Introduction
Disorders associated with individuals being
overweight and obese have become everyday issues over the past few
decades, and global statistics indicate that these disorders are on
a continuous rise (1). Obesity is
a metabolic syndrome that is associated with a cluster of
disorders, including type 2 diabetes mellitus, hypertension and
fatty liver disorder (2–5). Despite the increasing life expectancy
in the United States, babies born at the beginning of the
twenty-first century are predicted to be the first generation that
may have shorter life expectancies than their parents (6). Risk factors for becoming overweight
and obese are well known, but the underlying pathogenesis has not
yet been elucidated. At present, therapy is aimed at modifying the
risk factors, but there are no sustainable therapies for the
prevention or treatment of obesity.
Melatonin is a neurohormone synthesized in the
pineal gland (7). It exerts its
functions via membrane and nuclear receptors as well as
receptor-independent actions. Receptor-mediated neuroendocrine
functions of melatonin include the circadian rhythm, sleep, the
stress response, the process of aging and immunity (8,9). The
antioxidant effect of melatonin, along with the free radical
scavenging action, is a well-established receptor-independent
function (10).
Melatonin has various functions, including the
promotion and regulation of energy homeostasis (11). For instance, evidence has
demonstrated that melatonin is involved in the regulation of food
intake, energy storage and energy expenditure (12,13).
Melatonin treatment in drinking water or a liquid diet has been
revealed to reduce body weight and abdominal fat in rats
independently of food intake reduction (14–17).
In the zebrafish, melatonin inhibits appetite and stimulates
satiety signals in the central nervous system (18). A previous study has also indicated
that there is a synchronizing function of melatonin with the
metabolism in white adipocytes (19).
Although studies with exogenous administration of
melatonin demonstrate the ‘anti-obesity’ effect of melatonin, as
aforementioned, the effects of endogenous melatonin on obesity are
controversial and have not yet been established. Thus, the aim of
the current study was to determine the effects of endogenous
melatonin, especially pineal gland-derived, on obesity by employing
rat pinealectomy (Pnx) model.
Materials and methods
Animals
A total of 23 male Wistar rats, aged 3-weeks-old,
were purchased from the Laboratory Animal Center of Japan SLC, Inc.
The rats were housed in individual cages under a controlled
temperature (24±1°C), humidity and lighting (12-h light/dark
cycle). After 1-week of acclimation, rats were fed a high-fat diet
(HFD; cat. no. D12451; Research Diets, Inc.) for 10 weeks, which
contained 45% kcal as fat, 20% as protein and 35% as carbohydrates,
with access to water ad libitum prior to the operation. Rats
were randomized into two groups with a similar average body weight,
the sham group with 13 ats, and the Pnx group with 10 rats. After
the operation, rats were fed a normal chow diet (NCD) with access
to water ad libitum for 10 weeks. This study was approved by
the Institutional Animal Care and Use Committee of Keimyung
University, School of Medicine, Daegu, Korea (approval number.
KM-2014-16). Body weight, food intake and water intake were
measured twice a week. Peritoneal glucose tolerance tests (PGTT)
were performed before and after the operation on overnight-fasted
rats. After an overnight fast, rats were administrated with 1 g/kg
glucose [D-(+)-glucose; Sigma-Aldrich; Merck KGaA] by peritoneal
gavage. Blood samples were collected from the tip of the tail
immediately prior to and 30, 60, 90 and 120 min after injection.
Blood glucose was measured using a glucometer (Model GU, Accu-Chek
Active; Roche Diagnostics GmbH). Rats were sacrificed by isoflurane
[2% volume-to-volume (v/v) concentration] inhalation at 10 weeks
after operation followed by exsanguination. Tissues, including
brown adipose tissues (BAT) and white adipose tissues (WAT), and
serum were harvested for further analyses.
Surgical procedures
Rats were fasted overnight prior to surgery. Pnx was
performed as described previously (20). Anesthesia was induced with an
isoflurane concentration (v/v) of 5% (JW Pharmaceutical
Corporation) in 30% oxygen and 70% nitrous oxide and maintained
with 2% isoflurane (v/v) during the surgical procedure. In brief, a
sagittal opening was made in the scalp followed by exposure of the
lambda suture. The skull around the lambda suture was drilled and
carefully removed. The pineal gland was then removed with fine
forceps. The removed skull was placed back, and the scalp was
sutured. The procedure was completed within 30 min.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted with TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) according to
the manufacturer's protocol. For RT-qPCR, 5 µg total RNA was
reverse transcribed for 30 min at 37°C in a reaction mixture
containing the RNA, 40 U RNase inhibitor (Promega Corporation), 0.5
mM deoxynucleotide triphosphate (Promega Corporation), 2 µM random
hexamer primers, 5X AMV reverse transcriptase reaction buffer and
30 U AMV reverse transcriptase (Promega Corporation). RT-qPCR
analysis was performed using the SYBR Green PCR Master Mix (Toyobo
Life Science) and RT-qPCR thermocycling conditions were as follows:
Initial denaturation at 95°C for 30 sec, followed by amplification
for 45 cycles performed at 95°C for 5 sec, 60°C for 10 sec and 72°C
for 15 sec, 95°C for 10 sec and 10 sec each at 0.2°C increments to
62°C for a melting curve analysis, and 65°C for 1 min, followed by
cooling to 37°C for 10 min with gene-specific primers. The relative
expression of each gene was normalized against β-actin. The samples
were assayed on a LightCycler 480 (Roche Diagnostics) instrument
and the concentration was calculated as copies per µl using the
standard curve. Primer sequences (Macrogen, Inc.) are listed in
Table I.
 | Table I.Primer sequences. |
Table I.
Primer sequences.
| Gene | Sequence
(5′→3′) | Temperature,
°C | bp |
|---|
| CPT1A | F:
ATGACGGCTATGGTGTCTCC | 60 | 154 |
|
| R:
GTGAGGCCAAACAAGGTGAT |
|
|
| CPT1B | F:
CTGGACCGAGAAGAGATCAA | 60 | 175 |
|
| R:
CCTTGAAGAAGCGACCTTTG |
|
|
| Cyt C | F:
TCAATGATGCTGCCTTTCAC | 60 | 233 |
|
| R:
ACTCCCAATCAGGCATGAAC |
|
|
| DIO2 | F:
TCCCAATTCCAGTGTGGTGC | 61 | 181 |
|
| R:
GCGGAAGGCTGGCAGTTGCC |
|
|
| FASN | F:
TGGCTTCCGTTCAGTCTCTT | 60 | 180 |
|
| R:
CAGTGCCAAGGTCTCTAGCC |
|
|
| MCAD | F:
CCTGGACAGGAAAACATTTG | 60 | 167 |
|
| R:
CCTTCGCAATAGAGGCAAAG |
|
|
| MTNR1A | F:
CAGACCTCGTGGTGGCTATT | 60 | 237 |
|
| R:
TCAGGAACACGTAGCACAGG |
|
|
| MTNR1B | F:
ATTCCTGCACCTTCATCCAG | 60 | 224 |
|
| R:
TATGGCGAAAACCACAAACA |
|
|
| PGC1α | F:
TCGCCTTCTTGCTCTTCCTT | 59 | 175 |
|
| R:
ATCTACTGCCTGGGGACCTT |
|
|
| PRDM16 | F:
CCTAGCCCTGAGCGATACTG | 61 | 233 |
|
| R:
AGTAGCTACGTTGCGGGAGA |
|
|
| SREBP1c | F:
GTGGTCTTCCAGAGGCTGAG | 62 | 178 |
|
| R:
GGGTGAGAGCCTTGAGACAG |
|
|
| UCP1 | F:
GAAGGATTGCCGAAACTGTA | 57 | 155 |
|
| R:
CCAGCCGAGATCTTGCTTCC |
|
|
| β-actin | F:
AGCCATGTACGTAGCCATCC | 60 | 233 |
|
| R:
TTTCCCTCTCAGCTGTGGTG |
|
|
Western blot analysis
Harvested tissues were subjected to SDS-PAGE and
immunoblotted. Briefly, tissues were lysed in ice-cold lysis buffer
[50 mM Tris-HCl (pH, 7.4), 25 mM EDTA (pH 8.0), 650 mM NaCl, 5%
Triton X-100] containing protease inhibitors (200 mM
phenylmethylsulfonyl fluoride, 100 µg/ml leupeptin, 10 µg/ml
pepstatin, 1 µg/ml aprotinin and 2 mM EDTA). The lysates were
centrifuged for 20 min at 7,500 × g at 4°C and the supernatant was
collected. The concentrations of protein were determined using a
BCA protein assay kit (cat. no. 23227; Thermo Fisher Scientific,
Inc.) and densitometric analysis was performed using SPECTROstar
Nano software (version 5.5; BMG Labtech GmbH). Proteins (30 µg)
were separated by SDS-PAGE (8%) and transferred to nitrocellulose
membrane (GE Healthcare). The membrane was blocked at room
temperature (RT) for 30 min in 5% skimmed milk in TBS with 0.1%
Tween-20 (TBST) before incubation overnight at 4°C with the
following primary antibodies: Anti-uncoupling protein 1 (UCP1;
1:1,000; cat. no. ab10983; Abcam), anti-peroxisome
proliferator-activated receptor γ coactivator-1α (PGC1α; 1:500;
cat. no. ab54481; Abcam), anti-iodothyronine deiodinase 2 (DIO2;
1:1,000; cat. no. ab77779; Abcam), anti-p-Akt (1:1,000; cat. no.
9271S; Cell Signaling Technology, Inc.), MTNR1A (1:1,000; cat. no.
NBP1-7113; Novus Biologicals, LLC), MTNR1B (1:1,000; cat. no.
NLS932; Novus Biologicals) and anti-sirtuin 1 (SIRT1; 1:1,000; cat.
no. sc-15404; Santa Cruz Biotechnology, Inc.). β-actin (1:1,000;
cat. no. A5441; Sigma-Aldrich; Merck KGaA) was used as an internal
control. The membrane was then washed in TBST and incubated with
horseradish peroxidase-conjugated rabbit and mouse secondary
antibodies (both 1:1,000; cat. nos. sc-2004 and sc-2005,
respectively; both Santa Cruz Biotechnology, Inc.). Protein bands
were detected using Super Signal West Pico Chemiluminescent
Substrate (Thermo Fisher Scientific, Inc.). The protein-specific
signals were detected using LAS-3000 (FujiFilm Wako Pure Chemical
Corporation).
Histological assessment of BAT
The BAT of each rat was fixed at RT for 24 h with
10% formalin, embedded in paraffin and sectioned. The sections (4
µm-thick) were stained at RT with hematoxylin and eosin (H&E)
for 3 min and 30 sec, respectively, or immunohistochemistry was
performed for light microscopic examination. To observe UCP1
expression, 5-µm thick sections were permeabilized in PBS,
incubated in 10 mM sodium citrate buffer with pH 6.0 at 100°C for
10 min, and then incubated with anti-UCP1 (1:3,000 antibody
overnight at 4°C. Sections were then incubated with the secondary
UCP1 antibody (1:200; Santa Cruz Biotechnology, Inc.) for 1 h at
RT, followed by staining at RT for 1 min with diaminobenzidine
chromogen (Vector Laboratories, Inc.) and counterstaining with
hematoxylin at RT for 3 min. The stained sections were examined
under a microscope at magnification ×200 (Nikon Corporation); all
histological assessments were made by a pathologist.
Enzyme-linked immunosorbent assay
(ELISA)
Blood samples were allowed to clot for 2 h at RT
before centrifugation at 4°C for 20 min at 2,000 × g. Serum levels
of leptin (R&D System, Inc.) and melatonin (cat. no. CEA908GE;
Cloud-Clone Corp.) were measured using ELISA kits. Enzyme levels in
the supernatants were measured according to the manufacturer's
instructions. Serum samples were then diluted at 1:10 in a diluent
solution from the kit and incubated in the plates at RT for 2 h,
after which the plates were washed five times with washing buffer
from the kit. Horseradish peroxidase conjugated rat leptin (100 µl)
was added and then incubated for 2 h at RT. The plates were washed
five times and incubated with substrate solution for 30 min, after
which the stop solution was added. The absorbance values were
determined with an ELISA microplate reader (Biochrom, Ltd.) at a
wavelength of 450 nm and then 570 nm. Hepatic triglyceride (TG;
cat. no. 10010303; Cayman Chemical Company) levels were measured
using an ELISA kit. Briefly, the samples or TG standard were
incubated with TG enzyme mixture in plates for 15 min at RT. A
standard curve was constructed to assign arbitrary units. The
absorbance values were determined with an ELISA microplate reader
operating at 530–550 nm.
Statistical analysis
Data were analyzed using an unpaired Student's
t-test. Each experiment was performed at least three times in
duplicate. All data are given in terms of relative values and are
presented as the mean ± standard deviation. P<0.05 was
considered to indicate a statistically significant difference.
Results
Pnx increases insulin sensitivity
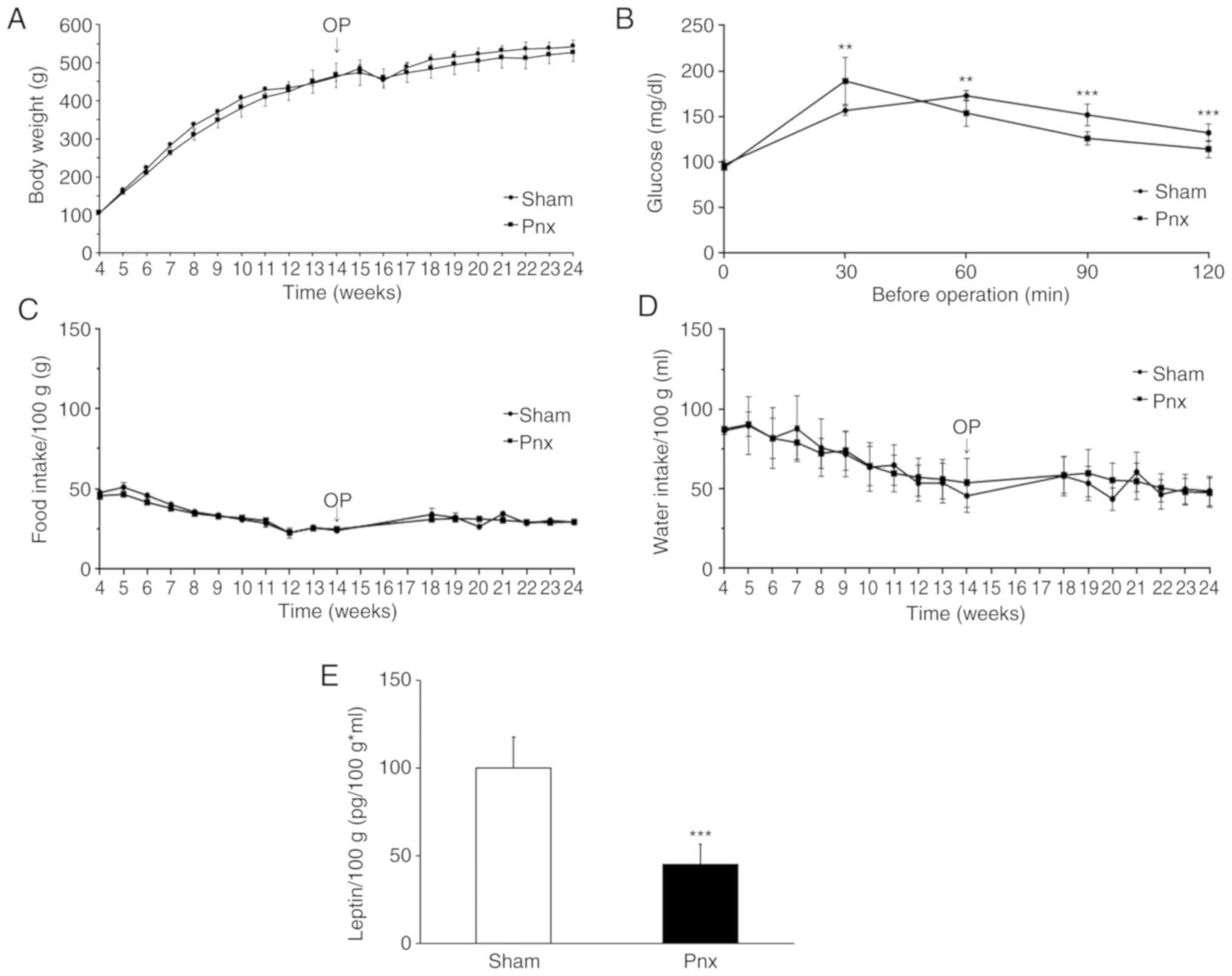
Before rats underwent Pnx, average body weight was
465±22.3 g. Average body weight at the end of the experiment was
542±17.5 g in the sham group and 529±28.1 g in the Pnx group.
Although not statistically different, throughout the experiment the
degree of increase in the body weight of the Pnx group was
consistently less than that observed in the sham group (Fig. 1A). PGTT before the operation
revealed impaired insulin responses. After the operation, PGTT
revealed that glucose levels at 60, 90 and 120 min in the Pnx group
were lower than those in the sham group, which indicated an
improvement of insulin sensitivity in the Pnx group (Fig. 1B). Food and water intake/100 g body
weight did not differ between the two groups (Fig. 1C and D). Since insulin resistance
is associated with elevated levels of plasma leptin, circulating
leptin levels in the sham and Pnx groups were investigated and
calculated as leptin levels per 100 g body weight (Fig. 1E). Circulating leptin levels in the
Pnx group significantly decreased (159.79±13.39 pg/ml; P<0.01)
compared with those in the sham group (344.67±48.64 pg/ml;
P<0.05), implicating increased insulin sensitivity following
Pnx. Serum insulin levels were also determined in sham and Pnx
groups and, although not statistically significant, decreased
insulin levels were found in the Pnx group (data not shown). Serum
levels of melatonin were also determined, but the measured values
were below the detectable levels of the kit (data not shown).
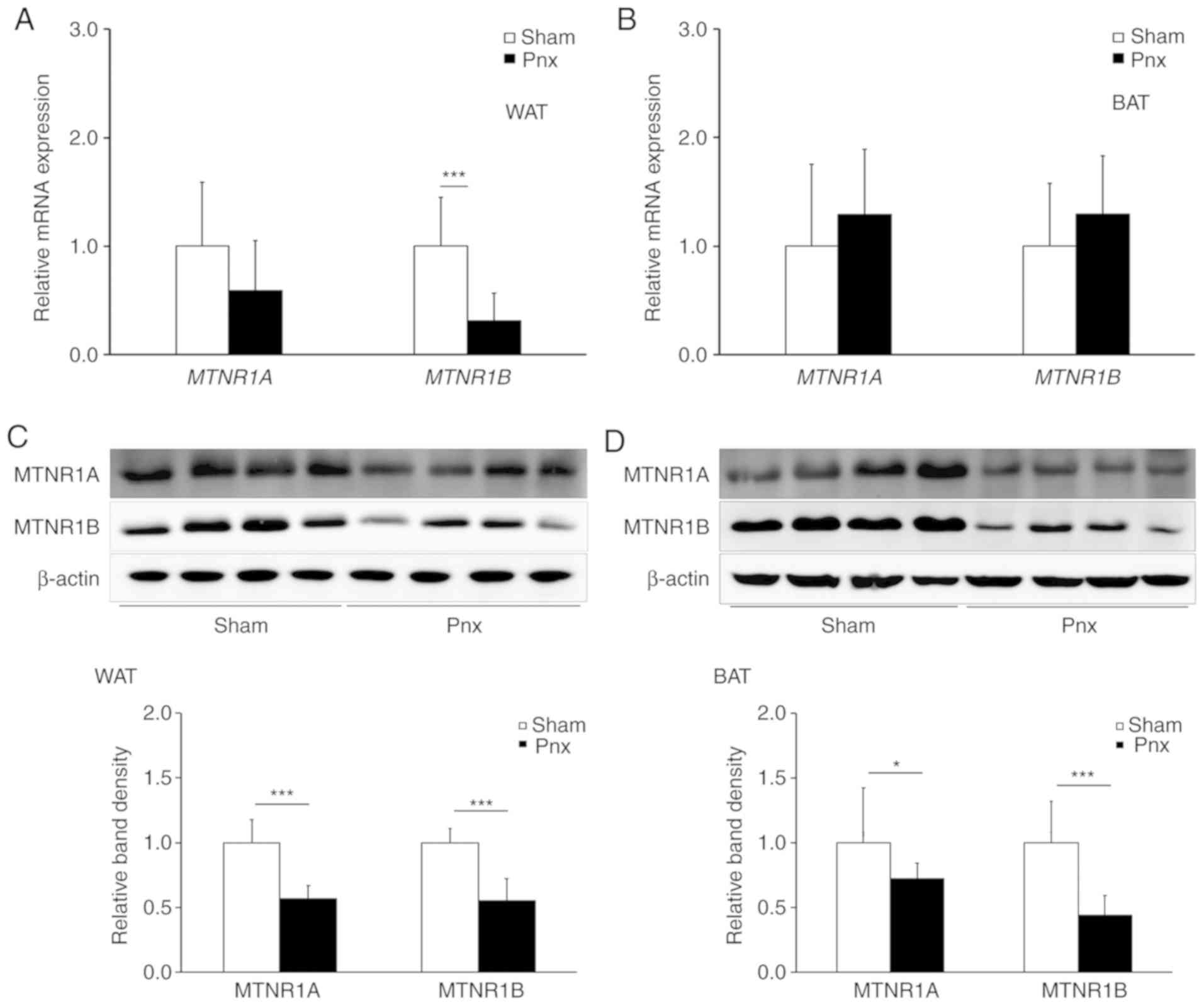
Pnx decreases melatonin receptor
(MTNR)1A and MTNR1B expression in WAT and BAT
To determine the effect of Pnx on the expression of
melatonin receptors, the expression levels of MTNR1A and MTNR1B
were examined in WAT and BAT. It was found that both mRNA (Fig. 2A and B) and protein (Fig. 2C and D) expression levels of MTNR1A
and MTNR1B decreased in WAT from the group that underwent Pnx
compared with those in the sham group. Although mRNA levels of
MTNR1A and MTNR1B were not different in BAT, proteins expression
levels of both MTNR1A and MTNR1B were significantly different,
which indicates decreased melatonin receptor activity in WAT and
BAT. Further research as to why and how expression levels of
melatonin receptor mRNA do not match those of melatonin receptor
proteins is required.
Pnx stimulates thermogenesis in
BAT
Based on the results that showed increased insulin
sensitivity, and decreased MTNR1A and MTNR1B expression, in WAT and
BAT in the Pnx group, it was hypothesized that Pnx improved insulin
sensitivity by stimulating thermogenesis in BAT and regulating
lipogenesis in WAT. To investigate this hypothesis, the expression
of various thermogenic genes were examined in BAT (Fig. 3A). Fig. 3A shows a notable increase in the
expression levels (upper panel) and semi-quantification (lower
panel) of the thermogenic genes (SIRT1, PGC1α, UCP1 and DIO2) in
the groups that underwent Pnx compared with those in the sham
group. The invisibility of the SIRT and PGC1α western bands in the
control may be due to short exposure time. Accordingly, increased
mRNA expression of PGC1α, UCP1, DIO2 and PR domain-containing 16
(PRDM16) were also found in the Pnx group (Fig. 3B and C). However, no differences
were observed in the expression levels of peroxisome
proliferator-activated receptor γ (data not shown), the master
regulator of adipogenesis (21,22).
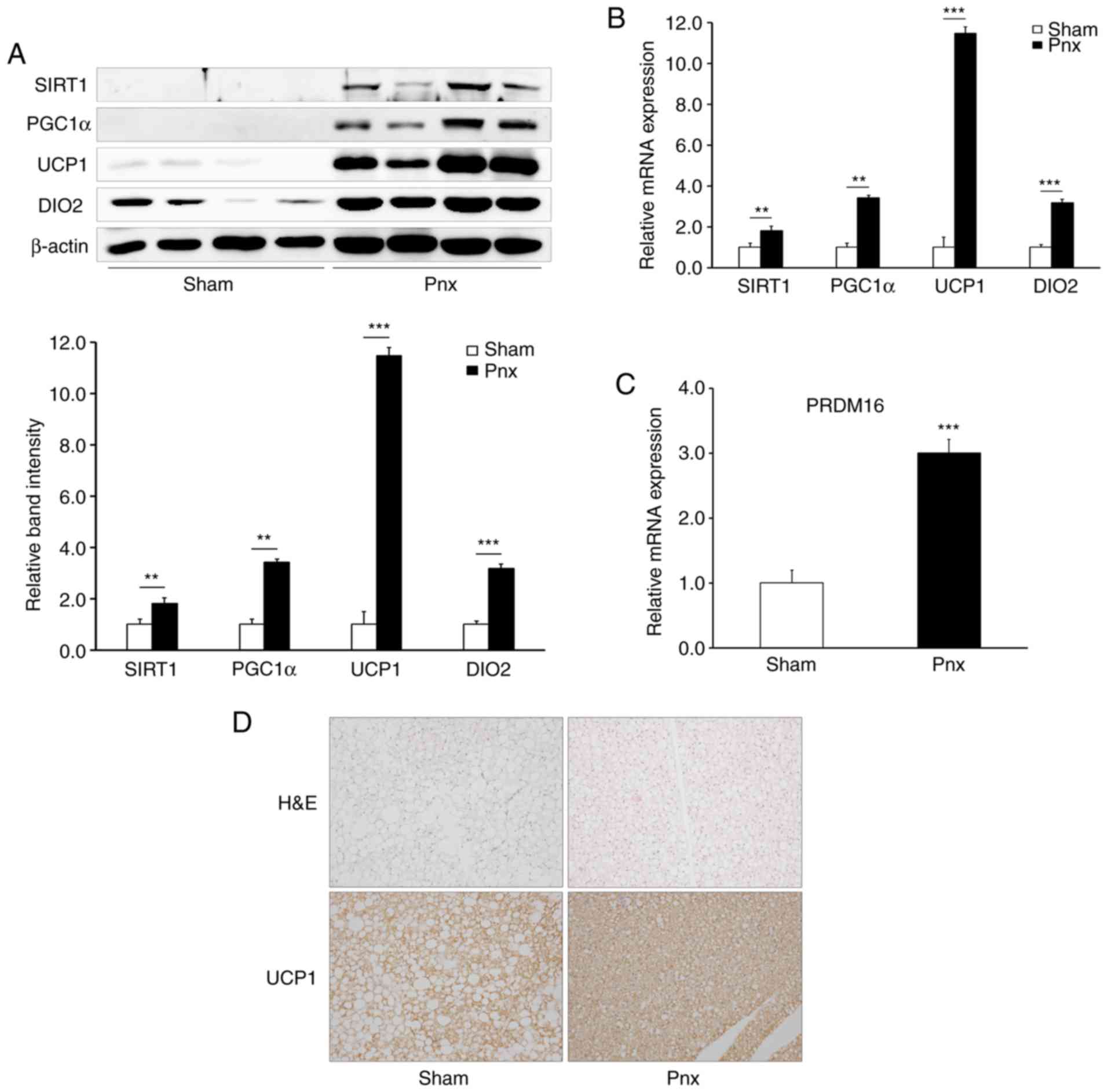 | Figure 3.Thermogenic genes and PRDM16 in BAT.
(A) Protein expression of SIRT1, PGC1α, UCP1 and DIO2 were examined
by western blot analysis (upper panel), and relative band density
was semi-quantified (lower panel). (B) mRNA expression levels of
PGC1α, UCP1, and DIO2 (C) PRDM16 was examined by reverse
transcription-quantitative PCR and normalized to β-actin. (D)
Changes in size of lipid droplets and UCP1 expression in BAT.
Representative images of BAT from the sham group and Pnx group
stained with H&E (upper panel) and immune-histochemical images
stained with UCP1 antibody (lower panel). Magnification, ×200.
Values are presented as the mean ± SD. **P<0.01, ***P<0.001
vs. the sham group. Pnx, pinealectomy; BAT, brown adipose tissue;
PGC1α, peroxisome proliferator-activated receptor γ coactivator
1-α; UCP1, uncoupling protein 1; PPARγ, peroxisome
proliferator-activated receptor γ; DIO2, deiodinase 2; PRDM16, PR
domain 16; H&E, hematoxylin and eosin; SIRT1, sirtuin 1. |
To further validate the thermogenic effect of Pnx,
the study proceeded to examine histological changes and UCP1
expression in BAT (Fig. 3D).
Histological analysis of BAT by H&E staining revealed a marked
decrease in the size of lipid droplets in the Pnx group (upper
right panel, Fig. 3D). Consistent
with the result in Fig. 3A and B,
immunohistochemical staining revealed a notable increase in the
expression of UCP1 in the Pnx group compared with the sham group
(lower right panel, Fig. 3D).
Pnx downregulates lipogenic genes in
WAT and the liver
Subsequently, the possible regulatory effects of Pnx
in WAT were examined by determining the expression levels of
various lipogenic genes (Fig. 4A).
It was found that Pnx led to a significant reduction in the
expression of sterol regulatory element binding protein 1c
(SREBP1c), the master regulatory transcription factor for
lipogenesis (21). Consistent with
the decreased expression of SREBP1c, the Pnx group was also found
to have a decreased expression of fatty acid synthase (FASN), a key
lipogenic enzyme in the first step of de novo lipogenesis
(21). Moreover, following Pnx the
expression of stearoyl-CoA desaturase (SCD1), which catalyzes the
unsaturation of fatty acids forming a double bond in stearoyl-CoA,
decreased in these rats. These results suggested that Pnx inhibited
the lipogenic pathway in WAT. Next, the effect of Pnx on the
expression of major mitochondrial biogenesis genes PGC1α and
cytochrome c (Cyt c) was investigated (21). Contrary to the expression in BAT
(Fig. 3A and B), the expression of
PGC1α in WAT decreased following Pnx, which might indicate
attenuated mitochondrial biogenesis. Decreased expression levels of
genes involved in fatty acid oxidation [carnitine
palmitoyltransferase (CPT)1b and medium-chain acyl-CoA
dehydrogenase], also supports this hypothesis (Fig. 4B) (21).
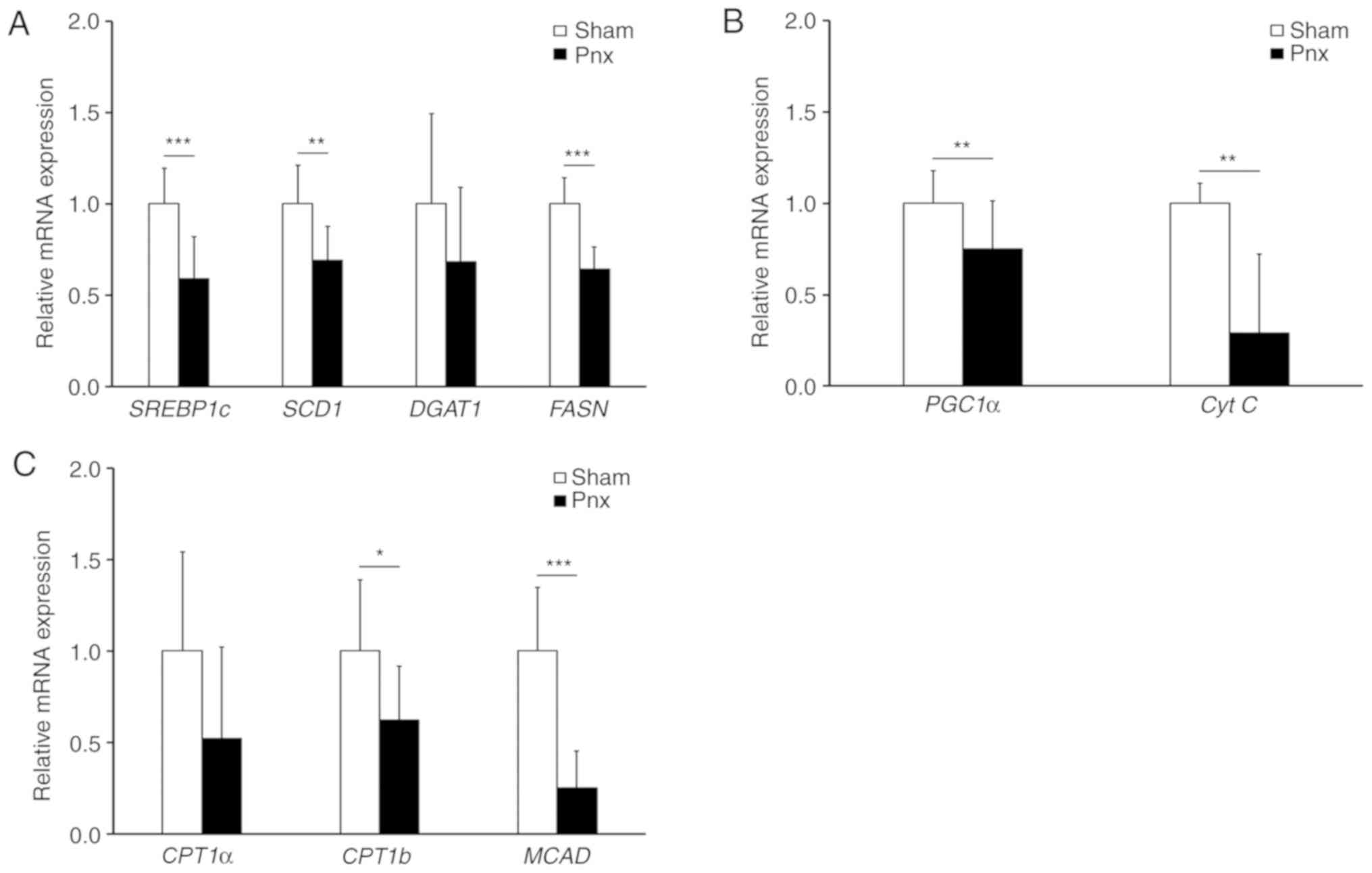 | Figure 4.Lipogenesis genes in WAT. mRNA
expression levels of (A) the lipogenic genes, SREBP1c, SCD1, DGAT1
and FASN, (B) the mitochondrial biogenesis genes, PGC1α and Cyt C,
and (C) fatty acid oxidation genes, CPT1a, CPT1b and MCAD, in WAT
were examined by reverse transcription-quantitative PCR and
normalized to β-actin. Values are presented as the mean ± SD.
*P<0.05, **P<0.01, ***P<0.001 vs. the sham group. Pnx,
pinealectomy; WAT, white adipose tissue; SREBP1c, sterol regulatory
element-binding protein 1; SCD1, stearoyl-CoA desaturase 1; DGAT1,
diacylglycerol O-acyltransferase 1; FASN, fatty acid synthase; CPT,
carnitine palmitoyltransferase; MCAD, medium-chain acyl-CoA
dehydrogenase; PGC1α, peroxisome proliferator-activated receptor γ
coactivator 1-α; Cyt C, cytochrome c. |
Since the liver is the organ primarily responsible
for lipogenesis, the expression levels of lipogenic genes were
further examined in the liver. Histological analysis of the liver
by H&E staining also demonstrated that lipid droplets decreased
in size and number in the Pnx group compared with the sham group
(Fig. 5A). Consistent with the
results of the expression levels in WAT, it was found that Pnx
induced a significant decrease in SREBP1c. Furthermore, the
expression levels of the SREPB1c target genes, SCD1, FASN and
DGAT2, although not statistically significant, decreased in the
group that underwent Pnx (Fig.
5B). Expression levels of genes involved in mitochondrial
biogenesis and fatty acid oxidation, with the exception of CPT1a,
showed no significant differences between the sham and Pnx groups
(Fig. 5C and D). The present study
also observed an increased expression of phosphorylated AKT in the
Pnx group (Fig. 5E).
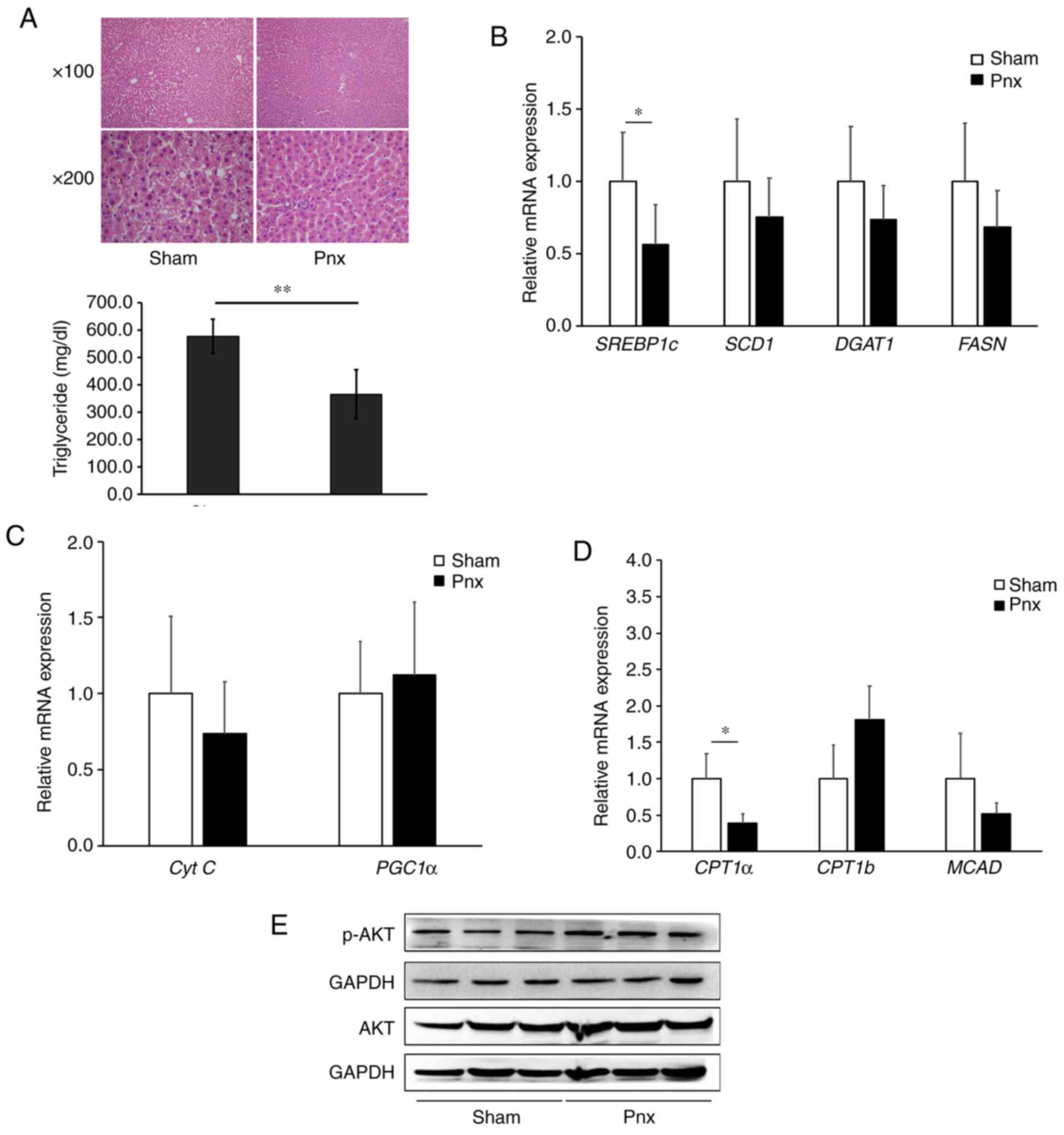 | Figure 5.Changes in hepatic histology and
lipogenesis genes in the liver. (A) Liver sections were stained
with H&E. Magnification at ×100 in the upper panel and ×200 in
the lower panel. mRNA expression levels of the (B) lipogenic genes,
SREBP1c, SCD1, DGAT1 and FASN, (C) mitochondrial biogenesis genes,
PGC1α and Cyt C, and (D) fatty acid oxidation genes, CPT1a, CPT1b
and MCAD, in the liver were examined by reverse
transcription-quantitative PCR and normalized to β-actin. (E)
Protein expression levels of p-AKT, AKT and GAPDH were examined by
western blotting. Values are presented as the mean ± SD.
*P<0.05, **P<0.01 vs. the sham group. Pnx, pinealectomy;
SREBP1c, sterol regulatory element-binding protein 1; SCD1,
stearoyl-CoA desaturase 1; DGAT1, diacylglycerol O-acyltransferase
1; FASN, fatty acid synthase; CPT, carnitine palmitoyltransferase;
MCAD, medium-chain acyl-CoA dehydrogenase; PGC1α, peroxisome
proliferator-activated receptor γ coactivator 1-α; Cyt C,
cytochrome c; H&E, hematoxylin and eosin; p-,
phosphorylated. |
Discussion
Numerous studies have demonstrated that exogenous
melatonin reduces body weight and food intake (14–17).
However, the current study has shown that Pnx stimulated
thermogenesis in BAT, and attenuated lipogenesis in WAT and the
liver of HFD-fed rats without any significant changes in body
weight compared with control rats.
No differences were observed in the body weight or
amount of food intake in the group that underwent Pnx compared with
those in the sham group, which may indicate that endogenously
synthesized pineal melatonin does not affect systemic energy
balance. To support this result, a recent study also demonstrated
that there was no difference in the body weight and food intake of
NCD-fed controls and Pnx rats (23). Buonfiglio et al (23) reported that exogenous melatonin
treatment following Pnx resulted in a decrease in body weight and
food intake, which suggests that exogenous, rather than endogenous,
melatonin is a possible contributing factor for the decrease in
body weight and food intake. Decreased protein expression levels of
MTNR1A and MTNR1B in WAT and BAT of the Pnx group were also
observed, which suggests reduced melatonin receptor activities in
WAT and BAT. Furthermore, it was found that mRNA expression levels
of MTNR1A and MTNR1B were not consistent with protein expression
levels of MTNR1A and MTNR1B in BAT, which may indicate that there
is a different response to compensate for decreased concentrations
of melatonin in WAT and BAT.
Notably, despite the lack of significant differences
in body weight changes, Pnx was observed to increase insulin
sensitivity, evidenced by faster disposal of plasma glucose and
lower levels of plasma leptin in the Pnx group. These results are
in contrast to a previous report, which found that Pnx induced
insulin resistance (24). Lima
et al (24) demonstrated
that Pnx causes glucose intolerance and decreases adipose cell
responsiveness to insulin in NCD-fed rats. Since HFD-fed rats were
employed in the present study, it was hypothesized that there was
an interaction between diet and the absence of pineal melatonin
that affected systemic metabolism. In the present study, decreased
expression levels of lipogenic genes were observed in WAT, which
indicated a reduction in lipogenesis. Whether the decreased
lipogenesis in WAT contributed to the decreased adiposity in the
Pnx group needs further study since lipogenesis in WAT under a HFD
may not be as tightly associated with the adiposity caused by a
NCD. It is commonly known that leptin levels increase in obesity
and are associated with insulin resistance (25–28),
so decreased leptin levels in the Pnx group compared with that in
the control HFD-fed group further validates Pnx-induced insulin
sensitivity.
UCP1, also known as thermogenin, is predominantly
expressed in BAT. UCP1 is a mitochondrial protein that regulates
dissipation of excess energy via the uncoupling of oxidative
phosphorylation (21). It is also
positively associated with insulin sensitivity (22). The present study found that Pnx
markedly increased the expression of UCP1 in BAT. Consistent with
this finding, increased expression levels of PGC1α, DIO2 and SIRT1
were also found in the Pnx group. UCP1 plays a role in the activity
of several transcription factors and genes, such as PGC1α, DIO2,
SIRT1 and PRDM16 (21). PGC1α is a
master nuclear transcription factor that controls the expression of
thermogenic genes (21). In the
present study, PGC1α expression was significantly increased in the
Pnx rats, which suggested that PGC1α-induced thermogenic genes are
upregulated following Pnx. DIO2 is another commonly known master
regulatory molecule that controls the expression of thermogenic
genes (29). The present study
also found that Pnx stimulated the expression of DIO2. As expected,
the expression of the thermogenic gene UCP1 was increased in the
BAT following Pnx. Furthermore, DIO2 activity has been demonstrated
to generate a significant fraction of the circulating hormone
triiodothyronine in rats, and enhance the cAMP-generated acute
increase in UCP1 mRNA via increased UCP1 gene transcription
(30). PRDM16 is a regulatory
molecule known to induce brown adipogenicity (31), and the expression of PRDM16 in the
BAT of the Pnx group was found to be increased, which indicated
that Pnx may induce BAT. Furthermore, the expression of SIRT1 in
the Pnx group was found to increase the expressions of SIRT1 in
BAT, which further indicated that the Pnx-induced insulin
sensitivity may be mediated via BAT activation (32). The present study is limited by the
absence of a control group of rats fed NCD. Furthermore, this study
lacks an investigation into the underlying mechanisms of the effect
of Pnx on BAT activation. Therefore, further studies are required
to elucidate this.
Overall, in the present study Pnx was found to
decrease lipogenesis in WAT and the liver. It was also demonstrated
that expression levels of lipogenic genes were decreased in both
the WAT and liver of the Pnx group compared with the sham group,
which indicated that Pnx attenuates lipogenesis via a currently
unidentified mechanism. These results indicate that Pnx may inhibit
lipogenic activity by inhibiting the expression of genes involved
in lipogenesis, as evidenced by the decrease in mRNA expression of
the mitochondrial biogenesis genes, PGC1α and Cyt C. In conclusion,
rats that underwent Pnx experience an increase in thermogenesis in
BAT, and a decrease in lipogenesis in WAT, leading to increased
insulin sensitivity.
Acknowledgements
Not applicable.
Funding
This work was supported by the Basic Science
Research Program through the National Research Foundation of Korea
funded by the Ministry of Education (grant no.
NRF-2018R1A2B6006175) and by the research fund of Hanyang
University (grant no. HY-201500000003026), Seoul, Republic of
Korea.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
This study was conceptualized by EH and JHS, data
collection was performed by MK and SML. Data analysis was conducted
by MK, SML, JJ and YJK, the experiments were performed by MK, SML,
JJ, KCM and EH. EH, JJ, KCM, JHS and TKH supervised the project,
and data processing was performed by KCM. This study was supported
by funding acquired by EH, JHS and TKH. The initial draft of the
manuscript was written by MK, SML and KCM, and the review and
editing were conducted by EH, JHS and TKH. TKH supervised
additional experiments, analyzed and interpreted the data, provided
funding and prepared the draft for revision. All authors read and
approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
Pnx
|
pinealectomy
|
|
HFD
|
high-fat diet
|
|
NCD
|
normal chow diet
|
|
MTNR
|
melatonin receptor
|
|
BAT
|
brown adipose tissue
|
|
WAT
|
white adipose tissue
|
|
UCP1
|
uncoupling protein 1
|
|
PGTT
|
peritoneal glucose tolerance test
|
|
SIRT1
|
sirtuin 1
|
|
PGC1α
|
peroxisome proliferator-activated
receptor γ coactivator-1α
|
|
PRDM16
|
PR domain-containing 16
|
|
SREBP1c
|
sterol regulatory element binding
protein 1c
|
|
FASN
|
fatty acid synthase
|
|
SCD1
|
stearoyl-CoA desaturase
|
|
CPT
|
carnitine palmitoyltransferase
|
|
MCAD
|
medium-chain acyl-CoA
dehydrogenase
|
|
Cyt c
|
cytochrome c
|
References
|
1
|
Ng M, Fleming T, Robinson M, Thomson B,
Graetz N, Margono C, Mullany EC, Biryukov S, Abbafati C, Ferede S,
et al: Global, regional, and national prevalence of overweight and
obesity in children and adults during 1980–2013: A systematic
analysis for the global burden of disease study 2013. Lancet.
384:766–781. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Berrington de Gonzalez A, Hartge P, Cerhan
JR, Flint AJ, Hannan L, MacInnis RJ, Moore SC, Tobias GS,
Anton-Culver H, Freeman LB, et al: Body-mass index and mortality
among 1.46 million white adults. N Engl J Med. 363:2211–2219. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Renehan AG, Tyson M, Egger M, Heller RF
and Zwahlen M: Body-mass index and incidence of cancer: A
systematic review and meta-analysis of prospective observational
studies. Lancet. 371:569–578. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Prospective Studies Collaboration, ;
Whitlock G, Lewington S, Sherliker P, Clarke R, Emberson J, Halsey
J, Qizilbash N, Collins R and Peto R: Body-mass index and
cause-specific mortality in 900 000 adults: Collaborative analyses
of 57 prospective studies. Lancet. 373:1083–1096. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Skrypnik D, Bogdański P, Zawiejska A and
Wender-Ożegowska E: Role of gestational weight gain, gestational
diabetes, breastfeeding, and hypertension in mother-to-child
obesity transmission. Pol Arch Intern Med. 129:267–275.
2019.PubMed/NCBI
|
|
6
|
Olshansky SJ, Passaro DJ, Hershow RC,
Layden J, Carnes BA, Brody J, Hayflick L, Butler RN, Allison DB and
Ludwig DS: A potential decline in life expectancy in the United
States in the 21st century. N Engl J Med. 352:1138–1145. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Stehle JH, Saade A, Rawashdeh O, Ackermann
K, Jilg A, Sebestény T and Maronde E: A survey of molecular details
in the human pineal gland in the light of phylogeny, structure,
function and chronobiological diseases. J Pineal Res. 51:17–43.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cardinali DP, Golombek DA, Rosenstein RE,
Cutrera RA and Esquifino AI: Melatonin site and mechanism of
action: Single or multiple? J Pineal Res. 23:32–39. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Galano A, Tan DX and Reiter RJ: Melatonin
as a natural ally against oxidative stress: A physicochemical
examination. J Pineal Res. 51:1–16. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Reiter RJ, Tan DX, Osuna C and Gitto E:
Actions of melatonin in the reduction of oxidative stress. A
review. J Biomed Sci. 7:444–458. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cipolla-Neto J, Amaral FG, Afeche SC, Tan
DX and Reiter RJ: Melatonin, energy metabolism, and obesity: A
review. J Pineal Res. 56:371–381. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Bartness TJ and Wade GN: Body weight, food
intake and energy regulation in exercising and melatonin-treated
siberian hamsters. Physiol Behav. 35:805–808. 1985. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bubenik G and Pang SF: The role of
serotonin and melatonin in gastrointestinal physiology: Ontogeny,
regulation of food intake, and mutual serotonin-melatonin feedback.
J Pineal Res. 16:91–99. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bojkova B, Orendáš P, Friedmanova L,
Kassayová M, Datelinka I, Ahlersová E and Ahlers I: Prolonged
melatonin administration in 6-month-old sprague-dawley rats:
Metabolic alterations. Acta Physiol Hung. 95:65–76. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Puchalski SS, Green JN and Rasmussen DD:
Melatonin effect on rat body weight regulation in response to
high-fat diet at middle age. Endocrine. 21:163–167. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Ríos-Lugo MJ, Jiménez-Ortega V,
Cano-Barquilla P, Mateos PF, Spinedi EJ, Cardinali DP and Esquifino
AI: Melatonin counteracts changes in hypothalamic gene expression
of signals regulating feeding behavior in high-fat fed rats. Horm
Mol Biol Clin Investig. 21:175–183. 2015.PubMed/NCBI
|
|
17
|
Wolden-Hanson T, Mitton DR, McCants RL,
Yellon SM, Wilkinson CW, Matsumoto AM and Rasmussen DD: Daily
melatonin administration to middle-aged male rats suppresses body
weight, intraabdominal adiposity, and plasma leptin and insulin
independent of food intake and total body fat. Endocrinology.
141:487–497. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Piccinetti CC, Migliarini B, Olivotto I,
Coletti G, Amici A and Carnevali O: Appetite regulation: The
central role of melatonin in danio rerio. Horm Behav. 58:780–785.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Alonso-Vale MIC, Andreotti S, Mukai PY,
Borges-Silva CDN, Peres SB, Cipolla-Neto J and Lima FB: Melatonin
and the circadian entrainment of metabolic and hormonal activities
in primary isolated adipocytes. J Pineal Res. 45:422–429. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Maganhin CC, Simões RS, Fuchs LFP,
Oliveira-Filho RM, de Jesus Simões M, Neto JE, Baracat EC and Maria
J: Rat pinealectomy: A modified direct visual approach. Acta Cir
Bras. 24:321–324. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Seale P, Kajimura S, Yang W, Chin S, Rohas
LM, Uldry M, Tavernier G, Langin D and Spiegelman BM:
Transcriptional control of brown fat determination by PRDM16. Cell
Metab. 6:38–54. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Chondronikola M, Volpi E, Børsheim E,
Porter C, Annamalai P, Enerbäck S, Lidell ME, Saraf MK, Labbe SM,
Hurren NM, et al: Brown adipose tissue improves whole body glucose
homeostasis and insulin sensitivity in humans. Diabetes.
63:4089–4099. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Buonfiglio D, Parthimos R, Dantas R, Silva
RC, Gomes G, Andrade-Silva J, Ramos-Lobo A, Amaral FG, Matos R,
Sinésio J Jr, et al: Melatonin absence leads to long-term leptin
resistance and overweight in rats. Front Endocrinol (Lausanne).
9:1222018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lima FB, Machado UF, Bartol I, Seraphim
PM, Sumida DH, Moraes SM, Hell NS, Okamoto MM, Saad MJ, Carvalho CR
and Cipolla-Neto J: Pinealectomy causes glucose intolerance and
decreases adipose cell responsiveness to insulin in rats. Am J
Physiol. 275:E934–E941. 1998.PubMed/NCBI
|
|
25
|
Considine RV, Sinha MK, Heiman ML,
Kriauciunas A, Stephens TW, Nyce MR, Ohannesian JP, Marco CC, McKee
LJ and Bauer TL: Serum immunoreactive-leptin concentrations in
normal-weight and obese humans. N Engl J Med. 334:292–295. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Handjieva-Darlenska T and Boyadjieva NJ:
The effect of high-fat diet on plasma ghrelin and leptin levels in
rats. J Physiol Biochem. 65:157–164. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Segal KR, Landt M and Klein S:
Relationship between insulin sensitivity and plasma leptin
concentration in lean and obese men. Diabetes. 45:988–991. 1996.
View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Skrypnik D, Mostowska A, Jagodziński PP
and Bogdański P: Association of rs699947 (−2578 C/A) and rs2010963
(−634 G/C) single nucleotide polymorphisms of the VEGF gene, VEGF-A
and leptin serum level, and cardiovascular risk in patients with
excess body mass: A case-control study. J Clin Med. 9:4692020.
View Article : Google Scholar
|
|
29
|
Bartelt A and Heeren J: Adipose tissue
browning and metabolic health. Nat Rev Endocrinol. 10:24–36. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
De Jesus LA, Carvalho SD, Ribeiro MO,
Schneider M, Kim SW, Harney JW, Larsen PR and Bianco AC: The type 2
iodothyronine deiodinase is essential for adaptive thermogenesis in
brown adipose tissue. J Clin Invest. 108:1379–1385. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Giralt M and Villarroya F: White, brown,
beige/brite: Different adipose cells for different functions?
Endocrinology. 154:2992–3000. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Boutant M, Joffraud M, Kulkarni SS,
García-Casarrubios E, García-Roves PM, Ratajczak J,
Fernández-Marcos PJ, Valverde AM, Serrano M and Cantó C: SIRT1
enhances glucose tolerance by potentiating brown adipose tissue
function. Mol Metab. 4:118–131. 2014. View Article : Google Scholar : PubMed/NCBI
|