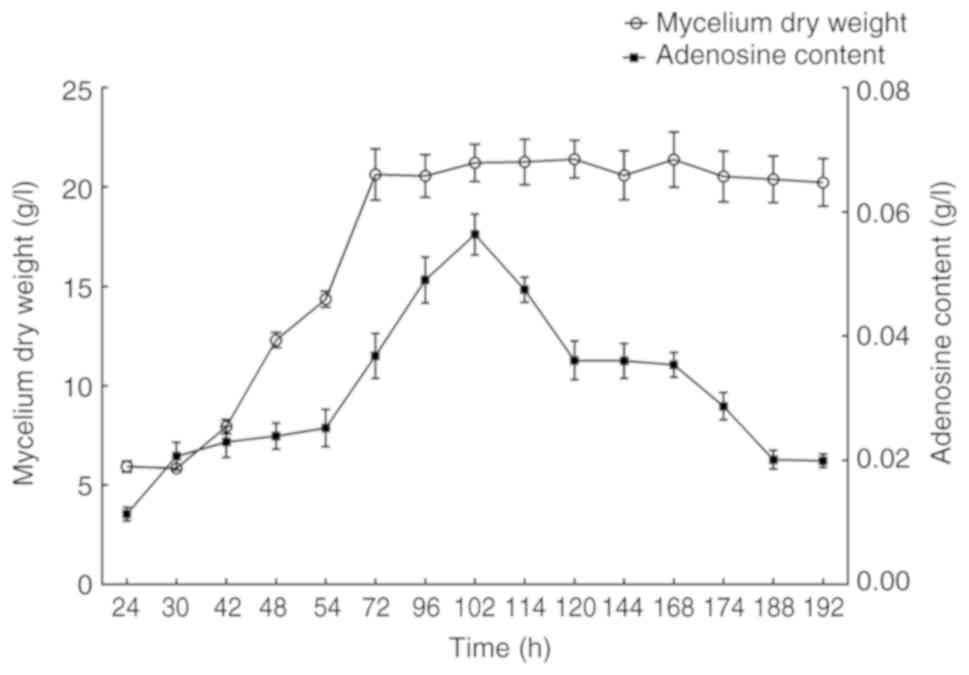
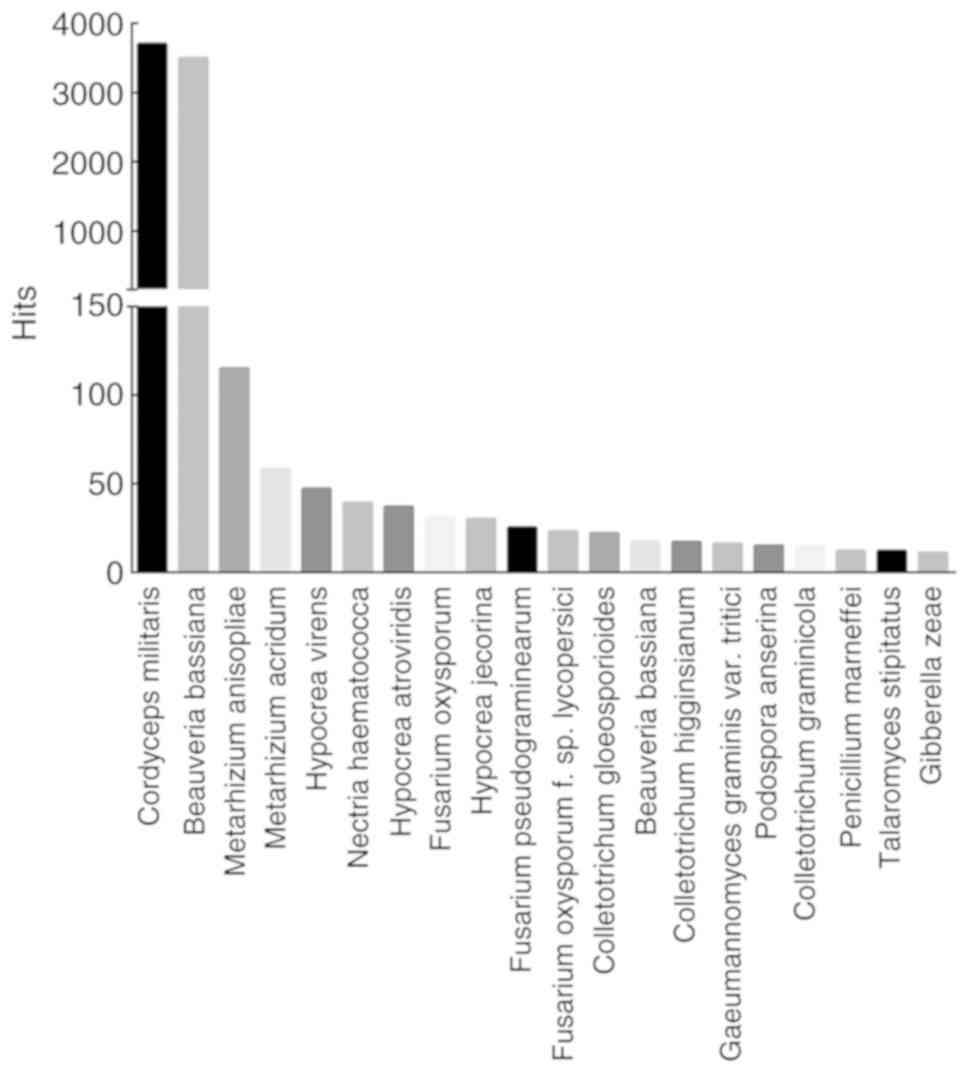
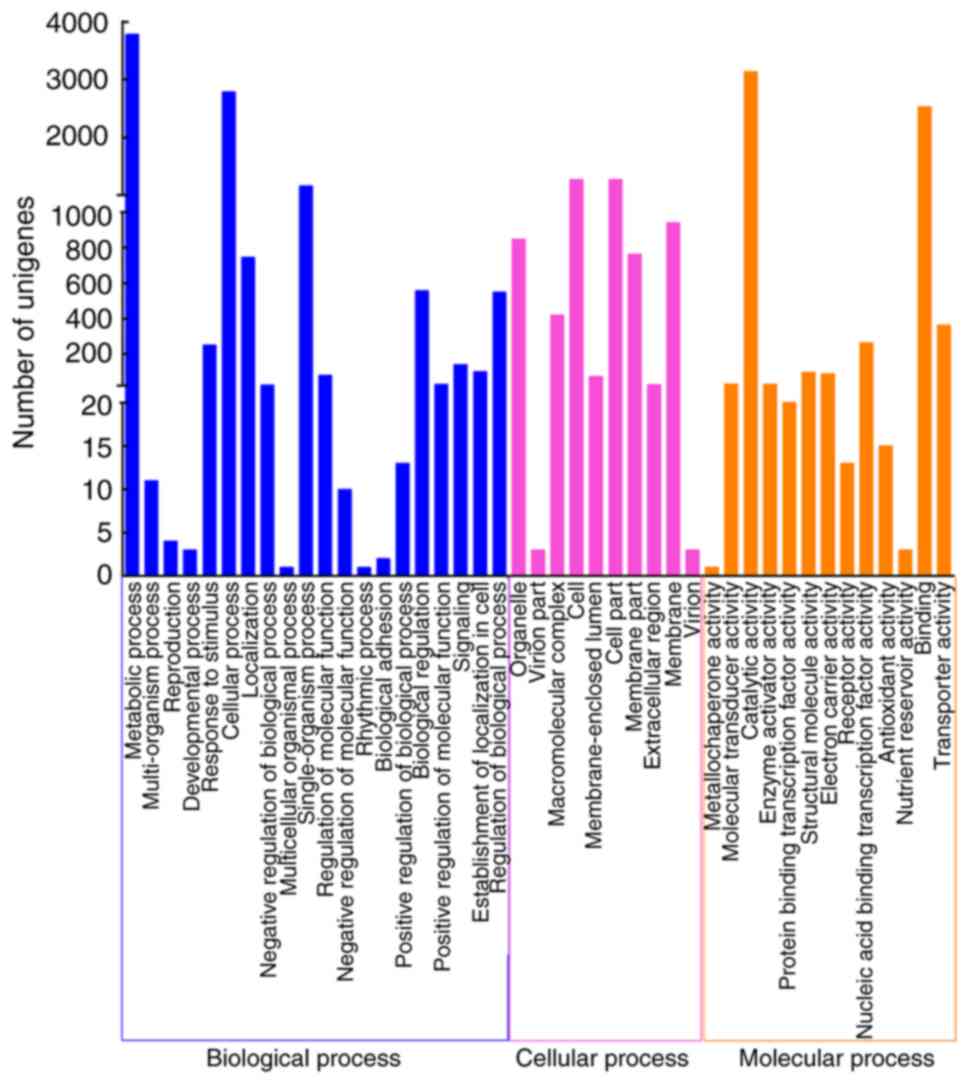
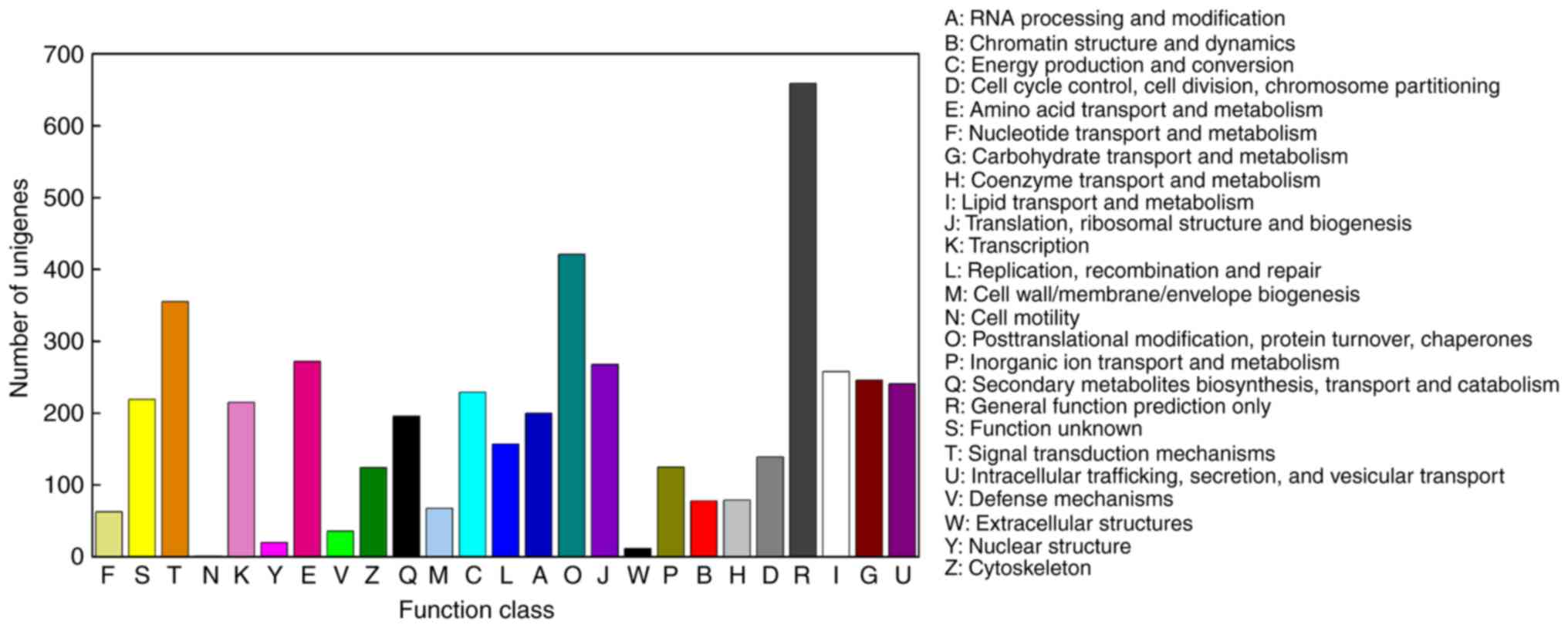
|
1
|
Wang M, Guan X, Chi Y, Robinson N and Liu
JP: Chinese herbal medicine as adjuvant treatment to chemotherapy
for multidrug-resistant tuberculosis (MDR-TB): A systematic review
of randomized clinical trials. Tuberculosis (Edinb). 95:364–372.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Chen SY, Lin YH, Huang JW and Chen YC:
Chinese herbal medicine network and core treatments for allergic
skin diseases: Implications from a nationwide database. J
Ethnopharmacol. 168:260–267. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Xu CP, Kim SW, Hwang HJ and Yun JW:
Production of exopolysaccharides by submerged culture of an
enthomopathogenic fungus, Paecilomyces tenuipes C240 in
stirred-tank and airlift reactors. Bioresource Technol. 97:770–777.
2006. View Article : Google Scholar
|
|
4
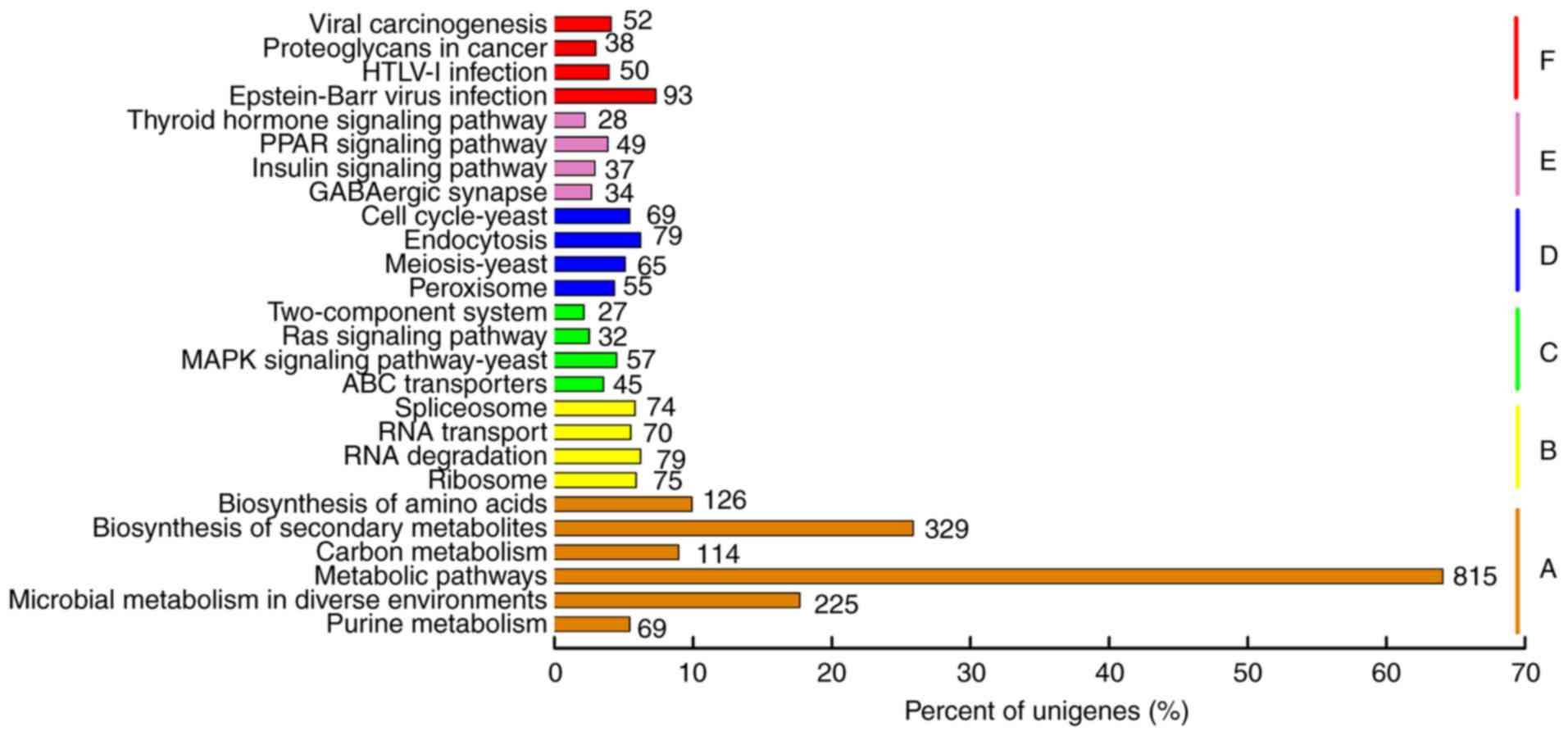
|
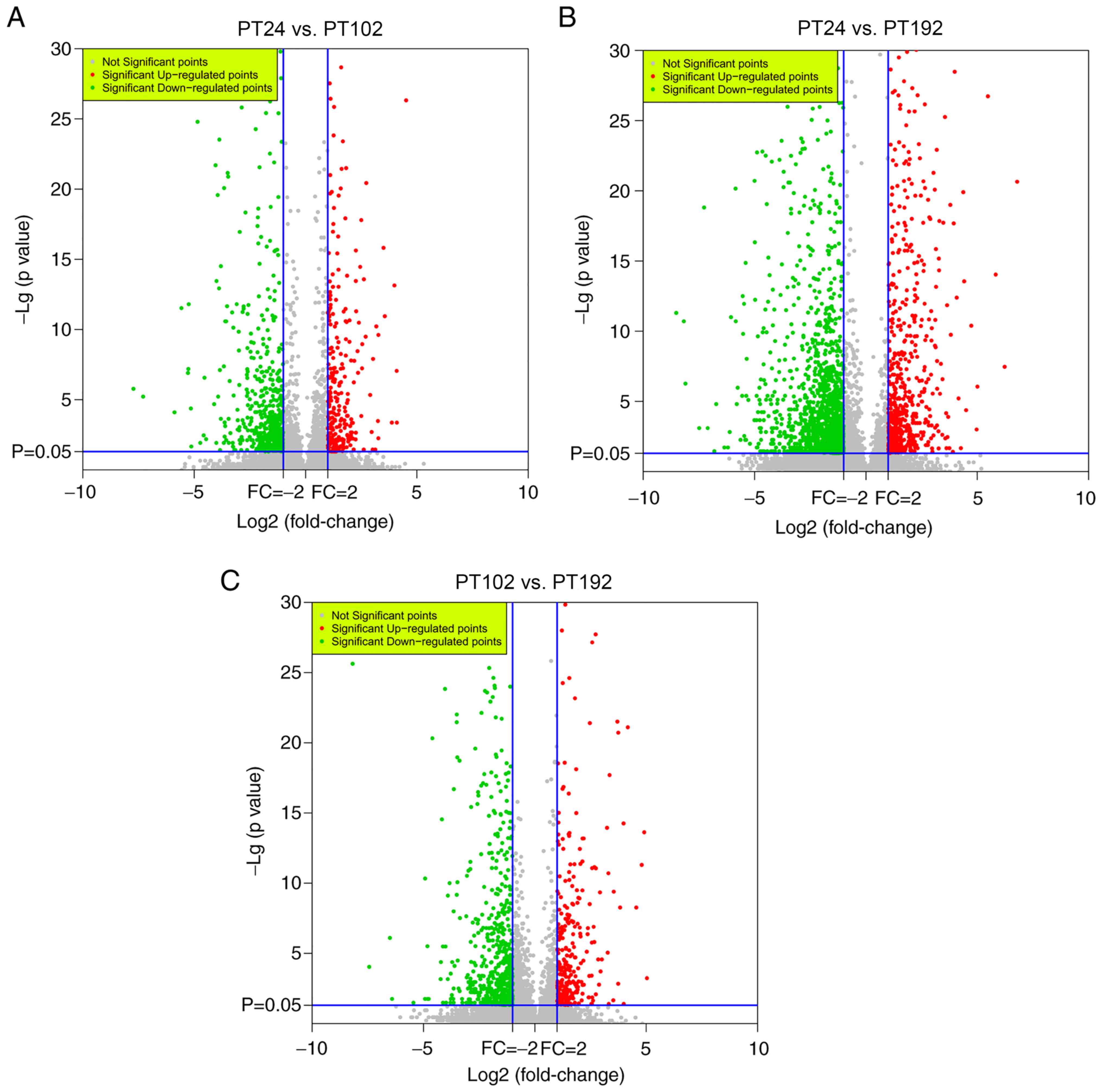
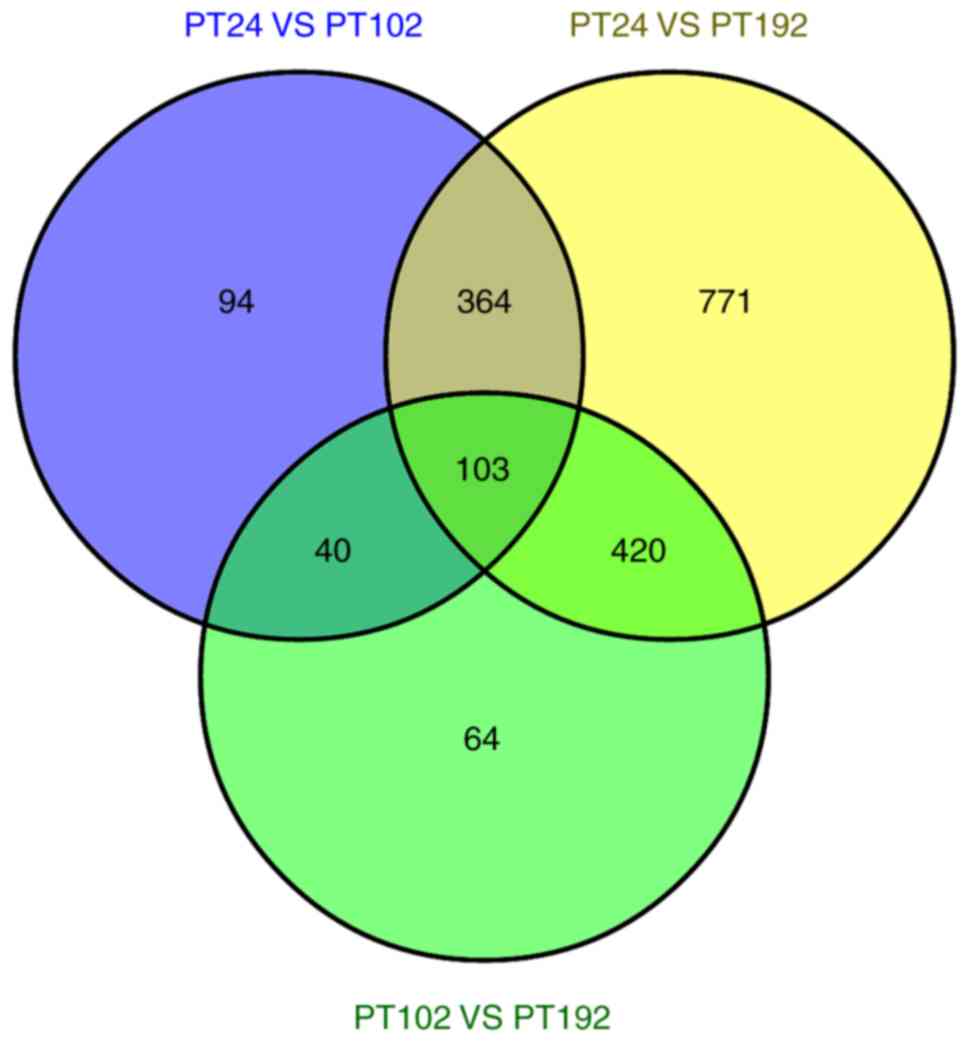
Du LN, Liu CG, Teng MY, Meng QF, Lu JH,
Zhou YL, Liu Y, Cheng YK, Wang D and Teng LS: Anti-diabetic
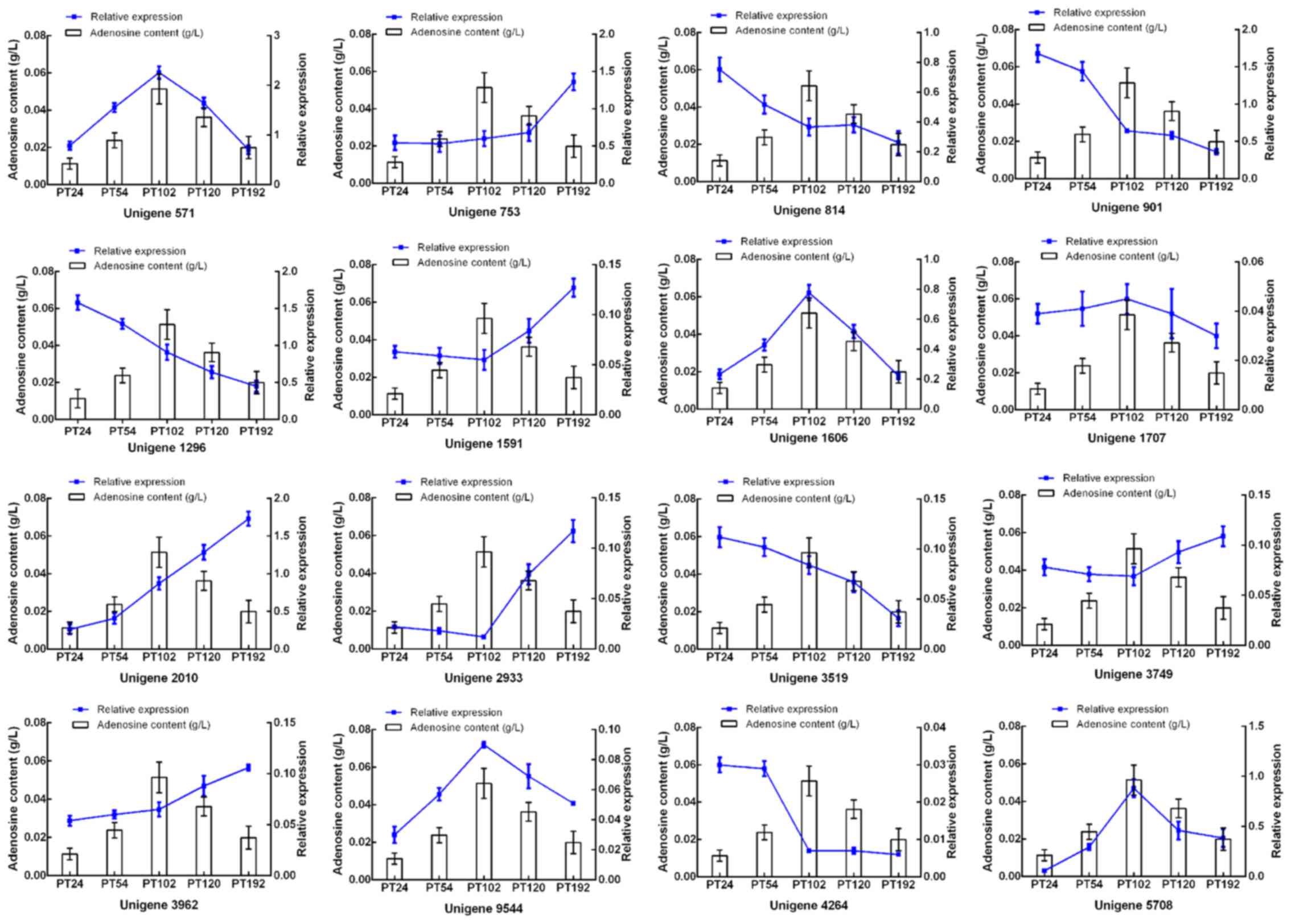
activities of Paecilomyces tenuipes N45 extract in
alloxan-induced diabetic mice. Mol Med Rep. 13:1701–1708. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sapkota K, Moon SM, Choi BS, Kim S, Kim YS
and Kim SJ: Enhancement of IL-18 expression by Paecilomyces
tenuipes. Mycoscience. 52:260–267. 2011. View Article : Google Scholar
|
|
6
|
Kim HC, Choi BS, Sapkota K, Kim S and Lee
HJ: Purification and characterization of a novel, highly potent
fibrinolytic enzyme from Paecilomyces tenuipes. Process
Biochem. 46:1545–1553. 2011. View Article : Google Scholar
|
|
7
|
Moezi L, Akbarian R, Niknahad H and
Shafaroodi H: The interaction of adenosine and morphine on
pentylenetetrazole-induced seizure threshold in mice.
Neuropharmacology. 72:1–8. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Huang CH, Tsai SK, Chiang SC, Lai CC and
Weng ZC: The role of adenosine in preconditioning by brief pressure
overload in rats. J Formos Med Assoc. 114:756–763. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ondrackova P, Kovaru H, Kovaru F, Leva L
and Faldyna M: Adenosine modulates LPS-induced cytokine production
in porcine monocytes. Cytokine. 61:953–961. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Yang DQ, Song JY, Wu LJ, Ma YF, Song CH,
Dovat S, Nishizaki T and Liu J: Induction of senescence by
adenosine suppressing the growth of lung cancer cells. Biochem
Biophys Res Commun. 440:62–67. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Magocha TA, Zabed H, Yang MM, Yun JH,
Zhang HH and Qi XH: Improvement of industrially important microbial
strains by genome shuffling: Current status and future prospects.
Bioresource Technol. 257:281–289. 2018. View Article : Google Scholar
|
|
12
|
Li DM, Wu W, Zhang D, Liu XR, Liu XF and
Lin YJ: Floral transcriptome analyses of four Paphiopedilum
Orchids with distinct flowering behaviors and development of
simple sequence repeat markers. Plant Mol Biol Rep. 33:1928–1952.
2015. View Article : Google Scholar
|
|
13
|
Cândido Ede S, Fernandes Gda R, de Alencar
SA, Cardoso M, Lima SM, Miranda Vde J, Porto WF, Nolasco DO, de
Oliveira-Júnior NG, Barbosa AE, et al: Shedding some light over the
floral metabolism by arum lily (Zantedeschia aethiopica)
spathe de novo transcriptome assembly. PLoS One. 9:e904872014.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Shakeel A, Zhan CS, Yang YY, Wang XK, Yang
TW, Zhao ZY, Zhang QY, Li XH and Hu XB: The transcript profile of a
traditional Chinese medicine, Atractylodes lancea, revealing
its sesquiterpenoid biosynthesis of the major active components.
PLoS One. 11:e01519752016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Sangwan RS, Tripathi S, Singh J, Narnoliya
LK and Sangwan NS: de novo sequencing and assembly of Centella
asiatica leaf transcriptome for mapping of structural,
functional and regulatory genes with special reference to secondary
metabolism. Gene. 525:58–76. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gao X, Han J, Lu Z, Li Y and He C: De novo
assembly and characterization of spotted seal Phoca largha
transcriptome using Illumina paired-end sequencing. Comp Comp
Biochem Physiol Part D Genomics Proteomics. 8:103–110. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bräutigam A, Mullick T, Schliesky S and
Weber APM: Critical assessment of assembly strategies for non-model
species mRNA-Seq data and application of next-generation sequencing
to the comparison of C3 and C4 species. J Exp
Bot. 62:3093–3102. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang Y, Jiang R, Wu H, Liu P, Xie J, He Y
and Pang H: Next-generation sequencing-based transcriptome analysis
of Cryptolaemus montrouzieri under insecticide stress
reveals resistance-relevant genes in laybirds. Genomics. 100:35–41.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Farlora R, Araya-Garay J and
Gallardo-Escárate C: Discovery of sex-related genes through
high-throughput transcriptome sequencing from the salmon louse
Caligus rogercresseyi. Mar Genom. 15:85–93. 2014. View Article : Google Scholar
|
|
20
|
Liu G, Wei X, Chen R, Zhou H, Li X, Sun Y,
Xie S, Zhu Q, Qu N, Yang G, et al: A novel mutation of the SLC25A13
gene in a Chinese patient with citrin deficiency detected by target
next-generation sequencing. Gene. 533:547–553. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhang S, Sui Z, Chang L, Kang K, Ma J,
Kong F, Zhou W, Wang JG, Guo L, Geng H, et al: Transcriptome de
novo assembly sequencing and analysis of the toxic dinoflagellate
Alexandrium catenella using the Illumina platform. Gene.
537:285–293. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Du L, Liu Y, Liu C, Meng Q, Song J, Wang
D, Lu J and Teng L, Zhou Y and Teng L: Acute and subchronic
toxicity studies on safety assessment of Paecilomyces
tenuipes N45 extracts. Comb Chem High Throughput Screen.
18:809–818. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Li G, Zhao Y, Liu Z, Gao C, Yan F, Liu B
and Feng J: De novo assembly and characterization of the spleen
transcriptome of common carp (Cyrinus carpio) using Illumina
paired-end sequencing. Fish Shellfish Immunol. 44:420–129. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
The Gene Ontology Consortium, . The gene
ontology resource: 20 years and still going strong. Nucleic Acids
Res. 47:D330–D338. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zheng Y, Xu QF, Chen HY, Chen QP, Gong ZJ
and Lai W: Transcriptome analysis of ultraviolet A-induced photo
aging cells with deep sequencing. J Dermatol. 45:175–181. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ye J, Fang L, Zheng HK, Zhang Y, Chen J,
Zhang ZJ and Wang J, Li ST, Li RQ, Bolund L and Wang J: WEGO: A web
tool for plotting GO annotations. Nucleic Acids Res. 34:W293–W297.
2006. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Sello C, Liu C, Sun Y, Msuthwana P, Hu J,
Sui Y, Chen S, Zhou Y, Lu H, Xu C, et al: De novo assembly and
comparative transcriptome profiling of Anser and Anser cygnodies
geese species' embryonic skin feather follicles. Genes (Basel).
10:3512019. View Article : Google Scholar
|
|
28
|
Kanehisa M, Sato Y, Furumichi M, Morishima
K and Tanabe M: New approach for understanding genome variations in
KEGG. Nucleic Acids Res. 47:D590–D595. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Anders S and Huber W: Differential
expression analysis for sequence count data. Genome Biol.
11:R1062010. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Li WT: Volcano plots in analyzing
differential expressions with mRNA microarrays. J Bioinf Comput
Biol. 10:12310032012. View Article : Google Scholar
|
|
31
|
Forconi M, Biscotti MA, Barucca M,
Buonocore F, Moro GD, Fausto AM, Fausto AM, Gerdol M, Pallavicini
G, Schartl M, et al: Characterization of purine catabolic pathway
genes in coelacanths. J Exp Zool B Mol Dev Evol. 322:334–341. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Losenkova K, Zuccarini M, Karikoski M,
Laurila J, Boison D, Jalkanen S and Yegutkin GG:
Compartmentalization of adenosine metabolism in cancer cells and
its modulation during acute hypoxia. J Cell Sci. 133:jcs2414632020.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Striepen B, Pruijssers AJ, Huang JL, Li C,
Gubbels MJ, Umejiego NN, Hedstrom L and Kissinger JC: Gene transfer
in the evolution of parasite nucleotide biosynthesis. Proc Natl
Acad Sci USA. 101:3154–3159. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Stasolla C, Katahira R, Thorpe TA and
Ashihara H: Purine and pyrimidine nucleotide metabolism in higher
plants. J Plant Physiol. 160:1271–95. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Layland J, Carrick D, Lee M, Oldroyd K and
Berry C: Adenosine: Physiology, pharmacology, and clinical
applications. JACC Cardiovasc Interv. 7:581–591. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pettersson J, Schrumpf ME, Raffel SJ,
Porcella SF, Guyard C, Lawrence K, Gherardini FC and Schwan TG:
Purine salvage pathways among Borrelia species. Infect Immun.
75:3877–3884. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Sasaki Y, Goto H, Wake T and Sasaki R:
Purine ribonucleotide homopolymer formation activity of RNA
polymerase from cauliflower. Biochim Biophys Acta. 366:443–453.
1974. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lunt SY, Muralidhar V, Hosios AM,
Israelsen WJ, Gui DY, Newhouse L, Ogrodzinski M, Hecht V, Xu K,
Acevedo PM, et al: Pyruvate kinase isoform expression alters
nucleotide synthesis to impact cell proliferation. Mol Cell.
57:95–107. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Wang JJ, Bai WW, Zhou W, Liu J, Chen J,
Liu XY, Xiang TT, Liu RH, Wang WH, Zhang BL and Wan YJ:
Transcriptomic analysis of two Beauveria bassiana strains
grown on cuticle extracts of the silkworm uncovers their different
metabolic response at early infection stage. J Invertebr Pathol.
145:45–54. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wang B, Zhou J, Liu HZ and Zheng FR:
Analysis of transcriptome profiling from the brain at maturation
and regression phases in starry flounder (Platichthys
stellatus). Gene Rep. 4:45–52. 2016. View Article : Google Scholar
|
|
41
|
Sun L, Wang Q, Wang Q, Dong K, Xiao Y and
Zhang YJ: Identification and characterization of odorant binding
proteins in the forelegs of Adelphocoris lineolatus (Goeze).
Front Physiol. 8:7352017. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li JY, Pan LQ, Miao JJ, Xu RY and Xu WJ:
De novo assembly and characterization of the ovarian transcriptome
reveal mechanisms of the final maturation stage in Chinese scallop
Chlamys farreri. Comp Biochem Physiol Part D Genomics
Proteomics. 20:118–124. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Fan XD, Wang JQ, Yang N, Dong YY, Liu L,
Wang FW, Wang N, Chen H, Liu WC, Sun YP, et al: Gene expression
profiling of soybean leaves and roots under salt, saline-alkali and
drought stress by high-throughput Illumina sequencing. Gene.
512:392–402. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Stutzer C, Mans BJ, Gaspar ARM, Neitz AWH
and Maritz-Olivier C: Ornithodoros savignyi: Soft tick
apyrase belongs to the 5′-nucleotidase family. Exp Parasitol.
122:318–327. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Santos CA, Saraiva AM, Toledo MAS, Beloti
LL, Crucello A, Favaro MTP, Horta MAC, Santiago AS, Mendes JS,
Souza AA and Souza AP: Initial biochemical and functional
characterization of a 5′-nucleotidase from Xylella
fastidiosa related to the human cytosolic 5′-nucleotidase I.
Microb Pathog. 59-60:1–6. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Fenckova M, Hobizalova R, Fric ZF and
Dolezal T: Functional characterization of ecto-5′-nucleotidases and
apyrases in Drosophila melanogaster. Insect Biochem Mol
Biol. 41:956–967. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hunsucker SA, Mitchell BS and Spychala J:
The 5′-nucleotidases as regulators of nucleotide and drug
metabolism. Pharmacol Ther. 107:1–30. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Covarrubias R, Chepurko E, Reynolds A,
Huttinger ZM, Huttinger R, Stanfill K, Wheeler DG, Novitskaya T,
Robson SC, Dwyer KM, et al: Role of the CD39/CD37 purinergic
pathway in modulating arterial thrombosis in mice. Arterioscler
Thromb Vasc Biol. 36:1809–1820. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Xiang L, Li Y, Zhu Y, Luo H, Li C, Xu X,
Sun C, Song J, Shi L, He L, et al: Transcriptome analysis of the
Ophiocordyceps sinensis fruiting body reveals putative genes
involved in fruiting body development and cordycepin biosynthesis.
Genomics. 103:154–159. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Zheng P, Xia Y, Xiao G, Xiong C, Hu X,
Zhang S, Zheng H, Huang Y, Zhou Y, Wang S, et al: Genome sequence
of the insect pathogenic fungus Cordyceps militaris, a
valued traditional Chinese medicine. Genome Biol. 12:R1162011.
View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Sakai Y, Tamao Y, Shimamoto T, Hama H,
Tsuda M and Tsuchiya T: Cloning and expression of the
5′-nucleotidase gene of Vibrio parahaemolyticus in Escherichia
coli and overproduction of the enzyme. J Biochem. 105:841–846.
1989. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Rampazzo C, Mazzon C, Reichard P and
Bianchi V: 5′-Nucleotidases: Specific assays for five different
enzymes in cell extracts. Biochem Biophys Res Commun. 293:258–263.
2002. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Ubeidat M, Eristi CM and Rutherford CL:
Expression pattern of 5′-nucleotidase in Dictyostelium. Mech
Dev. 110:237–239. 2002. View Article : Google Scholar : PubMed/NCBI
|