Cancer stem cells (CSCs), a population of cells with
similar characteristics to those of stem cells, are associated with
the occurrence, recurrence, metastasis and chemoradiation
resistance of cancer (1,2). The existence of CSCs is a challenge
for tumor treatment, but can also provide a novel direction for
clinical treatment. To date, there have been various speculations
about the occurrence of CSCs, but it remains unclear. Hypoxia in
the tumor microenvironment is usually found in advanced cancer and
is associated with low survival rates and poor prognosis (3,4). An
increasing number of studies have found that hypoxia can induce
cancer cell invasion, metastasis and epithelial-mesenchymal
transition (EMT), which can promote stem-like characteristics in
cancer cells (5,6). Hypoxia inducible factor-1 (HIF-1), as
a pivotal molecule in the regulation of CSCs by hypoxia,
participates in tumor growth, immune evasion, metabolic
reprogramming and drug resistance by regulating the transcription
of target genes (4,6,7). HIF-1
seems to play an important, or even core, role in the generation
and maintenance of CSCs, but the explicit mechanism remains to be
elucidated. This review attempted to summarize the role and
mechanism of HIF-1 in CSCs, in order to provide more targets to
solve the limitations of clinical tumor treatment.
Epigenetic modifications are reversible, heritable
changes in gene function that do not alter the DNA sequence
(26,27). HIF-1α can be used as a key regulator
of genomic methylation in hepatocellular carcinoma cells (28). The presence of HIF-1α binding sites
on methionine adenosyltransferase 2A (MAT2A) can promote the
transcription of MAT2A to maintain the genome of the
demethylation state (28).
Unfortunately, most currently available studies have focused on the
genetic modification of HIF-1 downstream target genes or key
enzymes (28). Research on the
epigenetic modification of HIF-1 in CSCs is rare.
Post-translational modification is one of the most
important regulatory mechanisms for dynamically and reversibly
regulating proteins that have biological functions (29). A previous study has found that
lysine methyltransferase G9a can mono- or di-methylate HIF-1α on
lysine 674, which reduces the transcriptional activity of HIF-1α
and expression of downstream genes by reducing the TAD activity of
HIF-1α; meanwhile G9a is reduced in glioblastoma cells, maintaining
the activity of HIF-1α and promoting HIF-1-dependent cell migration
(30). As a small ubiquitin-like
modifier (SUMO) protease, small ubiquitin-like modifier protease 1
(SENP1) forms a positive feedback loop with HIF-1α in hepatoma
cells, which causes HIF-1α deSUMOylation and stable expression
during hypoxia, and can promote the production of liver CSCs
(31). In addition, the
SENP1/HIF-1α positive feedback loop promotes hypoxia-induced cell
EMT and invasion in osteosarcoma cells (32). Kinase ERK-mediated phosphorylation
of HIF-1α increases its stability and accumulation in the nucleus
to promote the transcriptional activation of target genes (25,33).
Protein kinase A in HeLa cells can also phosphorylate threonine 63
and serine 692 of HIF-1α, inhibiting HIF-1α degradation and
increasing HIF-1α transcription (34). The knockout of Y-box binding protein
1 in gliomas inhibits cell proliferation and blocks the cell cycle
by downregulating the phosphorylation level of HIF-1, which may
affect the proliferation, differentiation and metastasis of glioma
cells (35,36). There is not enough information on
the epigenetic and post-translational modification of HIF-1, thus
further studies are needed to explore how the proliferation and
growth of CSCs can be inhibited via the expression of HIF.
Non-coding RNA (ncRNA) is a class of RNA that is
transcribed from DNA but is not translated into a protein, types of
ncRNA include small interfering RNA (siRNA), long ncRNA (lncRNA)
and microRNA (miRNA) (37,38). A previous study demonstrated that
the interaction between HIF-1 and ncRNA is significant in the
self-renewal and proliferation of CSCs. miR-124 can reverse the
resistance of breast CSCs to doxorubicin through the inhibition of
the STAT3/HIF-1α signaling pathway (39). miR-200b can target and inhibit the
anti-angiogenic Krüppel-like factor 2 gene during acute hypoxia,
thereby stabilizing HIF-1 signaling to promote angiogenesis
(40). miR-200c inhibits the
metastasis and invasion of lung cancer cells by inhibiting HIF-1α
expression (41). miR-18a targets
HIF-1α and inhibits the distant metastasis of breast cancer through
the HIF-1α-dependent hypoxic response; miR-18a-5p also increases
the radiotherapy sensitivity of lung CSCs by inhibiting HIF-1α
(42,43). The promoter of miR-302 responds to
HIF-1α, which is beneficial for enhancing stem-like characteristics
of hypoxic cancer cells (44). The
expression of miR-210, an important regulator that inhibits DNA
repair, is directly regulated by HIF-1α and promotes the
degradation of normoxic gene mRNA (45,46).
The knockout of miR-210 suppresses the self-renewal capacity and
resistance of glioma stem-like cells induced by hypoxia (47). miR-21 and HIF-1α are positively
correlated in multiple tumors, and miR-126 and HIF-1α are
significantly negatively correlated in colon cancer, thus
indicating that their expression could be used for the early
diagnosis and screening of cancer (46,48,49).
miR-21 and miR-130b activate EMT through the phosphatase and tensin
homolog/Akt/HIF-1α pathway and promote hepatocellular carcinoma
metastasis (50). HIF-1α binds to
the miR-1275 promoter, which promotes miR-1275 expression and
maintains the pluripotency of stem cells by activating the
β-catenin and Notch signaling pathways in lung adenocarcinoma
(51). miR-421 is upregulated by
HIF-1α and promotes gastric cancer metastasis and chemotherapy
resistance (52). miR-107 inhibits
Ewing sarcoma cell proliferation, blocks the cell cycle and
promotes apoptosis by targeting HIF-1β (53). miR-99a, which is reduced in breast
CSCs, suppresses the stem-like phenotype of breast cancer by
inhibiting the mTOR/HIF-1α signaling pathway (54). In addition, under normoxia, the
upregulated miR-31 in head and neck squamous cell carcinoma can
target the 3′ untranslated region of the FIH transcript to promote
the transactivation activity of HIF, leading to increased
tumorigenicity (55). Similar
results were also found in oral squamous cell carcinoma and
colorectal cancer (56,57). The upregulated lncRNA LOC554202 in
non-small cell lung cancer is positively correlated with miR-31,
thereby targeting FIH to promote the development of acquired
gefitinib resistance (58).
miR-31-5p, which is upregulated in lung cancer, induces glycolytic
gene expression by regulating the FIH/HIF pathway, and ultimately
promotes cell proliferation and tumor growth (59). miR-21 and miR-184, which are also
upregulated in head and neck squamous cell carcinoma, have similar
tumorigenic mechanisms to those of miR-31 (60).
Several hypoxia-related lncRNAs, such as HOTAIR,
H19, lncRNA-NUTF2P3-001, lncRNA-UCA1, lncRNA-EFNA3, ANRIL, HINCUTs
and GAPLINC, are directly regulated by HIF-1α, as their promoters
have hypoxic response elements (HREs), which are required for
HIF-1α-mediated transcriptional activation (61,62).
The low expression of LINC00996 in colorectal cancer cells may
participate in the occurrence and metastasis of colorectal cancer
by regulating the HIF-1 signaling pathway (63). HIF-1α can promote the transcription
of lincRNA-p21 (64). Conversely,
hypoxia-related lncRNA-p21 can bind to HIF-1α, which can prevent
the interaction of HIF-1α and Von Hippel-Lindau (VHL) and cause the
accumulation of HIF-1α in cells (64). Beyond that, the knockout of
lincRNA-p21 can also induce apoptosis through the
HIF-1α/Akt/mTOR/P70S6 kinase 1 (S6K)-pathway and increase the
sensitivity of radiotherapy (65).
By forming a complex with HIF-1α, lncHIFCAR/MIR31HG promotes the
binding of HIF-1α to the target promoters and increases the
sphere-forming and metastatic ability of oral cancer cells
(66). lncRNA PCGEM1 may be used as
a scaffold to form a complex with HIF-1α and transcription factor
Snail homolog 1 (SNAI1) and regulate the invasion and metastasis of
gastric cancer cells (67).
Hypoxia-induced lncRNA CRPAT4 is regulated by HIF-1α and plays a
carcinogenic role by promoting the growth and migration of cancer
cells (68). The presence of siRNA
targeting HIF-1α may provide a novel direction for specific
treatment against HIF-1α, and it could achieve molecular therapy by
inducing apoptosis and increasing the sensitivity of radiotherapy
(69,70).
In the regulation of HIF-1 expression, the
cooperation of miRNA and related lncRNA is equally important. The
significantly upregulated lncRNA TUG1 in osteosarcoma protects the
HIF-1α mRNA 3′ untranslated region from miR-143-5p, thereby
promoting osteosarcoma metastasis (71). lncRNA MIR31HG is the host gene of
miR-31, which is located in the first intron of MIR31HG and has
consistent transcriptional regulation (66,72).
Studies have found that MIR31HG is a hypoxia-inducible lncRNA and
acts as a HIF-1α co-activator to regulate the HIF-1 transcriptional
network (66). Mechanistically,
MIR31HG directly interacts with HIF-1α to promote the recruitment
of HIF-1α and p300 to target gene promoters (66). It is worth noting that, although the
expression of MIR31HG is positively correlated with miR-31 in
certain types of cancer, the knockout of MIR31HG has no effect on
miR-31, indicating that the tumor regulatory effect of MIR31HG may
be independent of miR-31 (73). A
variety of lncRNAs serve as competing endogenous RNAs (ceRNAs) to
inhibit the interaction of miRNAs with their targets, thereby
forming a lncRNA-miRNA-mRNA ceRNA network, which can regulate
multiple signaling pathways, including the HIF-1α pathway (74,75).
In addition, the HIF-1α-mediated hypoxia-induced upregulation of
lncRNA-NEAT1 in hepatocellular carcinoma regulates the expression
of the uridine-cytidine kinase 2 gene, which is associated with low
survival rates of patients with liver cancer, by suppressing
miR-199a-3p, and ultimately promotes tumor growth (76).
Although there are only a small number of studies on
HIF-1-related ncRNAs, the existing independent studies are
sufficient to illustrate HIF-1 as an important regulator or
participant in CSC-related ncRNA (Table
I), which may eradicate CSCs and prolong the life of patients
by targeting ncRNA or HIF-1.
Markers of CSCs induce the pluripotency of cancer
cells and are used to distinguish CSC subpopulations, some of which
have been found to be associated with metastasis (77,78).
Studies have found that HIF-1 is associated with the generation of
CSC markers. The data has indicated that HIF-1α can induce the
production of multiple stem cell markers, such as OCT4, SOX2, NANOG
and Krüppel-like factor 4 (KLF4) (44,79,80).
In addition, the silencing of HIF-1α can hinder the progression of
cancer by inhibiting the expression of stem cell markers (81). HIF-1 was found to bind directly to
the CD47 promoter to facilitate gene transcription, which
helps to escape phagocytosis of macrophages and maintain the stem
phenotype of breast CSCs (7,82).
Endogenous HIF-1α is recruited to the promoter of CD24,
which promotes CD24 expression, as well as tumor formation and
metastasis (83). HIF-1α appears to
bind to the CD133 promoter and promote the production of
CD133+ glioblastoma, and colon and pancreatic CSCs via
OCT4 and SOX2 (81,84–87).
In addition, a correlation has been found between HIF-1α and CD133
in the cytoplasm, rather than other parts of the cell, such as the
gland cavity (88). In turn, CD133
promotes HIF-1α expression and its translocation to the nucleus
under hypoxia (89). However, there
is a different opinion that hypoxia-induced HIF-1α expression leads
to a decrease in CD133 expression in gastrointestinal cancer cells
that overexpress CD133. Under normoxic conditions, the inhibition
of mTOR signaling in CD133-overexpressing gastrointestinal cancer
cells suppresses HIF-1α expression and promotes that of CD133
(90). In breast CSCs, HIF-1
increases the stability of NANOG mRNA through the
transactivation of RNA demethylase ALKBH5, which is involved in
encoding N6-methyladenosine demethylase (7). In prostate cancer samples, the
co-localization of HIF-1α, OCT4 and NANOG suggests that HIF-1α may
regulate the production of CSCs by regulating stem factors
(44). Surprisingly, OCT4 isoform
OCT4B in cervical cancer cells promotes neovascularization by
upregulating HIF-1α production (91). The subtype of aldehyde
dehydrogenase, 4-trimethylaminobutyraldehyde dehydrogenase
(ALDH1A1), which is associated with the self-renewal, metastasis
and resistance of cancer cells, is regulated by HIF-1α in breast
cancer (81). In turn, ALDH1A1
promotes HIF-1α expression via retinoic acid signaling (81,92).
In addition to promoting the production of mesenchymal or EMT
marker proteins, HIF-1α also inhibits the expression of epithelial
marker proteins, which can be confirmed by the use of HIF-1α
inhibitors (93–95). HIF-1α can be used as a malignant
marker of chondrosarcoma, due to its association with
neovascularization (96). In
conclusion, CSC markers can be used to isolate CSC subpopulations,
and have been demonstrated to be involved in the self-renewal of
CSCs, as well as cancer invasion and metastasis. Under hypoxic
conditions, HIF-1α, as a direct or indirect upstream regulator of
the marker protein, may become a novel target for the elimination
of CSCs.
Hypoxia not only regulates the production of CSCs,
but also participates in regulating the immune system. Hypoxia
promotes the B cell differentiation potential of lymphoid-primed
multipotent progenitors through HIF-1α, resulting in the production
of B cells (97). Under hypoxic
conditions, HIF-1α also regulates innate immune responses, induces
regulatory T cells (Tregs), and mediates immune escape from
cytotoxic T lymphocytes and other complex immune responses
(98–100). In previous years, HIF-1α-mediated
tumor immunity has been proposed as a direction to solve the
problems associated with tumor therapy (101–103), so the immune response of CSCs
mediated by HIF-1α is also worth exploring when investigating
antitumor therapies.
During EMT, in addition to the induction of cancer
stemness, immunosuppression is also observed, which can lead to
increased malignancy of the tumor, drug resistance and metastasis
(79). Studies have found that
hypoxia further increases the production of immunosuppressive
factors, inhibition of monocyte phagocytosis, inhibition of T cell
proliferation, and activation and induction of Tregs in
glioblastoma multiforme-related CSCs, which may be achieved via the
phoshphorylated STAT3/HIF-1α/vascular endothelial growth factor
(VEGF) pathway (104–106). During HIF-1α-induced EMT of liver
cancer cells, the upregulated cytokine CCL20 promotes the
expression of indoleamine 2,3-dioxygenase in monocyte-derived
macrophages, which inhibits the activation and proliferation of T
cells and induces Tregs by increasing the degradation of tryptophan
(107). Immune escape and
immunosuppression are equally important for the existence of CSCs
(108). One of the breast CSC
marker proteins, CD47, can bind to the signal regulatory protein α
on the surface of macrophages to escape macrophage phagocytosis,
and the induction of CD47 depends on the direct regulation of
HIF-1α (82). There are two types
of tumor-associated macrophages, M1 and M2, which inhibit or
promote tumor growth, respectively. The M2 type is more common in
the tumor microenvironment and promotes tumor invasion (108,109). Hypoxia induces nuclear factor-κB
(NF-κB) and HIF-1α successively, leading to the infiltration of M2
macrophages in the tumor microenvironment and tumorigenesis
(110). As compared with normal
cells, triple-negative breast cancer cells have more
HIF-1α-specific IgG, which indicates that HIF-1α is immunogenic
(111). Treatment using a HIF-1α
vaccine recruits type I T cells to the tumor tissue and effectively
inhibits basal-like breast CSCs, which can inhibit tumor metastasis
(111). In order to adapt to
chronic hypoxia, CD8+ T cells increase the expression of
active HIF-1α and increase their own effector functions (112). The production of NANOG in hypoxic
tumor cells depends on the expression of HIF-1α, and upregulated
NANOG reduces the sensitivity of hypoxic tumor cells to the lysis
of cytotoxic T lymphocytes; however, this process is not caused by
the increased phenotype of CSCs, but NANOG increased the viability
of the tumor cells (113). At the
same time, some studies have proposed that hypoxia-induced NANOG,
which also depends on HIF-1α, promotes the self-renewal of melanoma
cells and induces Tregs and macrophages by directly regulating
TGF-β1, but whether this immunosuppression is associated with CSCs
has not been reported (114). In
summary, HIF-1α-mediated immunosuppression and immune escape are
crucial in CSCs. This common regulatory mechanism in the tumor
microenvironment gives CSCs a new definition and provides novel
ideas for tumor immunotherapy.
It is commonly known that cancer cells can
metabolize and reprogram under hypoxic conditions to complete the
conversion of oxidative phosphorylation (OXPHOS) to glycolysis to
meet their own energy needs (7,115).
However, a study recently proposed the concept of metabolic
plasticity, that is, even within the same cancer cell population,
cancer cells can complete the conversion of glycolysis to OXPHOS,
while maintaining metabolic reprogramming and energy requirements
(116). Indeed, hypoxic cells do
not metabolize all glucose to lactic acid, and non-hypoxic cells do
not metabolize all glucose to carbon dioxide and water, which makes
metabolic balance particularly important (117). Among them, HIF-1 plays a crucial
role as a regulator in metabolic reprogramming, and the role of
HIF-1 in CSC metabolism is worthy of attention.
HIF-1, as the main regulator of several glycolytic
transporters and enzymes, including glucose transporter,
monocarboxylate transporter, hexokinase and lactate dehydrogenase,
regulates glycolytic transformation (95,118).
HIF-1α also promotes the production of carbonic anhydrases, which
interact with extracellular acidification to change the pH of the
intracellular and extracellular environment of the cell, thereby
affecting the absorption of anticancer drugs and producing drug
resistance (119,120). Colon CSC clones with liver
receptor homolog-1-overexpression, a target of GATA binding protein
6, were found to have increased intracellular hypoxia, HIF-1α and
reactive oxygen species (ROS); it was further proven that
glycolysis and OXPHOS co-exist in the clones but mainly
mitochondrial respiration (121).
The knockout of CSC marker protein CD44 induces glycolysis to
OXPHOS via a complex signaling pathway involving HIF-1α (115). The loss of ATP synthase,
especially the D subunit ATP5H, leads to the accumulation of ROS in
cells and the stabilization of normoxic HIF-1α and activation of
the HIF-1α pathway, affecting mitochondrial metabolic
reprogramming, the production of stem-like ability, and therapeutic
resistance (122). The α-KG
analogue dimethyl 2-ketoglutarate allows excess succinate/fumarate
to be transferred from the mitochondria to the cytoplasm, where it
can impair prolyl hydroxylase and thus stabilize and activate
HIF-1α, eventually leading to increased glycolysis and the
acquisition of the stem-like characteristics of breast cancer cells
(118). In addition, the
upregulation of HIF-1α, under hypoxia, inhibits the expression of
mitochondrial phosphoenolpyruvate carboxykinase, which leads to a
weakened tricarboxylic acid cycle and the enrichment of fumarate,
ultimately leading to increased ROS levels and breast CSC growth
(123). In conclusion, HIF-1
appears to mainly act as an intermediate participant regulating the
complex conversion mechanism of glycolysis and OXPHOS and the
generation and maintenance of stemness in CSCs.
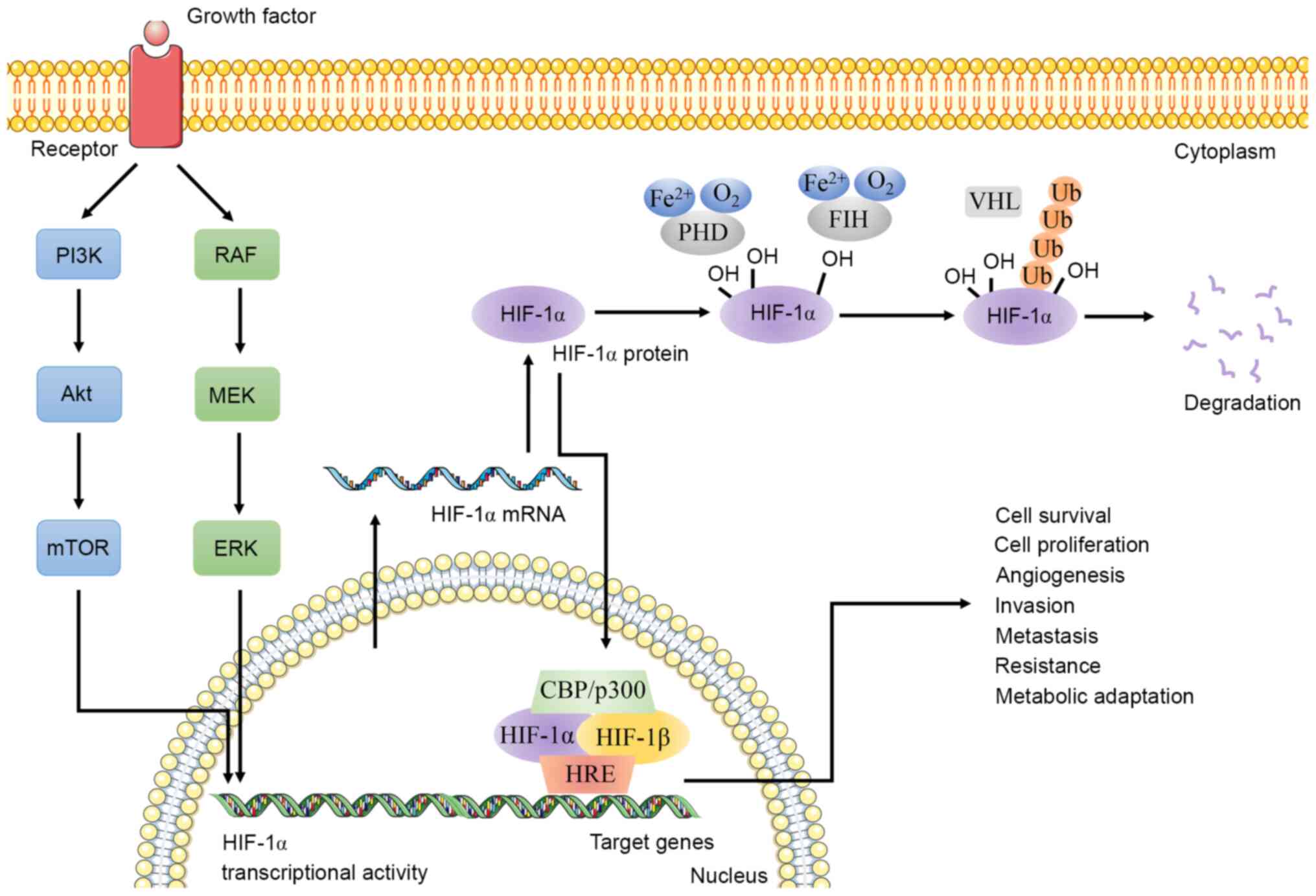
HIF-1 is regulated by multiple signaling pathways in
CSCs and also participates in regulating the characteristics of
CSCs through signaling pathways, such as the Notch, MAPK/ERK and
Wnt signaling pathways (81).
Studies have found that the PI3K/Akt/mTOR pathway maintains the
transcription, translation and biological activity of HIF-1α
(54,124). Hypoxia also induces tuftelin 1 in
a HIF-1α-dependent manner; subsequently, tuftelin1 activates the
Ca2+/PI3K/Akt pathway to induce EMT and metastasis in
hepatocellular carcinoma (50).
HIF-1α promotes the survival of prostate CSCs by inhibiting mTOR
and activating Akt phosphorylation, which may be accomplished by
the feedback regulation of PI3K via P70-S6K-mediated insulin
receptor substrate 1 phosphorylation (125). The mitochondrial autophagy
regulator NIX interacts with Ras homolog enriched in the brain to
activate mTOR/Akt/HIF signaling, and subsequently increase the
self-renewal capacity of glioma stem cells (126). The Wnt/β-catenin signaling pathway
activates the transcription of HIF-1α, inhibits the
apoptosis of hepatocellular carcinoma and promotes the occurrence
of EMT, and then triggers the metastasis of hepatocellular
carcinoma (50). Conversely, HIF-1α
maintains the stemness of esophageal squamous cell carcinoma by
activating the Wnt/β-catenin signaling pathway (127). HIF-1α induces the generation of
breast CSCs and the enhancement of self-renewal capacity by
upregulating the expression of yes-associated protein (YAP) and
tafazzin (TAZ) in the Hippo pathway (7,86).
HIF-1α, which can also function as a bidirectional co-activator of
TAZ, is recruited by TAZ to the promoter of encoding connective
tissue growth factor (CTGF) to activate the transcription of
CTGF, which is involved in promoting the onset of EMT and
maintaining the stem-like phenotype of breast CSCs (6,128).
In addition, high-mobility group box 1 (HMGB1) released from
injured or dying cells following X-ray radiation induces the
dedifferentiation of CD133- pancreatic cancer cells, and promotes
pancreatic CSC production and pancreatic cancer metastasis via the
HMGB1/toll-like receptor 2 (TLR2)/YAP/HIF-1α axis, in which
HMGB1-TLR2 promotes HIF-1α and YAP nuclear localization and HIF-1α
DNA binding ability (129). The
ROS-mediated transition of breast CSCs from a quiescent mesenchymal
state to a proliferative epithelial state is promoted by the
activation of the Notch pathway and AMP-activated protein
kinase/HIF-1α axis (130,131).
HIF-1α maintains the stemness of leukemia and
glioblastoma stem cells through the Notch signaling pathway
(81,86). Studies have also suggested that the
hypoxia/Notch1/SOX2 axis is essential for the development of
ovarian CSCs (81). The combination
of HIF-1α and notch intracellular domain activates Notch-responsive
promoters and increases the expression of Notch downstream genes,
such as Hes1 and Hey2, and Hes1 is important
in the stemness maintenance and self-renewal of leukemia stem cells
(50,81). In addition, studies have suggested
that there is negative feedback regulation of the Hes1 expression,
which is completed by the combination of Hes1 and N-boxes on the
Hes1 promoter (132).
Furthermore, HIF-1α may enhance the Notch pathway-induced Hes1
expression by inhibiting the negative feedback regulation of the
Hes1 gene, and ultimately promote the maintenance of the
stemness of mouse cholangiocarcinoma CSCs (132,133). STAT3 induced by HIF-1α through the
JAK or adenylate receptor 2B pathway can upregulate interleukin-6
and Nanog, which can maintain the CSC phenotype (81). Under hypoxic conditions, the
increased HIF-1α in glioma stem-like cells activates the
JAK1/2-STAT3 signaling pathway by promoting the production of VEGF,
a HIF-1α target, and ultimately enhances the self-renewal ability
of glioma stem-like cells (134).
HIF-1α inhibits the expression of E-cadherin and promotes EMT in
hepatocellular carcinoma via the SNAI1 signaling pathway, in which
HIF-1α binds to two HREs on the SNAI1 promoter to upregulate
SNAI1, a transcriptional inhibitor of E-cadherin (50,135).
At the same time, researchers found that HIF-1α promotes EMT in
gastric CSCs by activating the Snail signaling pathway, and the
same result was found in ovarian cancer (136,137). Dual-specificity phosphatases
(DUSPs) negatively regulate MAPK pathway activity (138). Chemotherapy induces an increase in
DUSP9 expression and a decrease in that of DUSP16 in a
HIF-1-dependent manner; it then upregulates NANOG and KLF4 through
the reduction of ERK activity and the increase of P38 activity,
respectively, finally promoting the development of breast CSCs
(138). Studies have found that
hypoxia activates the Sonic Hedgehog signaling pathway to induce
the production of CSC markers in cholangiocarcinoma cells in a
HIF-1α-dependent manner, which can be blocked by HIF-1α inhibition
(139). The expression level of
glioma-associated oncogene homolog 1 (Gli1) in the Hedgehog pathway
in prostate cancer was higher in the HIF-1α+ group,
indicating that hypoxia promotes the expression of Gli1 and that
the increased Gli1 expression is significantly associated with EMT
of prostate cancer cells (140).
Under hypoxia, upregulated HIF-1α promotes the expression of its
downstream target gene, sirtuin type 1 (SIRT1), by
activating the NF-κB signaling pathway, and increased SIRT1
promotes the maintenance of stem-like characteristics of ovarian
cancer cells (141). Of note,
studies have suggested that hypoxia-related factors switch HIF-1α
to HIF-2α by activating the NF-κB pathway to increase the
malignancy of bladder cancer and maintain the expression of stem
cell markers (142).
In addition, some signaling pathways also regulate
the HIF-1α-related complex. NF-κB-mediated inflammatory signaling
can prevent the HIF-1α transcription network by directly competing
for the binding of p300 to the promoter of HRE-encoding genes
(143). Raf Kinase Trapping to
Golgi in clear-cell renal cell carcinoma prevents the formation of
the HIF-1α/p300 complex and transactivation of HIF-1α by inhibiting
the MAPK pathway (144). Since
PI3K promotes the induction of HIF-1α levels and there is a protein
kinase B (PKB) phosphorylation site on p300, the PI3K/PKB pathway
can boost the binding of HIF-1α/phosphorylated p300 to glucokinase
gene (GK)-HRE, thereby promoting insulin-mediated GK
gene expression (145). LB-1, a
triptolide derivative, prevents the formation of the
HIF-1α/p300/p-STAT3 complex by downregulating the PI3K/Akt/mTOR
pathway that regulates HIF-1α at the translation level, and
ultimately exerts anti-tumor properties (146). In turn, the increase in the Wnt
signaling pathway mediated by CBP plays an important role in
hypoxia-induced leukemia stem cells (147,148). In fact, multiple factors in the
HIF-1α signaling pathway can also interfere with the formation of
the HIF-1α/p300 complex. CBP/p300-interacting transactivator with
an ED-rich tail 2 in hypoxia signaling was also found to prevent
p300 from recruiting to N-TAD and C-TAD, thereby inactivating
HIF-1α (149). The ferritin heavy
chain in the hypoxia signaling pathway has also been demonstrated
to interfere with p300 recruitment to HIF-1α by promoting
FIH-mediated hydroxylation of N803 (150). In prostate cancer, N-myc
downstream-regulated gene 3 regulates Akt-dependent HIF-1α hypoxia
signaling through dissociating the p300 from HIF-1α (151).
In conclusion, HIF-1α promotes self-expression by
interacting with multiple signaling pathways and participates in
the maintenance of stem-like characteristics of CSCs (Table II).
It is worth noting that the regulatory effect of
HIF-1 is also different in specific types of cancer (Table III). As HIF-1/HIF-1α is a key
factor regulating CSCs, its inhibitor may be used for adjuvant
therapy. Acriflavine, a HIF-1 inhibitor, prevents the dimerization
of HIF-1α and HIF-1β, leading to undimerized HIF-1α degradation and
inhibition of hypoxia-induced gene expression (82). Acriflavine can effectively inhibit
the stemness and growth of chronic myeloid leukemia cells (81). Digoxin, also a HIF-1 inhibitor, or
acriflavine can prevent glioblastoma stem cells from responding to
hypoxia by reducing the expression of receptor for advanced
glycation end products, which is a membrane receptor that senses
the necrotic stimulation of cells that die due to hypoxia (152). The targeted inhibition of
HIF-1 by siRNA can increase the radiotherapy sensitivity of
malignant gliomas, but the limited delivery efficiency of siRNA
still needs to be resolved (153,154). The HIF-1α vaccine is proposed to
inhibit breast cancer metastasis from the perspective of tumor
immunity (111). Not only that,
the combination of HIF-1α and co-activators can also become a
potential therapeutic target. Even under hypoxic conditions,
3-(5-hydroxymethyl-2-furyl)-1-benzyl indazole can stimulate FIH to
bind to C-TAD and reduce the recruitment of p300 to inhibit HIF-1α
in a N803 hydroxylation-independent manner (155). Bortezomib, which is used for
clinical testing of multiple tumors, has similar FIH-mediated
anticancer effects (156). Osmium
metal complex 1 can be used as an inhibitor to directly interfere
with the interaction between HIF-1α and p300 to stop the HIF-1α
expression and inhibit cell proliferation (157). Chetomin, discorhabdin L (2) and melatonin play a similar role
(153,158–160). The combination of enzalutamide, an
androgen receptor antagonist, and chetomin effectively inhibits the
growth of castration-resistant prostate cancer cells (158). TEL03, a perylene derivative, acts
as a dual targeted inhibitor of STAT3 and HIF-1α, interrupts the
phosphorylation of STAT3 and inhibits its transcription; it also
inhibits the combination of HIF-1α and CBP/p300 to induce HIF-1α
degradation, thereby inducing apoptosis and suppressing tumor
growth (161). LB-1 also prevents
the formation of the HIF-1α/p300/p-STAT3 complex by targeting both
the HIF-1α and STAT3 pathways to inhibit the growth of prostate
cancer cells (146). Minnelide, a
pro-drug of triptolide, has shown clinical prospects in a recent
phase I trial of advanced gastrointestinal malignancies, with the
phase II trial in preparation (162). In addition, the inhibition of key
enzymes and signaling pathways (Table
II), key gene knockout, as well as CSC-related immune and
metabolic regulation, all comprise potential targets (Table IV) for CSC therapy.
CSCs are a population with the potential for
differentiation and self-renewal, and participate in tumor
metastasis, recurrence and treatment resistance (81,163).
Understanding the mechanisms through which CSCs produce and
maintain stemness may help overcome the limitations of clinical
cancer treatment. Hypoxia regulates angiogenesis, tumorigenesis,
immune response, cancer recurrence and metastasis, and participates
in EMT progression and CSC production (81,82,164).
HIF-1 stably expressed under hypoxic conditions binds to HRE on the
promoter of key genes and regulates glycolysis, angiogenesis, cell
apoptosis, tissue invasion and PH regulation (165,166). HIF-1α, as the active subunit of
HIF-1, is a primary transcription regulator in hypoxic adaptive
responses (81,82). Therefore, the present review focused
more on the role of HIF-1α in CSCs instead of HIF-1, in order to
propose novel methods for the eradication of CSCs from various
perspectives, including epigenetic modification, immune response,
metabolic reprogramming, stem cell marker, and ncRNA and signaling
pathways associated with CSCs (Fig.
2).
The current tumor treatment methods, combined with
adjuvant therapy with HIF-1/HIF-1α as the core, may prevent the
recurrence and metastasis of cancer cells, and ultimately improve
the cure rate and prolong the life of patients.
Not applicable.
This review was supported by the National Natural
Science Foundation of China (grant nos. 81372835 and 81670143), and
National Key Research and Development Program of China (grant no.
2018YFA0106902).
Not applicable.
QZ wrote the manuscript. ZH, YZ, JC and WL reviewed
the manuscript critically. All authors read and approved the final
manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Lagunas-Rangel FA: Circular RNAs and their
participation in stemness of cancer. Med Oncol. 37:422020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Wang H, Cui G, Yu B, Sun M and Yang H:
Cancer stem cell niche in colorectal cancer and targeted therapies.
Curr Pharm Des. 26:1979–1993. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Sureshbabu SK, Chaukar D and Chiplunkar
SV: Hypoxia regulates the differentiation and anti-tumor effector
functions of γδT cells in oral cancer. Clin Exp Immunol. 201:40–57.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Zhang C, Samanta D, Lu H, Bullen JW, Zhang
H, Chen I, He X and Semenza GL: Hypoxia induces the breast cancer
stem cell phenotype by HIF-dependent and ALKBH5-mediated
m6A-demethylation of NANOG mRNA. Proc Natl Acad Sci USA.
113:E2047–E2056. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Xu QH, Xiao Y, Li XQ, Fan L, Zhou CC,
Cheng L, Jiang ZD and Wang GH: Resveratrol Counteracts
Hypoxia-Induced Gastric Cancer Invasion and EMT through Hedgehog
Pathway Suppression. Anticancer Agents Med Chem. 20:1105–1114.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ajduković J: HIF-1 - a big chapter in the
cancer tale. Exp Oncol. 38:9–12. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Schito L and Semenza GL: Hypoxia-inducible
factors: Master regulators of cancer progression. Trends Cancer.
2:758–770. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lu Y, Wang L, Ding W, Wang D, Wang X, Luo
Q and Zhu L: Ammonia mediates mitochondrial uncoupling and promotes
glycolysis via HIF-1 activation in human breast cancer MDA-MB-231
cells. Biochem Biophys Res Commun. 519:153–159. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Azimi I: The interplay between HIF-1 and
calcium signalling in cancer. Int J Biochem Cell Biol. 97:73–77.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hong M, Shi H, Wang N, Tan HY, Wang Q and
Feng Y: Dual effects of Chinese herbal medicines on angiogenesis in
cancer and ischemic stroke treatments: Role of HIF-1 network. Front
Pharmacol. 10:6962019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Albadari N, Deng S and Li W: The
transcriptional factors HIF-1 and HIF-2 and their novel inhibitors
in cancer therapy. Expert Opin Drug Discov. 14:667–682. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Garner KEL, Hull NJ, Sims AH, Lamb R and
Clarke RB: The milk protein alpha-casein suppresses triple negative
breast cancer stem cell activity via STAT and HIF-1alpha signalling
pathways in breast cancer cells and fibroblasts. J Mammary Gland
Biol Neoplasia. 24:245–256. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Masoud GN and Li W: HIF-1α pathway: Role,
regulation and intervention for cancer therapy. Acta Pharm Sin B.
5:378–389. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chan ST, Patel PR, Ransom TR, Henrich CJ,
McKee TC, Goey AK, Cook KM, Figg WD, McMahon JB, Schnermann MJ, et
al: Structural elucidation and synthesis of eudistidine A: An
unusual polycyclic marine alkaloid that blocks interaction of the
protein binding domains of p300 and HIF-1α. J Am Chem Soc.
137:5569–5575. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Freedman SJ, Sun ZY, Kung AL, France DS,
Wagner G and Eck MJ: Structural basis for negative regulation of
hypoxia-inducible factor-1alpha by CITED2. Nat Struct Biol.
10:504–512. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kasper LH, Boussouar F, Boyd K, Xu W,
Biesen M, Rehg J, Baudino TA, Cleveland JL and Brindle PK: Two
transactivation mechanisms cooperate for the bulk of
HIF-1-responsive gene expression. EMBO J. 24:3846–3858. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Henchey LK, Kushal S, Dubey R, Chapman RN,
Olenyuk BZ and Arora PS: Inhibition of hypoxia inducible factor
1-transcription coactivator interaction by a hydrogen bond
surrogate alpha-helix. J Am Chem Soc. 132:941–943. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Cai X, Huang Y, Zhang X, Wang S, Zou Z,
Wang G, Wang Y and Zhang Z: Cloning, characterization, hypoxia and
heat shock response of hypoxia inducible factor-1 (HIF-1) from the
small abalone Haliotis diversicolor. Gene. 534:256–264. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu CJ, Sataur A, Wang L, Chen H and Simon
MC: The N-terminal transactivation domain confers target gene
specificity of hypoxia-inducible factors HIF-1alpha and HIF-2alpha.
Mol Biol Cell. 18:4528–4542. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Soñanez-Organis JG, Peregrino-Uriarte AB,
Gómez-Jiménez S, López-Zavala A, Forman HJ and Yepiz-Plascencia G:
Molecular characterization of hypoxia inducible factor-1 (HIF-1)
from the white shrimp Litopenaeus vannamei and
tissue-specific expression under hypoxia. Comp Biochem Physiol C
Toxicol Pharmacol. 150:395–405. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Soni S and Padwad YS: HIF-1 in cancer
therapy: Two decade long story of a transcription factor. Acta
Oncol. 56:503–515. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Ju UI, Park JW, Park HS, Kim SJ and Chun
YS: FBXO11 represses cellular response to hypoxia by destabilizing
hypoxia-inducible factor-1α mRNA. Biochem Biophys Res Commun.
464:1008–1015. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Mandl M and Depping R: Hypoxia-inducible
aryl hydrocarbon receptor nuclear translocator (ARNT) (HIF-1β): Is
it a rare exception? Mol Med. 20:215–220. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Qiu Y, Shan W, Yang Y, Jin M, Dai Y, Yang
H, Jiao R, Xia Y, Liu Q, Ju L, et al: Reversal of sorafenib
resistance in hepatocellular carcinoma: Epigenetically regulated
disruption of 14-3-3η/hypoxia-inducible factor-1α. Cell Death
Discov. 5:1202019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Karagiota A, Kourti M, Simos G and Mylonis
I: HIF-1α-derived cell-penetrating peptides inhibit ERK-dependent
activation of HIF-1 and trigger apoptosis of cancer cells under
hypoxia. Cell Mol Life Sci. 76:809–825. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Singh A, Gupta S and Sachan M: Epigenetic
biomarkers in the management of ovarian cancer: Current
prospectives. Front Cell Dev Biol. 7:1822019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Urbano A, Smith J, Weeks RJ and Chatterjee
A: Gene-specific targeting of DNA methylation in the mammalian
genome. Cancers. 11:15152019. View Article : Google Scholar
|
|
28
|
Liu Q, Liu L, Zhao Y, Zhang J, Wang D,
Chen J, He Y, Wu J, Zhang Z and Liu Z: Hypoxia induces genomic DNA
demethylation through the activation of HIF-1α and transcriptional
upregulation of MAT2A in hepatoma cells. Mol Cancer Ther.
10:1113–1123. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Han ZJ, Feng YH, Gu BH, Li YM and Chen H:
The post-translational modification, SUMOylation, and cancer
(Review). Int J Oncol. 52:1081–1094. 2018.PubMed/NCBI
|
|
30
|
Bao L, Chen Y, Lai HT, Wu SY, Wang JE,
Hatanpaa KJ, Raisanen JM, Fontenot M, Lega B, Chiang CM, et al:
Methylation of hypoxia-inducible factor (HIF)-1α by G9a/GLP
inhibits HIF-1 transcriptional activity and cell migration. Nucleic
Acids Res. 46:6576–6591. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Cui CP, Wong CC, Kai AK, Ho DW, Lau EY,
Tsui YM, Chan LK, Cheung TT, Chok KS, Chan AC, et al: SENP1
promotes hypoxia-induced cancer stemness by HIF-1α deSUMOylation
and SENP1/HIF-1α positive feedback loop. Gut. 66:2149–2159. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Wang X, Liang X, Liang H and Wang B:
SENP1/HIF-1α feedback loop modulates hypoxia-induced cell
proliferation, invasion, and EMT in human osteosarcoma cells. J
Cell Biochem. 119:1819–1826. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mylonis I, Kourti M, Samiotaki M,
Panayotou G and Simos G: Mortalin-mediated and ERK-controlled
targeting of HIF-1α to mitochondria confers resistance to apoptosis
under hypoxia. J Cell Sci. 130:466–479. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Bullen JW, Tchernyshyov I, Holewinski RJ,
DeVine L, Wu F, Venkatraman V, Kass DL, Cole RN, Van Eyk J and
Semenza GL: Protein kinase A-dependent phosphorylation stimulates
the transcriptional activity of hypoxia-inducible factor 1. Sci
Signal. 9:ra562016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gong H, Gao S, Yu C, Li M, Liu P, Zhang G,
Song J and Zheng J: Effect and mechanism of YB-1 knockdown on
glioma cell growth, migration, and apoptosis. Acta Biochim Biophys
Sin (Shanghai). 52:168–179. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
El-Naggar AM, Veinotte CJ, Cheng H,
Grunewald TG, Negri GL, Somasekharan SP, Corkery DP, Tirode F,
Mathers J, Khan D, et al: Translational activation of HIF1α by YB-1
promotes sarcoma metastasis. Cancer Cell. 27:682–697. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Yu AM, Batra N, Tu MJ and Sweeney C: Novel
approaches for efficient in vivo fermentation production of
noncoding RNAs. Appl Microbiol Biotechnol. 104:1927–1937. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Panir K, Schjenken JE, Robertson SA and
Hull ML: Non-coding RNAs in endometriosis: A narrative review. Hum
Reprod Update. 24:497–515. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Liu C, Xing H, Guo C, Yang Z and Wang Y
and Wang Y: MiR-124 reversed the doxorubicin resistance of breast
cancer stem cells through STAT3/HIF-1 signaling pathways. Cell
Cycle. 18:2215–2227. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Bartoszewski R, Serocki M,
Janaszak-Jasiecka A, Bartoszewska S, Kochan-Jamrozy K, Piotrowski
A, Króliczewski J and Collawn JF: miR-200b downregulates Krüppel
Like Factor 2 (KLF2) during acute hypoxia in human endothelial
cells. Eur J Cell Biol. 96:758–766. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Byun Y, Choi YC, Jeong Y, Lee G, Yoon S,
Jeong Y, Yoon J and Baek K: MiR-200c downregulates HIF-1α and
inhibits migration of lung cancer cells. Cell Mol Biol Lett.
24:282019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen X, Wu L, Li D, Xu Y, Zhang L, Niu K,
Kong R, Gu J, Xu Z, Chen Z, et al: Radiosensitizing effects of
miR-18a-5p on lung cancer stem-like cells via downregulating both
ATM and HIF-1α. Cancer Med. 7:3834–3847. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Krutilina R, Sun W, Sethuraman A, Brown M,
Seagroves TN, Pfeffer LM, Ignatova T and Fan M: MicroRNA-18a
inhibits hypoxia-inducible factor 1α activity and lung metastasis
in basal breast cancers. Breast Cancer Res. 16:R782014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Mathieu J, Zhang Z, Zhou W, Wang AJ,
Heddleston JM, Pinna CM, Hubaud A, Stadler B, Choi M, Bar M, et al:
HIF induces human embryonic stem cell markers in cancer cells.
Cancer Res. 71:4640–4652. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Ho AS, Huang X, Cao H, Christman-Skieller
C, Bennewith K, Le QT and Koong AC: Circulating miR-210 as a novel
hypoxia marker in pancreatic cancer. Transl Oncol. 3:109–113. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sabry D, El-Deek SE, Maher M, El-Baz MA,
El-Bader HM, Amer E, Hassan EA, Fathy W and El-Deek HE: Role of
miRNA-210, miRNA-21 and miRNA-126 as diagnostic biomarkers in
colorectal carcinoma: Impact of HIF-1α-VEGF signaling pathway. Mol
Cell Biochem. 454:177–189. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Yang W, Wei J, Guo T, Shen Y and Liu F:
Knockdown of miR-210 decreases hypoxic glioma stem cells stemness
and radioresistance. Exp Cell Res. 326:22–35. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Han M, Wang Y, Liu M, Bi X, Bao J, Zeng N,
Zhu Z, Mo Z, Wu C and Chen X: MiR-21 regulates
epithelial-mesenchymal transition phenotype and hypoxia-inducible
factor-1α expression in third-sphere forming breast cancer stem
cell-like cells. Cancer Sci. 103:1058–1064. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Hermansen SK, Nielsen BS, Aaberg-Jessen C
and Kristensen BW: miR-21 is linked to glioma angiogenesis: A
co-localization study. J Histochem Cytochem. 64:138–148. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Guo Y, Xiao Z, Yang L, Gao Y, Zhu Q, Hu L,
Huang D and Xu Q: Hypoxia inducible factors in hepatocellular
carcinoma (Review). Oncol Rep. 43:3–15. 2020.PubMed/NCBI
|
|
51
|
Jiang N, Zou C, Zhu Y, Luo Y, Chen L, Lei
Y, Tang K, Sun Y, Zhang W, Li S, et al: HIF-1α-regulated miR-1275
maintains stem cell-like phenotypes and promotes the progression of
LUAD by simultaneously activating Wnt/β-catenin and Notch
signaling. Theranostics. 10:2553–2570. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Ge X, Liu X, Lin F, Li P, Liu K, Geng R,
Dai C, Lin Y, Tang W, Wu Z, et al: MicroRNA-421 regulated by HIF-1α
promotes metastasis, inhibits apoptosis, and induces cisplatin
resistance by targeting E-cadherin and caspase-3 in gastric cancer.
Oncotarget. 7:24466–24482. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Chen J, Zhou X, Xiao Q, Wang T, Shao G, Li
Y and Zhang Z: MiR-107 suppresses cell proliferation and tube
formation of Ewing sarcoma cells partly by targeting HIF-1β. Hum
Cell. 31:42–49. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Yang Z, Han Y, Cheng K, Zhang G and Wang
X: miR-99a directly targets the mTOR signalling pathway in breast
cancer side population cells. Cell Prolif. 47:587–595. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Liu CJ, Tsai MM, Hung PS, Kao SY, Liu TY,
Wu KJ, Chiou SH, Lin SC and Chang KW: miR-31 ablates expression of
the HIF regulatory factor FIH to activate the HIF pathway in head
and neck carcinoma. Cancer Res. 70:1635–1644. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Hung PS, Tu HF, Kao SY, Yang CC, Liu CJ,
Huang TY, Chang KW and Lin SC: miR-31 is upregulated in oral
premalignant epithelium and contributes to the immortalization of
normal oral keratinocytes. Carcinogenesis. 35:1162–1171. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Chen T, Yao LQ, Shi Q, Ren Z, Ye LC, Xu
JM, Zhou PH and Zhong YS: MicroRNA-31 contributes to colorectal
cancer development by targeting factor inhibiting HIF-1α (FIH-1).
Cancer Biol Ther. 15:516–523. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
He J, Jin S, Zhang W, Wu D, Li J, Xu J and
Gao W: Long non-coding RNA LOC554202 promotes acquired gefitinib
resistance in non-small cell lung cancer through upregulating
miR-31 expression. J Cancer. 10:6003–6013. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Zhu B, Cao X, Zhang W, Pan G, Yi Q, Zhong
W and Yan D: MicroRNA-31-5p enhances the Warburg effect via
targeting FIH. FASEB J. 33:545–556. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Kao SY, Tsai MM, Wu CH, Chen JJ, Tseng SH,
Lin SC and Chang KW: Co-targeting of multiple microRNAs on
factor-Inhibiting hypoxia-Inducible factor gene for the
pathogenesis of head and neck carcinomas. Head Neck. 38:522–528.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Shih JW and Kung HJ: Long non-coding RNA
and tumor hypoxia: New players ushered toward an old arena. J
Biomed Sci. 24:532017. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Dong J, Xu J, Wang X and Jin B: Influence
of the interaction between long noncoding RNAs and hypoxia on
tumorigenesis. Tumour Biol. 37:1379–1385. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Ge H, Yan Y, Wu D, Huang Y and Tian F:
Potential role of LINC00996 in colorectal cancer: A study based on
data mining and bioinformatics. OncoTargets Ther. 11:4845–4855.
2018. View Article : Google Scholar
|
|
64
|
Yang F, Zhang H, Mei Y and Wu M:
Reciprocal regulation of HIF-1α and lincRNA-p21 modulates the
Warburg effect. Mol Cell. 53:88–100. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Shen Y, Liu Y, Sun T and Yang W:
LincRNA-p21 knockdown enhances radiosensitivity of hypoxic tumor
cells by reducing autophagy through HIF-1/Akt/mTOR/P70S6K pathway.
Exp Cell Res. 358:188–198. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Shih JW, Chiang WF, Wu AT, Wu MH, Wang LY,
Yu YL, Hung YW, Wang WC, Chu CY, Hung CL, et al: Long noncoding RNA
LncHIFCAR/MIR31HG is a HIF-1α co-activator driving oral cancer
progression. Nat Commun. 8:158742017. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Zhang J, Jin HY, Wu Y, Zheng ZC, Guo S,
Wang Y, Yang D, Meng XY, Xu X and Zhao Y: Hypoxia-induced LncRNA
PCGEM1 promotes invasion and metastasis of gastric cancer through
regulating SNAI1. Clin Transl Oncol. 21:1142–1151. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Zhang W, Wang J, Chai R, Zhong G, Zhang C,
Cao W, Yan L, Zhang X and Xu Z: Hypoxia-regulated lncRNA CRPAT4
promotes cell migration via regulating AVL9 in clear cell renal
cell carcinomas. OncoTargets Ther. 11:4537–4545. 2018. View Article : Google Scholar
|
|
69
|
Yang W, Sun T, Cao J and Fan S:
Hypoxia-inducible factor-1α downregulation by small interfering RNA
inhibits proliferation, induces apoptosis, and enhances
radiosensitivity in chemical hypoxic human hepatoma SMMC-7721
cells. Cancer Biother Radiopharm. 26:565–571. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Staab A, Fleischer M, Loeffler J, Said HM,
Katzer A, Plathow C, Einsele H, Flentje M and Vordermark D: Small
interfering RNA targeting HIF-1alpha reduces hypoxia-dependent
transcription and radiosensitizes hypoxic HT 1080 human
fibrosarcoma cells in vitro. Strahlenther Onkol. 187:252–259. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Yu X, Hu L, Li S, Shen J, Wang D, Xu R and
Yang H: Long non-coding RNA Taurine upregulated gene 1 promotes
osteosarcoma cell metastasis by mediating HIF-1α via miR-143-5p.
Cell Death Dis. 10:2802019. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Augoff K, McCue B, Plow EF and
Sossey-Alaoui K: miR-31 and its host gene lncRNA LOC554202 are
regulated by promoter hypermethylation in triple-negative breast
cancer. Mol Cancer. 11:52012. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Sun Y, Jia X, Wang M and Deng Y: Long
noncoding RNA MIR31HG abrogates the availability of tumor
suppressor microRNA-361 for the growth of osteosarcoma. Cancer
Manag Res. 11:8055–8064. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Liu Y, Zhu J, Ma X, Han S, Xiao D, Jia Y
and Wang Y: ceRNA network construction and comparison of gastric
cancer with or without Helicobacter pylori infection. J Cell
Physiol. 234:7128–7140. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Sun J, Yan J, Yuan X, Yang R, Dan T, Wang
X, Kong G and Gao S: A computationally constructed ceRNA
interaction network based on a comparison of the SHEE and SHEEC
cell lines. Cell Mol Biol Lett. 21:212016. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Zhang Q, Cheng Q, Xia M, Huang X, He X and
Liao J: Hypoxia-induced lncRNA-NEAT1 sustains the growth of
hepatocellular carcinoma via regulation of miR-199a-3p/UCK2. Front
Oncol. 10:9982020. View Article : Google Scholar : PubMed/NCBI
|
|
77
|
van Schaijik B, Davis PF, Wickremesekera
AC, Tan ST and Itinteang T: Subcellular localisation of the stem
cell markers OCT4, SOX2, NANOG, KLF4 and c-MYC in cancer: A review.
J Clin Pathol. 71:88–91. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Wang Z and Zöller M: Exosomes, metastases,
and the miracle of cancer stem cell markers. Cancer Metastasis Rev.
38:259–295. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Lin YT and Wu KJ: Epigenetic regulation of
epithelial-mesenchymal transition: Focusing on hypoxia and TGF-β
signaling. J Biomed Sci. 27:392020. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Rankin EB, Nam JM and Giaccia AJ: Hypoxia:
Signaling the metastatic cascade. Trends Cancer. 2:295–304. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Hajizadeh F, Okoye I, Esmaily M, Ghasemi
Chaleshtari M, Masjedi A, Azizi G, Irandoust M, Ghalamfarsa G and
Jadidi-Niaragh F: Hypoxia inducible factors in the tumor
microenvironment as therapeutic targets of cancer stem cells. Life
Sci. 237:1169522019. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Zhang H, Lu H, Xiang L, Bullen JW, Zhang
C, Samanta D, Gilkes DM, He J and Semenza GL: HIF-1 regulates CD47
expression in breast cancer cells to promote evasion of
phagocytosis and maintenance of cancer stem cells. Proc Natl Acad
Sci USA. 112:E6215–E6223. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Thomas S, Harding MA, Smith SC, Overdevest
JB, Nitz MD, Frierson HF, Tomlins SA, Kristiansen G and Theodorescu
D: CD24 is an effector of HIF-1-driven primary tumor growth and
metastasis. Cancer Res. 72:5600–5612. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Ohnishi S, Maehara O, Nakagawa K, Kameya
A, Otaki K, Fujita H, Higashi R, Takagi K, Asaka M, Sakamoto N, et
al: hypoxia-inducible factors activate CD133 promoter through ETS
family transcription factors. PLoS One. 8:e662552013. View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Hashimoto O, Shimizu K, Semba S, Chiba S,
Ku Y, Yokozaki H and Hori Y: Hypoxia induces tumor aggressiveness
and the expansion of CD133-positive cells in a hypoxia-inducible
factor-1alpha-dependent manner in pancreatic cancer cells.
Pathobiology. 78:181–192. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Chiu DK, Zhang MS, Tse AP and Wong CC:
Assessment of stabilization and activity of the HIFs important for
hypoxia-induced signalling in cancer cells. Methods Mol Biol.
1928:77–99. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Soeda A, Park M, Lee D, Mintz A,
Androutsellis-Theotokis A, McKay RD, Engh J, Iwama T, Kunisada T,
Kassam AB, et al: Hypoxia promotes expansion of the CD133-positive
glioma stem cells through activation of HIF-1alpha. Oncogene.
28:3949–3959. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Hashimoto K, Aoyagi K, Isobe T, Kouhuji K
and Shirouzu K: Expression of CD133 in the cytoplasm is associated
with cancer progression and poor prognosis in gastric cancer.
Gastric Cancer. 17:97–106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Maeda K, Ding Q, Yoshimitsu M, Kuwahata T,
Miyazaki Y, Tsukasa K, Hayashi T, Shinchi H, Natsugoe S and Takao
S: CD133 modulate HIF-1alpha expression under hypoxia in EMT
phenotype pancreatic cancer stem-like cells. Int J Mol Sci.
17:10252016. View Article : Google Scholar
|
|
90
|
Matsumoto K, Arao T, Tanaka K, Kaneda H,
Kudo K, Fujita Y, Tamura D, Aomatsu K, Tamura T, Yamada Y, et al:
mTOR signal and hypoxia-inducible factor-1 alpha regulate CD133
expression in cancer cells. Cancer Res. 69:7160–7164. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
91
|
Li SW, Wu XL, Dong CL, Xie XY, Wu JF and
Zhang X: The differential expression of OCT4 isoforms in cervical
carcinoma. PLoS One. 10:e01180332015. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Ciccone V, Terzuoli E, Donnini S,
Giachetti A, Morbidelli L and Ziche M: Stemness marker ALDH1A1
promotes tumor angiogenesis via retinoic acid/HIF-1α/VEGF
signalling in MCF-7 breast cancer cells. J Exp Clin Cancer Res.
37:3112018. View Article : Google Scholar : PubMed/NCBI
|
|
93
|
Chen Y, Xu H, Shi Q, Gu M, Wan X, Chen Q
and Wang Z: Hypoxia-inducible factor 1α (HIF-1α) mediates the
epithelial-mesenchymal transition in benign prostatic hyperplasia.
Int J Clin Exp Pathol. 12:295–304. 2019.PubMed/NCBI
|
|
94
|
Abouhashem NS, Ibrahim DA and Mohamed AM:
Prognostic implications of epithelial to mesenchymal transition
related proteins (E-cadherin, Snail) and hypoxia inducible factor
1α in endometrioid endometrial carcinoma. Ann Diagn Pathol.
22:1–11. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
95
|
Lee SY, Ju MK, Jeon HM, Lee YJ, Kim CH,
Park HG, Han SI and Kang HS: Oncogenic metabolism acts as a
prerequisite step for induction of cancer metastasis and cancer
stem cell phenotype. Oxid Med Cell Longev. 2018:10274532018.
View Article : Google Scholar : PubMed/NCBI
|
|
96
|
Kouvaras E, Christoni Z, Siasios I,
Malizos K, Koukoulis GK and Ioannou M: Hypoxia-inducible factor
1-alpha and vascular endothelial growth factor in cartilage tumors.
Biotech Histochem. 94:283–289. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
97
|
Chabi S, Uzan B, Naguibneva I, Rucci J,
Fahy L, Calvo J, Arcangeli ML, Mazurier F, Pflumio F and Haddad R:
Hypoxia regulates lymphoid development of human hematopoietic
progenitors. Cell Rep. 29:2307–2320.e6. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
98
|
Barsoum IB, Smallwood CA, Siemens DR and
Graham CH: A mechanism of hypoxia-mediated escape from adaptive
immunity in cancer cells. Cancer Res. 74:665–674. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
99
|
Polke M, Seiler F, Lepper PM, Kamyschnikow
A, Langer F, Monz D, Herr C, Bals R and Beisswenger C: Hypoxia and
the hypoxia-regulated transcription factor HIF-1α suppress the host
defence of airway epithelial cells. Innate Immun. 23:373–380. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
100
|
Flück K, Breves G, Fandrey J and Winning
S: Hypoxia-inducible factor 1 in dendritic cells is crucial for the
activation of protective regulatory T cells in murine colitis.
Mucosal Immunol. 9:379–390. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
101
|
Sun X, Kanwar JR, Leung E, Lehnert K, Wang
D and Krissansen GW: Gene transfer of antisense hypoxia inducible
factor-1 alpha enhances the therapeutic efficacy of cancer
immunotherapy. Gene Ther. 8:638–645. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
102
|
Liu F, Wang P, Jiang X, Tan G, Qiao H,
Jiang H, Krissansen GW and Sun X: Antisense hypoxia-inducible
factor 1alpha gene therapy enhances the therapeutic efficacy of
doxorubicin to combat hepatocellular carcinoma. Cancer Sci.
99:2055–2061. 2008.PubMed/NCBI
|
|
103
|
Tartour E, Pere H, Maillere B, Terme M,
Merillon N, Taieb J, Sandoval F, Quintin-Colonna F, Lacerda K,
Karadimou A, et al: Angiogenesis and immunity: A bidirectional link
potentially relevant for the monitoring of antiangiogenic therapy
and the development of novel therapeutic combination with
immunotherapy. Cancer Metastasis Rev. 30:83–95. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
104
|
Wei J, Wu A, Kong LY, Wang Y, Fuller G,
Fokt I, Melillo G, Priebe W and Heimberger AB: Hypoxia potentiates
glioma-mediated immunosuppression. PLoS One. 6:e161952011.
View Article : Google Scholar : PubMed/NCBI
|
|
105
|
Wu A, Wei J, Kong LY, Wang Y, Priebe W,
Qiao W, Sawaya R and Heimberger AB: Glioma cancer stem cells induce
immunosuppressive macrophages/microglia. Neuro Oncol. 12:1113–1125.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
106
|
Wei J, Barr J, Kong LY, Wang Y, Wu A,
Sharma AK, Gumin J, Henry V, Colman H, Priebe W, et al:
Glioblastoma cancer-initiating cells inhibit T-cell proliferation
and effector responses by the signal transducers and activators of
transcription 3 pathway. Mol Cancer Ther. 9:67–78. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
107
|
Ye LY, Chen W, Bai XL, Xu XY, Zhang Q, Xia
XF, Sun X, Li GG, Hu QD, Fu QH, et al: Hypoxia-induced
epithelial-to-Mesenchymal transition in hepatocellular carcinoma
induces an immunosuppressive tumor microenvironment to promote
metastasis. Cancer Res. 76:818–830. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
108
|
Noman MZ, Hasmim M, Messai Y, Terry S,
Kieda C, Janji B and Chouaib S: Hypoxia: A key player in antitumor
immune response. A review in the theme: Cellular responses to
hypoxia. Am J Physiol Cell Physiol. 309:C569–C579. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
109
|
Guo X, Xue H, Shao Q, Wang J, Guo X, Chen
X, Zhang J, Xu S, Li T, Zhang P, et al: Hypoxia promotes
glioma-associated macrophage infiltration via periostin and
subsequent M2 polarization by upregulating TGF-beta and M-CSFR.
Oncotarget. 7:80521–80542. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
110
|
Yu X, Li Z, Zhang Y, Xu M, Che Y, Tian X,
Wang R, Zou K and Zou L: β-elemene inhibits radiation and
hypoxia-induced macrophages infiltration via Prx-1/NF-κB/HIF-1α
signaling pathway. OncoTargets Ther. 12:4203–4211. 2019. View Article : Google Scholar
|
|
111
|
Cecil DL, Slota M, O'Meara MM, Curtis BC,
Gad E, Dang Y, Herendeen D, Rastetter L and Disis ML: Immunization
against HIF-1α inhibits the growth of basal mammary tumors and
targets mammary stem cells in vivo. Clin Cancer Res. 23:3396–3404.
2017. View Article : Google Scholar : PubMed/NCBI
|
|
112
|
de Almeida PE, Mak J, Hernandez G,
Jesudason R, Herault A, Javinal V, Borneo J, Kim JM and Walsh KB:
Anti-VEGF treatment enhances CD8+ T-cell antitumor
activity by amplifying hypoxia. Cancer Immunol Res. 8:806–818.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
113
|
Hasmim M, Noman MZ, Lauriol J, Benlalam H,
Mallavialle A, Rosselli F, Mami-Chouaib F, Alcaide-Loridan C and
Chouaib S: Hypoxia-dependent inhibition of tumor cell
susceptibility to CTL-mediated lysis involves NANOG induction in
target cells. J Immunol. 187:4031–4039. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
114
|
Hasmim M, Noman MZ, Messai Y, Bordereaux
D, Gros G, Baud V and Chouaib S: Cutting edge: Hypoxia-induced
Nanog favors the intratumoral infiltration of regulatory T cells
and macrophages via direct regulation of TGF-β1. J Immunol.
191:5802–5806. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
115
|
Nam K, Oh S and Shin I: Ablation of CD44
induces glycolysis-to-oxidative phosphorylation transition via
modulation of the c-Src-Akt-LKB1-AMPKα pathway. Biochem J.
473:3013–3030. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
116
|
Moldogazieva NT, Mokhosoev IM and
Terentiev AA: Metabolic heterogeneity of cancer cells: An interplay
between HIF-1, GLUTs, and AMPK. Cancers (Basel). 12:8622020.
View Article : Google Scholar
|
|
117
|
Semenza GL: Hypoxia-inducible factors:
Coupling glucose metabolism and redox regulation with induction of
the breast cancer stem cell phenotype. EMBO J. 36:252–259. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
118
|
Kuo CY, Cheng CT, Hou P, Lin YP, Ma H,
Chung Y, Chi K, Chen Y, Li W, Kung HJ, et al: HIF-1-alpha links
mitochondrial perturbation to the dynamic acquisition of breast
cancer tumorigenicity. Oncotarget. 7:34052–34069. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
119
|
Deshmukh A, Deshpande K, Arfuso F,
Newsholme P and Dharmarajan A: Cancer stem cell metabolism: A
potential target for cancer therapy. Mol Cancer. 15:692016.
View Article : Google Scholar : PubMed/NCBI
|
|
120
|
Shen YA, Pan SC, Chu I, Lai RY and Wei YH:
Targeting cancer stem cells from a metabolic perspective. Exp Biol
Med (Maywood). 245:465–476. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
121
|
Lai HT, Chiang CT, Tseng WK, Chao TC and
Su Y: GATA6 enhances the stemness of human colon cancer cells by
creating a metabolic symbiosis through upregulating LRH-1
expression. Mol Oncol. 14:1327–1347. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
122
|
Song KH, Kim JH, Lee YH, Bae HC, Lee HJ,
Woo SR, Oh SJ, Lee KM, Yee C, Kim BW, et al: Mitochondrial
reprogramming via ATP5H loss promotes multimodal cancer therapy
resistance. J Clin Invest. 128:4098–4114. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
123
|
Tang K, Yu Y, Zhu L, Xu P, Chen J, Ma J,
Zhang H, Fang H, Sun W, Zhou L, et al: Hypoxia-reprogrammed
tricarboxylic acid cycle promotes the growth of human breast
tumorigenic cells. Oncogene. 38:6970–6984. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
124
|
Patra K, Jana S, Sarkar A, Mandal DP and
Bhattacharjee S: The inhibition of hypoxia-induced angiogenesis and
metastasis by cinnamaldehyde is mediated by decreasing HIF-1α
protein synthesis via PI3K/Akt pathway. Biofactors. 45:401–415.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
125
|
Marhold M, Tomasich E, El-Gazzar A, Heller
G, Spittler A, Horvat R, Krainer M and Horak P: HIF1α regulates
mTOR signaling and viability of prostate cancer stem cells. Mol
Cancer Res. 13:556–564. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
126
|
Jung J, Zhang Y, Celiku O, Zhang W, Song
H, Williams BJ, Giles AJ, Rich JN, Abounader R, Gilbert MR, et al:
Mitochondrial NIX promotes tumor survival in the hypoxic niche of
glioblastoma. Cancer Res. 79:5218–5232. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
127
|
Lv Z, Liu RD, Chen XQ, Wang B, Li LF, Guo
YS, Chen XJ and Ren XQ: HIF 1α promotes the stemness of oesophageal
squamous cell carcinoma by activating the Wnt/β catenin pathway.
Oncol Rep. 42:726–734. 2019.PubMed/NCBI
|
|
128
|
Xiang L, Gilkes DM, Hu H, Luo W, Bullen
JW, Liang H and Semenza GL: HIF-1α and TAZ serve as reciprocal
co-activators in human breast cancer cells. Oncotarget.
6:11768–11778. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
129
|
Zhang L, Shi H, Chen H, Gong A, Liu Y,
Song L, Xu X, You T, Fan X, Wang D, et al: Dedifferentiation
process driven by radiotherapy-induced HMGB1/TLR2/YAP/HIF-1α
signaling enhances pancreatic cancer stemness. Cell Death Dis.
10:7242019. View Article : Google Scholar : PubMed/NCBI
|
|
130
|
Qian J and Rankin EB: Hypoxia-induced
phenotypes that mediate tumor heterogeneity. Adv Exp Med Biol.
1136:43–55. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
131
|
Luo M, Shang L, Brooks MD, Jiagge E, Zhu
Y, Buschhaus JM, Conley S, Fath MA, Davis A, Gheordunescu E, et al:
Targeting breast cancer stem cell state equilibrium through
modulation of redox signaling. Cell Metab. 28:69–86.e6. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
132
|
Wang Y and Liu Y, Malek SN, Zheng P and
Liu Y: Targeting HIF1α eliminates cancer stem cells in
hematological malignancies. Cell Stem Cell. 8:399–411. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
133
|
Hirata H, Yoshiura S, Ohtsuka T, Bessho Y,
Harada T, Yoshikawa K and Kageyama R: Oscillatory expression of the
bHLH factor Hes1 regulated by a negative feedback loop. Science.
298:840–843. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
134
|
Almiron Bonnin DA, Havrda MC, Lee MC, Liu
H, Zhang Z, Nguyen LN, Harrington LX, Hassanpour S, Cheng C and
Israel MA: Secretion-mediated STAT3 activation promotes
self-renewal of glioma stem-like cells during hypoxia. Oncogene.
37:1107–1118. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
135
|
Zhang L, Huang G, Li X, Zhang Y, Jiang Y,
Shen J, Liu J, Wang Q, Zhu J, Feng X, et al: Hypoxia induces
epithelial-mesenchymal transition via activation of SNAI1 by
hypoxia-inducible factor −1α in hepatocellular carcinoma. BMC
Cancer. 13:1082013. View Article : Google Scholar : PubMed/NCBI
|
|
136
|
Yang SW, Zhang ZG, Hao YX, Zhao YL, Qian
F, Shi Y, Li PA, Liu CY and Yu PW: HIF-1α induces the
epithelial-mesenchymal transition in gastric cancer stem cells
through the Snail pathway. Oncotarget. 8:9535–9545. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
137
|
Du J, Sun B, Zhao X, Gu Q, Dong X, Mo J,
Sun T, Wang J, Sun R and Liu Y: Hypoxia promotes vasculogenic
mimicry formation by inducing epithelial-mesenchymal transition in
ovarian carcinoma. Gynecol Oncol. 133:575–583. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
138
|
Lu H, Tran L, Park Y, Chen I, Lan J, Xie Y
and Semenza GL: Reciprocal regulation of DUSP9 and DUSP16
expression by HIF1 controls ERK and p38 MAP kinase activity and
mediates chemotherapy-induced breast cancer stem cell enrichment.
Cancer Res. 78:4191–4202. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
139
|
Bhuria V, Xing J, Scholta T, Bui KC,
Nguyen MLT, Malek NP, Bozko P and Plentz RR: Hypoxia induced Sonic
Hedgehog signaling regulates cancer stemness,
epithelial-to-mesenchymal transition and invasion in
cholangiocarcinoma. Exp Cell Res. 385:1116712019. View Article : Google Scholar : PubMed/NCBI
|
|
140
|
Lv L, Yang Z, Ma T and Xuan Y: Gli1, a
potential cancer stem cell marker, is strongly associated with
prognosis in prostate cancer. Int J Clin Exp Pathol. 11:4957–4966.
2018.PubMed/NCBI
|
|
141
|
Qin J, Liu Y, Lu Y, Liu M, Li M, Li J and
Wu L: Hypoxia-inducible factor 1 alpha promotes cancer stem
cells-like properties in human ovarian cancer cells by upregulating
SIRT1 expression. Sci Rep. 7:105922017. View Article : Google Scholar : PubMed/NCBI
|
|
142
|
Guan Z, Ding C, Du Y, Zhang K, Zhu JN,
Zhang T, He D, Xu S, Wang X and Fan J: HAF drives the switch of
HIF-1α to HIF-2α by activating the NF-κB pathway, leading to
malignant behavior of T24 bladder cancer cells. Int J Oncol.
44:393–402. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
143
|
Mendonça DB, Mendonça G, Aragão FJ and
Cooper LF: NF-κB suppresses HIF-1α response by competing for P300
binding. Biochem Biophys Res Commun. 404:997–1003. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
144
|
Zhang Y, Jiang X, Qin X, Ye D, Yi Z, Liu
M, Bai O, Liu W, Xie X, Wang Z, et al: RKTG inhibits angiogenesis
by suppressing MAPK-mediated autocrine VEGF signaling and is
downregulated in clear-cell renal cell carcinoma. Oncogene.
29:5404–5415. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
145
|
Roth U, Curth K, Unterman TG and Kietzmann
T: The transcription factors HIF-1 and HNF-4 and the coactivator
p300 are involved in insulin-regulated glucokinase gene expression
via the phosphatidylinositol 3-kinase/protein kinase B pathway. J
Biol Chem. 279:2623–2631. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
146
|
Niu F, Li Y, Lai FF, Ni L, Ji M, Jin J,
Yang HZ, Wang C, Zhang DM and Chen XG: LB-1 exerts antitumor
activity in pancreatic cancer by inhibiting HIF-1α and Stat3
signaling. J Cell Physiol. 230:2212–2223. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
147
|
Kida A and Kahn M: Hypoxia selects for a
quiescent, CML stem/leukemia initiating-like population dependent
on CBP/catenin transcription. Curr Mol Pharmacol. 6:204–210. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
148
|
Bordonaro M and Lazarova DL: CREB-binding
protein, p300, butyrate, and Wnt signaling in colorectal cancer.
World J Gastroenterol. 21:8238–8248. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
149
|
Yoon H, Lim JH, Cho CH, Huang LE and Park
JW: CITED2 controls the hypoxic signaling by snatching p300 from
the two distinct activation domains of HIF-1α. Biochim Biophys
Acta. 1813:2008–2016. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
150
|
Jin P, Kang J, Lee MK and Park JW:
Ferritin heavy chain controls the HIF-driven hypoxic response by
activating the asparaginyl hydroxylase FIH. Biochem Biophys Res
Commun. 499:475–481. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
151
|
Lee GY, Shin SH, Shin HW, Chun YS and Park
JW: NDRG3 lowers the metastatic potential in prostate cancer as a
feedback controller of hypoxia-inducible factors. Exp Mol Med.
50:1–13. 2018. View Article : Google Scholar
|
|
152
|
Papale M, Buccarelli M, Mollinari C, Russo
MA, Pallini R, Ricci-Vitiani L and Tafani M: Hypoxia, inflammation
and necrosis as determinants of glioblastoma cancer stem cells
progression. Int J Mol Sci. 21:26602020. View Article : Google Scholar
|
|
153
|
Kessler J, Hahnel A, Wichmann H, Rot S,
Kappler M, Bache M and Vordermark D: HIF-1α inhibition by siRNA or
chetomin in human malignant glioma cells: Effects on hypoxic
radioresistance and monitoring via CA9 expression. BMC Cancer.
10:6052010. View Article : Google Scholar : PubMed/NCBI
|
|
154
|
Lo WL, Chien Y, Chiou GY, Tseng LM, Hsu
HS, Chang YL, Lu KH, Chien CS, Wang ML, Chen YW, et al: Nuclear
localization signal-enhanced RNA interference of EZH2 and Oct4 in
the eradication of head and neck squamous cell carcinoma-derived
cancer stem cells. Biomaterials. 33:3693–3709. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
155
|
Li SH, Shin DH, Chun YS, Lee MK, Kim MS
and Park JW: A novel mode of action of YC-1 in HIF inhibition:
Stimulation of FIH-dependent p300 dissociation from HIF-1{alpha}.
Mol Cancer Ther. 7:3729–3738. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
156
|
Shin DH, Chun YS, Lee DS, Huang LE and
Park JW: Bortezomib inhibits tumor adaptation to hypoxia by
stimulating the FIH-mediated repression of hypoxia-inducible
factor-1. Blood. 111:3131–3136. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
157
|
Yang C, Wang W, Li GD, Zhong HJ, Dong ZZ,
Wong CY, Kwong DW, Ma DL and Leung CH: Anticancer osmium complex
inhibitors of the HIF-1α and p300 protein-protein interaction. Sci
Rep. 7:428602017. View Article : Google Scholar : PubMed/NCBI
|
|
158
|
Fernandez EV, Reece KM, Ley AM, Troutman
SM, Sissung TM, Price DK, Chau CH and Figg WD: Dual targeting of
the androgen receptor and hypoxia-inducible factor 1α pathways
synergistically inhibits castration-resistant prostate cancer
cells. Mol Pharmacol. 87:1006–1012. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
159
|
Goradel NH, Asghari MH, Moloudizargari M,
Negahdari B, Haghi-Aminjan H and Abdollahi M: Melatonin as an
angiogenesis inhibitor to combat cancer: Mechanistic evidence.
Toxicol Appl Pharmacol. 335:56–63. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
160
|
Harris EM, Strope JD, Beedie SL, Huang PA,
Goey AK, Cook KM, Schofield CJ, Chau CH, Cadelis MM, Copp BR, et
al: Preclinical evaluation of discorhabdins in antiangiogenic and
antitumor models. Mar Drugs. 16:2412018. View Article : Google Scholar
|
|
161
|
Chen H, Guan Y, Yuan G, Zhang Q and Jing
N: A perylene derivative regulates HIF-1α and Stat3 signaling
pathways. Bioorg Med Chem. 22:1496–1505. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
162
|
McGinn O, Gupta VK, Dauer P, Arora N,
Sharma N, Nomura A, Dudeja V, Saluja A and Banerjee S: Inhibition
of hypoxic response decreases stemness and reduces tumorigenic
signaling due to impaired assembly of HIF1 transcription complex in
pancreatic cancer. Sci Rep. 7:78722017. View Article : Google Scholar : PubMed/NCBI
|
|
163
|
Wang T, Shigdar S, Gantier MP, Hou Y, Wang
L, Li Y, Shamaileh HA, Yin W, Zhou SF, Zhao X, et al: Cancer stem
cell targeted therapy: Progress amid controversies. Oncotarget.
6:44191–44206. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
164
|
Xiang L and Semenza GL: Hypoxia-inducible
factors promote breast cancer stem cell specification and
maintenance in response to hypoxia or cytotoxic chemotherapy. Adv
Cancer Res. 141:175–212. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
165
|
Yeo CD, Kang N, Choi SY, Kim BN, Park CK,
Kim JW, Kim YK and Kim SJ: The role of hypoxia on the acquisition
of epithelial-mesenchymal transition and cancer stemness: A
possible link to epigenetic regulation. Korean J Intern Med.
32:589–599. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
166
|
Lee JH, Hur W, Hong SW, Kim JH, Kim SM,
Lee EB and Yoon SK: ELK3 promotes the migration and invasion of
liver cancer stem cells by targeting HIF-1α. Oncol Rep. 37:813–822.
2017. View Article : Google Scholar : PubMed/NCBI
|