Introduction
Atrial natriuretic peptide (ANP) and brain
natriuretic peptide (BNP) are members of the NP family, a group of
structurally similar but genetically distinct peptides that are
involved in important processes, such as natriuresis, diuresis,
vasodilation, and metabolic regulation (1). ANP and BNP act by binding to the
natriuretic peptide receptor A (NPRA), also called NPR1 or guanylyl
cyclase A. Activation of NPRA could elevate the levels of the
second messenger cyclic guanosine monophosphate (cGMP), which, in
turn, mediates physiological and pathophysiological processes
through cGMP-dependent protein kinase (2). Thus, NPRA signalling represents a
classical signalling pathway mediating the functions of various
types of organs and tissues, including the heart and blood vessels.
Importantly, weakened signals of NPs were observed in
cardiometabolic diseases, which might represent molecular targets
for novel therapeutic approaches. To date, however, the underlying
mechanisms governing these phenomena remain unclear (3).
Circular RNA (circRNA) has long been considered to
be the outcome of ‘splicing errors’ (4). However, with advances in
high-throughput sequencing, circRNA is now recognised as a class of
RNA produced by the alternative shearing of pre-mRNA (5). Because circRNA molecules lack free
ends, they are resistant to exonucleases and can, as such, escape
normal RNA degradation. These characteristics imply that circRNA
molecules could serve many potential roles, possibly acting as
transcription regulators (6),
microRNA (miRNA) sponges, and protein sponges, thereby modulating
various biological processes (7–11).
Moreover, circRNA molecules may also play a critical role in
several cellular functions that participate in the pathogenesis and
progression of diseases. For example, it has been widely reported
that circRNA is a key regulatory factor of cardiovascular diseases,
such as myocardial infarction (12), atherosclerosis (13), cardiomyopathy (14), and cardiac fibrosis (15,16).
The results of previous studies have suggested that circRNA
molecules may represent new potential therapeutic targets and
biomarkers for cardiovascular diseases (17,18).
To the best of our knowledge, the functional
relationship between NPs and circRNA has not been documented. In
the present study, the expression profile of circRNA and mRNA
molecules in the myocardium of mice with cardiac-specific deletion
of NPRA was examined, in order to characterise novel underlying
mechanisms of heart disease induced by the inactivation of
natriuretic signalling, based on circRNA networks.
Materials and methods
Generation of mice with
cardiac-specific deficiency in NPRA
All experimental protocols included in this
manuscript were approved by The Institutional Animal Care and Use
Committee of Xi'an Medical University. The NPRA conditional
knockout mouse model (NprAflox/flox) was generated by
the Shanghai Model Organisms Centre, Inc. This model was developed
using the CRISPR/Cas9 system against the C57BL/6 mouse background,
as previously described (19,20).
An NprA donor vector containing flox sites flanking exon 2 of the
NprA gene was constructed. Two single guide RNA (sgRNA) sites
targeting intron 1 and intron 2 were transcribed in vitro.
The sequences of the sgRNA target sites were
5′-GAGACGAGGACCAAGACTGCAGG-3′ and 5′-GCACAGGGGCCGTCTCATGCAGG-3′.
The donor vector with two sgRNAs and Cas9 mRNA was microinjected
into C57BL/6 fertilized eggs. F0-generation mice with
positive homologous recombination were identified by long PCR. The
primers used for correct 5′ homology arm recombination were
5′-GACTTCAAATCCCAGCTCACAACC-3′ and 5′-CAAAGATCGAATGGAGCCCTGTGT-3′.
The primers used for correct 3′ homology arm recombination were
5′-TATCTTGCGGCCCTCTGACTGTAT-3′ and 5′-ACTGGCCCTCCGTGGTTAGCA-3′. The
identity of the PCR products were confirmed by sequencing. NprA
flox heterozygous mice were obtained by mating F0
generation mice with wild-type C57BL/6 mice. The genotypes of
F1-generation NprAflox/flox mice were
identified using the same genotyping methods as
F0-generation mice.
To achieve tissue-specific deletion of the NPRA
gene, NprAflox/flox mice were crossed with Myh6-Cre
transgenic mice (Shanghai Model Organisms Centre, Inc.) to obtain
NprAflox/flox/Myh6-Cre mice. Both the NPRA−/−
and NPRA−/+ mice were identified by agarose gel
electrophoresis and ethidium bromide staining (Fig. S1).
Isolation of myocardium
All mice were euthanised by inhaling oxygen with 5%
isoflurane at a rate of 1 l/min. The mice were confirmed to be
deeply anaesthetized if they were immobile for 1 min. A 25% volume
of CO2 gas was constantly flowed into the chamber to
bring the mice to the point of clinical death, which was confirmed
by cessation of the heartbeat and respiration. The hearts were
subsequently removed to obtain the myocardium. Briefly, the cardiac
tissue was isolated and placed in ice-cold PBS and all fat and
connective tissue were removed. The samples were stored in −80°C
until use.
Reverse transcription-quantitative
(RT-q) PCR validation
To measure cardiac mRNA expression of NPRA in mice,
the myocardium samples (n=5 in each group) were collected and
washed in PBS. Total RNA was then isolated from myocardium samples
using TRIzol® (Thermo Fisher Scientific, Inc.) from
myocardium samples. RNA concentration was then measured by the
NanoDrop™ ND-1000 (NanoDrop™ Technologies; Thermo Fisher
Scientific, Inc.). The RNA samples were reverse transcribed into
cDNA using SuperScript™ III Reverse Transcriptase (Invitrogen;
Thermo Fisher Scientific, Inc.). Briefly, the mixture contained
RNA, Oligo (dT)18 (Takara Bio, Inc.), Random 6 mers
(Takara Bio, Inc.), dNTPs Mix (HyTest Ltd.), RNase free
dH2O and was placed in a 65°C water bath for 5 min and
on ice for 2 min. Following a short centrifugation in a palm
centrifuge for 5–10 sec to make the mixture to gather at the bottom
of the centrifuge tube, the RT reaction solution including 5X
First-Strand Buffer (Invitrogen; Thermo Fisher Scientific, Inc.),
DTT (Beijing Solarbio Science & Technology Co., Ltd.), RNase
Inhibitor (Epicentre; Illumina, Inc.) and SuperScript™ III Reverse
Transcriptase (Invitrogen; Thermo Fisher Scientific, Inc.) was
sequentially added and the mixture was kept at 37°C for 1 min. The
solution was incubated at 50°C for 60 min, then the reaction was
terminated by incubation at 70°C for 15 min. The cDNA obtained was
diluted 1:10 and stored at −20°C until use. qPCR was performed
using TransStart™ SYBR Green qPCR Supermix (Takara Bio, Inc.) to
determine the expression levels of NPRA. β-actin was included as an
internal normalization reference gene for mRNA expression. All the
PCR products were subjected to electrophoresis on a 2% agarose gel
and visualized using ethidium bromide staining to confirm the
presence of a single band of the expected size. The amplification
efficiencies of qPCR for the target gene and the endogenous control
were approximately equal and were calculated through a dilution
series of cDNA. The qPCR reaction system included 2 µl cDNA, 5 µl
2C Master mix (Arraystar, Inc.) and 0.5 µl of each primer (10 µM
solution) and diethylpyrocarbonate-treated water was added for a
total volume of 10 µl. All qPCR reactions were performed in
triplicate, including template-free controls. The thermocycling
conditions consisted of an initial denaturation at 94°C for 3 min,
followed by 40 cycles of denaturation at 94°C for 10 sec, and
annealing at 56°C 20 sec, then extension at 72°C for 30 sec, and a
final extension at 72°C for 5 min The data were analysed using the
comparative Cq (ΔΔCq) method (21).
To confirm the expression of differentially
expressed genes (DEGs), including mRNAs and circRNAs, briefly, the
sequences of candidate mRNAs and circRNAs were obtained from the
CircInteractome database (http://circinteractome.nia.nih.gov/). The GAPDH and
β-actin housekeeping genes were used as internal controls. The
reverse transcription was performed as described above. The
thermocycling conditions were as follows: i) Initial denaturation
at 95°C for 10 min; and ii) 40 cycles of denaturation at 95°C for
10 sec, and annealing at 60°C for 1 min. A melting curve analysis
was performed after the amplification was complete by cooling the
reaction to 60°C and then heating slowly to 99°C. The amplification
products were analysed by 2% agarose gel electrophoresis and
ethidium bromide staining. The relative quantification of the
expression of the investigated genes was performed using a standard
curve.
All primers were designed using Primer 5.0 and
synthesized using a ViiA 7 Real-time PCR System (Applied
Biosystems; Thermo Fisher Scientific, Inc.). Primer sequences are
presented in Tables I and II.
 | Table I.Reverse transcription-quantitative
PCR primers used to confirm the expression of mRNAs. |
Table I.
Reverse transcription-quantitative
PCR primers used to confirm the expression of mRNAs.
| Target genes | Primer
sequences | Annealing
temperature, °C | Product length,
bp |
|---|
| β-actin
(mouse) | F: 5′
GCAGGAGTACGATGAGTCCG 3′ |
|
|
|
| R:
5′ACGCAGCTCAGTAACAGTCC3′ | 60 | 247 |
| NPRA | F: 5′
GGCTGTGAAACGTGTGAACC 3′ |
|
|
|
| R: 5′
GTCGGTACAAGCTCCCACAA 3′ | 60 | 121 |
| Rhobtb1 | F: 5′
AAAGCGCCAACCGTGAG 3′ |
|
|
|
| R: 5′
CTGCTTGGTGTAGAGGTATTCC 3′ | 60 | 83 |
| Zbtb4 | F: 5′
CTGTGAGAAGGTGTTTGCCC 3′ |
|
|
|
| R: 5′
GCTCCCTGACTGTAGGTCTTGT 3′ | 60 | 279 |
| Tfpi | F: 5′
GATTCGTGTACGGTGGCT 3′ |
|
|
|
| R: 5′
GGCACTTTGGGAGACTGG 3′ | 60 | 199 |
| S100a9 | F: 5′
TGGTGGAAGCACAGTTGG 3′ |
|
|
|
| R: 5′
TTGCCATCAGCATCATACAC 3′ | 60 | 135 |
| Hp | F: 5′
GAGGCAAGAGAGGTCCACGAT 3′ |
|
|
|
| R: 5′
CAAGTGCTCCACATAGCCGTT 3′ | 60 | 160 |
| Cdkn1a | F: 5′
TTGTCGCTGTCTTGCACTC 3′ |
|
|
|
| R: 5′
GTGGGCACTTCAGGGTTT 3′ | 60 | 159 |
| Cbfa2t2 | F: 5′
GATGCCAACGGGCTCTAA 3′ |
|
|
|
| R: 5′
TGCTTCCTGCCAACTTCA 3′ | 60 | 138 |
| Agpat2 | F: 5′
CAGATCGCCAAGCGTGAG 3′ |
|
|
|
| R: 5′
CCCATTGTCGTTGCGTGTA 3′ | 60 | 189 |
| Nutf2 | F: 5′
AACCCAACTAGGCGCAATT 3′ |
|
|
|
| R: 5′
GCCGTGATGCTATGCTGAA 3′ | 60 | 132 |
| Alg12 | F: 5′
AGGGCATATCTTGGTGAA 3′ |
|
|
|
| R: 5′
TCTTGTCATACCTCCAGTCA 3′ | 60 | 203 |
| Gm20521 | F: 5′
CTTTGGTGGGACAAGTGC 3′ |
|
|
|
| R: 5′
TGTAGCTCCTTTAGCTTCTCA 3′ | 60 | 151 |
| Gbp10 | F: 5′
TTGTTGGATGGTCCCGTACT 3′ |
|
|
|
| R: 5′
GTGATTCTGTCCCGCCAG 3′ | 60 | 62 |
 | Table II.Reverse transcription quantitative
PCR primers used to confirm the expression of circRNAs. |
Table II.
Reverse transcription quantitative
PCR primers used to confirm the expression of circRNAs.
| Target genes | Primer
sequences | Annealing
temperature, °C | Product length,
bp |
|---|
| β-actin
(mouse) | F: 5′
GTACCACCATGTACCCAGGC3′ | 60 | 247 |
|
| R:
5′AACGCAGCTCAGTAACAGTCC3′ |
|
|
| GAPDH (mouse) | F: 5′
CACTGAGCAAGAGAGGCCCTAT 3′ | 60 | 144 |
|
| R: 5′
GCAGCGAACTTTATTGATGGTATT 3′ |
|
|
|
mmu_circRNA_43449 | F: 5′
CCTTCAGGGACAAAAAGGACAT 3′ | 60 | 159 |
|
| R: 5′
CAGCTTATCCGTTGCTCCAAT 3′ |
|
|
|
mmu_circRNA_36265 | F: 5′
AGCAAGTCGGCAAAAGGC 3′ | 60 | 129 |
|
| R: 5′
TCCACAGACACTGAAAGCTGAT 3′ |
|
|
|
mmu_circRNA_30261 | F: 5′
CTCACGCTACCCTACACTCTGG 3′ | 60 | 85 |
|
| R: 5′
CACACACCTGGACACATCTGAAT 3′ |
|
|
|
mmu_circRNA_36266 | F: 5′
GAATAAAAACAAAAAGTTAGAGAAGC 3′ | 60 | 139 |
|
| R: 5′
ATGAGATAAGAAATCAGGCCCT 3′ |
|
|
|
mmu_circRNA_32945 | F: 5′
TCTACCAGATCAACGTCCTCC 3′ | 60 | 94 |
|
| R: 5′
CAGCACACATTTAAGCACCAG 3′ |
|
|
|
mmu_circRNA_22217 | F: 5′
AAGGAGGAAAAGCCCCAGACT 3′ | 60 | 111 |
|
| R: 5′
GACATCAGAGAGCACACACCGT 3′ |
|
|
|
mmu_circRNA_005865 | F: 5′
GAAGCCAGGCTGTGTGTGTTA 3′ | 60 | 84 |
|
| R: 5′
GGTGATTCTCTTGGACCCTTG 3′ |
|
|
|
mmu_circRNA_42481 | F: 5′
TGGCATGTACAGTTTCTGAGTTTT 3′ | 60 | 71 |
|
| R: 5′
CTTGATGGAGGAGCAGGTTTG 3′ |
|
|
|
mmu_circRNA_19474 | F: 5′
AAAAACTCATTAATTGGGGTGGT 3′ | 60 | 55 |
|
| R: 5′
CAGCCGTCACGCATCTCAT 3′ |
|
|
|
mmu_circRNA_19519 | F: 5′
TAGACCATTCCAGTTTCCACAG 3′ | 60 | 128 |
|
| R: 5′
TTACACCCTTCAACCTACCCAT 3′ |
|
|
|
mmu_circRNA_19029 | F: 5′
ATGCCTGCTTCCTCAAAAACC 3′ | 60 | 134 |
|
| R: 5′
TACCTTACCTGGAACCAAACTCTC 3′ |
|
|
|
mmu_circRNA_29619 | F: 5′
TGGTGGTGTTTGTGTCTGTGAT 3′ | 60 | 108 |
|
| R: 5′
CATGACCAGTTCTTGGGCAGT 3′ |
|
|
|
mmu_circRNA_25320 | F: 5′
AAGAGAGTATAATGATTTTCTGGAAG 3′ | 60 | 103 |
|
| R: 5′
TCCCACACTCAGGACAGTTTC 3′ |
|
|
|
mmu_circRNA_26033 | F: 5′
GGATGGCTTCAAAGTGTGTATT 3′ | 60 | 85 |
|
| R: 5′
TCCTGTGATTCCACCTGTGC 3′ |
|
|
Western blotting
For the preparation of tissue lysates, hearts from
mice were dissected and washed with ice-cold PBS, then homogenized
with RIPA buffer (Beyotime Institute of Biotechnology) containing
150 mmol/l NaCl as previously described (22). Cell lysates were prepared for
western blotting as previously described (23). Protein concentrations were
determined using a BCA protein assay kit (Beyotime Institute of
Biotechnology). After adjustment of protein concentration, the
lysates were boiled in SDS loading buffer for 5 min then resolved
by 10% SDS-PAGE and 20 µg of protein was loaded per lane. Gels were
then transferred to an Immobilon-P PVDF membrane (EMD Millipore),
which was blocked by 5% non-fat dried milk in TBST (Tris Buffer
Saline with 0.1% Tween 20) buffer for 2 h at room temperature and
incubated with primary antibodies against mouse NPRA (cat. no.
LS-C264634; 1:500; LifeSpan BioSciences, Inc.) and GAPDH (cat. no.
2118, 1:1,000; Cell Signaling Technology, Inc.) in TBS + 0.1%
Tween-20 (TBST) and 5% non-fat dry milk overnight at 4°C. Membranes
were subsequently washed in 5% milk/TBST and incubated with
HRP-conjugated secondary antibody for 2 h at room temperature.
Protein bands were visualised using a chemiluminescence detection
kit (EMD Millipore).
RNA sequencing (RNA-seq)
Total RNA from each sample was quantified by agarose
gel electrophoresis and further verified and quantified by Nanodrop
ND-1000 spectrophotometer (NanoDrop Technologies; Thermo Fisher
Scientific, Inc.). Agarose electrophoresis was used to determine
the integrity of total RNA samples. A total of 1–2 µg total RNA
from each sample was used for RNA-seq library preparation. Briefly,
rRNA was removed from the total RNA with a RiboZero Magnetic Gold
kit (cat. no. MRZG12324; Epicentre; Illumina, Inc.). The
rRNA-depleted samples were then prepared for sequencing using the
KAPA-stranded RNA-Seq Library Prep kit for Illumina (cat. no.
KK8400; Kapa Biosystems; Roche Diagnostics). The library
preparation procedure included the following steps: i) RNA
molecules fragmentation; ii) first strand cDNA synthesis by reverse
transcription; iii) second-strand cDNA synthesis dUTP instead of
dTTP (with no Uracil-DNA glycosylase treatment); iv) end repair and
3′ adenylation of the double-stranded cDNA; v) ligation of the
Illumina adapter; vi) PCR amplification; and vii) purification
using magnetic beads (cat. no. KK8000; Kapa Biosystems; Roche
Diagnostics).
The quality of the libraries was evaluated using an
Agilent 2100 Bioanalyzer, in order to check the concentration, the
fragment size distribution (400–600 bp), and levels of adapter
dimer contamination. The amount was quantified using the absolute
qPCR method (24). For sequencing,
the barcoded libraries were mixed in equimolar amounts. Sequencing
was carried out using TruSeq SR Cluster kit v3-cBot-HS (150 cycles;
paired end, cat. no. GD-401-3001; Illumina, Inc.) on an Illumina
HiSeq 4000 platform according to the manufacturer's instructions.
The sequencing data are available in the Gene Expression Omnibus
(GEO) database (https://www.ncbi.nlm.nih.gov/geo) under accession no.
GEO140678.
Bioinformatics analysis
As previously reported (25), image analysis and base calling were
performed using the Off-Line Base Caller software v1.8 (Illumina,
Inc.). Sequence quality was examined using FastQC software v0.11.7
(26). The cutadapt software
(v1.2.1; http://code.google.com/p/cutadapt/) was then used to
trim the fragments with a 5′,3′-adaptor and to filter the reads ≤20
bp. The next step was alignment of the reads to the reference
genome using Hisat 2 software (2.1.0; http://daehwankimlab.github.io/hisat2/) (27). The average reads was 35,871,134.2
and 88.9% of the clean reads were aligned. According to a
previously published method (28),
the transcript abundances were estimated with StringTie v1.3.3
(http://ccb.jhu.edu/software/stringtie/), and the
number of identified genes and transcripts per group was calculated
based on the mean of fragments per kilobase of exon model million
reads mapped (FPKM) in each group, and the transcripts with FPKM
≥0.5 were retained. The ballgown R package v 3.5.0 (www.bioconductor.org/packages/release/bioc/html/ballgown.html)
was used to analyse the differentially expressed genes (DEGs) and
transcripts by comparing the NPRA−/− group to the
NPRA+/+ group. The adjusted P-values of DEGs were
obtained by multiple checking and correction using the Benjamini
and Hochberg method (29).
Filtering was based on an absolute fold change cut-off >1.5,
adjusted P≤0.05 and mean FPKM ≥0.5. CPAT v1.2.4 (http://lilab.research.bcm.edu/cpat/) was then
used to determine whether transcript sequences were coding or
non-coding. Principal component analysis (PCA) and Pearson
correlation analysis were carried out on gene expression levels.
Hierarchical clustering, Gene Ontology (GO) (30), Kyoto Encyclopedia of Genes and
Genomes (KEGG) (31) pathway,
scatter plot and volcano plot analysis were performed with the
differentially expressed genes in the R v3.5.0 (http://www.r-project.org/), Python v2.7 or shell
environment for statistical computing and graphics.
circRNA array analysis
Total RNA from the samples of both groups was
quantified. As described previously (32,33),
sample preparation and microarray hybridization were performed
according to the manufacturer's protocols (Arraystar, Inc.).
Briefly, total RNA from each sample was digested with RNase R
(Epicentre, Inc.) to remove linear RNA. The resulting
circRNA-enriched samples were then amplified and labelled with
fluorescently labelled nucleotides using an Arraystar Super RNA
labeling kit (Arraystar, Inc.) utilizing a random priming method
according to Arraystar Super RNA labeling protocol. The labelled
circRNA molecules were hybridized to the Arraystar Mouse circRNA
Array v2 (8×15 K, Arraystar). Microarray slides were scanned with a
G2505C Agilent Scanner (Agilent Technologies, Inc.).
The array images were analysed by Agilent Feature
Extraction software (version 11.0.1.1; Agilent Technologies, Inc.).
Quantile normalization and subsequent data processing were
performed using with the limma package (version 3.11; http://bioconductor.org/packages/release/bioc/html/limma.html).
Differentially expressed circRNA molecules (DE-circRNAs) with
statistical significance in expression levels between the two
groups were identified using a volcano plot. The circRNAs that were
differentially expressed between the two groups were filtered by
fold change filtering (FC>2). Hierarchical clustering was
subsequently conducted to illustrate the distinguishable circRNA
expression profiles among samples. The raw data of microarray data
are available in the GEO (https://www.ncbi.nlm.nih.gov/geo/) under accession no.
GSE140798.
Prediction of circRNA-miRNA-mRNA
network
As described previously (34,35),
the circRNA-miRNA interaction was predicted using the Arraystar
miRNA target prediction software (version 1.0, Arraystar Inc.)
based on TargetScan44 and miRanda, and the DE-circRNAs were
annotated in detail with circRNA-miRNA interaction information.
TargetScan (version 7.2; http://www.targetscan.org/), miRanda (http://www.microrna.org/), and miRDB (http://www.mirdb.org/) were used to predict the
microRNA-mRNA interactions that match the seed region of mouse
miRNA sequences obtained from miRBase (http://www.mirbase.org/). In addition, if a miRNA
molecule could bind to both the circRNA and target mRNA, the
targeted circRNA was defined as a candidate competing endogenous
RNA (ceRNA) for the gene, and the circRNA-miRNA-mRNA interactions
thus represent candidate ceRNA pairs (circRNA connected with miRNA
and miRNA connected with mRNA). In addition, circRNA and genes in
candidate ceRNA pairs are required to have the same direction of
change of expression level (upregulation or downregulation) in DEG
analysis because of a positive correlation in the expression of
ceRNA pairs (36). Then, the
DE-circRNAs and differentially expressed mRNA transcripts
(DE-mRNAs) that were validated by RT-qPCR were included within the
ceRNA network, while all others were excluded. The
circRNA-miRNA-mRNA interaction network was constructed and visually
displayed using Cytoscape (version 3.8.1; http://www.cytoscape.org/) based on the data analysis
results.
Statistical analysis
Statistical analysis was performed using R
statistical package (37) (v3.5.0;
http://www.R-project.org/), SPSS v19.0 (SPSS,
Inc.) and Prism 6 (GraphPad Software). A total of five samples were
included in each group. DEGs and DE-circRNAs were obtained by
comparing the NPRA−/− group and the NPRA+/+
group. Student's t-test was used to obtain P-values. The adjusted
P-values of DEGs were obtained by multiple checking and correction
based on the Benjamini and Hochberg method (29). Filtering was based on absolute FC
>1.5 in DEGs and FC >2 in DE-circRNAs, adjusted P-value ≤0.05
and mean FPKM ≥0.5 were employed for filtering. RT-qPCR,
statistical significance was assessed with Student's t-test. Data
are presented as the mean ± SEM of three experimental repeats.
P<0.05 was considered to indicate a statistically significant
difference.
Results
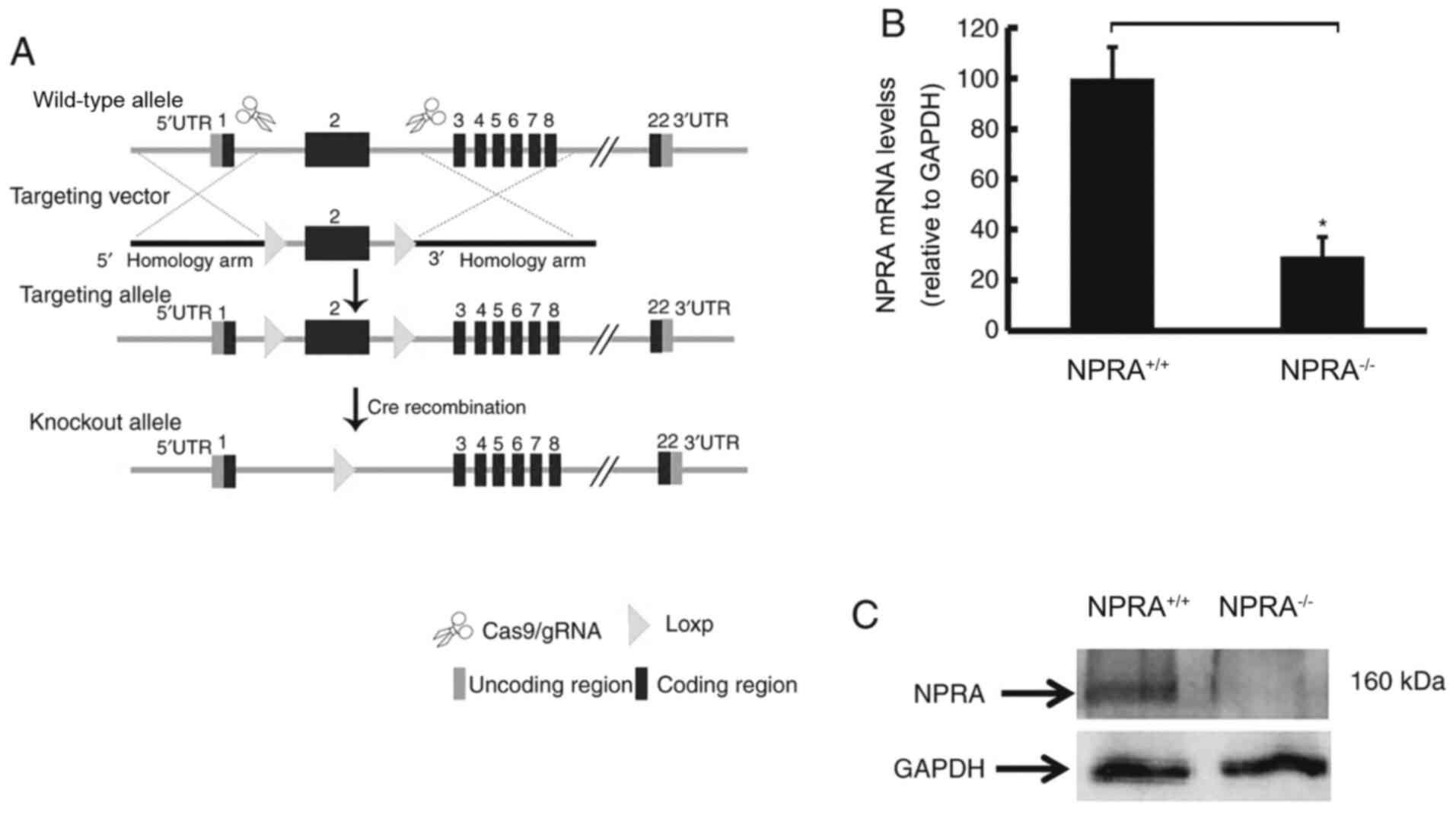
NPRA was specifically knocked out in
the myocardium of mice
To obtain a Cre recombinase-mediated, loxP-directed
deletion of the NprA gene, a targeting construct was designed in
which the second exon of NprA was flanked by two loxP sites
(Fig. 1A). In
NprAflox/flox/Myh6-Cre mice the NPRA gene was
specifically disrupted in the myocardium. Following Cre
recombination, exon 2 of NprA was deleted, leading to gene reading
frame transcoding and protein function loss. The loss of cardiac
expression of NprA was determined by RT-qPCR (Fig. 1B) and western blotting (Fig. 1C). Homozygous knockout
(NPRA−/−) and heterozygous (NPRA−/+) mice
were identified by PCR genotyping and agarose gel electrophoresis
staining (Fig. S1).
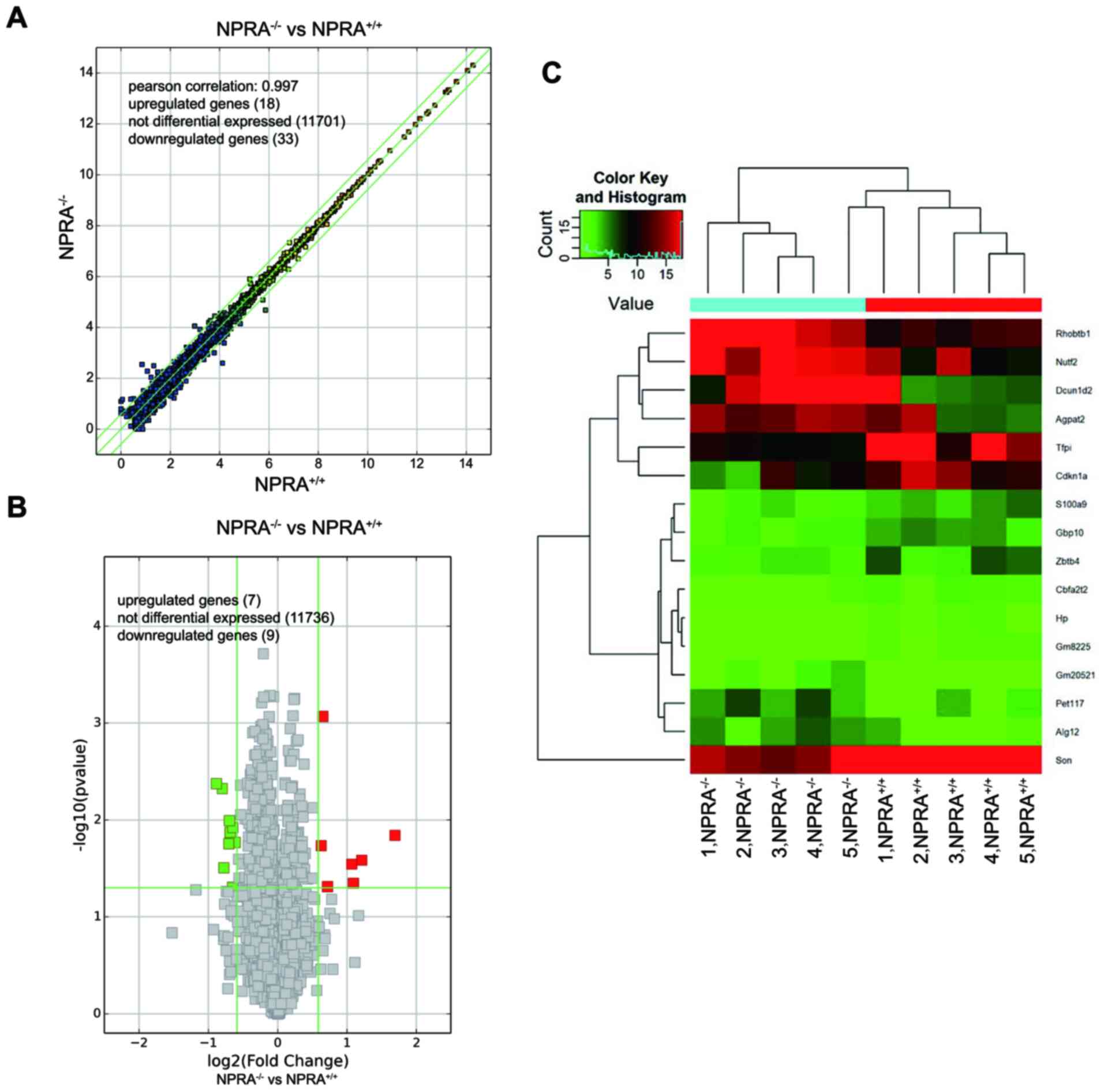
Differentially expressed mRNAs
associated with metabolic processes are present in cardiac-specific
NPRA−/− mice
The transcriptional profiles of NPRA−/−
and NPRA+/+ mice were determined using RNA-seq. The
resulting scatter plot (Fig. 2A),
volcano plot (Fig. 2B), and
hierarchical clustering map (Fig.
2C) highlighted significant (DEGs) between NPRA−/−
and NPRA+/+ mice (n=5 in each group). Scatter plots are
commonly used to display differences in gene expression between two
datasets and to assess trends in population distribution. The
values corresponding to the × and y-axes in the scatter plot are
the normalized, log2-scaled FPKM of NPRA+/+
and NPRA−/− mice, respectively. Symbols above the upper
green line and below the lower green line indicate fold change
>1.5 between the between NPRA−/− and
NPRA+/+ mice. The volcano plot displays the relationship
between the fold change of each DEG and the associated P-value in
each sample group. Data clusterisation successfully discriminated
NPRA−/− from NPRA+/+ samples. All 10 samples
were included for further analysis. However, only very few of these
genes were considered to be differentially expressed with an
adjusted P<0.05 and fold change >1.5. Indeed, seven were
upregulated, and nine were downregulated.
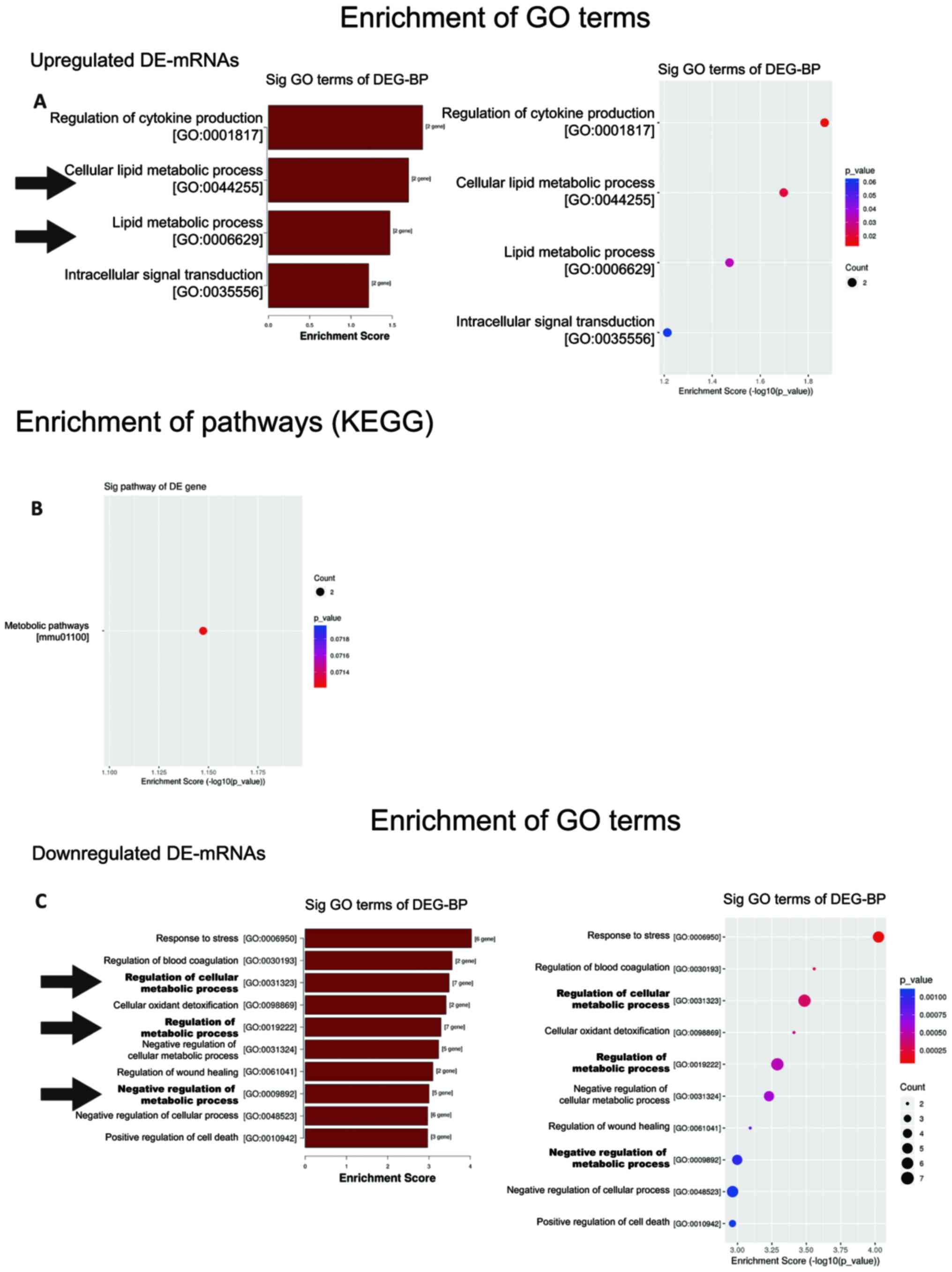
GO enrichment analysis was used to identify
significantly enriched GO terms associated with the potential
functional roles of DEGs through three categories: Biological
process (BP), molecular function (MF), and cellular component (CC).
Enriched terms with significant differences (P<0.05) were
selected and ranked by enrichment score (-log10-scaled
P-value). A total of 4 BP terms, 2 MF terms, and 11 CC terms were
significantly enriched in NPRA−/− mice, relative to
NPRA+/+ mice (Fig.
S2A). Intriguingly, 2 out of 4 significantly enriched BP terms
were related to the ‘lipid metabolic process’ (Fig. 3A). In KEGG pathway analysis, only a
single significant pathway, ‘metabolic pathways’, was observed
(Fig. 3B).
Among the downregulated genes, 99 BP terms, 13 MF
terms, and 11 CC terms were found by GO analysis (Fig. S2B). The top 10 BP terms in the
NPRA-deficient group ranked by enrichment score are shown in
Fig. 3C. Notably, three of the top
10 BP terms were related to ‘metabolic process’. These three terms
are regulation of cellular metabolic process (GO: 0031323),
regulation of metabolic process (GO: 0019222) and negative
regulation of metabolic process (GO: 0009892).
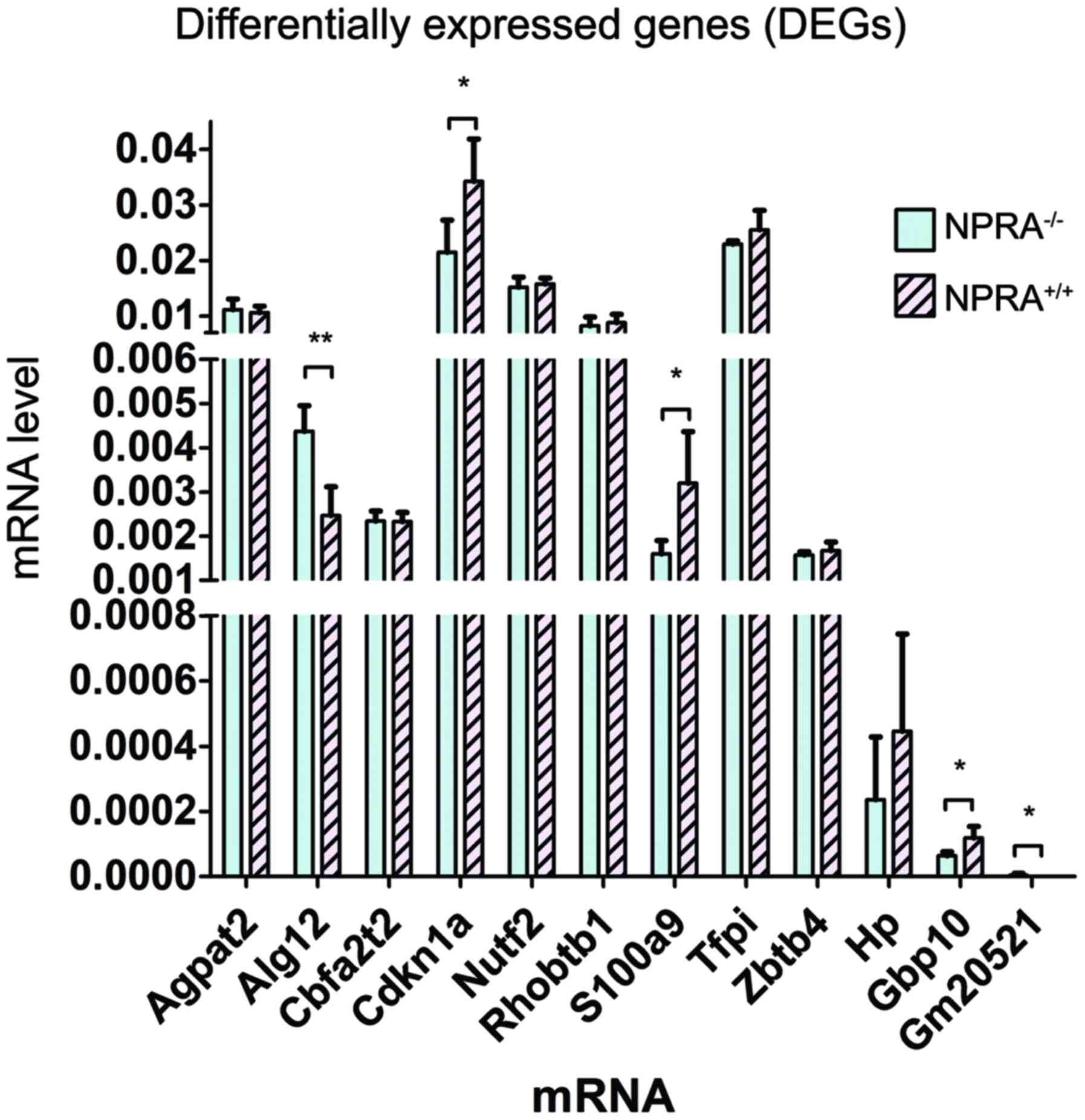
To validate the RNA-seq results and confirm the
differential gene expression profile, a subset of 12 genes which
involving with metabolic process terms we got in GO analysis,
including Agpat12, Alg12, Cbfa2t2, Cdkn1a, Nutf2, Rhobtb1, S100a9,
Tfpi, Zbtb4, Hp, Gbp10, and Gm20521, were selected out of all 16
DEGs, and their expressions were measured using RT-qPCR. Among
these genes, Alg12, Cdkn1a, S100a9, Gbp10 and Gm20521 were
significantly differentially expressed in NPRA−/− mice,
compared with NPRA+/+ mice (Fig. 4). Thus, these five genes were
selected for subsequent experiments.
circRNA-miRNA-mRNA interaction network
is activated to regulate cardiac metabolism under the condition of
NPRA deletion
circRNA can act as a miRNA ‘sponge’ that regulates
the expression of mRNA, and such circRNA-miRNA-mRNA interaction
network plays important roles in the genesis and development of
cardiovascular diseases (38).
Therefore, it was hypothesized that a circRNA-miRNA-mRNA network
might be at play in the context of cardiac-specific NPRA deletion,
thus inducing metabolic dysfunction in the myocardium.
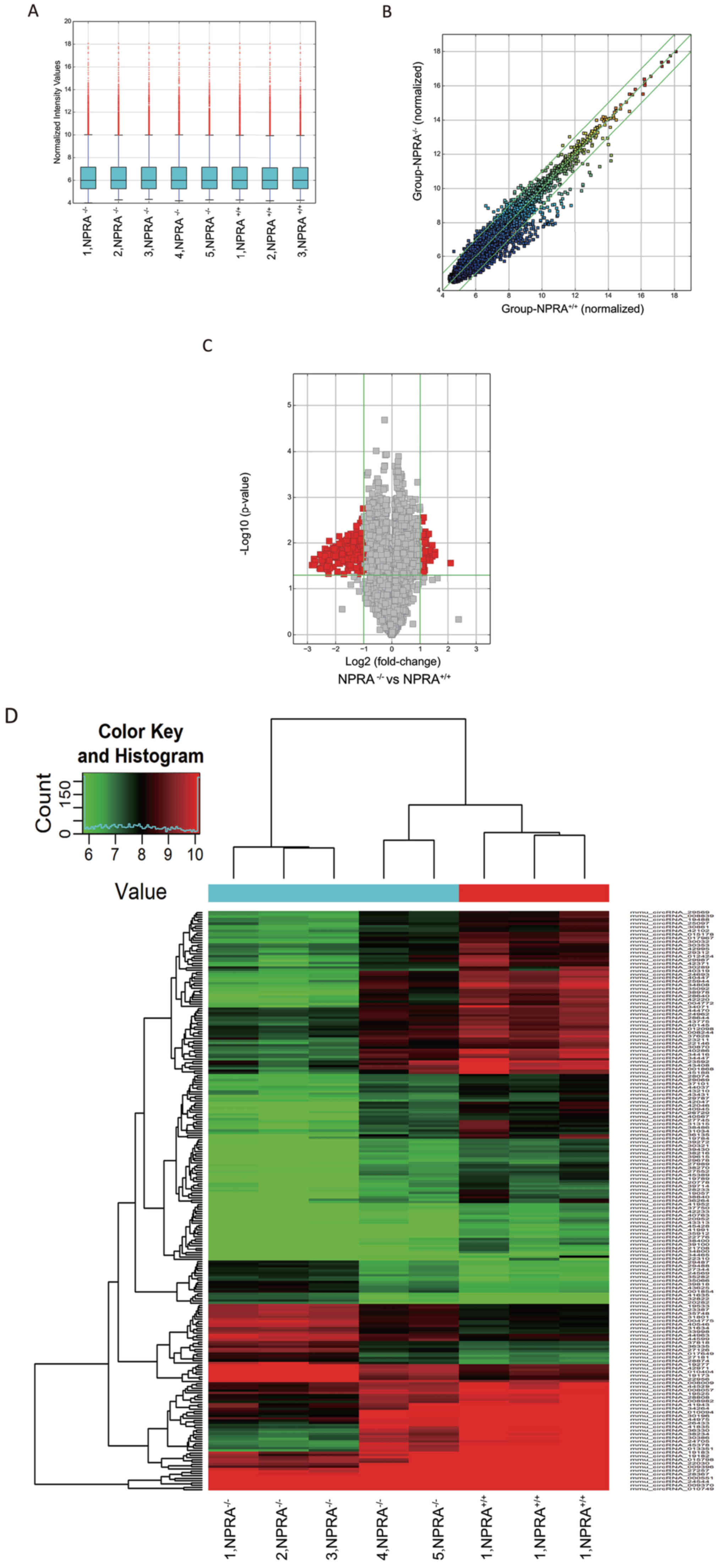
A murine circRNA microarray was used to analyse the
circRNA profiles of the myocardium from NPRA−/− mice,
compared with NPRA+/+ mice. The normalized intensity for
the NPRA+/+ and NPRA−/− groups was nearly the
same (Fig. 5A). DE-circRNA
molecules between NPRA−/− and NPRA+/+ mice
are displayed in a hierarchical clustering map (Fig. 5D), as well as a scatter plot
(Fig. 5B) and a volcano plot
(Fig. 5C). Clusterisation
successfully discriminated successfully discriminated
NPRA−/− from NPRA+/+ samples. In total, 55
upregulated and 197 downregulated DE-circRNAs were identified.
To further examine the regulatory mechanism
underlying DEG profiles, all global changes in the expression
pattern of circRNAs were predicted according to the complementary
miRNA matching sequence. Using ceRNA analysis, a circRNA-miRNA-mRNA
interaction network was constructed. To confirm the ceRNA network,
the predicted circRNAs were compared to the DE-circRNAs profiles
from the circRNA microarray, and the circRNAs involved in both the
predicted circRNA profiles and DE-circRNA profiles were selected
for validation. In addition, because of a positive correlation in
the expression of ceRNA pairs, candidate circRNAs were required to
be oriented in the same direction direction of change of expression
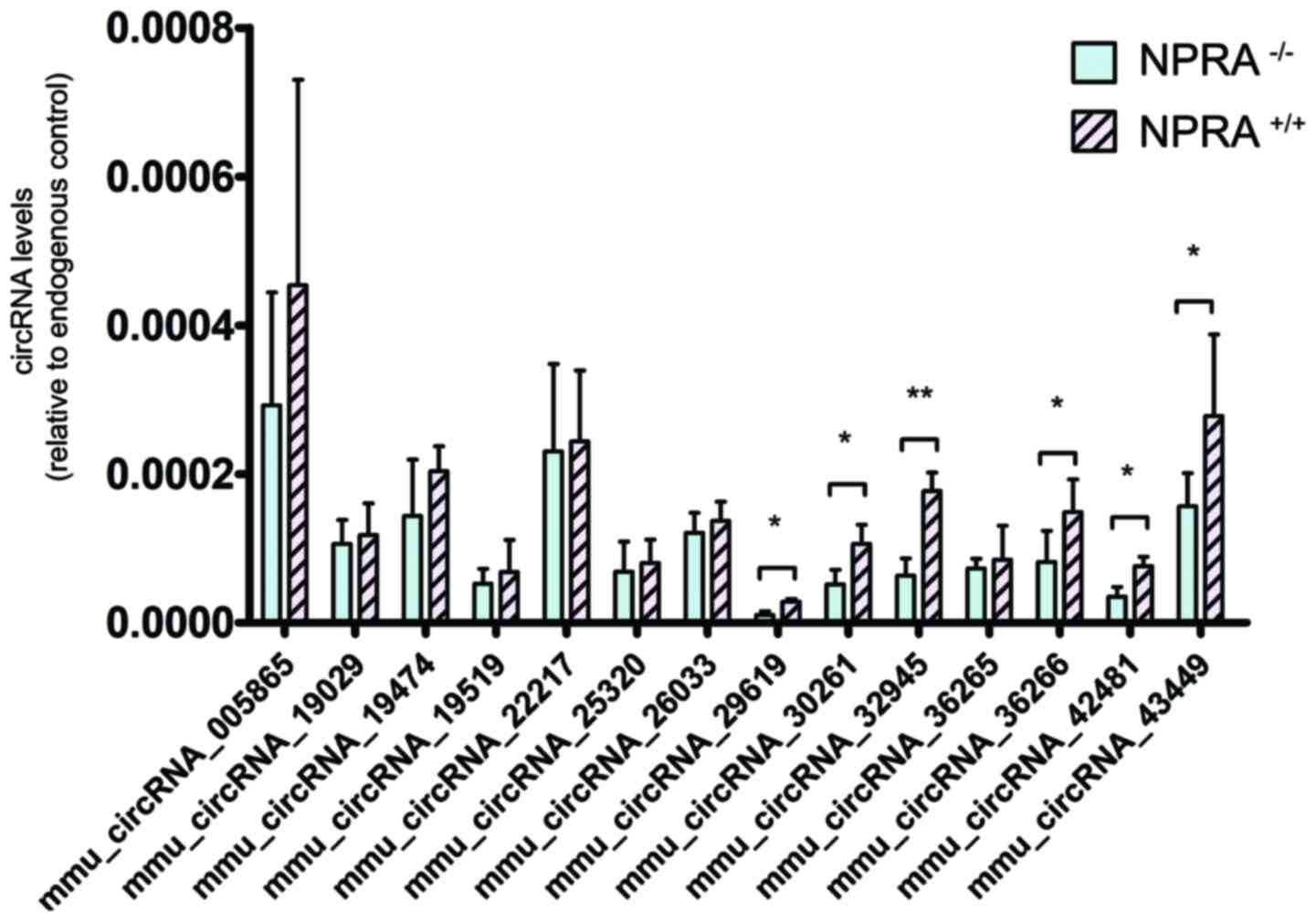
level in DEG analysis. Using this approach, 14 downregulated
circRNAs (mmu_circRNA_005865, mmu_circRNA_19029, mmu_circRNA_19474,
mmu_circRNA_19519, mmu_circRNA_22217, mmu_circRNA_25320,
mmu_circRNA_26033, mmu_circRNA_29619, mmu_circRNA_30261,
mmu_circRNA_32945, mmu_circRNA_36265, mmu_circRNA_36266,
mmu_circRNA_42481 and mmu_circRNA_43449) were selected for
validation by RT-qPCR. With RT-qPCR analysis, 6 circRNAs were
significantly downregulated in the NPRA−/− mice,
compared with NPRA+/+ mice (Fig. 6).
Thus, the DE-circRNAs and DEGs that have been
validated by RT-qPCR assay were included within the ceRNA network,
and all others were excluded. Thus, with the majority of possible
pathways excluded, only three target mRNA molecules (Alg12, Cdkn1a,
and Gbp10) were included within the ceRNA network. The entire
circRNA-miRNA-mRNA interaction network was constructed and visually
displayed by Cytoscape. The top five miRNA binding sites predicted
by ceRNA analysis for the six validated circRNAs in the ceRNA
network are presented in Table
III. The top 5 predicted miRNA response elements (MREs) in the
3′ UTR for each validated circRNA are presented in Fig. S3.
 | Table III.Top five miRNA binding elements for
validated circRNAs. |
Table III.
Top five miRNA binding elements for
validated circRNAs.
| CircRNA | MRE1 | MRE2 | MRE3 | MRE4 | MRE5 |
|---|
|
mmu_circRNA_29619 |
mmu-miR-1231-5p |
mmu-miR-449a-5p |
mmu-miR-6905-5p | mmu-miR-493-3p |
mmu-miR-7034-3p |
|
mmu_circRNA_30261 | mmu-miR-185-3p | mmu-miR-5120 |
mmu-miR-669e-5p |
mmu-miR-3100-3p |
mmu-miR-7048-5p |
|
mmu_circRNA_32945 | mmu-miR-8100 | mmu-miR-5110 |
mmu-miR-1966-5p | mmu-miR-6405 |
mmu-miR-6987-5p |
|
mmu_circRNA_42481 |
mmu-miR-3098-3p | mmu-miR-683 |
mmu-miR-7069-5p | mmu-miR-3552 |
mmu-miR-6990-3p |
|
mmu_circRNA_36266 |
mmu-miR-103-1-5p |
mmu-miR-103-2-5p | mmu-miR-335-3p | mmu-miR-107-5p | mmu-miR-6400 |
|
mmu_circRNA_43449 |
mmu-miR-7092-3p | mmu-miR-1187 | mmu-miR-466f |
mmu-miR-669a-5p |
mmu-miR-669p-5p |
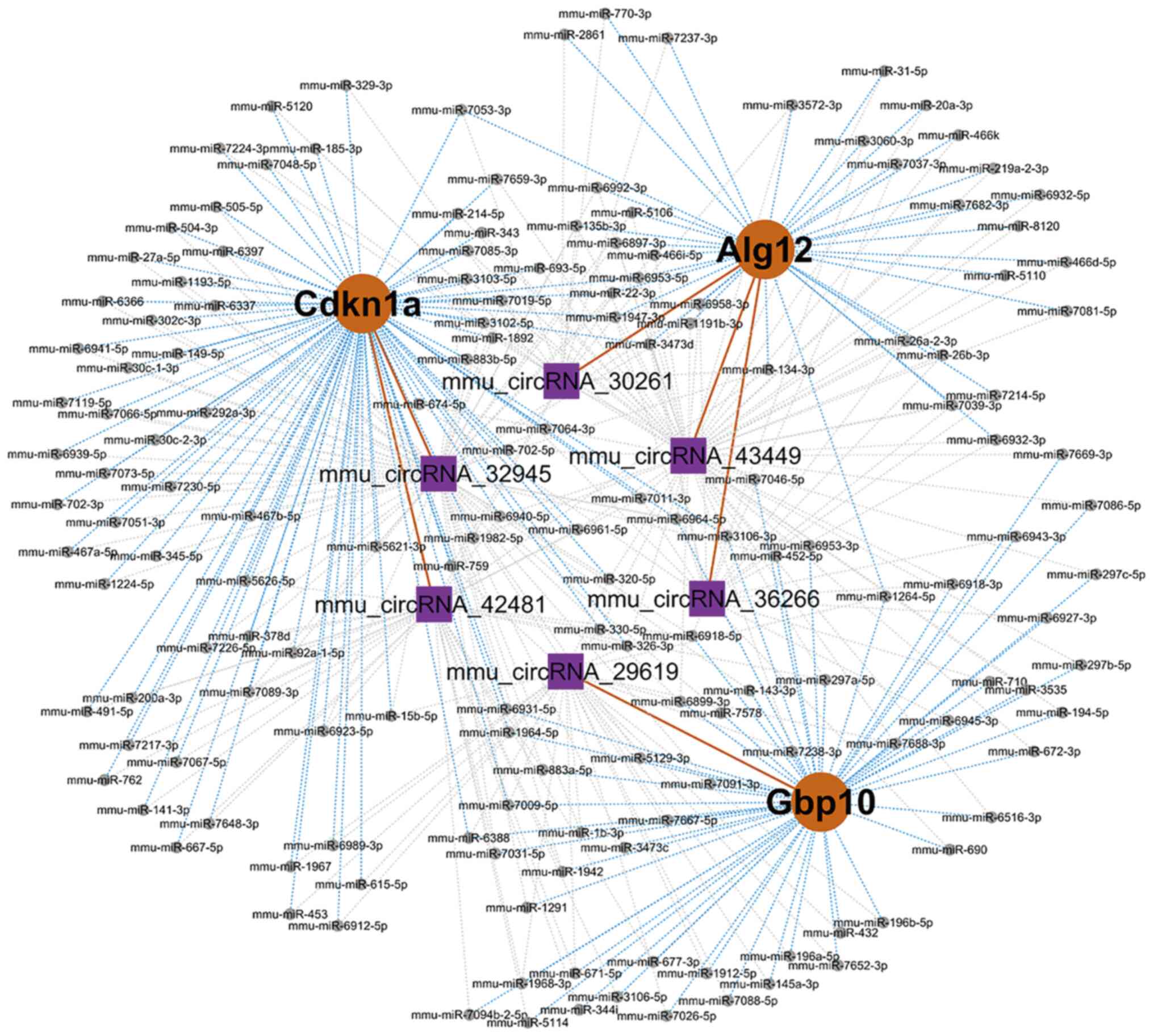
A circRNA-miRNA-mRNA interaction network was
constructed, which included three genes (Alg12, Cdkn1a, and Gbp10)
and six circRNAs (mmu_circRNA30261, mmu_circRNA43449,
mmu_circRNA36266, mmu_circRNA32945, mmu_circRNA42481, and
mmu_circRNA29619). Moreover, 162 miRNAs were predicted in this
network (Fig. 7; Table IV). This network may provide
insight into the potential interactions between circRNA candidates
and their target genes.
 | Table IV.Details of the ceRNA network. |
Table IV.
Details of the ceRNA network.
| Gene symbol | CeName | CeSymbol | CeType | Common miRNAs |
|---|
| Cdkn1a |
mmu_circRNA_32945 |
mmu_circRNA_32945 | circRNA | mmu-miR-1193-5p,
mmu-miR-1224-5p, mmu-miR-149-5p, mmu-miR-15b-5p, mmu-miR-1892,
mmu-miR-22-3p, mmu-miR-27a-5p, mmu-miR-292a-3p, mmu-miR-302c-3p,
mmu-miR-30c-1-3p, mmu-miR-30c-2-3p, mmu-miR-3102-5p,
mmu-miR-3103-5p, mmu-miR-320-5p, mmu-miR-345-5p, mmu-miR-378d,
mmu-miR-467a-5p, mmu-miR-467b-5p, mmu-miR-504-3p, mmu-miR-505-5p,
mmu-miR-5621-3p, mmu-miR-5626-5p, mmu-miR-6337, mmu-miR-6366,
mmu-miR-6397, mmu-miR-6923-5p, mmu-miR-6931-5p, mmu-miR-6939-5p,
mmu-miR-6941-5p, mmu-miR-6964-5p, mmu-miR-7019-5p, mmu-miR-702-3p,
mmu-miR-702-5p, mmu-miR-7051-3p, mmu-miR-7064-3p, mmu-miR-7066-5p,
mmu-miR-7073-5p, mmu-miR-7089-3p, mmu-miR-7119-5p, mmu-miR-7226-5p,
mmu-miR-7230-5p, mmu-miR-883b-5p, mmu-miR-92a-1-5p |
| Alg12 |
mmu_circRNA_43449 |
mmu_circRNA_43449 | circRNA | mmu-miR-1191b-3p,
mmu-miR-135b-3p, mmu-miR-1947-3p, mmu-miR-20a-3p,
mmu-miR-219a-2-3p, mmu-miR-26a-2-3p, mmu-miR-26b-3p, mmu-miR-31-5p,
mmu-miR-3473d, mmu-miR-3572-3p, mmu-miR-466d-5p, mmu-miR-466i-5p,
mmu-miR-466k, mmu-miR-5106, mmu-miR-5110, mmu-miR-6897-3p,
mmu-miR-6932-3p, mmu-miR-6932-5p, mmu-miR-6953-5p, mmu-miR-6958-3p,
mmu-miR-6992-3p, mmu-miR-7039-3p, mmu-miR-7081-5p, mmu-miR-7214-5p,
mmu-miR-7682-3p, mmu-miR-8120 |
| Cdkn1a |
mmu_circRNA_42481 |
mmu_circRNA_42481 | circRNA | mmu-miR-141-3p,
mmu-miR-1964-5p, mmu-miR-200a-3p, mmu-miR-22-3p, mmu-miR-326-3p,
mmu-miR-330-5p, mmu-miR-378d, mmu-miR-491-5p, mmu-miR-5626-5p,
mmu-miR-667-5p, mmu-miR-6923-5p, mmu-miR-6940-5p, mmu-miR-6961-5p,
mmu-miR-7011-3p, mmu-miR-7067-5p, mmu-miR-7089-3p, mmu-miR-7217-3p,
mmu-miR-7226-5p, mmu-miR-762, mmu-miR-7648-3p,
mmu-miR-92a-1-5p |
| Gbp10 |
mmu_circRNA_29619 |
mmu_circRNA_29619 | circRNA |
mmu-miR-1264-5p,mmu-miR-145a-3p,
mmu-miR-1912-5p, mmu-miR-1968-3p, mmu-miR-196a-5p, mmu-miR-196b-5p,
mmu-miR-1b-3p, mmu-miR-432, mmu-miR-6388, mmu-miR-671-5p,
mmu-miR-6899-3p, mmu-miR-7009-5p, mmu-miR-7026-5p, mmu-miR-7088-5p,
mmu-miR-7238-3p, mmu-miR-7652-3p, mmu-miR-7688-3p |
| Alg12 |
mmu_circRNA_30261 |
mmu_circRNA_30261 | circRNA | mmu-miR-134-3p,
mmu-miR-135b-3p, mmu-miR-2861, mmu-miR-3572-3p, mmu-miR-7053-3p,
mmu-miR-7237-3p, mmu-miR-770-3p |
| Alg12 |
mmu_circRNA_36266 |
mmu_circRNA_36266 | circRNA | mmu-miR-1947-3p,
mmu-miR-26a-2-3p, mmu-miR-26b-3p, mmu-miR-3060-3p, mmu-miR-3473d,
mmu-miR-6932-3p, mmu-miR-6953-5p, mmu-miR-6958-3p, mmu-miR-7037-3p,
mmu-miR-7039-3p, mmu-miR-7214-5p |
Discussion
The present study suggested that deficiency in NP
signalling may result in metabolic dysfunction involving a
circRNA-miRNA-mRNA interaction network. This conclusion is based on
the following findings: i) The expression profiles of mRNA and
circRNA molecules were different between the NPRA−/− and
the NPRA+/+ mice; ii) GO analysis demonstrated that DEGs
were associated with BP terms related to metabolism, especially
‘lipid metabolic processes’; iii) KEGG analysis also revealed a
single ‘metabolic pathway’; and iv) a circRNA-miRNA-mRNA
interaction network involved with cardiac metabolism was
constructed by ceRNA analysis.
NPs are a group of peptide hormones that are mainly
secreted from the heart and signal through c-GMP-coupled receptors
(39). The most well-known
biological functions of NPs are their renal and cardiovascular
functions, reducing arterial blood pressure, as well as sodium
reabsorption. Recently, a number of studies have indicated that NPs
may play a pivotal role in metabolic functions, including the
activation of lipolysis, lipid oxidation, and mitochondrial
respiration (3,40). Despite high circulating
concentrations of immunoreactive peptides, functional natriuretic
peptide deficiency is observed in type-2 diabetes mellitus and
cardio-metabolic complications, suggesting the involvement of
weakened NP signalling in the pathophysiology of metabolic
disorders (41). The presence of an
NP deficiency often occur with metabolic disease, this phenomenon
is generally accepted, as acknowledged by large cohorts (although
challenged by some smaller cohorts), but the cause has not been
fully elucidated (42). However, to
the best of our knowledge, no previous study has investigated the
expression of mRNAs in NPRA-deficient mice. In this study, using
RNA-seq, the differential expression of mRNA transcriptional
profiles between NPRA−/− myocardium and wild-type
myocardium was determined. The differential expression of mRNA
determined a profile of possible effectors downstream of the
NPRA/cGMP signalling pathway. Notably, the number of DEGs was
relatively low. Moreover, the results from GO analysis identified
genes associated with metabolism. Similarly, although KEGG
enrichment analysis only obtained a single pathway, it also clearly
pointed to metabolic pathways, indicating that NP deficiency may
play a pivotal role in metabolic disease.
Recent studies and developments in genome-wide
analyses and RNA-seq technologies have demonstrated that small
non-coding RNA molecules, such as miRNAs, linear long noncoding
RNAs (lncRNAs) and circRNAs, can function in the regulation of
biological processes (43,44). circRNA molecules are genetic
products that are generated by back-splicing of a single pre-mRNA
transcript (17). A growing number
of circRNA molecules have been identified, and their
pathophysiological functions have been determined. In particular,
it has been demonstrated that circRNA are involved in
cardiovascular disease initiation and progression, including
atherosclerosis, cardiomyopathy, myocardial infarction, and cardiac
fibrosis (17). In the present
study, to determine whether the NPRA-related cardiometabolic
profile (a term to describe a group of metabolic factors in cardiac
associated with cardiovascular disease, metabolic syndrome and type
II diabetes) (45) is associated
with circRNAs, high-throughput screening of the DE-circRNAs was
carried out using circRNA microarray analysis. To the best of our
knowledge, the present study the first to use circRNA microarray
analysis in NPRA−/− and NPRA+/+ mice. A total
of 55 significantly upregulated circRNAs and 197 significantly
downregulated circRNAs were identified. Thus, deficiency in NP
signalling resulted in altered expression of circRNAs, suggesting
that circRNA molecules might represent important downstream
effectors of NPRA/cGMP signalling. However, the underlying
mechanism governing this phenomenon warrants requires further
study.
Functionally, cytoplasmic circRNAs act as miRNA
sponges by binding to miRNAs as competing endogenous RNAs (ceRNAs);
according to the ceRNA hypothesis, the expression of cirRNA and
miRNA should be negatively correlated (46). In the study, a circRNA-miRNA-mRNA
network was constructed. Each circRNA and its potential
complementary binding miRNAs were illustrated by the network, and
specific interactions, such as
mmu_circRNA_32945/mmu-miR-5626-5p/Alg12 were identified in the
ceRNA network. Three genes, including Alg12, Cdkn1a, and Gbp10,
were predicted to be associated with cardio-metabolic processes. In
particular, Alg12 participates in asparagine N-linked
glycosylation, metabolic pathways, metabolism of proteins, N-glycan
biosynthesis and dolichyl-diphospho-oligosaccharide biosynthesis in
both mice and humans (www.ncbi.nlm.nih.gov/gene/).
In conclusion, NP signalling plays an important role
in the regulation of cardiac metabolism. The underlying mechanism
governing this role involves a circRNA-miRNA-mRNA interacting
network. This finding may provide insight into a possible novel
pathway that is regulated by downstream signals of NPRA and
modulates cardiac metabolism. The components of this network may
represent candidate targets of cardiac metabolic disorders and
warrant further investigation.
There were some limitations in the present study.
First, the sample size in this study was small. Second, as circRNA
may act as miRNA sponge, the circRNA-miRNA-mRNA networks were
constructed. However, the molecular functions of the majority of
identified circRNAs remain unknown, and several reports have shown
that circRNAs can have multiple other functions, such as regulation
of transcription, alternative splicing and translation (47). For this reason, functional
experimental studies are still necessary to validate the roles
played by these circRNAs in NPRA−/− myocardium. Also,
further research should be designed and conducted to verify the
validity of the whole of interaction network. For instance, this
may include measurement of target gene expression, identification
of circRNA molecules, pull-down assays with biotinylated circRNA
probes, dual luciferase reporter gene experiments and other
experiments for myocardial function and morphological changes, in
order to examine the regulatory effect of NPs on cardiac function
through this proposed ceRNA mechanism at the post-transcriptional
level. Then whether one certain stable circRNA could formed by back
splicing mechanism, whether one specific miRNA can be sponged by
the circRNA, and certain target genes regulated by NPs will be
identified. Future work should also focus on specific metabolic
pathways and their regulation by NP signalling.
Supplementary Material
Supporting Data
Acknowledgements
The authors thank Dr Jin-Song Chen (The Second
Affiliated Hospital, Xi'an Medical University, Shaanxi, China) for
his valuable suggestions for the present study.
Funding
The present study was supported by the National
Natural Science Foundation of China (grant no. 81870172), the
Shaanxi Provincial Key Research and Development Project (grant nos.
2018ZDXM-SF-068 and 2018SF-114), and Scientific Research Project of
Xi'an Health Commission (grant no. 2020yb60).
Availability of data and materials
The sequence data were uploaded to the Gene
Expression Omnibus database (https://www.ncbi.nlm.nih.gov/geo/). The GEO accession
number is GEO: GSE140678. Microarray data were uploaded into the
GEO database. The GEO accession number is GEO: GSE140798.
Authors' contributions
JY contributed to conception and design, funding
acquisition and manuscript review. BC contributed to study design,
provision of methodology and project administration. PC contributed
to experimental operation and acquisition of data. XS contributed
to data collection, statistical analysis, interpretation of data
and manuscript preparation. XZ, JZ and XW contributed to formal
analysis. All authors read and approved the final version of the
manuscript.
Ethics approval and consent to
participate
All experimental protocols included in this
manuscript were approved by the Institutional Animal Care and Use
Committee of Xi'an Medical University.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Boerrigter G, Costello-Boerrigter LC and
Burnett JC Jr: Natriuretic peptides in the diagnosis and management
of chronic heart failure. Heart Fail Clin. 5:501–514. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Schipke J, Roloff K, Kuhn M and Mühlfeld
C: Systemic, but not cardiomyocyte-specific, deletion of the
natriuretic peptide receptor guanylyl cyclase A increases
cardiomyocyte number in neonatal mice. Histochem Cell Biol.
144:365–375. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cannone V, Cabassi A, Volpi R and Burnett
JC Jr: Atrial natriuretic peptide: A molecular target of novel
therapeutic approaches to cardio-metabolic disease. Int J Mol Sci.
20:32652019. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Hsu MT and Coca-Prados M: Electron
microscopic evidence for the circular form of RNA in the cytoplasm
of eukaryotic cells. Nature. 280:339–340. 1979. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Ren S, Xin Z, Xu Y, Xu J and Wang G:
Construction and analysis of circular RNA molecular regulatory
networks in liver cancer. Cell Cycle. 16:2204–2211. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Jeck WR, Sorrentino JA, Wang K, Slevin MK,
Burd CE, Liu J, Marzluff WF and Sharpless NE: Circular RNAs are
abundant, conserved, and associated with ALU repeats. RNA.
19:141–157. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Hansen TB, Wiklund ED, Bramsen JB,
Villadsen SB, Statham AL, Clark SJ and Kjems J: MiRNA-dependent
gene silencing involving Ago2-mediated cleavage of a circular
antisense RNA. EMBO J. 30:4414–4422. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Hansen TB, Jensen TI, Clausen BH, Bramsen
JB, Finsen B, Damgaard CK and Kjems J: Natural RNA circles function
as efficient microRNA sponges. Nature. 495:384–388. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Ashwal-Fluss R, Meyer M, Pamudurti NR,
Ivanov A, Bartok O, Hanan M, Evantal N, Memczak S, Rajewsky N and
Kadener S: circRNA biogenesis competes with pre-mRNA splicing. Mol
Cell. 56:55–66. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Guo JU, Agarwal V, Guo H and Bartel DP:
Expanded identification and characterization of mammalian circular
RNAs. Genome Biol. 15:4092014. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Huang S, Li X, Zheng H, Si X, Li B, Wei G,
Li C, Chen Y, Chen Y, Liao W, et al: Loss of
super-enhancer-regulated circRNA Nfix induces cardiac regeneration
after myocardial infarction in adult mice. Circulation.
139:2857–2876. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Zhang F, Zhang R, Zhang X, Wu Y, Li X,
Zhang S, Hou W, Ding Y, Tian J, Sun L and Kong X: Comprehensive
analysis of circRNA expression pattern and circRNA-miRNA-mRNA
network in the pathogenesis of atherosclerosis in rabbits. Aging
(Albany NY). 10:2266–2283. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Yang F, Li A, Qin Y, Che H, Wang Y, Lv J,
Li Y, Li H, Yue E, Ding X, et al: A novel Circular RNA mediates
pyroptosis of diabetic cardiomyopathy by functioning as a competing
endogenous RNA. Mol Ther Nucleic Acids. 17:636–643. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Zhou B and Yu JW: A novel identified
circular RNA, circRNA_010567, promotes myocardial fibrosis via
suppressing miR-141 by targeting TGF-β1. Biochem Biophys Res
Commun. 487:769–775. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Altesha MA, Ni T, Khan A, Liu K and Zheng
X: Circular RNA in cardiovascular disease. J Cell Physiol.
234:5588–5600. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Bayoumi AS, Aonuma T, Teoh JP, Tang YL and
Kim IM: Circular noncoding RNAs as potential therapies and
circulating biomarkers for cardiovascular diseases. Acta Pharmacol
Sin. 39:1100–1109. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Wang K, Long B, Liu F, Wang JX, Liu CY,
Zhao B, Zhou LY, Sun T, Wang M, Yu T, et al: A circular RNA
protects the heart from pathological hypertrophy and heart failure
by targeting miR-223. Eur Heart J. 37:2602–2611. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang H, Wang H, Shivalila CS, Cheng AW,
Shi L and Jaenisch R: One-Step generation of mice carrying reporter
and conditional alleles by CRISPR/Cas-Mediated genome engineering.
Cell. 154:1370–1379. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Feng GK, Ye JC, Zhang WG, Mei Y, Zhou C,
Xiao YT, Li XL, Fan W, Wang F and Zeng MS: Integrin α6 targeted
positron emission tomography imaging of hepatocellular carcinoma in
mouse models. J Control Release. 310:11–21. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Yu J, Bulk E, Ji P, Hascher A, Tang M,
Metzger R, Marra A, Serve H, Berdel WE, Wiewroth R, et al: The
EPHB6 receptor tyrosine kinase is a metastasis suppressor that is
frequently silenced by promoter DNA hypermethylation in non-small
cell lung cancer. Clin Cancer Res. 16:2275–2283. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Zhao HK, Chen BY, Chang R, Wang JB, Ni F,
Yang L, Dong XC, Sun SH, Zhao G, Fang W, et al: Vasonatrin peptide,
a novel protector of dopaminergic neurons against the injuries
induced by n-methyl-4-phenylpyridiniums. Peptides. 49:117–122.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Lu Y, Xie L and Chen J: A novel procedure
for absolute real-time quantification of gene expression patterns.
Plant Methods. 8:92012. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Wan QQ, Wu D and Ye QF: Candidate genes as
biomarkers in Lipopolysaccharide-Induced acute respiratory distress
syndrome based on mRNA expression profile by Next-Generation
RNA-Seq analysis. Biomed Res Int. 2018:43847972018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Babraham Bioinformatics-FastQC A Quality
Control tool for High Throughput Sequence Data. PubMed/NCBI
|
|
27
|
Kim D, Langmead B and Salzberg SL: HISAT:
A fast spliced aligner with low memory requirements. Nat Methods.
12:357–360. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Poole CJ, Lodh A, Choi JH and van Riggelen
J: MYC deregulates TET1 and TET2 expression to control global DNA
(hydroxy)methylation and gene expression to maintain a neoplastic
phenotype in T-ALL. Epigenetics Chromatin. 12:412019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Benjamini Y and Hochberg Y: Controlling
the false discovery rate: A practical and powerful approach to
multiple testing. J R Stat Soc Ser B (Methodological). 57:289–300.
1995.
|
|
30
|
Aebersold R and Mann M: Mass-spectrometric
exploration of proteome structure and function. Nature.
537:347–355. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Kanehisa M, Furumichi M, Tanabe M, Sato Y
and Morishima K: KEGG: New perspectives on genomes, pathways,
diseases and drugs. Nucleic Acids Res. 45:D353–D361. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
32
|
He R, Liu P, Xie X, Zhou Y, Liao Q, Xiong
W, Li X, Li G, Zeng Z and Tang H: circGFRA1 and GFRA1 act as ceRNAs
in triple negative breast cancer by regulating miR-34a. J Exp Clin
Cancer Res. 36:1452017. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Lv C, Sun L, Guo Z, Li H, Kong D, Xu B,
Lin L, Liu T, Guo D, Zhou J and Li Y: Circular RNA regulatory
network reveals cell-cell crosstalk in acute myeloid leukemia
extramedullary infiltration. J Transl Med. 16:3612018. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lin SP, Ye S, Long Y, Fan Y, Mao HF, Chen
MT and Ma QJ: Circular RNA expression alterations are involved in
OGD/R-induced neuron injury. Biochem Biophys Res Commun. 471:52–56.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhou J, Xiong Q, Chen H, Yang C and Fan Y:
Identification of the spinal expression profile of non-coding rnas
involved in neuropathic pain following spared nerve injury by
sequence analysis. Front Mol Neurosci. 10:912017. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Li JH, Liu S, Zhou H, Qu LH and Yang JH:
StarBase v2.0: Decoding miRNA-ceRNA, miRNA-ncRNA and protein-RNA
interaction networks from large-scale CLIP-Seq data. Nucleic Acids
Res. 42:D92–D97. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Team R: R Core Team. R A Language and
Environment for Statistical Computing 2014. R Foundation for
Statistical Computing. 2008.
|
|
38
|
Li M, Duan L, Li Y and Liu B: Long
noncoding RNA/circular noncoding RNA-miRNA-mRNA axes in
cardiovascular diseases. Life Sci. 233:1164402019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Schlueter N, de Sterke A, Willmes DM,
Spranger J, Jordan J and Birkenfeld AL: Metabolic actions of
natriuretic peptides and therapeutic potential in the metabolic
syndrome. Pharmacol Ther. 144:12–27. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Wu W, Shi F, Liu D, Ceddia RP, Gaffin R,
Wei W, Fang H, Lewandowski ED and Collins S: Enhancing natriuretic
peptide signaling in adipose tissue, but not in muscle, protects
against diet-induced obesity and insulin resistance. Sci Signal.
10:eaam68702017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Zois NE, Bartels ED, Hunter I, Kousholt
BS, Olsen LH and Goetze JP: Natriuretic peptides in cardiometabolic
regulation and disease. Nat Rev Cardiol. 11:403–412. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Verboven K, Hansen D, Jocken JWE and Blaak
EE: Natriuretic peptides in the control of lipid metabolism and
insulin sensitivity. Obes Rev. 18:1243–1259. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Ding B, Lou W, Xu L and Fan W: Non-coding
RNA in drug resistance of hepatocellular carcinoma. Biosci Rep.
38:BSR201809152018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Xuan L, Qu L, Zhou H, Wang P, Yu H, Wu T,
Wang X, Li Q, Tian L, Liu M and Sun Y: Circular RNA: A novel
biomarker for progressive laryngeal cancer. Am J Transl Res.
8:932–939. 2016.PubMed/NCBI
|
|
45
|
Fernández-Cao JC and Doepking C: Role of
ethnicity and environment on lifestyle and cardiometabolic profile
in the Native American Mapuche population: Protocol for a
systematic review and meta-analysis. Medicine (Baltimore).
97:e133542018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Salmena L, Poliseno L, Tay Y, Kats L and
Pandolfi PP: A ceRNA hypothesis: The Rosetta Stone of a hidden RNA
language? Cell. 146:353–358. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Wilusz JE: A 360° view of circular RNAs:
From biogenesis to functions. Wiley Interdiscip Rev RNA.
9:e14782018. View Article : Google Scholar : PubMed/NCBI
|