Introduction
Kidney cancer is prevalent in both males and
females, and renal cell carcinoma (RCC) is the most common form of
kidney malignancy (1). Although
treatment options for RCC have improved over the last decade from a
non-specific immune approach (2),
to therapies targeting vascular endothelial growth factor (VEGF) or
tyrosine-protein kinase receptor UFO precursor (AXL) (3), the overall survival rate remains low.
Although the five-year disease specific survival in patients with
stage I–II reaches 80%, it is only 60% in stage III and <10% in
stage IV patients (4). Therefore,
understanding the molecular mechanisms of RCC development and
progression is essential.
Long non-coding RNAs (lncRNAs) are non-protein
coding transcripts of >200 nucleotides in length (5). Previous studies have suggested that
lncRNAs have a prominent role in the competing endogenous RNA
(ceRNA) network, in which they serve as microRNA (miRNA/miR)
sponges, thus controlling the expression of genes associated with
cancer development (6–8). For instance, the lncRNA HOX
transcript antisense RNA (HOTAIR) was discovered to affect the
proliferation and tumorigenesis of RCC by inhibiting the
miR-217/hypoxia-inducible factor-1 (HIF-1)α/AXL pathway (9). Additionally, lncRNA H19 was suggested
to potentially mediate breast cancer cell plasticity by
differentially sponging miR-200b/c and let-7b (10). Thus, these findings indicated that
lncRNAs may be crucial regulators of cancer progression and
metastasis (11–13). Nonetheless, their function in RCC
remains poorly understood.
The expression levels of small nucleolar RNA host
gene 12 (SNHG12), located on chromosome 1p35.3, were discovered to
be upregulated in several types of cancer, including non-small cell
lung cancer, glioma, cervical cancer and bladder cancer (14–17).
In previous studies, SNHG12 was also reported to function as an
miRNA sponge and influence several tumor-related pathways, such as
the Wnt/β-catenin signaling pathway and the Jak/STAT3 signaling
pathway (15,17–19).
In addition, SNHG12 was identified to serve as an oncogene, where
it had a crucial role in tumor progression (20,21).
SNHG12 has also been associated with the unfavorable prognosis of
nasopharyngeal carcinoma and gastric cancer, where it served as a
potential biomarker for cancer progression (22,23).
However, to the best of our knowledge, the role of SNHG12 in RCC
remains unknown.
In the present study, SNHG12 expression levels were
investigated in samples obtained from patients with RCC. SNHG12
knockdown significantly inhibited the viability and invasion of RCC
cell lines, while increasing apoptosis. Notably, SNHG12 was
discovered to directly interact with miR-200c-5p, which indicated
that it may serve as a sponge and control collagen type XI α1 chain
(COL11A1) expression levels. In conclusion, the findings of the
present study suggested that the SNHG12/miR-200c-5p/COL11A1 axis
may be involved in RCC progression.
Materials and methods
Clinical samples
A total of 58 patients (36 males and 22 females;
age, 31–72 years) diagnosed with RCC by pathological examination
after surgery at The Department of Urology, Hainan General Hospital
(Haikou, China) (August 2014-August 2017) were enrolled in the
present study. Patients who received chemotherapy, radiotherapy or
any other therapy were excluded from the present study. Tumor
tissues and paired adjacent non-tumor renal tissues were collected
and stored at −80°C for subsequent RNA extraction. Patients were
classified as RCC according to the World Health Organization
criteria (24) and tumor staging
was determined using the tumor-node-metastasis (TNM) classification
(4). The study was approved by the
Medical Ethics Committee of Hainan General Hospital (Haikou,
China). Written, informed consent was obtained from all 58
patients.
TCGA database
RNA sequencing and clinical data of patients with
RCC were downloaded from Genomic Data Commons Data Portal
(portal.gdc.cancer.gov). Patient data
were excluded if >75% lncRNA data was missing or if survival
information was not complete. Patients >18 years old were
included in the study. After screening, a total of 278 patients
with RCC and 33 non-tumor kidney samples were used in the present
study (25). TCGA data was
analyzed using RStudio version 0.99.896 software (RStudio, Inc.).
Survival analysis were conducted using Kaplan-Meier plots by SPSS
version 22.0 software (IBM, Corp.).
Cell culture
Human renal cell carcinoma cell lines A498, 786O,
Caki-1, Caki-2 and ACHN, and the normal renal epithelial cell line
HK-2 as well as 293T cell line were purchased from the American
Type Culture Collection. Cells were cultured in RPMI-1640 medium,
supplemented with 10% FBS and 1% penicillin/streptomycin (all from
Gibco; Thermo Fisher Scientific, Inc.). All cells were maintained
in a humidified incubator with 5% CO2 at 37°C.
Cell transfection
Small interfering RNAs (siRNAs/si) targeting SNHG12
and negative control (NC; si-NC) were synthesized by Guangzhou
RiboBio Co., Ltd. using the following sequences: Si-SNHG12 sense,
5′-GCAGUGUGCUACUGAACUUTT-3′ and antisense,
5′-AAGUUCAGUAGCACACUGCTT-3′; and si-NC sense,
5′-UUCUCCGAACGUGUCACGUTT-3′ and antisense,
5′-ACGUGACACGUUCGGAGAATT-3′. Si-COL11A1 was purchased from
Sigma-Aldrich; Merck KGaA. si-NC for si-COL11A1 was the same with
si-SNHG12. The miR-200c mimics, miR-NC, miR-200c inhibitor,
inhibitor-NC, pcDNA SNHG12 and pcDNA3.1 empty vector were purchased
from GeneCopoeia, Inc. The sequences were as follows: Si-COL11A1
sense, 5′-UUCUAAAUUUGAUGGUUUGCGTT-3′ and antisense,
5′-CAAACCAUCAAAUUUAGAAGA-3′; miR-200c-5p mimics,
5′-UAAUACUGCCGGGUAAUGAUGGA-3′; miR-mimics-NC,
5′-UCACAACCUCCUAGAAAGAGUAGA-3′; and miR-200c-5p inhibitor,
5′-UAAUACUGCCGGGUAAUGAUGGA-3′; inhibitor-NC,
5′-CGAACUUAUCUUAGGUACUC-3′. Briefly, 1×106 A498 or 786O
cells were seeded into six-well plates and transfected with 50 nM
siRNAs, 30 nM mimics, 30 nM inhibitors, 2 µg pcDNA SNHG12 or pcDNA
3.1 empty vector using Lipofectamine® 2000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's protocol. After 48 h of transfection at 37°C, the
transfection efficiency was determined by reverse
transcription-quantitative PCR (RT-qPCR). Untransfected RCC cells
were used as the ‘Blank’ control, and these cells were treated with
nothing.
RT-qPCR
Total RNA was extracted from cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.) and reverse transcribed into cDNA using PrimeScript Reverse
Transcriptase (Takara Biotechnology Co., Ltd.). RT was performed
using the following procedure: 30°C for 10 min followed by 42°C for
45 min. qPCR was subsequently performed using the SYBR Premix Taq
kit (Takara Biotechnology Co., Ltd.) on a LightCycler Real-Time PCR
instrument [Roche Diagnostics (Shanghai) Co., Ltd.]. qPCR was
performed using the following conditions: Enzyme activation at 95°C
for 2 min, 1 cycle; 40 cycles of denaturation at 95°C for 10 sec
and annealing/extension 60°C for 60 sec. U6 was used as the
endogenous control for miR-200c-5p and SNHG12, while GAPDH was used
as the loading control for COL11A1. The following primer sequences
were used for the qPCR: SNHG12 forward, 5′-AGGGCCATGTAACCAGTGAA-3′
and reverse, 5′-GCTGGCCTTAATCTGACTGC-3′; U6 forward,
5′-GCTTCGGCAGCACATATACTAAAAT-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′; miR-200c-5p forward,
5′-TACATCATAATACTGCCGGGTAA-3′ and reverse,
5′-GGATTGGATGTTCTCCACAGTCTC-3′; COL11A1 forward,
5′-AGTGGCATCGGGTAGCAATCA-3′ and reverse,
5′-TGTCCCCCTCAAAAACTTCTTCAT-3′; and GAPDH forward,
5′-CGCTCTCTGCTCCTCCTGTTC-3′ and reverse,
5′-ATCCGTTGACTCCGACCTTCAC-3′. The relative expression levels of
each target were quantified using the 2−ΔΔCq method
(26).
Western blotting
Cells were washed with PBS and total protein was
extracted using RIPA lysis buffer (Invitrogen; Thermo Fisher
Scientific, Inc.) containing a protease inhibitor cocktail (Roche
Diagnostics Co., Ltd.). Total protein was quantified using a
bicinchoninic acid protein assay kit (Pierce; Thermo Fisher
Scientific, Inc.) and 40 µg protein/well was separated via 10%
SDS-PAGE. The separated proteins were subsequently transferred onto
PVDF membranes (EMD Millipore) and blocked with 5% non-fat milk at
room temperature for 1 h. The membranes were then incubated with
the following primary antibodies overnight at 4°C: Anti-COL11A1
(cat. no. ab64883; Abcam) and anti-β-actin (cat. no. ab6276;
Abcam). Antibodies were used at 1:1,000 dilution. Following the
primary antibody incubation, the membranes were washed with
TBS-0.05% Tween-20 (TBST) 4–6 times for 1 h, then incubated with a
horseradish peroxidase-conjugated secondary antibody for 1 h at
room temperature (Goat anti-rabbit and anti-mouse IgG, cat. nos.
ab205718 and ab205719, respectively; both Abcam), secondary
antibodies were used at 1:5,000 dilution. The membranes were
subsequently washed with TBST 4–6 times for 1 h and protein bands
were visualized using an ECL substrate kit (Abcam). ImageJ software
(version 1.8.0; National Institutes of Health) was used for
densitometry.
Transwell invasion assay
Transwell invasion assays were performed using 8-µm
pore Matrigel®-precoated (37°C for 3 h and room
temperature overnight) polycarbonate membranes (BD Biosciences).
Briefly, 3×104 cells were seeded into the upper chambers
of Transwell plates in serum-free DMEM (Gibco; Thermo Fisher
Scientific, Inc.). The lower chambers contained complete DMEM
(Gibco; Thermo Fisher Scientific, Inc.) supplemented with 10% FBS.
Following incubation at 37°C for 48 h, cells in the upper chamber
were removed and the invasive cells on the lower surface of the
membranes were fixed in 4% paraformaldehyde at room temperature for
15 min, stained with 0.5% crystal violet at room temperature for 20
min and visualized using a light microscope (magnification, ×100)
(Olympus Corporation). ImageJ software (version 1.8.0; National
Institutes of Health) was used for data analysis.
Flow cytometric analysis of
apoptosis
Early and late (top and lower right corner of the
flow dot plot, respectively) stage apoptosis were detected by flow
cytometric analysis. Briefly, 0.5×106 A498 or 786O cells
were seeded to six-well plates and transfected with the
transfectants for 48 h at 37°C. The cells were subsequently
harvested (Eppendorf centrifuge 5418R) and washed twice with PBS.
The cells were then resuspended in binding buffer (cat. no.
BDB556454; BD Biosciences), and stained with Annexin V-FITC and
7-Aminoactinomycin D (AAD) using an Annexin V-FITC/7-AAD Apoptosis
Detection kit (BD Biosciences), according to the manufacturer's
protocol. Apoptotic cells were subsequently analyzed using a BD
FACSVerse flow cytometer (BD Biosciences) and analyzed using FlowJo
version 10.0.7 software (FlowJo LLC).
MTT assay
A total of 2×104 cells were seeded/well
into a 96-well plate and incubated for 24, 48 or 72 h at 37°C. Cell
viability was subsequently analyzed in triplicate using an MTT
assay [Roche Diagnostics (Shanghai) Co., Ltd.], according to the
manufacturer's protocol. Then, 10 µl MTT solution was added to each
well and DMSO was used to dissolve purple formazan. Cell viability
was analyzed at an absorbance at 570 nm using an iMark microplate
absorbance reader (Bio-Rad Laboratories, Inc.).
Luciferase reporter assay
TargetScan (targetscan.org/mamm_31) and miRcode (mircode.org) were used for the prediction of miRNA
targets. The putative miR-200c-5p binding site of the SNHG12 and
COL11A1 3′-untranslated region (UTR) sequences were amplified and
cloned into the pmirGLO vector (Promega Corporation) to synthesize
SNHG12-WT and COL11A1-WT, respectively. Mutant (MUT) 3′-UTR
fragments were generated by site-directed mutagenesis (InFusion HD
Cloning Plus; Takara Biotechnology Co., Ltd.) and then were cloned
into the pmirGLO plasmid (referred to as SNHG12-MUT and
COL11A1-MUT, respectively). Then, 2×105 293T cells were
seeded into 24-well plates and cultured for 24 h at 37°C. The cells
were then co-transfected with 5 µg SNHG12- or COL11A1-WT/MUT
luciferase constructs, and miR-200c-5p mimics (Lipofectamine 2000;
Thermo Fisher Scientific, Inc.). Following transfection for 48 h at
37°C, cells were collected and luciferase activity was detected
using a Dual-Luciferase Reporter Assay system (Promega
Corporation). Renilla Luciferase was used for
normalization.
Statistical analysis
Statistical analysis was performed using SPSS
Statistics 22.0 software (IBM Corp.). Pairwise comparisons were
assessed using an unpaired Student's t-test or a paired Student's
t-test when the data were paired (Fig.
1B). Multigroup comparisons were performed using two-way ANOVA,
followed by a Bonferroni's post hoc test. A Kaplan-Meier curve was
used for survival analysis and the P-value was obtained using the
log-rank test. A χ2 test was used to determine the
association between SNHG12 expression levels and
clinicopathological variables, whereas a Spearman's correlation
analysis test was used to determine the correlation between SNHG12
and miR-200c-5p expression levels. All experiments were conducted
in triplicate. Data are presented as the mean ± SD. P<0.05 was
considered to indicate a statistically significant difference.
Results
SNHG12 expression levels are
upregulated in RCC patient samples and RCC cell lines
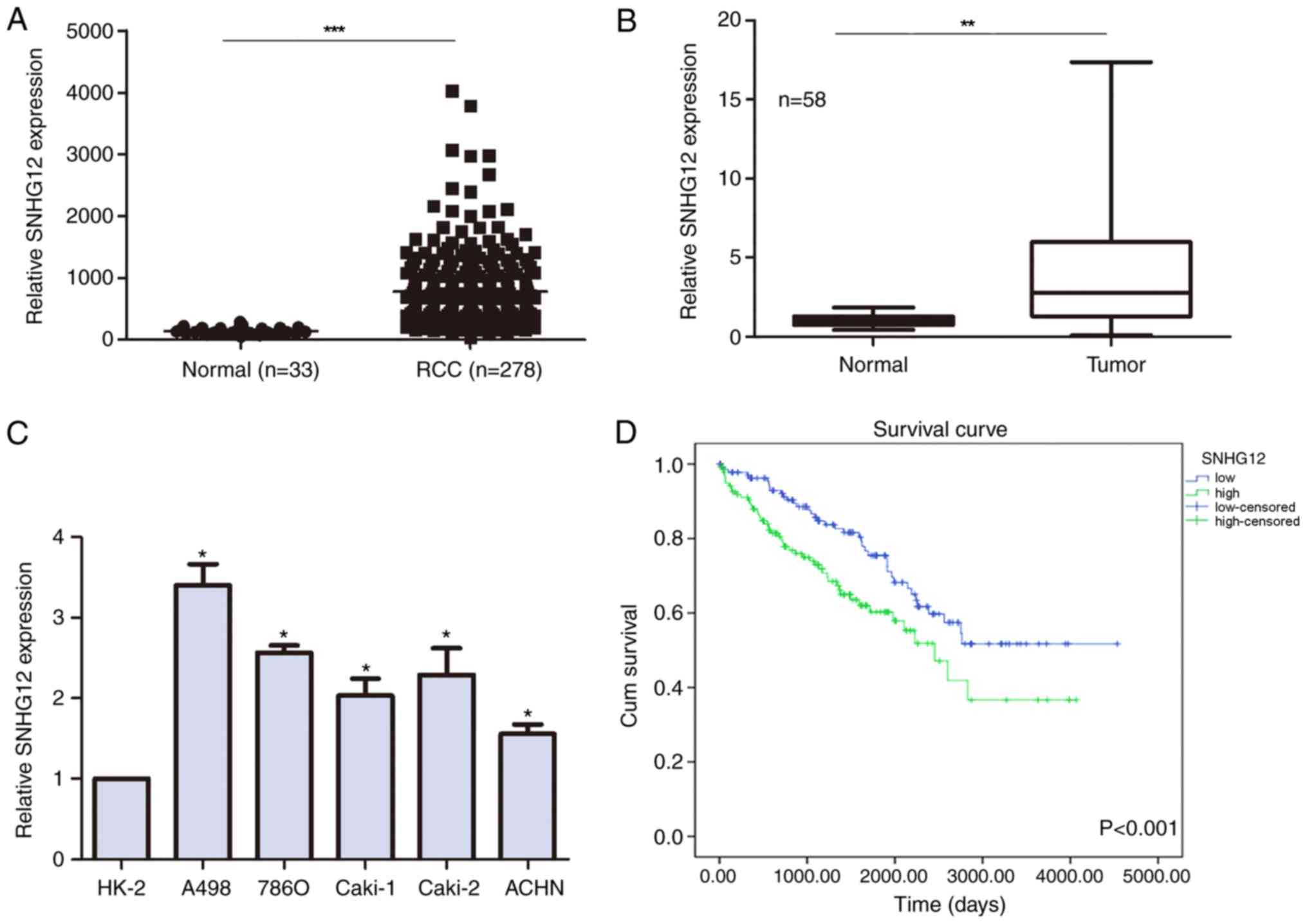
The expression levels of SNHG12 were compared
between RCC and non-tumor samples from TCGA database; SNHG12
expression levels were discovered to be significantly upregulated
in patients with RCC compared with in non-tumor patient samples
(799.9±35.41 vs. 141±8.625 normalized counts, respectively;
Fig. 1A). To validate this
finding, SNHG12 expression levels were also analyzed in clinical
samples and similarly, SNHG12 expression levels were significantly
upregulated in the RCC tumor tissues compared with the adjacent
non-tumor tissues (Fig. 1B). The
expression levels of SNHG12 were also evaluated in RCC cell lines,
all of which demonstrated upregulated expression levels of SNHG12
compared with the normal kidney epithelial cell line HK-2 (Fig. 1C).
Association between SNHG12 expression
levels and clinical characteristics in patients with RCC
The clinical characteristics analyzed in the present
study included age, sex, histological differentiation, TNM stage,
tumor size and lymph node and distant metastases (Table I). The median expression levels of
SNHG12 were used to categorize patients into ‘high’ and ‘low’
expression groups. High SNHG12 expression levels were significantly
associated with a lower histological differentiation advanced TNM
stage and further lymph node and distant metastases. TCGA datasets
were then used to further investigate whether SNHG12 expression
levels influenced the survival of patients with RCC; high SNHG12
expression levels in patients with RCC were associated with a
shorter overall survival rate compared with low expression levels
(Fig. 1D).
 | Table I.Association between SNHG12 expression
levels and clinicopathological characteristics in patients with
renal cell carcinoma (n=58). |
Table I.
Association between SNHG12 expression
levels and clinicopathological characteristics in patients with
renal cell carcinoma (n=58).
|
| SNHG12 expression
levels |
|
|---|
|
|
|
|
|---|
| Parameter | Low, n | High, n | P-value |
|---|
| Age, years |
|
| 0.905 |
|
>60 | 7 | 15 |
|
|
≤60 | 12 | 24 |
|
| Sex |
|
| 0.455 |
|
Male | 16 | 20 |
|
|
Female | 12 | 10 |
|
| Histological
differentiation |
|
| <0.001 |
|
High | 23 | 4 |
|
|
Low | 9 | 22 |
|
| Lymph node
metastasis |
|
| 0.018 |
|
N0-N1 | 20 | 9 |
|
|
N2-NX | 11 | 18 |
|
| Distant
metastasis |
|
| 0.018 |
| M0 | 18 | 11 |
|
|
M1-MX | 9 | 20 |
|
| Tumor node
metastasis stage |
|
| 0.001 |
|
T1-T2 | 19 | 10 |
|
|
T3-T4 | 6 | 23 |
|
| Tumor size |
|
| 0.001 |
| ≤4
cm | 20 | 9 |
|
| >4
cm | 5 | 24 |
|
SNHG12 promotes cell viability and
invasion and inhibits apoptosis in RCC cell lines
To determine the function of SNHG12 in RCC cells,
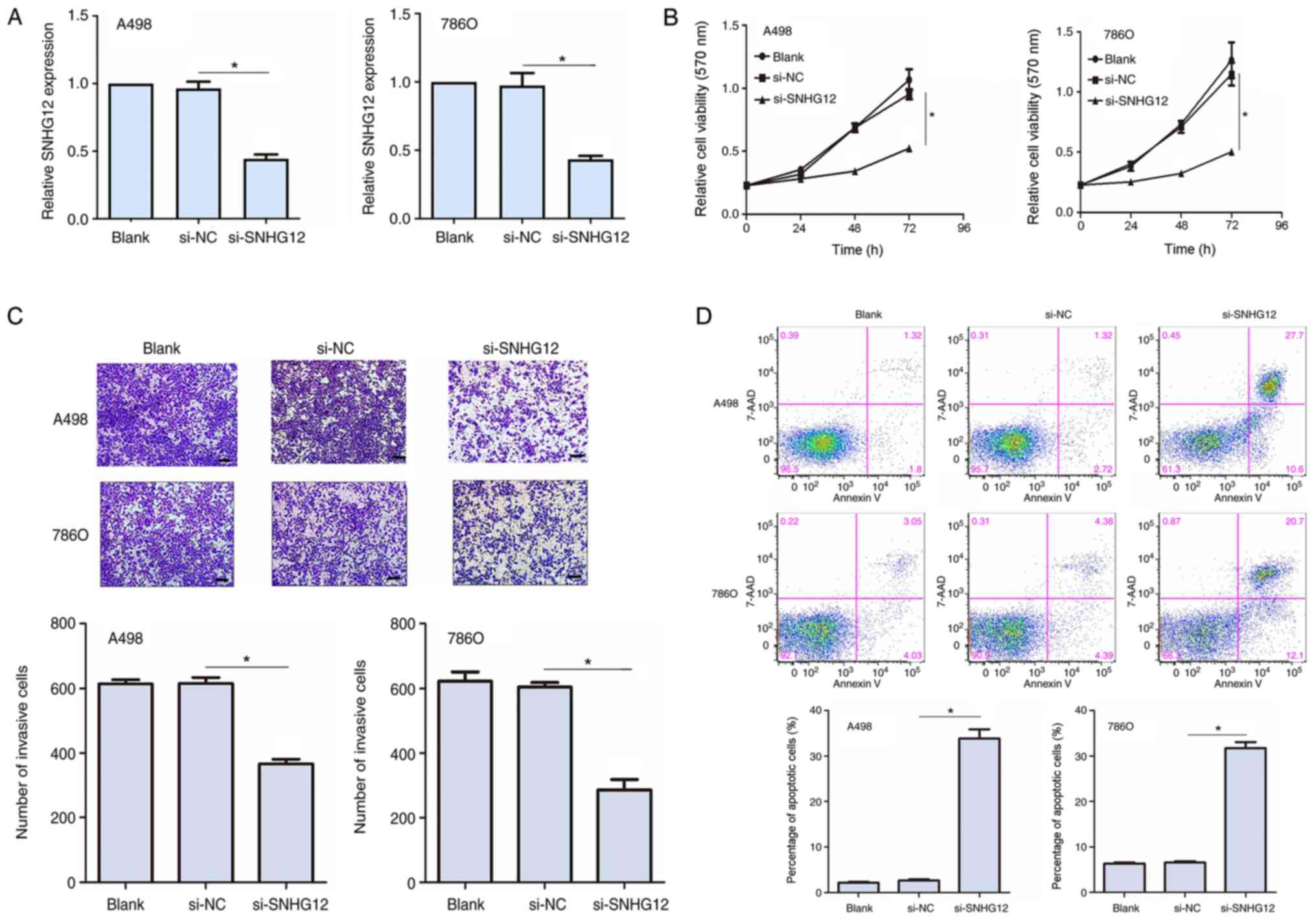
siRNA was used to knockdown SNHG12 expression levels in the A498
and 786O cell lines. In both cell lines, the transfection with
si-SNHG12 significantly downregulated SNHG12 expression levels
compared with the si-NC groups (Fig.
2A). An MTT assay was subsequently performed to determine the
cell viability in the transfected RCC cells. SNHG12 knockdown
significantly reduced cell viability at 48 and 72 h post
transfection compared with the si-NC group in both cell lines
(Fig. 2B). To investigate the
invasive properties of the transfected RCC cells, a Transwell
invasion assay was used. The knockdown of SNHG12 expression levels
significantly inhibited the invasive ability of A498 and 786O cells
(P<0.05; Fig. 2C). Flow
cytometric analysis also demonstrated that the knockdown of SNHG12
expression levels significantly increased the percentage of
apoptotic cells in both cell lines compared with the si-NC group
(Fig. 2D). Altogether, these
results indicated that SNHG12 may promote cell viability and
invasion, whilst inhibiting cell apoptosis in RCC cell lines.
SNHG12 downregulates the expression
levels of miR-200c-5p
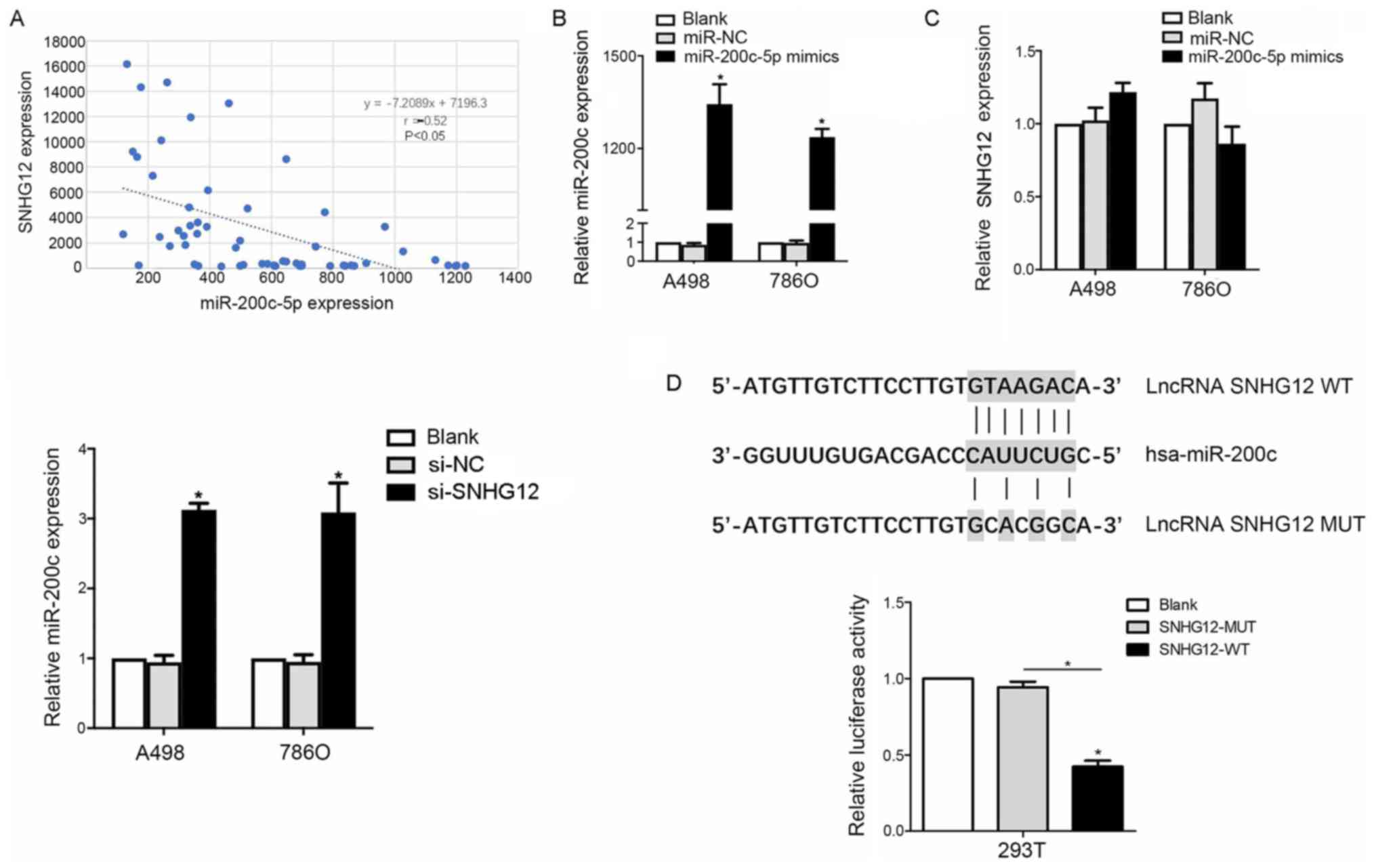
To further investigate the mechanism of SNHG12 in
the development of RCC, miRcode was used to screen for miRNAs
predicted to bind to SNHG12, and SNHG12 was discovered to contain
putative binding sites for miR-200c-5p. To validate this result, a
correlation analysis was conducted between SNHG12 and miR-200c-5p
expression levels in clinical RCC patient samples (n=58). The
expression levels of miR-200c-5p were revealed to be inversely
correlated with SNHG12 expression levels (r=−0.52; P=0.032;
Fig. 3A).
Thus, to determine the interaction between
miR-200c-5p and SNHG12, RCC cell lines were transfected with
miR-200c-5p mimics and si-SNHG12; the transfection efficiency of
the miR-200c-5p mimics was successful, as the expression levels
were significantly upregulated in the miR-200c-5p mimics group
(Fig. 3B). Subsequently, SNHG12
knockdown markedly upregulated miR-200c-5p expression levels, as
indicated by RT-qPCR. Notably, no significant differences were
observed in the SNHG12 expression levels following the
overexpression of miR-200c-5p compared with the two control groups
(Fig. 3C). Subsequently, a
luciferase reporter assay revealed that the relative luciferase
activity was decreased in 293T cells co-transfected with
pmirGLO-SNHG12-WT and miR-200c-5p mimics compared with
pmirGLO-SNHG12-MUT and miR-200c-5p mimics co-transfected cells
(P<0.05; Fig. 3D), which
further supported an interaction between SNHG12 and
miR-200c-5p.
SNHG12 exerts an oncogenic role
through the regulation of miR-200c-5p
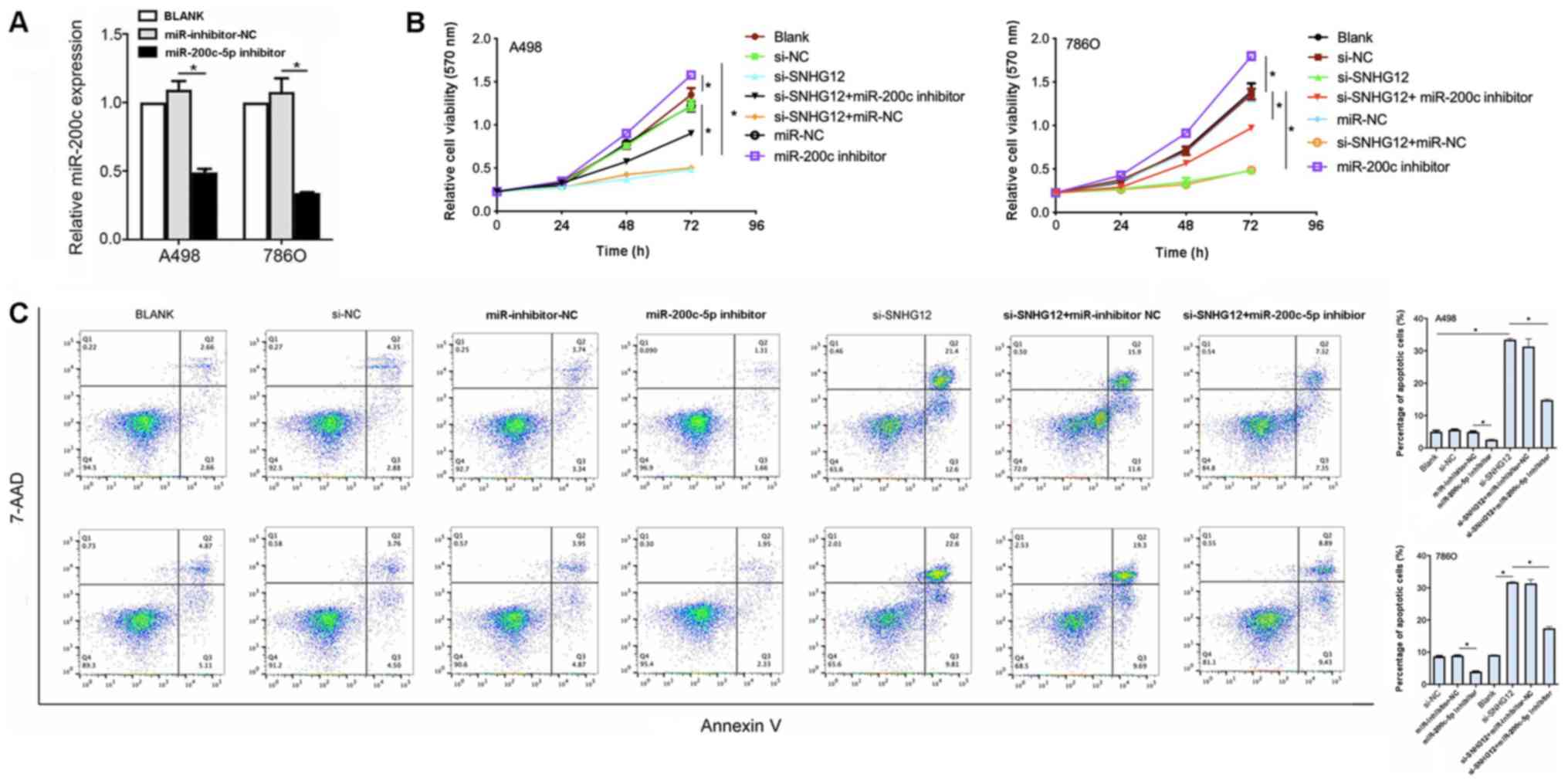
To determine whether SNGH12 exerted its effects
through miR-200c-5p, A498 and 786O cell lines were co-transfected
with si-SNHG12 and a miR-200c-5p inhibitor. The transfection of the
miR-200c-5p inhibitor into both cell lines was successful, as the
expression levels were significantly downregulated compared with
inhibitor NC (Fig. 4A). The cell
viability was significantly increased in both cell lines
co-transfected with si-SNHG12 and the miR-200c-5p inhibitor
compared with si-SNHG12 alone (Fig.
4B). In addition, in both cell lines, the percentage of
apoptotic cells in the si-SNHG12 + miR-200c-5p inhibitor group was
significantly decreased compared with the si-SNHG12 group
(P<0.05; Fig. 4C). Altogether,
these observations suggested that SNHG12 may exert its oncogenic
role through downregulating miR-200c-5p.
COL11A1 is a direct target of
miR-200c-5p
TargetScan and miRcode were used to determine target
genes of miR-200c-5p, which identified COL11A1 as one of the
predicted targets. COL11A1 is associated with adhesion and
extracellular matrix remodeling (27), which are both important processes
in RCC.
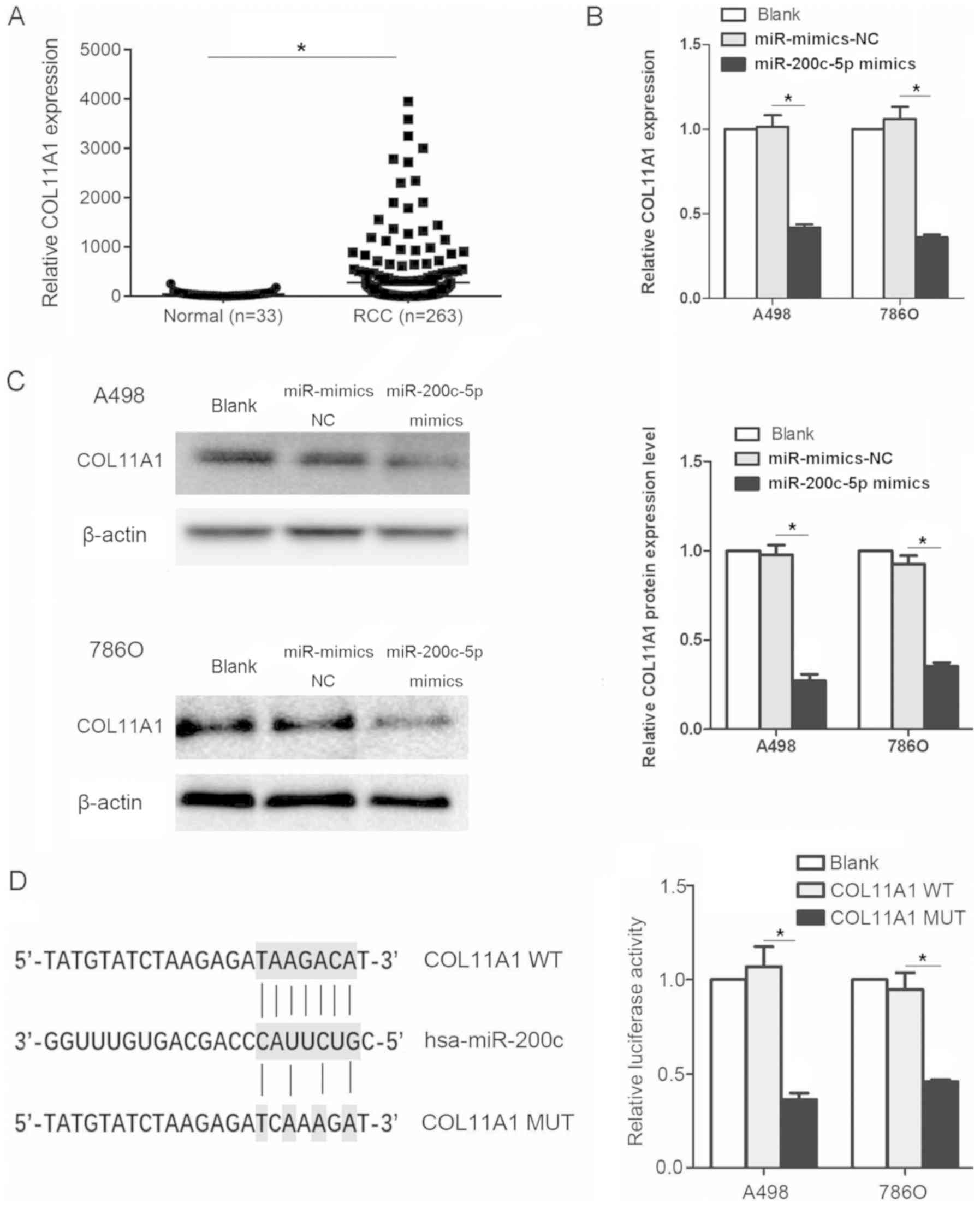
The relative expression levels of COL11A1 were
evaluated in datasets from patients with RCC and normal kidney
tissues obtained from TCGA database. The expression levels of
COL11A1 were significantly upregulated in the tissues from patients
with RCC compared with the normal tissues (Fig. 5A). Subsequently, to determine
whether COL11A1 was affected by miR-200c-5p, A498 and 786O cell
lines were transfected with miR-200c-5p mimics. The mRNA expression
levels of COL11A1 were significantly downregulated following the
transfection with the miR-200c-5p mimics (Fig. 5B). A similar trend was observed in
the protein expression levels of COL11A1 (Fig. 5C). Moreover, the relative
luciferase activity was significantly decreased in cells
transfected with the COL11A1-WT 3′-UTR construct compared with the
COL11A1-MUT 3′-UTR construct, which indicated that COL11A1 was a
direct target of miR-200c-5p (Fig.
5D). The binding sites of COL11A1 and miR-200c-5p was
discovered using miRanda database.
SNHG12 regulates COL11A1 expression
levels through miR-200c-5p
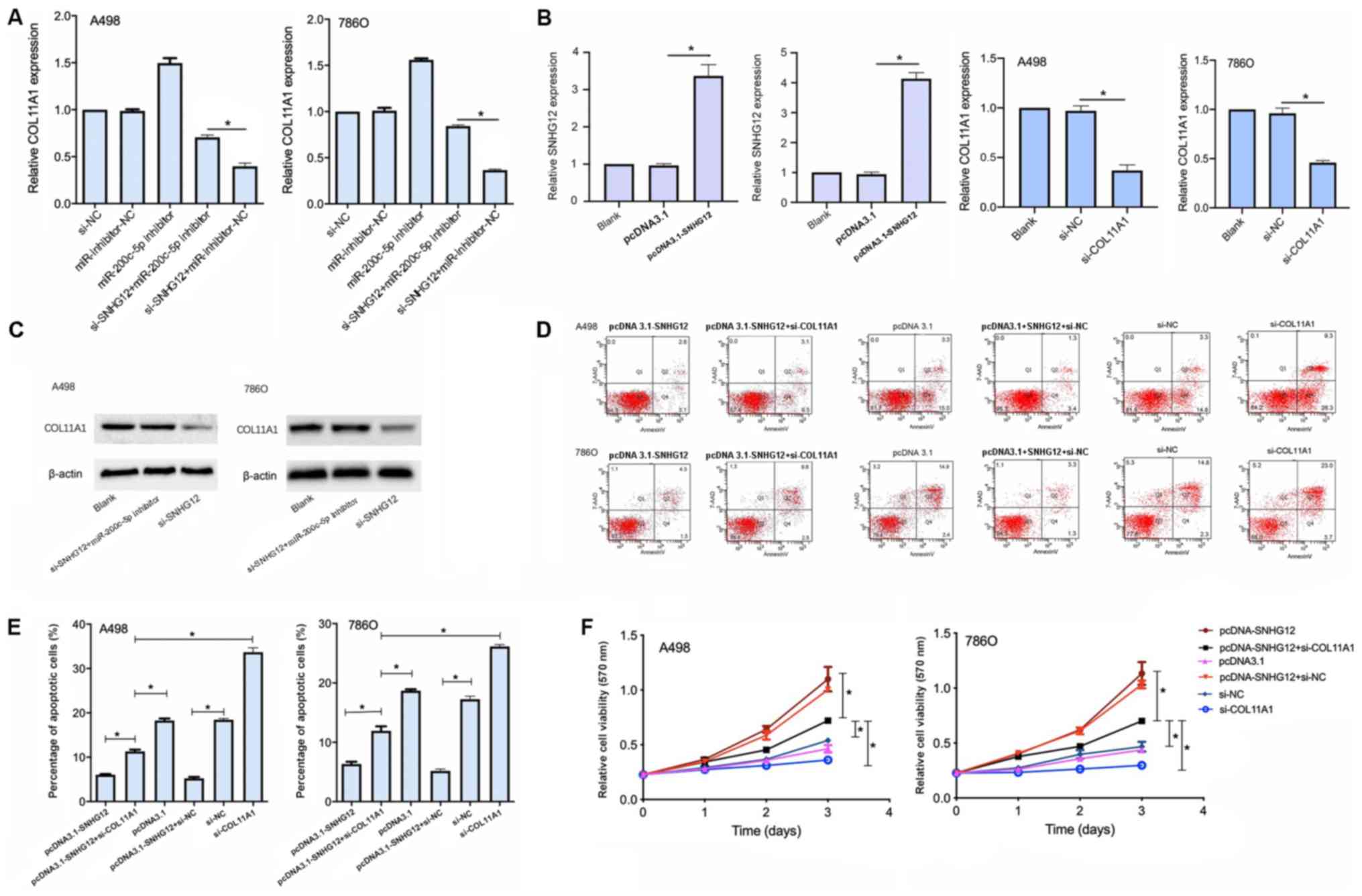
To investigate whether SNHG12 regulated COL11A1
expression levels by interacting with miR-200c-5p, COL11A1 mRNA
expression levels were analyzed following the transfection with
si-SNHG12 and the miR-200c-5p inhibitor. The transfection with
si-SNHG12 significantly downregulated the expression levels of
COL11A1 compared with the si-SNHG12 + miR-200c-5p inhibitor
transfection (Fig. 6A). Thus,
these findings indicated that the inhibition of miR-200c-5p may
partially reverse COL11A1 downregulation induced by si-SNHG12,
indicating that SNHG12 may regulate COL11A1 expression levels
through miR-200c-5p.
To confirm whether SNHG12 exerted its role through
COL11A1, A498 and 786O cell lines were transfected with the
pcDNA3.1-SNHG12 overexpression vector or si-COL11A1. The
transfection efficiency was proven to be successful (Fig. 6B and C), and MTT and apoptotic
assays were subsequently performed. The cell viability was
significantly decreased and the apoptotic rate was significantly
increased in cells co-transfected with both pcDNA3.1-SNHG12 and
si-COL11A1 compared with the cells transfected with pcDNA3.1-SNHG12
alone (Fig. 6D and E). These
findings indicated that the effect of SNHG12 on the growth of RCC
cell lines may be mediated, at least in part, by COL11A1.
Discussion
Previous studies have indicated that lncRNAs were
vital to physiological and pathological processes (28–30).
In fact, several lncRNAs have been associated with RCC; for
example, in one study, the lncRNA metastasis associated lung
adenocarcinoma transcript 1 interacted with miR-205 to promote
aggressive RCC (31), whereas in
another study, the lncRNA HOTAIR regulated HIF-α/AXL signaling by
inhibiting miR-217 (9).
Nevertheless, to the best of our knowledge, the role of SNHG12 in
RCC remains unclear.
SNHG12 has been extensively investigated in several
types of cancer, including papillary thyroid carcinoma, cervical
cancer, lung cancer, osteosarcoma and hepatocellular carcinoma
(15,16,18,32,33).
A previous study has reported that SNHG12 promoted tumor
progression in non-small cell lung carcinoma by binding to miR-218
and miR-181a, which regulated Slug/zinc finger E-box binding
homeobox (ZEB)2 and mitogen activated protein kinase/Slug signaling
(34).
The present study demonstrated that SNHG12
expression levels were upregulated in the tissues from patients
with RCC compared with the normal kidney tissues. This finding was
further validated in the A498 and 786O RCC cell lines and patient
samples. In addition, it was observed that SNHG12 knockdown
significantly suppressed cell viability and invasion and promoted
apoptosis in vitro. Moreover, analysis of TCGA data
indicated that high SNHG12 expression levels were associated with
poor clinical outcomes, a lower histological differentiation,
advanced clinical stages and metastasis. Overall, these results
suggested that SNHG12 may serve as an oncogene in RCC.
The ceRNA theory is widely accepted in the study of
lncRNAs. According to the ceRNA theory, lncRNAs regulate gene
expression by functioning as miRNA sponges, thereby interfering in
the binding between miRNAs and target genes (35). Several studies have suggested that
miR-200c-5p may function as a tumor suppressor gene in RCC
(36–40). Indeed, miR-200c-5p inhibited RCC
cell migration and invasion and participated in demethylation
drug-induced decreased cell migration and invasion (41). Moreover, miR-200c-5p was reported
to serve as a sponge for the lncRNA HOTAIR in the ceRNA network,
whereby it regulated RCC progression through several oncogenes,
including VEGFA, ZEB1 and ZEB2 (40). In the present study, miR-200c-5p
was also discovered to interact with SNHG12, as validated by
luciferase reporter assay, thus the findings suggested that SNHG12
may function as an oncogene in RCC through regulating COL11A1
expression levels by sponging miR-200c-5p. This was validated using
a luciferase reporter assay, which demonstrated that the 3′-UTR of
both SNH12 and COL11A1 were strongly bound to miR-200c-5p.
COL11A1, a primary interstitial extracellular matrix
constituent, is one of the most frequently upregulated genes
identified in chemoresistant tumors relative to normal tissues
(42). Notably, COL11A1 was
previously discovered to be associated with poor survival in
patients with RCC, owing to its high expression levels in the tumor
tissues compared with the normal kidney tissue (27). COL11A1 also promoted tumor
progression in ovarian cancer (43) and it was associated with poor
survival in multiple other types of cancer (e.g. pancreatic and
colon cancer) (44–46). In addition, a previous study
suggested that COL11A1 may be a highly specific biomarker and
molecular signature of cancer-associated fibroblasts, as it was
detected in several epithelial cancers, including lung cancer,
ovarian cancer, head and neck cancer, brain cancer, glioma, colon
cancer, liver cancer, bladder cancer, adrenocortical cancer, kidney
cancer and mesothelioma, according to data from TCGA database
(44). Thus, targeting COL11A1 or
COL11A1-associated genes may serve as an attractive therapeutic
target for RCC.
The function of miR-200c-5p and COL11A1 was further
analyzed in RCC cells using rescue assays, which indicated that
miR-200c-5p upregulation increased the rate of cell apoptosis and
decreased cell viability. COL11A1 knockdown also led to the same
results, which was consistent with previous findings (27).
In conclusion, the findings of the present study
suggested that SNHG12 may contribute to RCC progression by serving
as an oncogene, sponging miR-200c-5p and thereby upregulating the
expression levels of COL11A1. Thus, the present study provided
further insight into the role of SNHG12 in RCC development.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets analyzed during the current study are
available in the TCGA database (portal.gdc.cancer.gov/).
Authors' contributions
CX, YW, SL and XW conceived and designed the study,
performed experiments, and drafted and revised the manuscript. HL
and LS performed experiments and collected data. JZ performed
experiments. XK funded the study and interpreted the data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Medical Ethics
Committee of Hainan General Hospital (Haikou, China). Written
informed consent was obtained from all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Barata PC and Rini BI: Treatment of renal
cell carcinoma: Current status and future directions. CA Cancer J
Clin. 67:507–524. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Choueiri TK and Motzer RJ: Systemic
therapy for metastatic renal-cell carcinoma. N Engl J Med.
376:354–366. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Garcia JA and Rini BI: Recent progress in
the management of advanced renal cell carcinoma. CA Cancer J Clin.
57:112–125. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Jonasch E, Gao J and Rathmell WK: Renal
cell carcinoma. BMJ. 349:g47972014. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chi Y, Wang D, Wang J, Yu W and Yang J:
Long non-coding RNA in the pathogenesis of cancers. Cells.
8:10152019. View Article : Google Scholar
|
|
6
|
Schmitt AM and Chang HY: Long noncoding
RNAs in cancer pathways. Cancer Cell. 29:452–463. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zhan Y, Chen Z, Li Y, He A, He S, Gong Y,
Li X and Zhou L: Long non-coding RNA DANCR promotes malignant
phenotypes of bladder cancer cells by modulating the miR-149/MSI2
axis as a ceRNA. J Exp Clin Cancer Res. 37:2732018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Chen DL, Lu YX, Zhang JX, Wei XL, Wang F,
Zeng ZL, Pan ZZ, Yuan YF, Wang FH, Pelicano H, et al: Long
non-coding RNA UICLM promotes colorectal cancer liver metastasis by
acting as a ceRNA for microRNA-215 to regulate ZEB2 expression.
Theranostics. 7:4836–4849. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Hong Q, Li O, Zheng W, Xiao WZ, Zhang L,
Wu D, Cai GY, He JC and Chen XM: LncRNA HOTAIR regulates HIF-1α/AXL
signaling through inhibition of miR-217 in renal cell carcinoma.
Cell Death Dis. 8:e27722017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Zhou W, Ye XL, Xu J, Cao MG, Fang ZY, Li
LY, Guan GH, Liu Q, Qian YH and Xie D: The lncRNA H19 mediates
breast cancer cell plasticity during EMT and MET plasticity by
differentially sponging miR-200b/c and let-7b. Sci Signal.
10:eaak95572017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Trimarchi T, Bilal E, Ntziachristos P,
Fabbri G, Dalla-Favera R, Tsirigos A and Aifantis I: Genome-wide
mapping and characterization of Notch-regulated long noncoding RNAs
in acute leukemia. Cell. 158:593–606. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Atianand MK, Hu W, Satpathy AT, Shen Y,
Ricci EP, Alvarez-Dominguez JR, Bhatta A, Schattgen SA, McGowan JD,
Blin J, et al: A long noncoding RNA lincRNA-EPS acts as a
transcriptional brake to restrain inflammation. Cell.
165:1672–1685. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Kopp F and Mendell JT: Functional
classification and experimental dissection of long noncoding RNAs.
Cell. 172:393–407. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Tamang S, Acharya V, Roy D, Sharma R,
Aryaa A, Sharma U, Khandelwal A, Prakash H, Vasquez KM and Jain A:
SNHG12: An LncRNA as a potential therapeutic target and biomarker
for human cancer. Front Oncol. 9:9012019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Jin XJ, Chen XJ, Zhang ZF, Hu WS, Ou RY,
Li S, Xue JS, Chen LL, Hu Y and Zhu H: Long noncoding RNA SNHG12
promotes the progression of cervical cancer via modulating
miR-125b/STAT3 axis. J Cell Physiol. 234:6624–6632. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dong J, Wang Q, Li L and Xiao-Jin Z:
Upregulation of long non-coding RNA small nucleolar RNA host gene
12 contributes to cell growth and invasion in cervical cancer by
acting as a sponge for MiR-424-5p. Cell Physiol Biochem.
45:2086–2094. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Jiang B, Hailong S, Yuan J, Zhao H, Xia W,
Zha Z, Bin W and Liu Z: Identification of oncogenic long noncoding
RNA SNHG12 and DUXAP8 in human bladder cancer through a
comprehensive profiling analysis. Biomed Pharmacother. 108:500–507.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhou B, Li L, Li Y, Sun H and Zeng C: Long
noncoding RNA SNHG12 mediates doxorubicin resistance of
osteosarcoma via miR-320a/MCL1 axis. Biomed Pharmacother.
106:850–857. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun Y, Liu J, Chu L, Yang W, Liu H, Li C
and Yang J: Long noncoding RNA SNHG12 facilitates the tumorigenesis
of glioma through miR-101-3p/FOXP1 axis. Gene. 676:315–321. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Chen Q, Zhou W, Du SQ, Gong DX, Li J, Bi
JB, Li ZH, Zhang Z, Li ZL, Liu XK and Kong CZ: Overexpression of
SNHG12 regulates the viability and invasion of renal cell carcinoma
cells through modulation of HIF1α. Cancer Cell Int. 19:1282019.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang O, Yang F, Liu Y, Lv L, Ma R, Chen C,
Wang J, Tan Q, Cheng Y, Xia E, et al: C-MYC-induced upregulation of
lncRNA SNHG12 regulates cell proliferation, apoptosis and migration
in triple-negative breast cancer. Am J Transl Res. 9:533–545.
2017.PubMed/NCBI
|
|
22
|
Liu ZB, Tang C, Jin X, Liu SH and Pi W:
Increased expression of lncRNA SNHG12 predicts a poor prognosis of
nasopharyngeal carcinoma and regulates cell proliferation and
metastasis by modulating Notch signal pathway. Cancer Biomark.
23:603–613. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Xian HP, Zhuo ZL, Sun YJ, Liang B and Zhao
XT: Circulating long non-coding RNAs HULC and ZNFX1-AS1 are
potential biomarkers in patients with gastric cancer. Oncol Lett.
16:4689–4698. 2018.PubMed/NCBI
|
|
24
|
Moch H, Cubilla AL, Humphrey PA, Reuter VE
and Ulbright TM: The 2016 WHO classification of tumours of the
urinary system and male genital organs-part a: Renal, penile, and
testicular tumours. Eur Urol. 70:93–105. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ricketts CJ, De Cubas AA, Fan H, Smith CC,
Lang M, Reznik E, Bowlby R, Gibb EA, Akbani R, Beroukhim R, et al:
The cancer genome atlas comprehensive molecular characterization of
renal cell carcinoma. Cell Rep. 23:313–326.e315. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Boguslawska J, Kedzierska H, Poplawski P,
Rybicka B, Tanski Z and Piekielko-Witkowska A: Expression of genes
involved in cellular adhesion and extracellular matrix remodeling
correlates with poor survival of patients with renal cancer. J
Urol. 195:1892–1902. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Bhan A, Soleimani M and Mandal SS: Long
noncoding RNA and cancer: A new paradigm. Cancer Res. 77:3965–3981.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Fang Y and Fullwood MJ: Roles, functions,
and mechanisms of long non-coding RNAs in cancer. Genomics
Proteomics Bioinformatics. 14:42–54. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Yarani R, Mirza AH, Kaur S and Pociot F:
The emerging role of lncRNAs in inflammatory bowel disease. Exp Mol
Med. 50:1–14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Hirata H, Hinoda Y, Shahryari V, Deng G,
Nakajima K, Tabatabai ZL, Ishii N and Dahiya R: Long noncoding RNA
MALAT1 promotes aggressive renal cell carcinoma through Ezh2 and
interacts with miR-205. Cancer Res. 75:1322–1331. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lan T, Ma W, Hong Z, Wu L, Chen X and Yuan
Y: Long non-coding RNA small nucleolar RNA host gene 12 (SNHG12)
promotes tumorigenesis and metastasis by targeting miR-199a/b-5p in
hepatocellular carcinoma. J Exp Clin Cancer Res. 36:112017.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Ding S, Qu W, Jiao Y, Zhang J, Zhang C and
Dang S: LncRNA SNHG12 promotes the proliferation and metastasis of
papillary thyroid carcinoma cells through regulating wnt/β-catenin
signaling pathway. Cancer Biomark. 22:217–226. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Wang P, Chen D, Ma H and Li Y: LncRNA
SNHG12 contributes to multidrug resistance through activating the
MAPK/Slug pathway by sponging miR-181a in non-small cell lung
cancer. Oncotarget. 8:84086–84101. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Qi X, Zhang DH, Wu N, Xiao JH, Wang X and
Ma W: ceRNA in cancer: Possible functions and clinical
implications. J Med Genet. 52:710–718. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Maolakuerban N, Azhati B, Tusong H, Abula
A, Yasheng A and Xireyazidan A: MiR-200c-3p inhibits cell migration
and invasion of clear cell renal cell carcinoma via regulating
SLC6A1. Cancer Biol Ther. 19:282–291. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Jiang J, Yi BO, Ding S, Sun J, Cao W and
Liu M: Demethylation drug 5-Aza-2′-deoxycytidine-induced
upregulation of miR-200c inhibits the migration, invasion and
epithelial-mesenchymal transition of clear cell renal cell
carcinoma in vitro. Oncol Lett. 11:3167–3172. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Wang X, Chen X, Han W, Ruan A, Chen L,
Wang R, Xu Z, Xiao P, Lu X, Zhao Y, et al: miR-200c targets CDK2
and suppresses tumorigenesis in renal cell carcinoma. Mol Cancer
Res. 13:1567–1577. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Gao C, Peng FH and Peng LK: MiR-200c
sensitizes clear-cell renal cell carcinoma cells to sorafenib and
imatinib by targeting heme oxygenase-1. Neoplasma. 61:680–689.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ding J, Yeh CR, Sun Y, Lin C, Chou J, Ou
Z, Chang C, Qi J and Yeh S: Estrogen receptor β promotes renal cell
carcinoma progression via regulating LncRNA
HOTAIR-miR-138/200c/204/217 associated CeRNA network. Oncogene.
37:5037–5053. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ying G, Wu R, Xia M, Fei X, He QE, Zha C
and Wu F: Identification of eight key miRNAs associated with renal
cell carcinoma: A meta-analysis. Oncol Lett. 16:5847–5855.
2018.PubMed/NCBI
|
|
42
|
Wu YH, Chang TH, Huang YF, Chen CC and
Chou CY: COL11A1 confers chemoresistance on ovarian cancer cells
through the activation of Akt/c/EBPβ pathway and PDK1
stabilization. Oncotarget. 6:23748–23763. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Wu YH, Chang TH, Huang YF, Huang HD and
Chou CY: COL11A1 promotes tumor progression and predicts poor
clinical outcome in ovarian cancer. Oncogene. 33:3432–3440. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Jia D, Liu Z, Deng N, Tan TZ, Huang RY,
Taylor-Harding B, Cheon DJ, Lawrenson K, Wiedemeyer WR, Walts AE,
et al: A COL11A1-correlated pan-cancer gene signature of activated
fibroblasts for the prioritization of therapeutic targets. Cancer
Lett. 382:203–214. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yang H, Wu J, Zhang J, Yang Z, Jin W, Li
Y, Jin L, Yin L, Liu H and Wang Z: Integrated bioinformatics
analysis of key genes involved in progress of colon cancer. Mol
Genet Genomic Med. 7:e005882019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Feldmann G, Habbe N, Dhara S, Bisht S,
Alvarez H, Fendrich V, Beaty R, Mullendore M, Karikari C, Bardeesy
N, et al: Hedgehog inhibition prolongs survival in a genetically
engineered mouse model of pancreatic cancer. Gut. 57:1420–1430.
2008. View Article : Google Scholar : PubMed/NCBI
|