med J
(Engl). 129:2567–2575. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Shahi M, Peymani A and Sahmani M:
Regulation of bone metabolism. Rep Biochem Mol Biol. 5:73–82.
2017.PubMed/NCBI
|
|
19
|
Long F, Zhang XM, Karp S, Yang Y and
McMahon AP: Genetic manipulation of hedgehog signaling in the
endochondral skeleton reveals a direct role in the regulation of
chondrocyte proliferation. Development. 128:5099–5108.
2001.PubMed/NCBI
|
|
20
|
American Veterinary Medical Association, .
AVMA Guidelines for the Euthanasia of Animals: 2013 Edition.
https://www.avma.org/KB/Policies/Documents/euthanasia.pdf
|
|
21
|
Zhao GZ, Zhang LQ, Liu Y, Fang J, Li HZ,
Gao KH and Chen YZ: Effects of platelet-derived growth factor on
chondrocyte proliferation, migration and apoptosis via regulation
of GIT1 expression. Mol Med Rep. 14:897–903. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Estensoro I, Pérez-Cordón G,
Sitjà-Bobadilla A and Piazzon MC: Bromodeoxyuridine DNA labelling
reveals host and parasite proliferation in a fish-myxozoan model. J
Fish Dis. 41:651–662. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Bourgine A, Pilet P, Diouani S, Sourice S,
Lesoeur J, Beck-Cormier S, Khoshniat S, Weiss P, Friedlander G,
Guicheux J and Beck L: Mice with hypomorphic expression of the
sodium-phosphate cotransporter PiT1/Slc20a1 have an unexpected
normal bone mineralization. PLoS One. 8:e659792013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang Z, Wei X, Gao J, Zhao Y, Zhao Y, Guo
L, Chen C, Duan Z, Li P and Wei L: Intra-articular injection of
cross-linked hyaluronic acid-dexamethasone hydrogel attenuates
osteoarthritis: An experimental study in a rat model of
osteoarthritis. Int J Mol Sci. 17:4112016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Liu F, Fang F, Yuan H, Yang D, Chen Y,
Williams L, Goldstein SA, Krebsbach PH and Guan JL: Suppression of
autophagy by FIP200 deletion leads to osteopenia in mice through
the inhibition of osteoblast terminal differentiation. J Bone Miner
Res. 28:2414–2430. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Harris L, Zalucki O and Piper M: BrdU/EdU
dual labeling to determine the cell-cycle dynamics of defined
cellular subpopulations. J Mol Histol. 49:229–234. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Fan L, Bi T, Wang L and Xiao W: DNA-damage
tolerance through PCNA ubiquitination and sumoylation. Biochem J.
477:2655–2677. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Wang M, Sampson ER, Jin H, Li J, Ke QH, Im
HJ and Chen D: MMP13 is a critical target gene during the
progression of osteoarthritis. Arthritis Res Ther. 15:R52013.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Komori T: Runx2, an inducer of osteoblast
and chondrocyte differentiation. Histochem Cell Biol. 149:313–323.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Chen D, Liu Y, Liu Z and Wang P: OPG is
required for the postnatal maintenance of condylar cartilage.
Calcif Tissue Int. 104:461–474. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Eid AA, Lee DY, Roman LJ, Khazim K and
Gorin Y: Sestrin 2 and AMPK connect hyperglycemia to Nox4-dependent
endothelial nitric oxide synthase uncoupling and matrix protein
expression. Mol Cell Biol. 33:3439–3460. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Roux CH, Pisani DF, Gillet P, Fontas E,
Yahia HB, Djedaini M, Ambrosetti D, Michiels JF, Panaia-Ferrari P,
Breuil V, et al: Oxytocin controls chondrogenesis and correlates
with osteoarthritis. Int J Mol Sci. 21:39662020. View Article : Google Scholar
|
|
33
|
Michigami T: Wnt signaling and skeletal
dysplasias. Clin Calcium. 29:323–328. 2019.(In Japanese).
PubMed/NCBI
|
|
34
|
Han Q, Lin Q, Huang P, Chen M, Hu X, Fu H,
He S, Shen F, Zeng H and Deng Y: Microglia-derived IL-1β
contributes to axon development disorders and synaptic deficit
through p38-MAPK signal pathway in septic neonatal rats. J
Neuroinflammation. 14:522017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zelzer E, Mamluk R, Ferrara N, Johnson RS,
Schipani E and Olsen BR: VEGFA is necessary for chondrocyte
survival during bone development. Development. 131:2161–2171. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Zhou J, Li P, Chen Q, Wei X, Zhao T, Wang
Z and Wei L: Mitogen-activated protein kinase p38 induces HDAC4
degradation in hypertrophic chondrocytes. Biochim Biophys Acta.
1853:370–376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhou Y, Kipps TJ and Zhang S: Wnt5a
signaling in normal and cancer stem cells. Stem Cells Int.
2017:52952862017. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nusse R and Clevers H: Wnt/β-catenin
signaling, disease, and emerging therapeutic modalities. Cell.
169:985–999. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Kumawat K and Gosens R: WNT-5A: Signaling
and functions in health and disease. Cell Mol Life Sci. 73:567–587.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Bouaziz W, Sigaux J, Modrowski D, Devignes
CS, Funck-Brentano T, Richette P, Ea HK, Provot S, Cohen-Solal M
and Haÿ E: Interaction of HIF1α and β-catenin inhibits matrix
metalloproteinase 13 expression and prevents cartilage damage in
mice. Proc Natl Acad Sci USA. 113:5453–5458. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Nguyen L, Sharma A, Chakraborty C, Saibaba
B, Ahn ME and Lee SS: Review of prospects of biological fluid
biomarkers in osteoarthritis. Int J Mol Sci. 18:6012017. View Article : Google Scholar
|
|
43
|
Chen H, Ghori-Javed FY, Rashid H, Adhami
MD, Serra R, Gutierrez SE and Javed A: Runx2 regulates endochondral
ossification through control of chondrocyte proliferation and
differentiation. J Bone Miner Res. 29:2653–2665. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Walzer SM, Cetin E, Grübl-Barabas R,
Sulzbacher I, Rueger B, Girsch W, Toegel S, Windhager R and Fischer
MB: Vascularization of primary and secondary ossification centres
in the human growth plate. BMC Dev Biol. 14:362014. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Filipowska J, Tomaszewski KA, Niedźwiedzki
Ł, Walocha JA and Niedźwiedzki T: The role of vasculature in bone
development, regeneration and proper systemic functioning.
Angiogenesis. 20:291–302. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Hassanein S, Nasr Eldin MH, Amer HA,
Abdelhamid AE, El Houssinie M and Ibrahim A: Human umbilical cord
blood CD34-positive cells as predictors of the incidence and
short-term outcome of neonatal hypoxic-ischemic encephalopathy: A
pilot study. J Clin Neurol. 13:84–90. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xuan F, Yano F, Mori D, Chijimatsu R,
Maenohara Y, Nakamoto H, Mori Y, Makii Y, Oichi T, Taketo MM, et
al: Wnt/β-catenin signaling contributes to articular cartilage
homeostasis through lubricin induction in the superficial zone.
Arthritis Res Ther. 21:2472019. View Article : Google Scholar : PubMed/NCBI
|