Introduction
Gastric cancer (GC) is one of the most common
malignancies and is the third leading cause of cancer-related
mortality worldwide (1). In 2018,
there were 1,033,701 new cases of GC diagnosed, which accounted for
5.7% of the total worldwide number of cancer diagnoses (2). Patients with GC often present with
non-specific symptoms in the early stages and the majority of GC
cases are diagnosed in the advanced stages, which generally
indicates a poor prognosis (3).
Clinically, despite advancements in the therapy of GC, the
treatment effect remains unsatisfactory and the 5-year survival
rate is ≤30% (4,5). The improvement in the prognosis of
patients with GC requires the identification of novel and effective
therapeutic targets. It has been reported that the pathogenesis of
GC is caused by the altered expression of multiple genes (6). Therefore, the identification of tumor
suppressor genes and oncogenes is of great importance for the
diagnosis and treatment of GC.
Long non-coding RNAs (lncRNAs) are a class of RNA
transcripts that are >200 nucleotides in length (7). lncRNAs lack protein-coding function,
but exert physiological effects at the epigenetic, transcriptional
and post-transcriptional levels (8). Previous studies have reported that a
number of lncRNAs serve as crucial regulators in the progression of
GC. For instance, Wu et al (9) revealed that lncRNA FEZF1 antisense RNA
1 (FEZF1-AS1) was highly expressed in GC tissues and cells, and was
associated with poor prognosis in patients with GC. Therefore,
lncRNA FEZF1-AS1 may serve as an independent prognostic factor in
GC, which promotes the tumorigenesis of GC by activating the Wnt
signaling pathway. By contrast, Nie et al (10) observed that lncRNA MIR31 host gene
(MIR31HG) was downregulated in GC, which indicated poor prognosis
in patients with GC. In vitro studies have also revealed
that decreased lncRNA MIR31HG expression could promote GC cell
proliferation (10). Similarly, GC
associated transcript 2, transmembrane protein 238 like and
maternally expressed 3 are considered as tumor suppressor genes in
GC (11–13), whereas GC associated transcript 3,
BC005927, urothelial cancer associated 1 and colon cancer
associated transcript 1 were identified as cancer-promoting genes
(14–17). Thus, the discovery of lncRNAs
provides important therapeutic targets for GC.
lncRNA JPX transcript, XIST activator (JPX) is a
member of lncRNA family that, to the best of our knowledge, has not
been investigated in GC. In the present study, JPX expression was
detected in GC and the effects of JPX on the development of GC were
investigated. Several studies have revealed that microRNA
(miRNA/miR)-197 acts as a tumor suppressor in the progression of
GC. For example, Liao et al (18) reported that miR-197 suppressed GC
progression by regulating metadherin, while Chen et al
(19) indicated that
hsa_circ_0092306 facilitated GG tumorigenesis by targeting miR-197.
Moreover, increasing evidence has suggested that lncRNA could serve
as a competing endogenous RNA against miRNA to regulate the
progression of various types of cancer, such as cervical cancer, GC
and ovarian cancer (20–22). However, whether JPX regulates
miR-197 in GC requires further investigation. It has also been
observed that C-X-C motif chemokine receptor 6 (CXCR6) is involved
in the development of GC.
Therefore, the present study investigated whether
JPX regulated the development of GC via the miR-197/CXCR6 axis,
with the aim of identifying a novel potential target and
theoretical basis for the treatment of GC.
Materials and methods
The Cancer Genome Atlas (TCGA)
analysis
JPX and miR-197 expression levels in tumor and
healthy tissues of patients with GC were downloaded from TCGA
database (https://tcga-data.nci.nih.gov/tcga/). Differences in
JPX and miR-197 expression levels between healthy and tumor tissues
were analyzed.
Tissue specimens
A total of 32 paired GC and adjacent non-tumor
tissues (3–5 cm adjacent to tumor) were collected from 32 patients
with GC (mean age, 62 years; age range, 43–76 years; 17 male
patients and 15 female patients) at The Second Affiliated Hospital
of Nanjing Medical University between January 2016 and April 2018.
The diagnoses of these samples were verified by experienced
pathologists. Written informed consent was obtained from all
patients. The present study was approved by the Ethics Committee of
The Second Affiliated Hospital of Nanjing Medical University.
Cell culture
GC cell lines (NCI-N87 and MKN-45) and normal
gastric mucosa epithelial cells (GES-1) were purchased from The
Cell Bank of Type Culture Collection of the Chinese Academy of
Sciences. Cells were cultured in 3 ml DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo Fisher
Scientific, Inc.), 100 U/ml penicillin (Thermo Fisher Scientific,
Inc.) and 100 µg/ml streptomycin (Thermo Fisher Scientific, Inc.)
in sterilized culture flasks at 37°C with 5% CO2.
Transfection
The short hairpin RNA (shRNA) targeting JPX (shJPX;
5′-GUGCGUACAGUGCUGUACAGCAU-3′) with its negative control (NC) shRNA
(shNC; 5′-UACGCUCAGCAUGUGUCACUC-3′), miR-197 mimic
(5′-UCGUUCGUGAGCACUUGCGACG-3′) with its NC mimics
(5′-UCGUCGGAUCGACUGAGAUCU-3′) and miR-197 inhibitor
(5′-AGCCUUGCUGCAGGUGCGCAU-3′) with its NC inhibitor
(5′-UGCCUUACUGACGGUCGGAGA-3′) were purchased from Shanghai
GenePharma Co., Ltd. NCI-N87 and MKN-45 cells were transfected with
10 nM shJPX and 10 nM shNC. NCI-N87 and MKN-45 cells were
transfected with 10 nM miR-197 mimic and 10 nM NC to form the
miR-197 mimic and NC groups, respectively.
CXCR6 and Beclin1 overexpression plasmids
(pcDNA3.1-CXCR6 and pcDNA3.1-Beclin1, respectively) were
established by inserting the full-length CXCR6 or Beclin1 sequence
into the pcDNA3.1 vector (Thermo Fisher Scientific, Inc.). An empty
pcDNA3.1 vector (pcDNA3.1) was used as the control.
NCI-N87 and MKN-45 cells were co-transfected with 10
nM shJPX and 10 nM miR-197 inhibitor (shJPX + miR-197 inhibitor
group), miR-197 mimic and CXCR6 overexpression vector (miR-197
mimic + CXCR6 group) or miR-197 mimic and Beclin1 overexpression
vector (miR-197 mimic + Beclin1 group).
All transfections were performed using
Lipofectamine® 3000 reagent (Thermo Fisher Scientific,
Inc.). Cells were maintained at 37°C and 5% CO2 for 8 h.
Successfully transfected cells were screened and maintained in DMEM
supplemented with 10% FBS at 37°C and 5% CO2. At 48 h
post-transfection, cells were used for subsequent
experimentation.
Luciferase reporter gene assay
StarBase 2.0 (http://starbase.sysu.edu.cn) was used to predict the
potential miRNAs that can bind to JPX. Wild-type (WT) and mutant
(MUT) JPX sequences, as well as WT and MUT CXCR6 sequences, were
designed by Shanghai GenePharma Co., Ltd. and cloned into the
psiCHECK2 luciferase reporter (Promega Corporation). NCI-N87 cells
transfected with 10 nM miR-197 mimics and 10 nM NC were prepared as
single cell suspensions in serum-free DMEM at a density of
1×105 cells/ml. Cells were seeded in 6-well plates (1 ml
cell suspensions per well). NCI-N87 cells were transfected with WT
and MUT JPX psiCHECK2 luciferase reporters, as well as WT and MUT
CXCR6 psiCHECK2 luciferase reporters. Transfection was performed
using Lipofectamine 2000. At 48 h post-transfection, the luciferase
activity of each well was determined using a Dual-luciferase
reporter assay system (Promega Corporation) according to the
manufacturer's protocol. The relative luciferase activity of each
well was normalized to Renilla luciferase activity.
Reverse transcription-quantitative PCR
(RT-qPCR)
Tissue specimens were ground into powder in liquid
nitrogen prior to RNA extraction. Total RNA was isolated from
tissue specimens and cells using TRIzol® reagent (Thermo
Fisher Scientific, Inc.). Total RNA (200 ng) was reverse
transcribed into cDNA using SuperScript III (Invitrogen; Thermo
Fisher Scientific, Inc.) at 37°C for 15 min. Subsequently, qPCR was
performed using an ABI PRISM 7500 Sequence Detection system (Thermo
Fisher Scientific, Inc.) and SYBR Select Master mix (Thermo Fisher
Scientific, Inc.). U6 or GAPDH were used as the internal reference
genes. The following thermocycling conditions were used for qPCR:
Initial denaturation at 95°C for 5 min; followed by 40 cycles of
95°C for 20 sec, 58°C for 30 sec and 74°C for 30 sec and a final
extension step at 72°C for 5 min. The primers used for qPCR were as
follows: JPX forward, 5′-TGCAGTCAGAAGGGAGCAAT-3′ and reverse,
5′-CACCGTCATCAGGCTGTCTT-3′; miR-197 forward,
5′-GTTCACCACCTTCTCCAC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′; CXCR6
forward, 5′-ATGCCATGACCAGCTTTCACT-3′ and reverse,
5′-TTAAGGCAGGCCCTCAGG-3′; Beclin1 forward,
5′-TAGGATCCATGGAAGGGTCTAAGAC-3′ and reverse,
5′-GCGAAGCTTTCATTTGTTATAAAAT-3′; GAPDH forward,
5′-GTCGATGGCTAGTCGTAGCATCGAT-3′ and reverse,
5′-TGCTAGCTGGCATGCCCGATCGATC-3′; and U6 forward,
5′-GCTTCGGCAGCACATATACTAAAA-3′ and reverse,
5′-CGCTTCACGAATTTGCGTGTCAT-3′. miRNA and mRNA expression levels
were quantified using the 2−ΔΔCq method (23).
MTT assay
NCI-N87 and MKN-45 cells were re-suspended in DMEM
supplemented with 10% FBS at a density of 1×105
cells/ml. Cell suspension (100 µl) was added into 96-well plates
for 0, 24, 48 or 72 h at 37°C with 5% CO2. At each time
point, 20 µl MTT solution (5 mg/ml; Sigma-Aldrich; Merck KGaA) was
added into each well for 4 h incubation at 37°C. Subsequently, 150
µl DMSO was added to each well and the plated were gently agitated
for 10 min at room temperature to dissolve the formazan crystal.
The optical density of each well was measured at a wavelength of
490 nm using a microplate reader (Biotek Instruments, Inc.).
Wound healing assay
The wound healing assay was performed to determine
cell migration. Briefly, NCI-N87 and MKN-45 cells re-suspended in
DMEM supplemented with 1% FBS were inoculated into 6-well plates (1
ml cell suspension per well; 1×105 cells/ml). Cells were
incubated at 37°C with 5% CO2 for 24 h. Subsequently, a
straight line was made through the center of each well using a 200
µl pipette tip. The residual liquid in each well was discarded and
replaced with fresh DMEM supplemented with 10% FBS (24). Cells were incubated for 24 h at 37°C
with 5% CO2. The original wound width and wound width at
24 h was observed under a light microscope (magnification, ×200)
and analyzed using ImageJ software (version 1.48; National
Institutes of Health).
Transwell experiment
NCI-N87 and MKN-45 cell invasion was assessed using
Transwell chambers (pore size, 8 µm; EMD Millipore) precoated with
100 µl Matrigel (BD Biosciences) for 1 h at room temperature. Cells
(5×104) were plated in the upper chamber with 200 µl
serum-free DMEM. Into the lower chamber, 600 µl DMEM containing 10%
FBS was plated. The upper chambers were pre-coated with Matrigel
for 1 h at room temperature. Following incubation for 24 h at 37°C
with 5% CO2, the Transwell chamber was removed and cells
remaining on the upper chamber were gently removed using a cotton
swab. Invading cells were fixed with 4% formaldehyde and stained
with 0.1% crystal violet, both for 20 min at room temperature.
Invading cells were observed using a light microscope
(magnification, ×200) in five non-overlapping random fields of
view.
Western blotting
Total protein was extracted from NCI-N87 cells using
RIPA cell lysis (Thermo Fisher Scientific, Inc.) and quantified
using a BCA kit (Thermo Fisher Scientific, Inc.). A total of 10 µg
proteins were separated via 10% SDS-PAGE and transferred to PVDF
membranes, which were blocked with 5% skimmed milk for 2 h at room
temperature. The membranes were incubated for 12 h at 4°C with
primary antibodies targeted against: CXCR6 (1:1,000; cat. no.
ab137134; Abcam), Beclin1 (1:1,000; cat. no. 3738; Cell Signaling
Technology, Inc.), p62 (1:1,000; cat. no. 88588; Cell Signaling
Technology, Inc.), LC3B (1:1,000; cat. no. ab51520; Abcam;
containing LC3-I and LC3-II) and GAPDH (1:1,000; cat. no. ab8245;
Abcam). The membranes were washed three times with TBS-0.1%
Tween-20 and subsequently incubated with horseradish
peroxidase-conjugated secondary antibodies (1:1,000; goat
anti-mouse IgG; cat. no. ab205719; and goat anti-rabbit IgG; cat.
no. ab205718; Abcam) for 2 h at room temperature. Proteins were
visualized using an enhanced chemiluminescence detection system
(Amersham; Cytiva). Image-Pro® Plus software (version
6.0; Media Cybernetics, Inc.) was used to quantify the protein
expression.
Statistical analysis
All experiments were repeated three times
independently. Data are presented as the mean ± SD. Statistical
analyses were performed using SPSS software (version 19.0; IBM
Corp). Comparisons between tumor and adjacent healthy tissue
samples were performed using a paired Student's t-test, while
comparisons between the experimental and control groups were
conducted using an unpaired Student's t-test. Comparisons among
multiple groups were analyzed using one-way ANOVA followed by
Tukey's test. Kaplan-Meier survival curve analysis and the log-rank
test were conducted to analyze the overall survival rate of
patients. Patients were divided into high and low groups, as
determined by the mean expression value. The correlation between
JPX and miR-197 expression in patients with GC was analyzed using
Pearson's correlation analysis. P<0.05 was considered to
indicate a statistically significant difference.
Results
High JPX expression and low miR-197
expression in patients with GC indicates poor prognosis
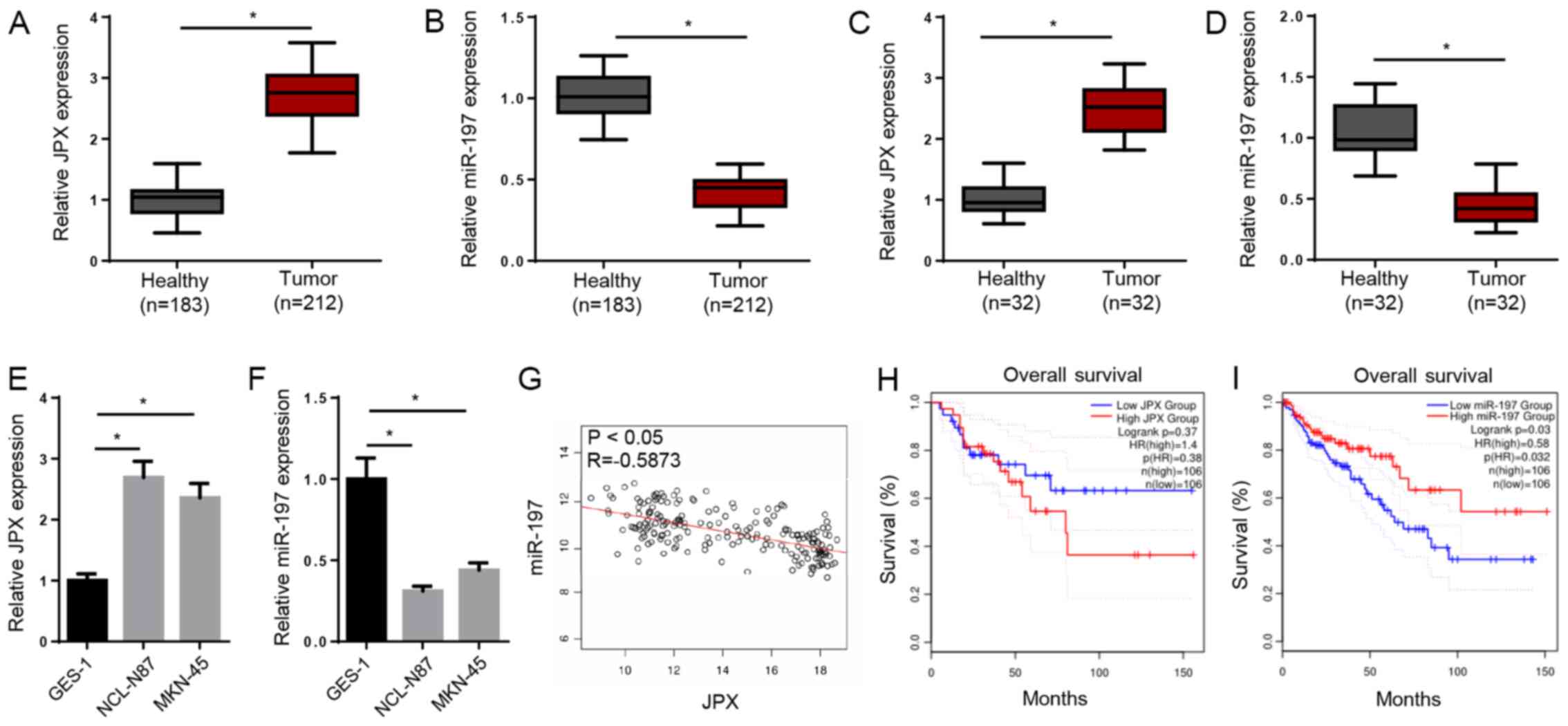
TCGA analysis demonstrated significantly upregulated
JPX expression and downregulated miR-197 expression in patients
with GC (Fig. 1A and B).
Furthermore, JPX and miR-197 expression levels were measured in
clinical GC tissues and adjacent non-tumor tissues. Consistently,
the expression of JPX was significantly increased, whereas miR-197
was decreased in GC tissues (Fig. 1C
and D). JPX expression was also higher and miR-197 expression
was lower in GC cell lines (NCI-N87 and MKN-45) compared with a
normal gastric mucosa epithelial cell line (GES-1; Fig. 1E and F).
The Pearson's correlation analysis indicated that
JPX and miR-197 expression levels were moderately, negatively
correlated in GC tissues (Fig. 1G).
The Kaplan-Meier analysis suggested that high JPX expression or low
miR-197 expression in patients were associated with a shorter
overall survival time (Fig. 1H and
I). Therefore, the results demonstrated that high JPX
expression and low miR-197 expression in patients with GC indicated
poor prognosis.
Knockdown of JPX inhibits GC cell
activity, migration and invasion by promoting miR-197
expression
The StarBase results indicated that JPX may bind to
miR-197 (Fig. 2A). The luciferase
reporter gene assay further suggested that miR-197 overexpression
significantly decreased the relative luciferase activity of WT JPX,
but did not obviously alter the relative luciferase activity of MUT
JPX (Fig. 2B). Moreover, miR-197
mimic significantly increased miR-197 expression, but markedly
decreased JPX expression in NCI-N87 cells (Fig. 2C). RT-qPCR indicated that the
expression levels of JPX or miR-197 were decreased in GC cells
transfected with shJPX or miR-197 inhibitor, respectively (Fig. 2D and E). Compared with the shNC
group, the shJPX group had higher miR-197 expression levels.
However, compared with the shJPX group, the shJPX + miR-197
inhibitor group displayed significantly lower miR-197 expression
(Fig. 2F). The results indicated
that JPX bound to miR-197 and suppressed miR-197 expression.
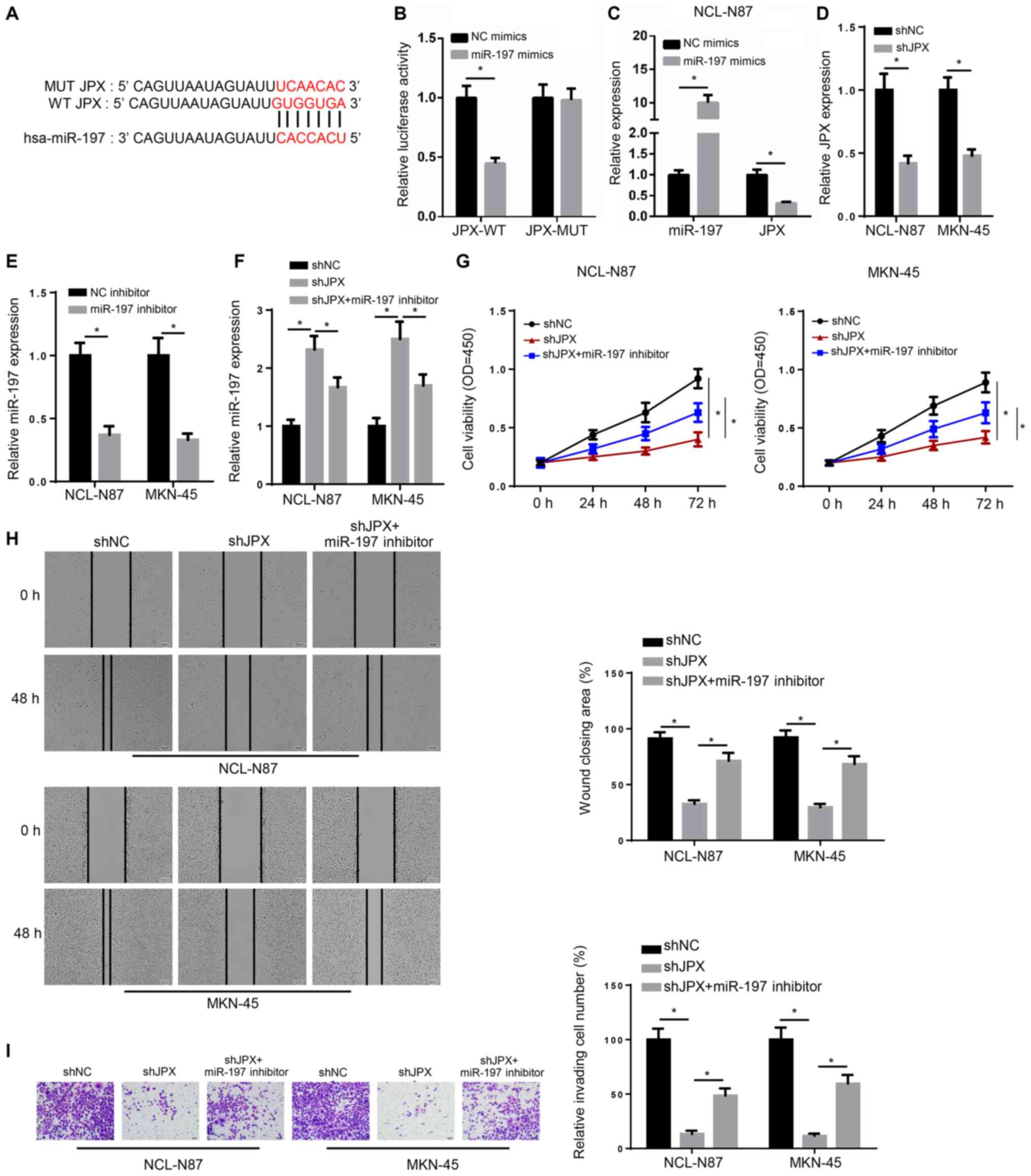 | Figure 2.Knockdown of JPX inhibits the
activity, migration and invasion of GC cells by promoting miR-197
expression. (A) Starbase predicted the targeted binding site
between JPX and miR-197. (B) Luciferase reporter gene assay was
used to confirm the relationship between JPX and miR-197. (C)
RT-qPCR was performed to detect JPX and miR-197 expression levels
in NCL-N87 cells. (D) RT-qPCR identified the expression of JPX in
NCL-N87 and MKN-45 transfected with shNC and shJPX. (E) RT-qPCR
demonstrated the expression of miR-197 in NCL-N87 and MKN-45
transfected with NC inhibitor and miR-197 inhibitor. (F) miR-197
expression in NCL-N87 and MKN-45 cells was assessed via RT-qPCR.
(G) Viability of NCL-N87 and MKN-45 cells was determined using a
Cell Counting Kit-8 assay. (H) Migration of cells was measured via
a wound healing test (magnification, ×200). (I) Invasion of cells
was evaluated with Transwell assays (magnification, ×200).
*P<0.05. miR, microRNA; JPX, long non-coding RNA JPX transcript,
XIST activator; RT-qPCR, reverse transcription-quantitative PCR;
NC, negative control; shRNA, short hairpin RNA; WT, wild-type; MUT,
mutant; OD, optical density. |
The shJPX group demonstrated lower cell viability at
72 h, a wider scratch width and decreased invasive abilities
compared with the shNC group. However, compared with the shJPX
group, the shJPX + miR-197 inhibitor group had a higher cell
viability at 72 h, a narrower scratch width and increased invasive
abilities (Fig. 2G-I). Therefore,
the results suggested that JPX knockdown inhibited GC cell
activity, migration and invasion by promoting miR-197
expression.
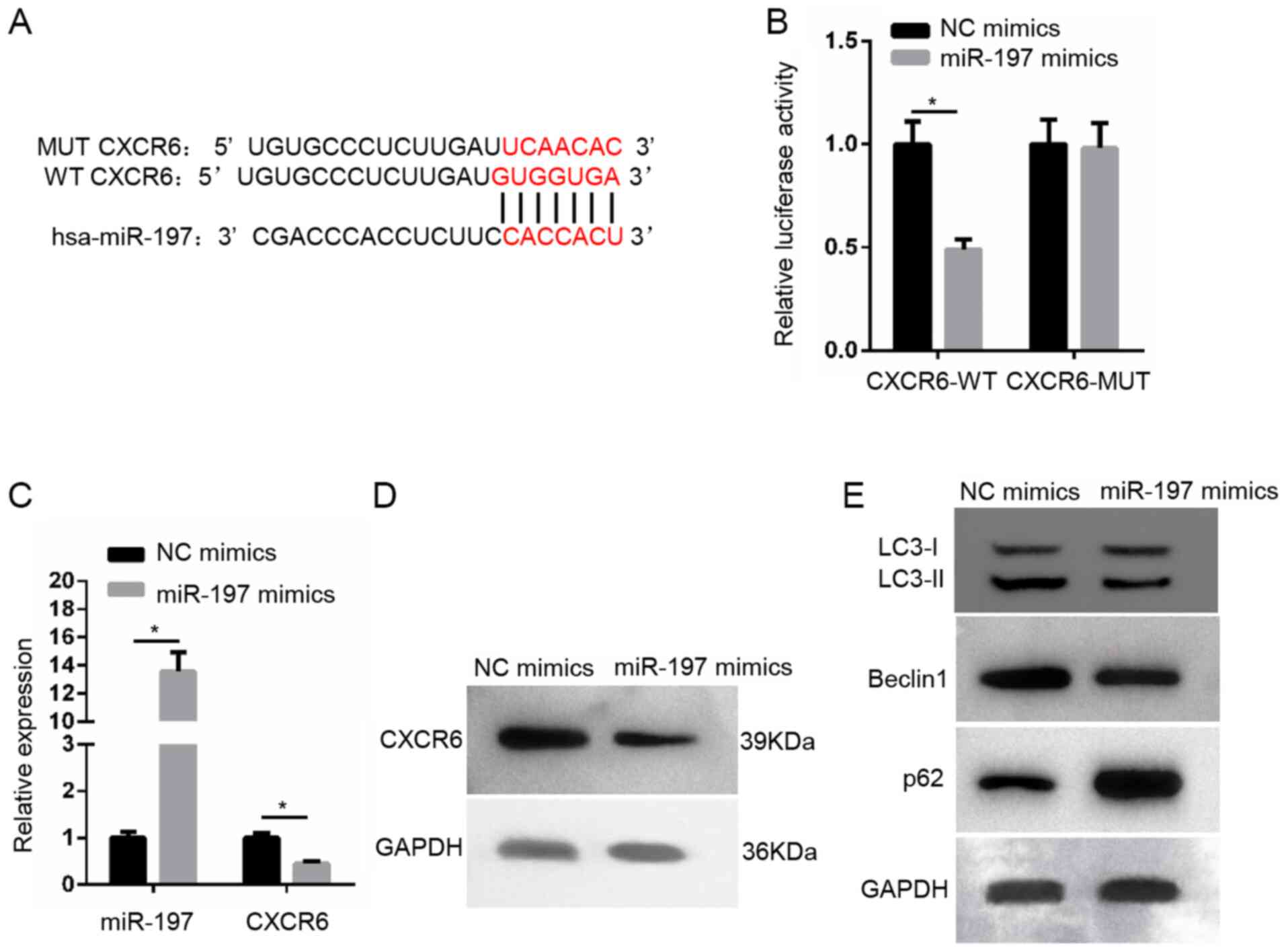
miR-197 inhibits CXCR6 expression and
autophagy
StarBase predicted that miR-197 possessed the
binding site for CXCR6 (Fig. 3A).
According to the luciferase reporter gene assay, miR-197
overexpression significantly inhibited the relative luciferase
activity of WT CXCR6, but did not notably alter the relative
luciferase activity of MUT CXCR6 (Fig.
3B). The miR-197 mimic group had higher miR-197 expression and
lower CXCR6 expression (Fig. 3C).
The western blotting results indicated that, compared with NC
mimics, miR-197 overexpression decreased CXCR6 expression, the
ratio of LC3-II/LC3-I and Beclin1 protein expression levels, but
increased p62 protein expression in NCI-N87 cells (Fig. 3D and E).
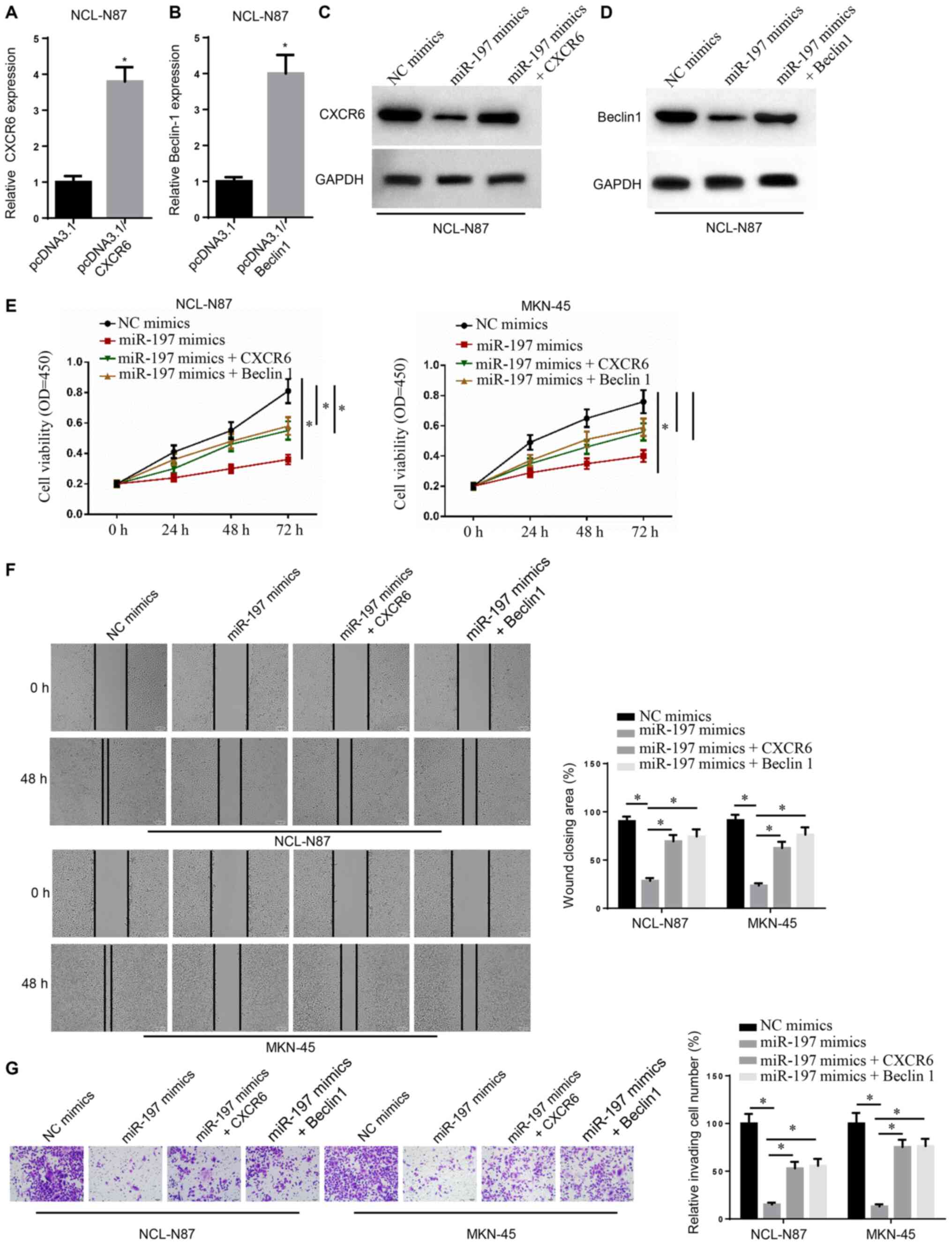
miR-197 decreases GC cell activity,
migration and invasion by inhibiting CXCR6 and Beclin1 expression
levels
The expression levels of CXCR6 and Beclin1 were
significantly upregulated in NCL-N87 cells transfected with
pcDNA3.1/CXCR6 or pcDNA3.1/Beclin1 (Fig. 4A and B) Following miR-197 mimic
transfection, NCI-N87 cells displayed markedly lower CXCR6 and
Beclin1 expression levels. However, compared with the miR-197 mimic
group, the miR-197 mimic + CXCR6 group displayed markedly higher
CXCR6 expression and the miR-197 mimic + Beclin1 group displayed
higher Beclin1 expression (Fig. 4C and
D).
NCI-N87 and MKN-45 cell viability, migration and
invasion were detected by performing CCK-8, wound healing and
Transwell assays, respectively. The miR-197 mimic group displayed
lower cell viability at 72 h, a wider scratch width and declined
invasive abilities compared with the NC group (Fig. 4E-G). However, compared with the
miR-197 mimic group, the miR-197 mimic + CXCR6 and miR-197 mimic +
Beclin1 groups had higher cell viability at 72 h, a narrower
scratch width and increased invasive abilities. Therefore, the
results indicated that miR-197 inhibited GC cell activity,
migration and invasion by inhibiting CXCR6 and Beclin1 expression
levels.
Discussion
The initiation and progression of human malignant
tumors are associated with various factors, such as etiological
factors, and genetic and epigenetic factors (25,26).
As important tumor regulators, lncRNAs can regulate the development
of tumors at the epigenetic, transcriptional and
post-transcriptional levels (27,28).
At present, the main mechanisms underlying lncRNAs affecting the
development of GC are generally as follows: Some lncRNAs promote GC
cell proliferation by inducing cell cycle arrest and inhibiting
apoptosis, thereby affecting the progression of GC (29); moreover, several lncRNAs can inhibit
or accelerate GC cell migration and invasion by affecting the
epithelial-mesenchymal transition (30); lncRNAs also serve as endogenous
competitive RNAs, which compete with miRNAs for binding to target
genes, thereby regulating the downstream signaling pathways and the
progression of GC (31); and
finally, lncRNAs can promote the progression of GC via autophagy,
metabolic stress and hypoxia (15).
The present study suggested that JPX served as an endogenous
competitive RNA, which promoted GC progression by regulating CXCR6
expression and promoting autophagy via sponging miR-197. To the
best of our knowledge, the present study was the first to indicate
that JPX served as a cancer-promoting gene in GC. Therefore, JPX
may be a novel potential target for the treatment of GC.
The present study demonstrated that miR-197 was
sponged and directly inhibited by JPX. Currently, the impact of
miR-197 on the development of GC is not completely understood.
Previous studies have investigated the effects of miR-197 on other
human diseases. For instance, miR-197 serves as a cancer-promoting
gene in bladder cancer, as indicated by the effect of miR-197
overexpression enhancing bladder cancer cell invasion and migration
(32). Chen and Yang (33) revealed that miR-197-3p expression
was significantly elevated in lung adenocarcinoma, and miR-197-3p
inhibition suppressed lung adenocarcinoma cell proliferation and
enhanced cell apoptosis. By contrast, TCGA analysis in the present
study indicated that miR-197-3p expression in patients with GC was
downregulated. Therefore, suggesting that miR-197-3p served as a
tumor suppressor gene in GC, as evidenced by miR-197-3p
overexpression attenuating cell proliferation, migration and
invasion.
In the present study, the mechanism underlying
miR-197-mediated inhibition of GC progression was suggested to
occur via regulation of CXCR6 and autophagy. CXCR6 was identified
to be a downstream gene of miR-197 and its expression was directly
suppressed by miR-197. CXCR6 is a chemokine receptor of CXCL16
(34). CXCR6 is abnormally
upregulated in multiple tumor tissues and cells, and the expression
of CXCR6 is increased with the severity of tumor malignancy
(35). In addition, CXCR6 has been
reported to serve as a cancer-promoting gene in human tumors. For
example, Ma et al (36)
indicated that CXCR6 accelerated osteosarcoma cell proliferation
and metastasis via the Akt signaling pathway. Gao et al
(37) also demonstrated that CXCR6
upregulation contributed to a proinflammatory tumor
microenvironment, which lead to metastasis and poor prognosis in
hepatocellular carcinoma. Moreover, Jin et al (38) reported that CXCR6 expression in GC
tissues was aberrantly increased, which was markedly associated
with lymph node metastases and advanced clinical stage. In
vitro studies have revealed that CXCR6 overexpression enhances
GC cell migration and invasion (37). Li et al (39) also demonstrated that CXCR6 was
significantly upregulated in patients with GC and GC cells.
Moreover, the aforementioned study indicated that blocking CXCR6
signaling weakened GC cell migration and invasion. Consistent with
the previous studies, the present findings also suggested that
CXCR6 was a cancer-promoting gene in GC. It was identified that
CXCR6 overexpression in GC cells reversed the inhibitory effect of
miR-197 on GC cell proliferation, migration and invasion.
Beclin1 and p62 are two important autophagy-related
proteins (40,41). Masuda et al (42) reported that autophagy was
prominently associated with poor survival in patients with GC,
especially for patients with stage I GC. Moreover, Geng et
al (43) revealed that Beclin1
was upregulated in GC tissues and cells, and Beclin1 expression was
an independent prognostic factor for patients with positive lymph
node metastasis. Valencia et al (44) also observed that the expression of
p62 was declined in the stroma of several tumors, and as an
anti-inflammatory tumor suppressor, p62 could inhibit tumor
development by modulating the metabolism in the tumor stroma. In
addition, low p62 expression is closely associated with poorer
outcomes in patients with colorectal cancer, as patients with low
p62 expression display the most aggressive tumor features (45). Thus, p62 is considered as a
candidate for autophagy-modulating therapies to treat tumors
(46). In the present study,
miR-197 overexpression decreased Beclin1 expression and increased
p62 expression levels. Furthermore, Beclin1 overexpression reversed
the inhibitory effect of miR-197 on GC cell viability, migration
and invasion. Therefore, miR-197 may inhibit the development of GC
by inhibiting autophagy.
However, there are limitations in the current study.
Firstly, 10% FBS was used in the wound healing assay; however,
cells should preferably be serum starved during wound healing
assays. Secondly, all findings were based on in vitro
assays; thus, in vivo experiments should be considered in
future studies to elucidate the role of JPX in GC.
In conclusion, the present study investigated the
effects of JPX in GC development and the results suggested that JPX
was a cancer-promoting gene in GC. Regarding the underlying
mechanism, JPX may promote the development of GC by regulating
CXCR6 and enhance autophagy via inhibiting miR-197. Therefore, JPX
may serve as a novel therapeutic target for GC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XH and ZL designed the present study, performed the
experiments, analyzed the data and prepared the figures, and
drafted the initial manuscript. ZL reviewed and revised the
manuscript. All authors read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of The Second Affiliated Hospital of Nanjing Medical
University. Written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Li A, Li J, Lin J, Zhuo W and Si J:
COL11A1 is overexpressed in gastric cancer tissues and regulates
proliferation, migration and invasion of HGC-27 gastric cancer
cells in vitro. Oncol Rep. 37:333–340. 2017. View Article : Google Scholar
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar
|
|
3
|
Maimaiti Y, Maimaitiming M, Li Y, Aibibula
S, Ainiwaer A, Aili A, Sun Z and Abudureyimu K: SSH1 expression is
associated with gastric cancer progression and predicts a poor
prognosis. BMC Gastroenterol. 18:122018. View Article : Google Scholar
|
|
4
|
Wei GH and Wang X: lncRNA MEG3 inhibit
proliferation and metastasis of gastric cancer via p53 signaling
pathway. Eur Rev Med Pharmacol Sci. 21:3850–3856. 2017.
|
|
5
|
Dai F, Xuan Y, Jin JJ, Yu S, Long ZW, Cai
H, Liu XW, Zhou Y, Wang YN, Chen Z and Huang H: CtBP2
overexpression promotes tumor cell proliferation and invasion in
gastric cancer and is associated with poor prognosis. Oncotarget.
8:28736–28749. 2017. View Article : Google Scholar
|
|
6
|
Tsukamoto Y, Uchida T, Karnan S, Noguchi
T, Nguyen LT, Tanigawa M, Takeuchi I, Matsuura K, Hijiya N and
Nakada C: Genome-wide analysis of DNA copy number alterations and
gene expression in gastric cancer. J Pathol. 216:471–482. 2008.
View Article : Google Scholar
|
|
7
|
Ruan X, Li P and Cao H: Identification of
transcriptional regulators that bind to long noncoding RNAs by RNA
pull-down and RNA immunoprecipitation. Methods Mol Biol.
1783:185–191. 2018. View Article : Google Scholar
|
|
8
|
Liu K, Mao X, Chen Y, Li T and Ton H:
Regulatory role of long non-coding RNAs during reproductive
disease. Am J Transl Res. 10:1–12. 2018.
|
|
9
|
Wu X, Zhang P, Zhu H, Li S, Chen X and Shi
L: Long noncoding RNA FEZF1-AS1 indicates a poor prognosis of
gastric cancer and promotes tumorigenesis via activation of Wnt
signaling pathway. Biomed Pharmacother. 96:1103–1108. 2017.
View Article : Google Scholar
|
|
10
|
Nie FQ, Ma S, Xie M, Liu YW, De W and Liu
XH: Decreased long noncoding RNA MIR31HG is correlated with poor
prognosis and contributes to cell proliferation in gastric cancer.
Tumour Biol. 37:7693–7701. 2016. View Article : Google Scholar
|
|
11
|
Shao Y, Chen H, Jiang X, Chen S, Li P, Ye
M, Li Q, Sun W and Guo J: Low expression of lncRNA-HMlincRNA717 in
human gastric cancer and its clinical significances. Tumour Biol.
35:9591–9595. 2014. View Article : Google Scholar
|
|
12
|
Zeng S, Xie X, Xiao YF, Tang B, Hu CJ,
Wang SM, Wu YY, Dong H, Li BS and Yang SM: Long noncoding RNA
LINC00675 enhances phosphorylation of vimentin on Ser83 to suppress
gastric cancer progression. Cancer Lett. 412:179–187. 2018.
View Article : Google Scholar
|
|
13
|
Sun M, Xia R, Jin F, Xu T, Liu Z, De W and
Liu X: Downregulated long noncoding RNA MEG3 is associated with
poor prognosis and promotes cell proliferation in gastric cancer.
Tumour Biol. 35:1065–1073. 2014. View Article : Google Scholar
|
|
14
|
Shen W, Yuan Y, Zhao M, Li J, Xu J, Lou G,
Zheng J, Bu S, Guo J and Xi Y: Novel long non-coding RNA GACAT3
promotes gastric cancer cell proliferation through the IL-6/STAT3
signaling pathway. Tumor Biol. 37:14895–14902. 2016. View Article : Google Scholar
|
|
15
|
Liu X, Wang Y, Sun L, Min J, Liu J, Chen
D, Zhang H, Zhang H, Zhang H, Zhou Y and Liu L: Long noncoding RNA
BC005927 upregulates EPHB4 and promotes gastric cancer metastasis
under hypoxia. Cancer Sci. 109:988–1000. 2018. View Article : Google Scholar
|
|
16
|
Wang ZQ, He CY, Hu L, Shi HP, Li JF, Gu
QL, Su LP, Liu BY, Li C and Zhu Z: Long noncoding RNA UCA1 promotes
tumour metastasis by inducing GRK2 degradation in gastric cancer.
Cancer Lett. 408:10–21. 2017. View Article : Google Scholar
|
|
17
|
Wang P, Liu X, Han G, Dai S, Ni Q, Xiao S
and Huang J: Downregulated lncRNA UCA1 acts as ceRNA to adsorb
microRNA-498 to repress proliferation, invasion and epithelial
mesenchymal transition of esophageal cancer cells by decreasing
ZEB2 expression. Cell Cycle. 18:2359–2376. 2019. View Article : Google Scholar
|
|
18
|
Liao Z, Li Y, Zhou Y, Huang Q and Dong J:
MicroRNA-197 inhibits gastric cancer progression by directly
targeting metadherin. Mol Med Rep. 17:602–611. 2018.
|
|
19
|
Chen Z, Ju H, Zhao T, Yu S, Li P, Jia J,
Li N, Jing X, Tan B and Li Y: hsa_circ_0092306 targeting miR-197-3p
promotes gastric cancer development by regulating PRKCB in MKN-45
cells. Mol Ther Nucleic Acids. 18:617–626. 2019. View Article : Google Scholar
|
|
20
|
Luan X and Wang Y: LncRNA XLOC_006390
facilitates cervical cancer tumorigenesis and metastasis as a ceRNA
against miR-331-3p and miR-338-3p. J Gynecol Oncol. 29:e952018.
View Article : Google Scholar
|
|
21
|
Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G,
Zhao Q, Wu D, Gong W, Du M, et al: LncRNA MT1JP functions as a
ceRNA in regulating FBXW7 through competitively binding to
miR-92a-3p in gastric cancer. Mol Cancer. 17:872018. View Article : Google Scholar
|
|
22
|
Liang H, Yu T, Han Y, Jiang H, Wang C, You
T, Zhao X, Shan H, Yang R, Yang L, et al: LncRNA PTAR promotes EMT
and invasion-metastasis in serous ovarian cancer by competitively
binding miR-101-3p to regulate ZEB1 expression. Mol Cancer.
17:1192018. View Article : Google Scholar
|
|
23
|
Livak K and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
24
|
Yang Q, Yu Y, Sun Z and Pan Y: Long
non-coding RNA PVT1 promotes cell proliferation and invasion
through regulating miR-133a in ovarian cancer. Biomed Pharmacother.
106:61–67. 2018. View Article : Google Scholar
|
|
25
|
Giovannucci E: Modifiable risk factors for
colon cancer. Gastroenterol Clin North Am. 31:925–943. 2002.
View Article : Google Scholar
|
|
26
|
Cavatorta O, Scida S, Miraglia C, Barchi
A, Nouvenne A, Leandro G, Meschi T, De'Angelis GL and Di Mario F:
Epidemiology of gastric cancer and risk factors. Acta Biomed.
89:82–87. 2018.
|
|
27
|
Tan H, Zhang S, Zhang J, Zhu L, Chen Y,
Yang H, Chen Y, An Y and Liu B: Long non-coding RNAs in gastric
cancer: New emerging biological functions and therapeutic
implications. Theranostics. 10:8880–8902. 2020. View Article : Google Scholar
|
|
28
|
Sun DE and Ye SY: Emerging roles of long
noncoding RNA regulator of reprogramming in cancer treatment.
Cancer Manag Res. 12:6103–6112. 2020. View Article : Google Scholar
|
|
29
|
Pan L, Liang W, Gu J, Zang X, Huang Z, Shi
H, Chen J, Fu M, Zhang P, Xiao X, et al: Long noncoding RNA DANCR
is activated by SALL4 and promotes the proliferation and invasion
of gastric cancer cells. Oncotarget. 9:1915–1930. 2017. View Article : Google Scholar
|
|
30
|
Liu YW, Sun M, Xia R, Zhang EB, Liu XH,
Zhang ZH, Xu TP, De W, Liu BR and Wang ZX: LincHOTAIR
epigenetically silences miR34a by binding to PRC2 to promote the
epithelial-to-mesenchymal transition in human gastric cancer. Cell
Death Dis. 6:e18022015. View Article : Google Scholar
|
|
31
|
Mao Y, Liu R, Zhou H, Yin S, Zhao Q, Ding
X and Wang H: Transcriptome analysis of miRNA-lncRNA-mRNA
interactions in the malignant transformation process of gastric
cancer initiation. Cancer Gene Ther. 24:267–275. 2017. View Article : Google Scholar
|
|
32
|
Wang YY, Wu ZY, Wang GC, Liu K, Niu XB, Gu
S and Meng JS: LINC00312 inhibits the migration and invasion of
bladder cancer cells by targeting miR-197-3p. Tumor Biol.
37:14553–14563. 2016. View Article : Google Scholar
|
|
33
|
Chen Y and Yang C: miR-197-3p-induced
downregulation of lysine 63 deubiquitinase promotes cell
proliferation and inhibits cell apoptosis in lung adenocarcinoma
cell lines. Mol Med Rep. 17:3921–3927. 2018.
|
|
34
|
Darash-Yahana M, Gillespie JW, Hewitt SM,
Chen YY, Maeda S, Stein I, Singh SP, Bedolla RB, Peled A, Troyer
DA, et al: The chemokine CXCL16 and its receptor, CXCR6, as markers
and promoters of inflammation-associated cancers. PLoS One.
4:e66952009. View Article : Google Scholar
|
|
35
|
Deng L, Chen N, Li Y, Zheng H and Lei Q:
CXCR6/CXCL16 functions as a regulator in metastasis and progression
of cancer. Biochim Biophys Acta. 1806:42–49. 2010.
|
|
36
|
Ma Y, Xu X and Luo M: CXCR6 promotes tumor
cell proliferation and metastasis in osteosarcoma through the Akt
pathway. Cell Immunol. 311:80–85. 2017. View Article : Google Scholar
|
|
37
|
Gao Q, Zhao YJ, Wang XY, Qiu SJ, Shi YH,
Sun J, Yi Y, Shi JY, Shi GM, Ding ZB, et al: CXCR6 upregulation
contributes to a proinflammatory tumor microenvironment that drives
metastasis and poor patient outcomes in hepatocellular carcinoma.
Cancer Res. 72:3546–3556. 2012. View Article : Google Scholar
|
|
38
|
Jin JJ, Dai FX, Long ZW, Cai H, Liu XW,
Zhou Y, Hong Q, Dong QZ, Wang YN and Huang H: CXCR6 predicts poor
prognosis in gastric cancer and promotes tumor metastasis through
epithelial-mesenchymal transition. Oncol Rep. 37:3279–3286. 2017.
View Article : Google Scholar
|
|
39
|
Li Y, Fu LX, Zhu WL, Shi H, Chen LJ and Ye
B: Blockade of CXCR6 reduces invasive potential of gastric cancer
cells through inhibition of AKT signaling. Int J Immunopathol
Pharmacol. 28:194–200. 2015. View Article : Google Scholar
|
|
40
|
Zhai QL, Hu XD, Xiao J and Yu DQ:
Astragalus polysaccharide may increase sensitivity of cervical
cancer HeLa cells to cisplatin by regulating cell autophagy.
Zhongguo Zhong Yao Za Zhi. 43:805–812. 2018.(In Chinese).
|
|
41
|
Xu Z, Yan Y, Zeng S, Qian L, Dai S, Xiao
L, Wang L, Yang X, Xiao Y and Gong Z: Reducing autophagy and
inducing G1 phase arrest by aloperine enhances radio-sensitivity in
lung cancer cells. Oncol Rep. 43:1541–1548. 2017.
|
|
42
|
Masuda G, Yashiro M, Kitayama K, Miki Y,
Kasashima H, Kinoshita H, Morisaki T, Fukuoka T, Hasegawa T,
Sakurai K, et al: Clinicopathological correlations of
autophagy-related proteins LC3, Beclin 1 and p62 in gastric cancer.
Anticancer Res. 36:129–136. 2016.
|
|
43
|
Geng QR, Xu DZ, He LJ, Lu JB, Zhou ZW,
Zhan YQ and Lu Y: Beclin-1 expression is a significant predictor of
survival in patients with lymph node-positive gastric Cancer. PLoS
One. 7:e459682012. View Article : Google Scholar
|
|
44
|
Valencia T, Kim JY, Abu-Baker S,
Moscat-Pardos J, Ahn CS, Reina-Campos M, Duran A, Castilla EA,
Metallo CM, Diaz-Meco MT and Moscat J: Metabolic reprogramming of
stromal fibroblasts through p62-mTORC1 signaling promotes
inflammation and tumorigenesis. Cancer Cell. 26:121–135. 2014.
View Article : Google Scholar
|
|
45
|
Zhang J, Yang S, Xu B, Wang T, Zheng Y,
Liu F, Ren F, Jiang J, Shi H, Zou B, et al: p62 functions as an
oncogene in colorectal cancer through inhibiting apoptosis and
promoting cell proliferation by interacting with the vitamin D
receptor. Cell Prolif. 52:e125852019. View Article : Google Scholar
|
|
46
|
Adams O, Dislich B, Berezowska S, Schläfli
AM, Seiler CA, Kröll D, Tschan MP and Langer R: Prognostic
relevance of autophagy markers LC3B and p62 in esophageal
adenocarcinomas. Oncotarget. 7:39241–39255. 2016. View Article : Google Scholar
|