Introduction
Gastric cancer (GC) is one of the most frequent
types of cancer and the second leading cause of cancer-associated
deaths worldwide (1). According to
recorded statistics from 2012, >70% of new cases of GC occured
in developing countries, primarily in Asia, Central Europe, Eastern
Europe and Latin America (2,3). GC
results in >951,000 new cases and 723,000 deaths per year
worldwide (3). Tumor metastasis,
which is responsible for 90% of all cancer-associated deaths, is a
multi-step process of biological cascades that ultimately leads to
the spread of tumor cells to different tissues (4,5). Early
surgical treatment, targeted chemotherapy and immunotherapy are
widely applied in managing the metastasis and progression of GC,
but the results are often unsatisfactory (6–8). Tumor
cells will develop multi-drug resistance to chemotherapy, which is
a key factor affecting the therapeutic effect on numerous types of
cancer, including GC (9).
Therefore, further exploration of the pathogenesis of GC and
development of a more effective treatment is necessary.
The human genome consists of >98% non-coding
regions and therefore the remaining <2% of the human genome are
translated into proteins; most non-coding regions are directly
transcribed into RNAs (10–12). These RNAs cannot be translated into
proteins and are therefore considered as non-coding RNAs (ncRNAs)
(10–12). The ncRNA family members are numerous
and highly diverse, and can be easily divided into short ncRNAs
(<200 nucleotides in length) and long ncRNAs (lncRNAs; >200
nucleotides in length) according to RNA length. The most
representative and the most studied ncRNAs are microRNAs
(miRNAs/miRs) and lncRNAs (13–17). A
number of studies have demonstrated a mutual regulation between
lncRNAs and miRNAs, and different regulatory forms of the two in
interaction affect various diseases, including GC (13–17).
Li et al (18) demonstrated that the lncRNA small
nucleolar RNA host gene 4 (SNHG4) can sponge miR-148a-3p to exert a
cancer-promoting function in cervical cancer. Chen et al
(19) demonstrated that miR-204-5p
acts as an antitumor factor in GC, suppressing GC progression. The
current study aimed to explore the association between SNHG4 and
miR-148a-3p in GC cells.
Materials and methods
Ethics approval
All experiments were approved by the Ethics Board of
Zhuji People's Hospital (Shaoxing, China). A total of 53 patients
diagnosed with GC by gastroscopy and pathological examination were
recruited in the current study. All patients in the present study
provided written informed consent and their pathophysiological
characteristics (age, sex, histology differentiation, borrmann
type, tumour location, lymph node metastasis, tumour invasion and
TNM stage) (20) were collected
(Table I). None of the enrolled
patients received chemotherapy, radiotherapy or targeted therapy
before radical surgery. Samples from GC tissues and adjacent
non-tumor tissues (≥2 cm from the tumour margin) were collected
from the 53 patients with GC (age range, 38–85 years; mean age, 61
years) who underwent radical gastrectomy at Zhuji People's Hospital
between April 2018 and June 2019. All the samples were frozen in
liquid nitrogen immediately after the resection and stored at −80°C
until use.
 | Table I.Association between SNHG4 expression
and clinical characteristics. |
Table I.
Association between SNHG4 expression
and clinical characteristics.
|
| Relative SNHG4
expression, n |
|
|
|---|
|
|
|
|
|
|---|
| Variable | Low (n=27) | High (n=26) | χ2 | P-value |
|---|
| Age, years |
|
| 0.573 | 0.449 |
|
≥60 | 16 | 18 |
|
|
|
<60 | 11 | 8 |
|
|
| Sex |
|
| 0.172 | 0.678 |
|
Male | 14 | 12 |
|
|
|
Female | 13 | 14 |
|
|
| Histological
differentiation |
|
| 3.627 | 0.163 |
|
Well | 9 | 2 |
|
|
|
Moderate | 11 | 10 |
|
|
|
Poor | 7 | 14 |
|
|
| Borrmann type |
|
| 2.051 | 0.359 |
| Early
stage | 5 | 2 |
|
|
| I+II
type | 9 | 7 |
|
|
| III+IV
type | 13 | 17 |
|
|
| Tumor location |
|
| 1.672 | 0.433 |
| Upper
stomach | 6 | 5 |
|
|
| Middle
stomach | 9 | 10 |
|
|
| Lower
stomach | 7 | 5 |
|
|
|
Mixed | 5 | 6 |
|
|
| Lymph node
metastasis |
|
| 8.578 | 0.003 |
|
Yes | 10 | 20 |
|
|
| No | 17 | 6 |
|
|
| Tumor invasion
(AJCC) |
|
| 8.312 | 0.004 |
|
Tis-T2 | 19 | 8 |
|
|
|
T3-T4 | 8 | 18 |
|
|
| TNM stage
(AJCC) |
|
| 13.746 | <0.001 |
|
I–II | 21 | 7 |
|
|
|
III–IV | 6 | 19 |
|
|
Bioinformatics
The expression levels of SNHG4 in GC and normal
tissues was predicted using starBase v3.0 (starbase.sysu.edu.cn). The patients were divided into
high SNHG4 and low SNHG4 groups according to the median (median
value, 41). The overall survival rate of patients with low or high
SNHG4 expression was plotted using Kaplan-Meier (21). The interaction between SNHG4 and
miR-204-5p was predicted using TargetScan v7.2 (www.targetscan.org).
Cell culture
Human gastric epithelial cell line GES-1 and GC cell
lines (SNU719, AGS and HGC-27) were purchased from The Cell Bank of
Type Culture Collection of the Chinese Academy of Sciences. GES-1
and HGC-27 cells were cultured in RPMI-1640 medium (cat. no.
21875091; Thermo Fisher Scientific, Inc.), SNU719 cells were
cultured in DMEM (cat. no. D0819; Sigma-Aldrich; Merck KGaA) and
AGS cells were cultured in Ham's F-12K (Kaighn's) medium (cat. no.
21127022; Thermo Fisher Scientific, Inc.). The cells were
supplemented with 10% FBS (cat. no. F8192) and
penicillin-streptomycin reagent (cat. no. V900929) (both from
Sigma-Aldrich; Merck KGaA), and cultured at 37°C with 5%
CO2.
Experimental design
According to the expression levels of SNHG4, AGS and
HGC-27 cell lines were used in subsequent experiments. To explore
the association between SNHG4 and miR-204-5p, AGS and HGC-27 cells
were divided into different groups depending on their treatment.
Cells were treated with short interfering (si)SNHG4 (Shanghai
GenePharma Co., Ltd.) plus miR-204-5p inhibitor control (IC;
non-targeting scrambled control; Shanghai GenePharma Co., Ltd.),
siSNHG4 plus miR-204-5p inhibitor (I; Shanghai GenePharma Co.,
Ltd.), siSNHG4 negative control (siNC; non-targeting scrambled
control; Shanghai GenePharma Co., Ltd.) plus miR-204-5p IC and siNC
plus miR-204-5p I, corresponding to the siSNHG4+IC, siSNHG4+I,
siNC+IC and siNC+I groups, respectively. The sequences of siRNAs
and miRNAs were as follows: siSNHG4, 5′-UAUUUCCUCCCUUCAGAUGGG-3′;
siNC, 5′-UUCUCCGAACGUGUCACGUTT-3′; miR-204-5p I,
5′-AGGCAUAGGAUGACAAAGGGAA-3′; and miR-204-5p IC,
5′-UCUACUCUUUCUAGGAGGUUGUGA-3′. For transfection, AGS and HGC-27
cells (2×105 cells) were seeded in 6-well plates and
incubated at 37°C for 24 h. When cell confluence reached 80–90%,
the cells were transfected with siSNHG4 (50 nM), siNC (50 nM),
miR-204-5p I (100 nM) and miR-204-5p IC (100 nM) using
Lipofectamine® 3000 (cat. no. L3000015; Thermo Fisher
Scientific, Inc.) according to the manufacturer's protocol.
Transfection efficiency was monitored using quantitative (q)PCR
after 48 h. At 48 h post-transfection, cells were used for
subsequent experiments.
Reverse transcription (RT)-qPCR
Following the collection of GC tissues from the
patients, total RNA was extracted from samples and GC cells
(5×106) using TRIzol® reagent (cat. no.
15596018; Thermo Fisher Scientific, Inc.) according to the
manufacturer's protocol and used to detect the expression levels of
SNHG4 and miR-204-5p in adjacent normal tissues (ANTs), GC tissues
and transfected cells. The same procedures were performed in GES-1,
SNU719, AGS and HGC-27 cells. cDNA was synthesized using
PrimeScript RT reagent kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's protocols. qPCR was performed in a
IQ5 thermocycler (Bio-Rad Laboratories, Inc.) under the following
conditions: 90 sec at 95°C, 30 sec at 95°C, 20 sec at 65°C and 30
sec at 72°C, for 40 cycles. The reaction system was composed of 8
µl 2X SYBR Green master mix [cat. no. 4913850001; Roche Diagnostics
(Shanghai) Co., Ltd.], 1 µl forward primer (10 µM), 1 µl reverse
primer (10 µM), 10 µl cDNA template and 5 µl double-distilled
H2O. Relative expression levels of each sample were
calculated using the 2−ΔΔCq method (22). miRNA and mRNA expression levels were
normalized to the internal reference genes U6 and GAPDH,
respectively. The primers are listed in Table II. The experiment was repeated
three times independently.
 | Table II.Primers used in the present
study. |
Table II.
Primers used in the present
study.
| Primer name | Sequence (5–3) |
|---|
| miR-204-5p | F
CCTTTGTCATCCTATGCC |
| miR-204-5p | R
GAACATGTCTGCGTATCTC |
| lncRNA SNHG4 | F
GCAGGTGACAGTCTGCATGT |
| lncRNA SNHG4 | R
TTTTAAGTCCCCTACCCCCATC |
| GAPDH | F
CGCTTCACGAATTTGCGTGTCAT |
| GAPDH | R
GAAGATGGTGATGGGATTTC |
| U6 | F
GCTTCGGCAGCACATATACTAAAAT |
| U6 | R
CGCTTCACGAATTTGCGTGTCAT |
Dual-luciferase reporter assay
The verification of the interaction between SNHG4
and miR-204-5p was performed using a dual-luciferase reporter
assay. SNHG4 mutation was created using the Quick-Change
Site-Directed Mutagenesis kit (Agilent Technologies, Inc.). The
pRL-TK plasmid (Promega Corporation) was transfected with
miR-204-5p I using Lipofectamine 3000. At 48 h post-transfection,
the dual-Glo luciferase assay kit (Promega Corporation) was used to
determine the relative luciferase activities of AGS and HGC-27
cells. Relative luciferase activity was normalized to
Renilla luciferase activity.
Cell Counting Kit (CCK)-8
Following cell transfection, AGS and HGC-27 cells
(1×106 cells/well) were cultured for 24, 48 and 72 h in
6-well plates and incubated with 10 µl CCK-8 reagent (cat. no.
96992-100TESTS-F; Sigma-Aldrich; Merck KGaA) at 37°C for 2 h.
Finally, the optical density (OD) at 450 nm was read and recorded
using a Multiskan microplate reader (Thermo Fisher Scientific,
Inc.).
Colony formation assay
Following cell transfection, the cells were
harvested at 1×106 cells/ml and seeded onto plates for
14 days at 37°C with 5% CO2 to assess colony formation.
After 100–120 clones were formed, the colonies were stained with
0.2% crystal violet for 30 min at room temperature after fixation
with 1 ml methanol for 15 min at room temperature. Finally, the
number of colonies (cell clusters containing ≥5 cells) were
observed and counted under a light microscope (magnification, ×1).
Colony formation rate (%) was calculated according to the following
formula: (number of clones/cells inoculated) ×100.
Flow cytometry
At 48 h post-transfection, cell apoptosis was
determined following transfection. AGS and HGC-27 cells were
diluted to 1×106 cells/ml after 48 h culture at 37°C,
washed with PBS and then mixed with 200 µl binding buffer.
Following the instructions of the Annexin V-FITC kit
(Sigma-Aldrich; Merck KGaA), 5 µl PI and 10 µl FITC-labeled Annexin
V was added to cells for 15 min at room temperature. The cells were
then resuspended in 300 µl binding buffer and then subjected to
flow cytometry (BD FACSVerse™; BD Biosciences) and analyzed using
FlowJo software (version 10.0; FlowJo LLC) to determine cell
apoptosis. The rate of apoptosis was calculated as the sum of early
and late apoptosis.
Wound-healing assay
At 48 h post-transfection, AGS and HGC-27 cells
(1×106 cells/ml) were harvested into the dishes to
create a monolayer. At 90–100% confluence, When the monolayers of
cells were grown to ~90–100% confluency, a gap in the center of the
layer was made using a 200-µl pipette tip and the cells were
maintained in serum-free RPMI-1640 medium with 5% CO2 at
37°C. After 48 h, the floating cells were removed by washing the
cells with PBS. The distance of cell migration was determined by
the mean value of the width of the gap between the top, middle and
bottom of the wound. The wounds were observed in five fields of
view using a light microscope (magnification, ×100).
Transwell assay
A Transwell assay was performed for determining cell
invasion. As aforementioned, AGS and HGC-27 cells (1×106
cells/ml) were harvested and seeded with serum-free
RPMI-1640-medium into the upper chamber of a 8-µm Transwell insert
pre-coated with Matrigel (BD Biosciences). In the bottom chamber,
300 µl RPMI-1640 medium with 10% FBS was added. The cells were
incubated for 48 h with 5% CO2 at 37°C for the invasion
test. Finally, the cells that had invaded into the bottom chamber
were washed with PBS and fixed with 4% paraformaldehyde for 15 min
at room temperature and then stained with 0.2% crystal violet for
10 min at room temperature for observation under a light microscope
(magnification, ×250).
Western blot analysis
The protein expression levels of E-cadherin (cad),
N-cad and Snail of the cells transfected with siSNHG4 or miR-204-5p
I were detected. A total of 1×106 cells/ml (AGS and
HGC-27 cells) were harvested and lysed using RIPA lysis buffer
(cat. no. R0278; Sigma-Aldrich; Merck KGaA) plus protease inhibitor
(cat. no. S8830; Sigma-Aldrich; Merck KGaA) to obtain the total
protein. Protein concentration was measured using BCA regent
(Sigma-Aldrich; Merck KGaA). Protein samples (40 µg/lane) were
separated via 12% SDS-PAGE at 90 V for 2 h, then transferred to
PVDF membranes and blocked using 5% skimmed milk for 1 h at room
temperature. Antibodies against proliferating cell nuclear antigen
(PCNA; cat. no. ab92552; Abcam; 1:1,000; 29 kDa), cyclin D1 (cat.
no. ab16663; Abcam; 1:200; 33 kDa), E-cad (cat. no. ab40772; Abcam;
1:1,000; 97 kDa), N-cad (cat. no. ab76057; Abcam; 1:1,000; 100
kDa), Snail (cat. no. ab216347; Abcam; 1:1,000; 29 kDa) and GAPDH
(cat. no. ab181602; Abcam; 1:1,000; 36 kDa) were mixed in 5%
skimmed milk and incubated with the membrane overnight at 4°C.
After washing with PBS-Tween (0.1% Tween), primary antibodies were
detected using an HRP-conjugated goat anti-rabbit secondary
antibody (cat. no. ab205718; Abcam; 1:2,000) at room temperature
for 2 h, and the protein signals were developed using SignalFire™
ECL Reagent (cat. no. 6883; Cell Signaling Technology, Inc.).
Protein expression levels were quantified using ImageJ software
(v1.8.0; National Institutes of Health), with GAPDH as the loading
control.
Statistical analysis
Data are presented as the mean ± SD of three
independent experiments. The data were analyzed using GraphPad
Prism 8.0 (GraphPad Software, Inc.). The statistical differences
with regard to the overall survival rate and pathophysiological
characteristics among patients with low or high SNHG4 expression
were analyzed using the log-rank and χ2 tests,
respectively. The correlation between the expression levels of
SNHG4 and miR-204-5p was analyzed using Pearson's correlation
analysis. An unpaired Student's t-test was used to analyze the
statistical difference between two groups. One-way ANOVA followed
by Tukey's post hoc test was conducted to measure the statistical
difference among multiple groups, and paired Student's t-test was
used to compare the data between tumor and adjacent non-tumor
tissues. P<0.05 was considered to indicate a statistically
significant difference.
Results
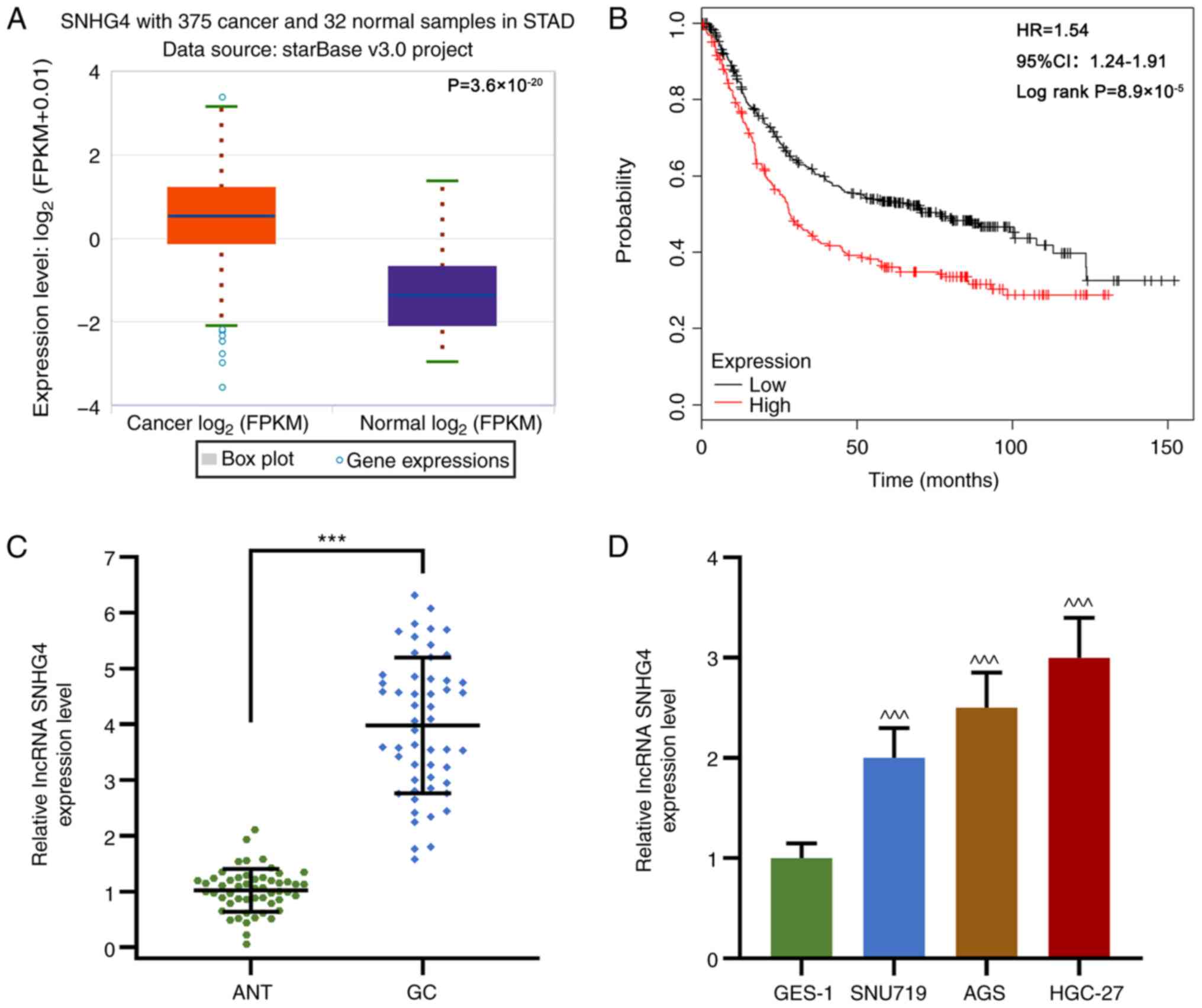
SNHG4 is upregulated in GC tissues and
cell lines
To examine the role of SNHG4 in GC, SNHG4 expression
was measured in GC tissues and cell lines. In GC tissues, SNHG4 was
highly expressed and high SNHG4 expression was associated with a
lower survival rate compared with low SNHG4 expression (P<0.001;
Fig. 1A-C). Similarly, SNHG4
expression in SNU719, AGS and HGC-27 cells was significantly
upregulated compared with that in GES-1 cells, which are normal
gastric epithelial cells (P<0.001; Fig. 1D). In addition, as shown in Table I, SNHG4 expression was significantly
associated with lymph node metastasis, tumor invasion and TNM
stage. Therefore, the data suggested that SNHG4 may promote GC
development.
miR-204-5p expression is suppressed in
GC tissues and cell lines, and is negatively correlated with SHNG4
expression
miR-204-5p was predicted and verified to interact
with SNHG4 by bioinformatics and a dual-luciferase reporter assay,
and the expression levels of miR-204-5p in GC were investigated.
Notably, miR-204-5p shared a complementary sequence with SNHG4, and
the dual-luciferase reporter assay demonstrated that miR-204-5p
inhibitor increased the relative luciferase activities of SNHG4-WT
in AGS and HGC-27 cells compared with the miR-204-5p inhibitor
control group (P<0.001; Fig.
2A-C). In addition, miR-204-5p expression in GC tissues and
cell lines (SNU719, AGS and HGC-27) was significantly decreased
compared with that in ANTs or GES-1 cells, respectively
(P<0.001; Fig. 2D and G).
Notably, in ANTs and GC tissues, the expression levels of
miR-204-5p and SNHG4 were found to be negatively correlated
(Fig. 2E and F). The results
indicated that SNHG4 may exert its regulatory function via
suppressing miR-204-5p expression.
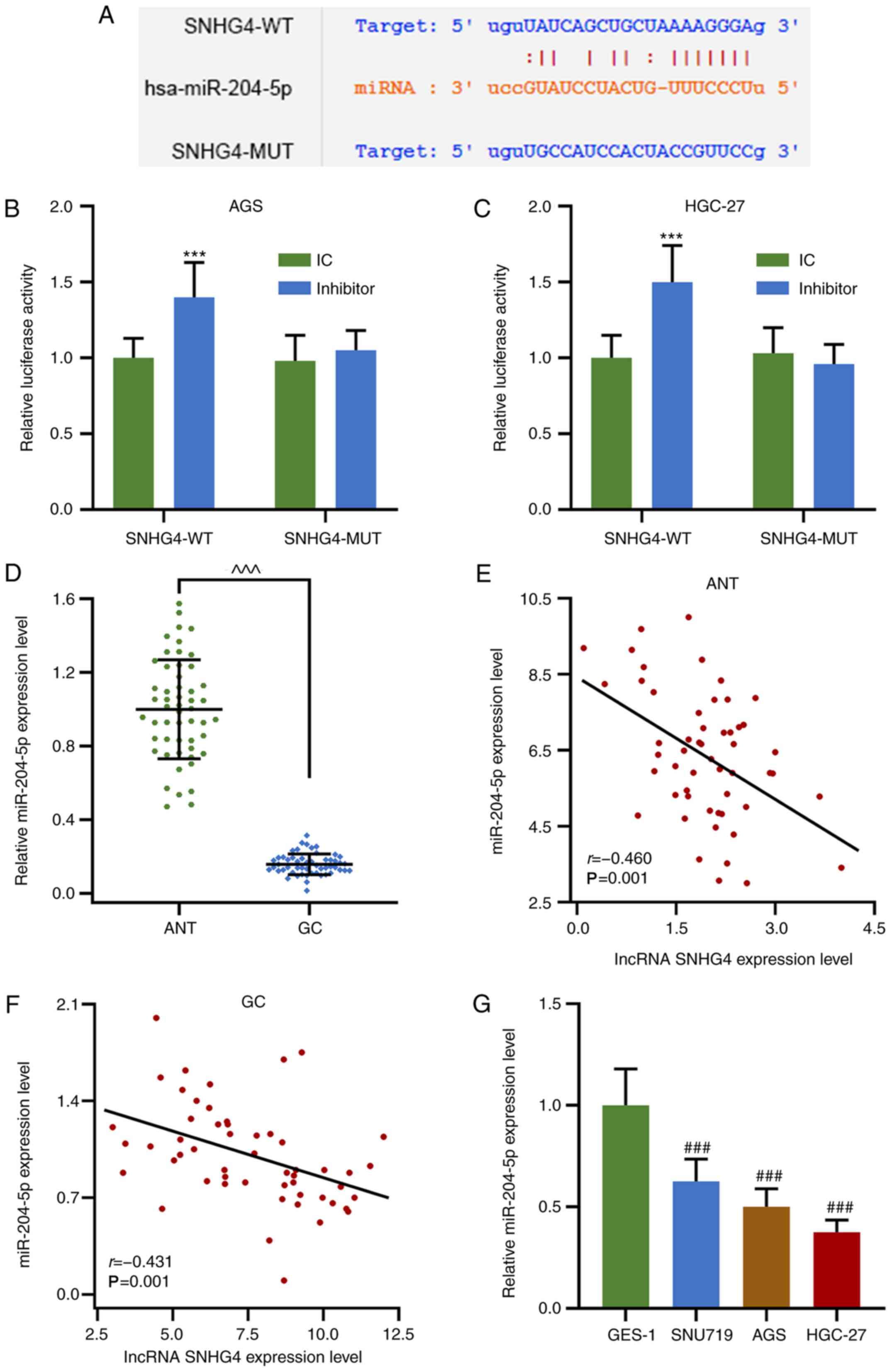 | Figure 2.miR-204-5p expression is downregulated
in GC tissues and cell lines, and its expression levels are
negatively correlated with those of SNHG4. (A) Possible
complementary sequences in SNHG4-WT and miR-204-5p. Relative
luciferase activity in (B) AGS and (C) HGC-27 cells treated with or
without miR-204-5p inhibitor when SNHG4 sequence was WT or MUT. (D)
Relative miR-204-5p expression levels in ANTs or GC tissues.
Correlation between the expression levels of SNHG4 and miR-204-5p
in (E) ANT and (F) GC tissues. (G) Relative miR-204-5p expression
levels in GES-1, SNU719, AGS and HGC-27 cell lines. Bars indicate
the mean ± standard deviation. ***P<0.001 vs. IC;
^^^P<0.001 vs. ANT; ###P<0.001 vs.
GES-1. SNHG4, small nucleolar RNA host gene 4; GC, gastric cancer;
miR, microRNA; WT, wild-type; MUT, mutant; ANT, adjacent normal
tissues; IC, inhibitor control. |
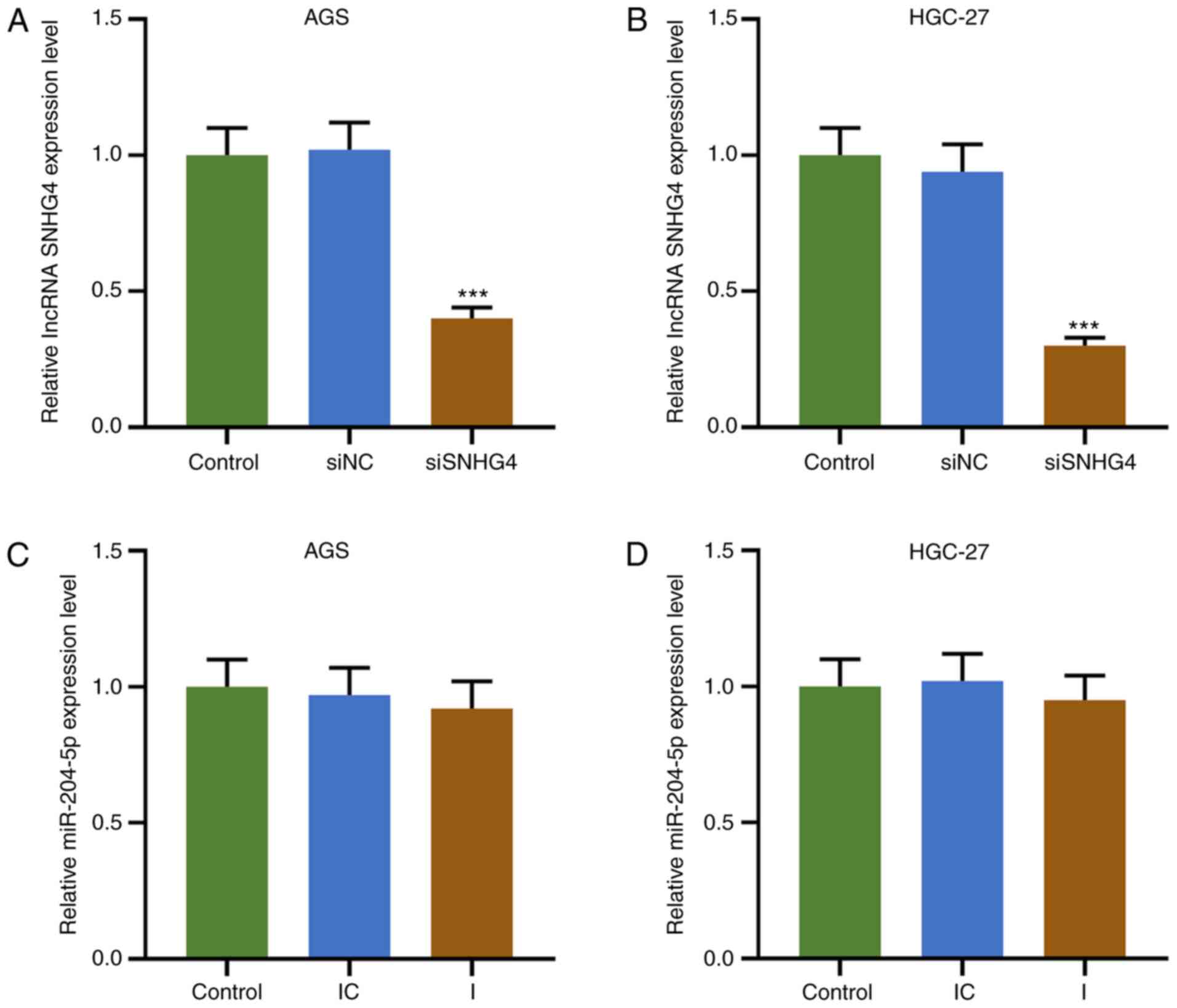
Downregulation of SNHG4 expression
reverses the effects of miR-204-5p inhibitor on the proliferation
and apoptosis of GC cells
To understand the association between SNHG4 and
miR-204-5p in GC, cell viability, colony formation and apoptosis
were evaluated when SNHG4 or miR-204-5p expression was
downregulated in AGS or HGC-27 cells. Firstly, the transfection
efficiency of SNHG4 and miR-204-5p was analyzed, and the results
demonstrated that SNHG4 expression was significantly decreased
after GC cells (AGS and HGC-27 cells) were transfected with siSNHG4
(P<0.001; Fig. 3A and B). No
significant difference was observed in miR-204-5p expression in
each group using the miR-204-5p inhibitor (Fig. 3C and D). The principle use of the
miR-204-5p inhibitor was to competitively bind to miR-204-5p with
SNHG4, resulting in a weakened effect of miR-204-5p despite
displaying no effect on miR-204-5p expression. In AGS and HGC-27
cells, the relative SNHG4 expression was significantly higher in
the siNC+I and lower in the siSNHG4+IC groups compared with that in
the siNC+IC group; additionally, SNHG4 expression was significantly
higher in the siSNHG4+I group compared with that in the siSNHG4+IC
group and lower compared with that in the siNC+I group (P<0.001;
Fig. 4A and B). Furthermore, the OD
value of AGS and HGC-27 cells was significantly higher in the
siNC+I group and lower in siSNHG4+IC groups compared with that in
the siNC+IC group, but it was significantly higher in the siSNHG4+I
group compared with that in the siSNHG4+IC and lower compared with
that in the siNC+I group at 48 h and 72 h (P<0.05 and P<0.01;
Fig. 4C and D). As expected, the
same trend was observed from the colony formation ability of both
AGS and HGC-27 cells (P<0.001; Fig.
4E and F). Additionally, the expression levels of
cycle-associated molecules were analyzed, and the results
demonstrated that the protein expression levels of PCNA and cyclin
D1 were significantly increased in the siNC+I group, while they
were significantly decreased in the siSNHG4+IC group, compared with
those in the siNC+IC group; miR-204-5p inhibitor reversed the
inhibitory effect of siSNHG4 on the PCNA and cyclin D1 expression
levels (P<0.001; Fig. 4G and H).
In addition, apoptosis was decreased in the siNC+I group
(P<0.01; Fig. 4I and J), but
significantly increased in the siSNHG4+IC group (P<0.001;
Fig. 4I and J) compared with the
siNC+IC group. Moreover, miR-204-5p inhibitor reversed
siSNHG4-mediated promotion of apoptosis (Fig. 4I and J). Therefore, downregulation
of miR-204-5p was able to promote the development of GC via
increasing GC cell proliferation and decreasing apoptosis. However,
downregulation of SNHG4 expression may reverse this trend caused by
miR-204-5p.
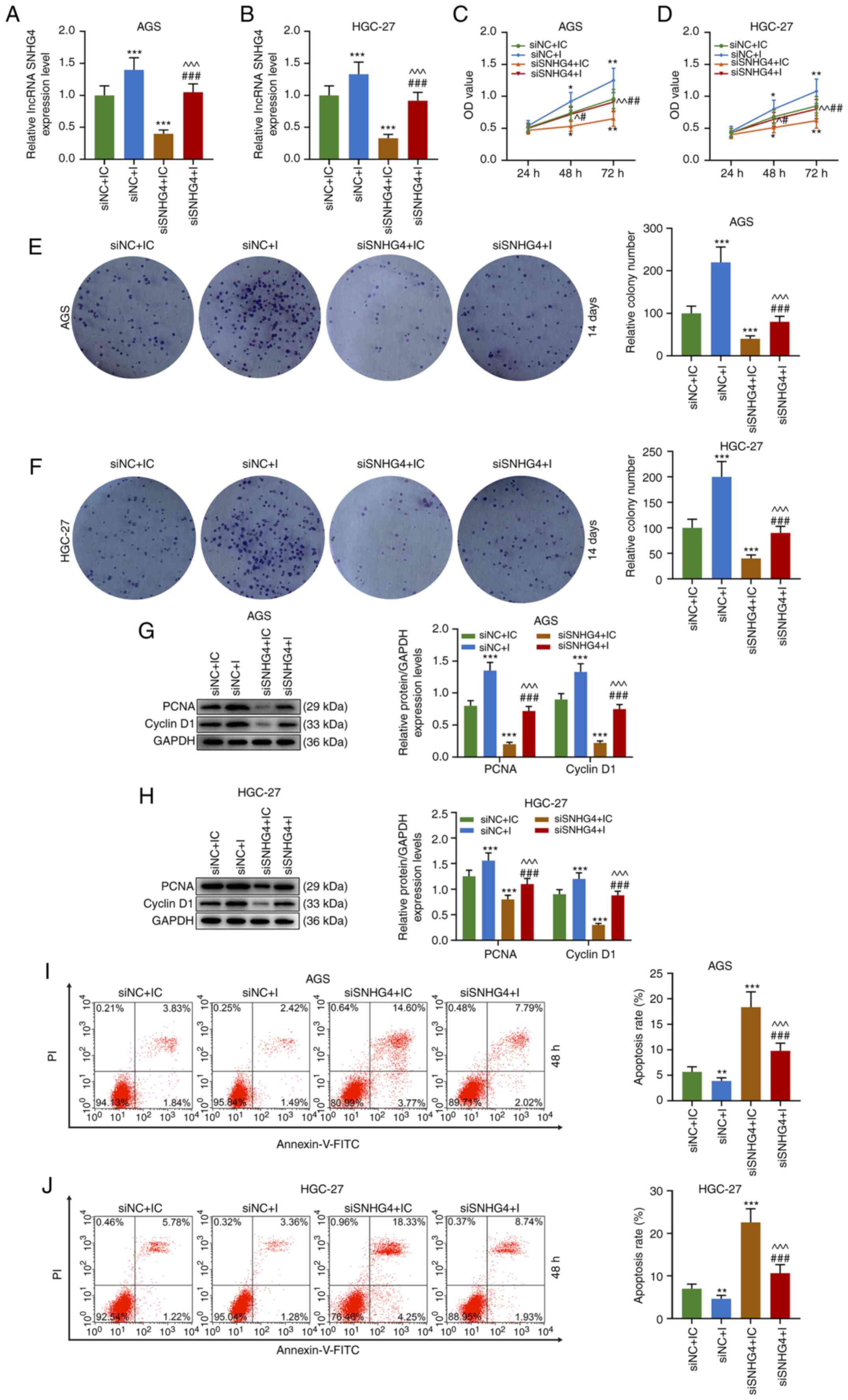 | Figure 4.Downregulation of SNHG4 expression
decreases the effects of miR-204-5p inhibition on the proliferation
and apoptosis of gastric cancer cells. Relative SNHG4 expression
levels in siNC+IC, siNC+I, siSNHG4+IC and siSNHG4+I groups in (A)
AGS or (B) HGC-27 cells. OD in each group in (C) AGS or (D) HGC-27
cells. Relative colony number in each group in (E) AGS or (F)
HGC-27 cells. Protein expression levels of PCNA and cyclin D1 in
(G) AGS or (H) HGC-27 cells in siNC+IC, siNC+I, siSNHG4+IC and
siSNHG4+I groups. Apoptosis rate of (I) AGS or (J) HGC-27 cells in
each group. Bars indicate the mean ± standard deviation.
*P<0.05, **P<0.01 and ***P<0.001 vs. siNC+IC;
^P<0.05, ^^P<0.01 and
^^^P<0.001 vs. siNC+I; #P<0.05,
##P<0.01 and ###P<0.001 vs. siSNHG4+IC.
SNHG4, small nucleolar RNA host gene 4; miR, microRNA; si, short
interfering; NC, negative control; IC, inhibitor control; I,
inhibitor; OD, optical density; PCNA, proliferating cell nuclear
antigen; lncRNA, long non-coding RNA. |
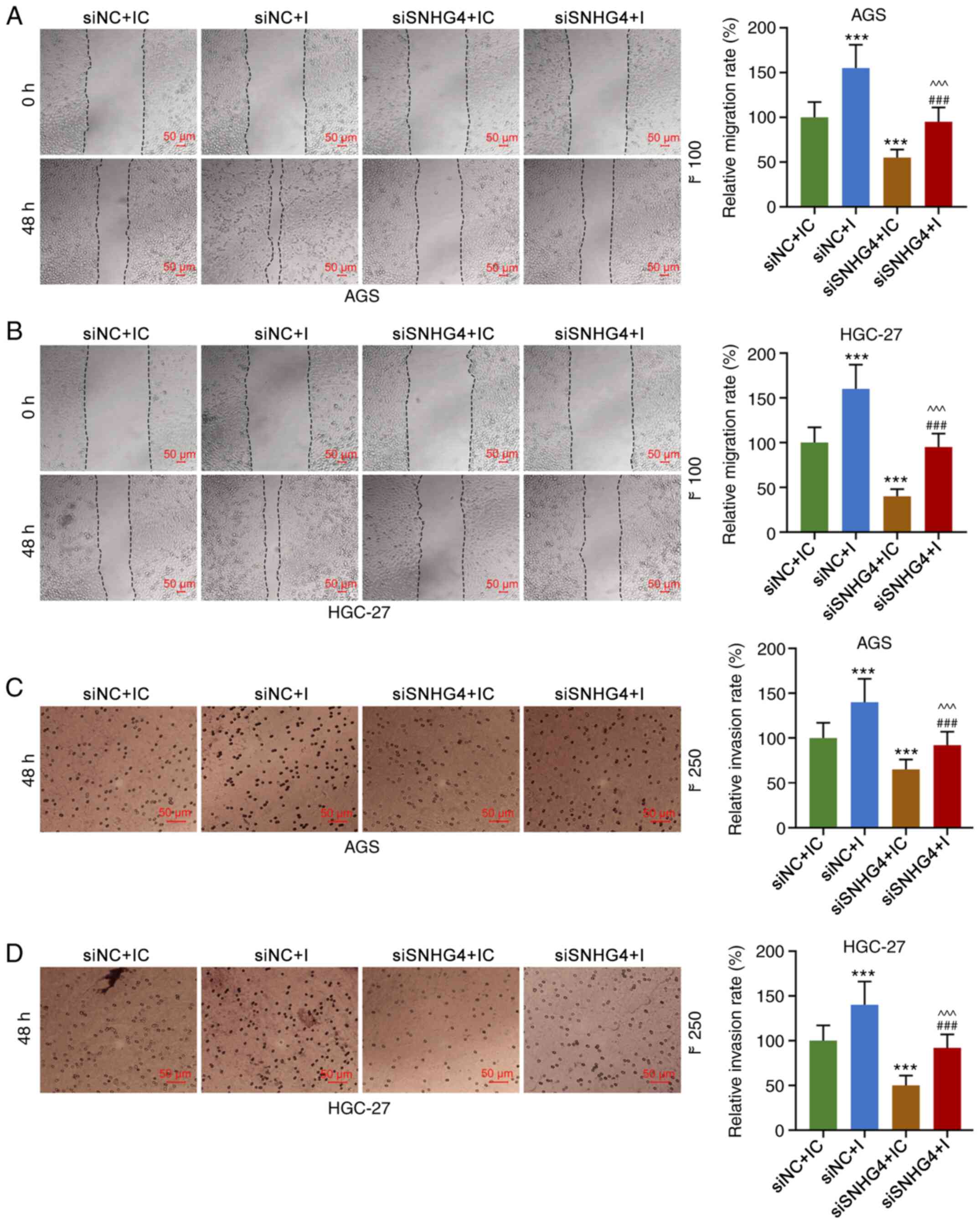
Downregulation of SNHG4 expression
reverses the effects of the miR-204-5p inhibitor on the migration
and invasion of GC cells
The migration and invasion of AGS and HGC-27 cells
were investigated, and the results revealed that the migration and
invasion of the cells were significantly higher in the siNC+I group
and lower in the siSNHG4+IC group compared with those in the
siNC+IC group, but they were significantly higher in the siSNHG4+I
group compared with those in the siSNHG4+IC and lower compared with
those in the siNC+I group (P<0.001; Fig. 5A-D). The data indicated that
downregulation of SNHG4 expression may decrease the migration and
invasion increased by miR-204-5p inhibitor in GC cells.
Downregulation of SNHG4 expression
reverses the effect of the miR-204-5p inhibitor on the
epithelial-mesenchymal transition (EMT) of GC cells
The EMT of AGS and HGC-27 cells was detected after
the treatment with siSNHG4 or miR-204-5p inhibitor. The results
demonstrated that in both AGS and HGC-27 cells, the expression
levels of N-cad and Snail were significantly higher in the siNC+I
and lower in the siSNHG4+IC groups compared with those in the
siNC+IC group, but they were significantly higher in the siSNHG4+I
group compared with those in the siSNHG4+IC and lower compared with
those in the siNC+I group (P<0.001; Fig. 6A and B). Overall, E-cad expression
displayed the opposite trend compared with the expression levels of
N-cad or Snail (Fig. 6A and B).
Therefore, SNHG4 downregulation may eliminate the effect of the
miR-204-5p inhibitor on the EMT of GC cells.
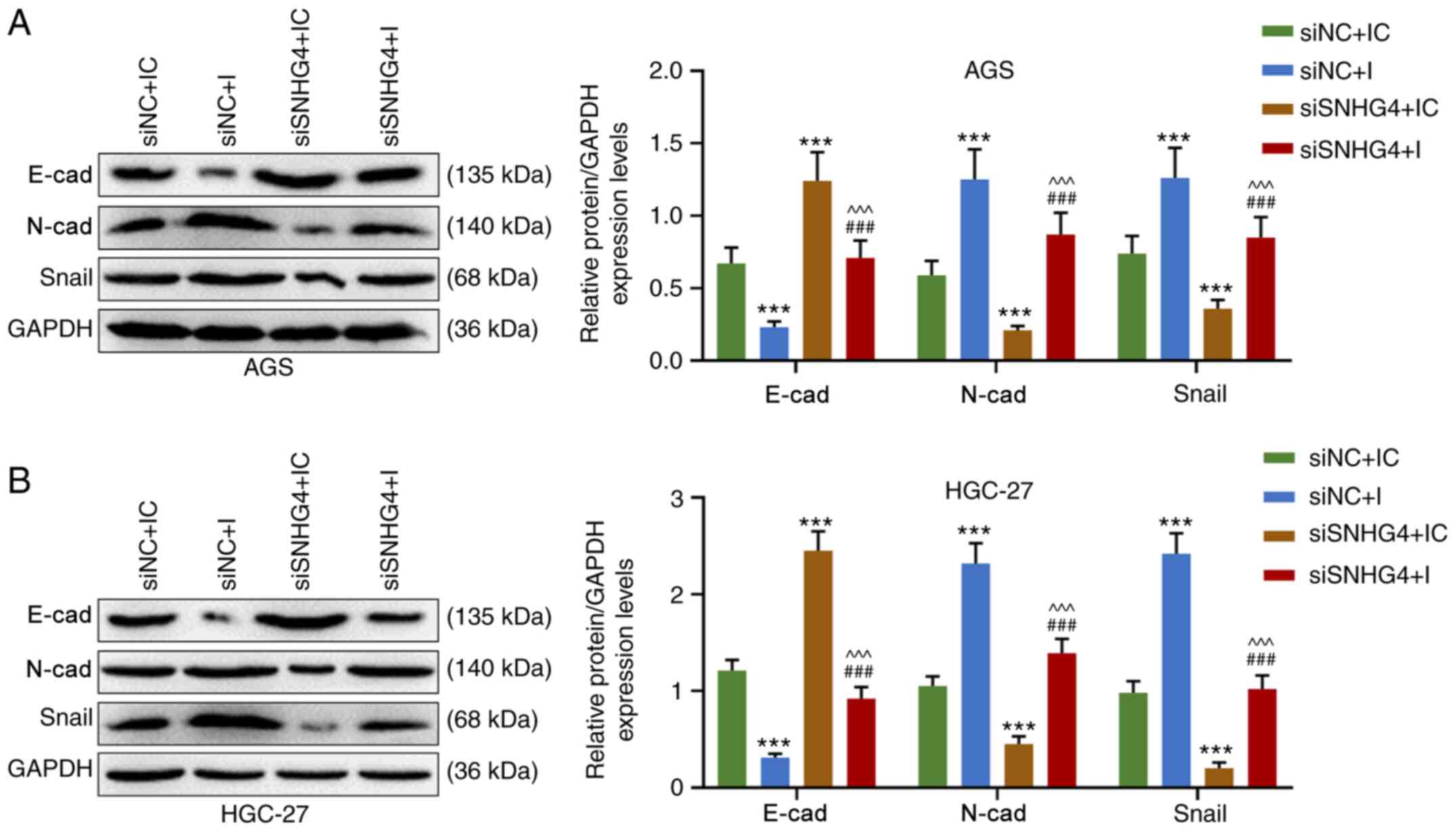 | Figure 6.Downregulation of SNHG4 expression
decreases the effect of miR-204-5p inhibition on the
epithelial-mesenchymal transition of gastric cancer cells. Protein
expression levels of E-cad, N-cad and Snail in (A) AGS or (B)
HGC-27 cells in siNC+IC, siNC+I, siSNHG4+IC and siSNHG4+I groups.
Bars indicate the mean ± standard deviation. ***P<0.001 vs.
siNC+IC; ^^^P<0.001 vs. siNC+I;
###P<0.001 vs. siSNHG4+IC. SNHG4, small nucleolar RNA
host gene 4; miR, microRNA; cad, cadherin; si, short interfering;
NC, negative control; IC, inhibitor control; I, inhibitor. |
Discussion
The present study demonstrated that SNHG4 expression
was upregulated in GC and resulted in a lower survival rate.
miR-204-5p expression was downregulated in GC tissues and cell
lines, and was predicted to be able to interact with SNHG4; in
addition, the expression levels of the two RNAs were found to be
negatively correlated. Furthermore, the proliferation, apoptosis,
migration, invasion and EMT increased by miR-204-5p inhibition in
GC cells was counteracted by the downregulation of SNHG4
expression. The data revealed a potential mechanism explaining the
development of GC.
The present study revealed that SNHG4 was highly
expressed in GC tissues and cell lines, and was associated with a
poor survival rate in patients with GC. Further exploration
demonstrated that the downregulation of SNHG4 expression decreased
the proliferation, migration and invasion of GC cells compared with
those of cells without treatment; in addition, it decreased the
expression levels of N-cad and Snail, and increased E-cad
expression. EMT is a process during which epithelial cells lose
polarity, tight junctions, adhesion and the morphology and
characteristics of cytoplasmic cells, thus gaining the ability to
invade and migrate (23,24). EMT serves a crucial role in tumor
formation and metastasis, especially in the invasion and metastasis
of tumors (25–28). In the process of EMT, the expression
levels of various cell adhesion factors, such as E-cad and
α-catenin, are downregulated, while others, such as Twist, Snail,
Slug and TGF-β, are upregulated (26–28).
Therefore, the increased expression levels of E-cad, along with
decreased expression levels of N-cad and Snail, reflect decreased
EMT. Xu et al (29) revealed
that, via miR-224-3p, SNHG4 acts as a promoter in osteosarcoma
development and causes poor survival rates. Tang et al
(30) noted that SNHG4 promoted the
proliferation, migration, invasiveness and epithelial-mesenchymal
transition of lung cancer cells by regulating miR-98-5p.
Collectively, the aforementioned studies indicated a promoting role
of SNHG4 in cancer; therefore, the present study further
investigated the role of SNHG4 in gastric cancer.
The present study demonstrated that miR-204-5p could
interact with SNHG4 and that miR-204-5p expression was
downregulated in GC tissues and cell lines; in addition, the
expression levels of the two RNAs were negatively correlated in
ANTs and GC tissues. Bian et al (31) demonstrated that lncRNA UCA1 promotes
the tumorigenesis of colorectal cancer via targeting miR-204-5p to
inhibit its expression. Similarly, Yin et al (32) identified that miR-204-5p could
target RAB22A and further restrain the biological characteristics
of colorectal cancer. In papillary thyroid carcinoma, miR-204-5p
inhibits the proliferation of tumor cells (33). Therefore, miR-204-5p may act as a
suppressor in GC development. However, the present study revealed
that SNHG4 may interact with miR-204-5p by negatively regulating
miR-204-5p expression and counteracting the antitumor effects of
miR-204-5p on the proliferation, apoptosis, colony formation,
migration, invasion and EMT of GC cells. The results of the current
study suggested that SNHG4 exerted its function via targeting and
interacting with miR-204-5p, thus promoting the progression of GC.
However, the results of the present study require validation by
performing in vivo experiments. In addition, the signaling
pathway regulation underlying SNHG4/miR-204-5p-mediated promotion
of GC progression requires further investigation.
In conclusion, GC progression may result from a loss
of regulation of SNHG4; specifically, SNHG4 upregulation may
promote GC by inhibiting miR-204-5p expression. The discovery of
the mechanism of this SNHG4-miR-204-5p pathway may contribute to
the development of drugs against the growth of GC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SW and WZ made substantial contributions to the
conception and design of the present study. JQ and FC performed
data acquisition, data analysis and interpretation. SW and WZ
drafted the manuscript and critically revised it for important
intellectual content. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
All experiments were approved by the Ethics Board of
Zhuji People's Hospital (approval no. ZJ-20180215) and were in
accordance with the 1964 Helsinki declaration and its later
amendments or comparable ethical standards. All patients in the
present study provided written informed consent.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016. View Article : Google Scholar
|
|
2
|
Cai J, Niu X, Chen Y, Hu Q, Shi G, Wu H,
Wang J and Yi J: Emodin-induced generation of reactive oxygen
species inhibits RhoA activation to sensitize gastric carcinoma
cells to anoikis. Neoplasia. 10:41–51. 2008. View Article : Google Scholar
|
|
3
|
Ferlay J, Soerjomataram I, Dikshit R, Eser
S, Mathers C, Rebelo M, Parkin DM, Forman D and Bray F: Cancer
incidence and mortality worldwide: Sources, methods and major
patterns in GLOBOCAN 2012. Int J Cancer. 136:E359–E386. 2015.
View Article : Google Scholar
|
|
4
|
Sun Y and Ma L: The emerging molecular
machinery and therapeutic targets of metastasis. Trends Pharmacol
Sci. 36:349–359. 2015. View Article : Google Scholar
|
|
5
|
Turajlic S and Swanton C: Metastasis as an
evolutionary process. Science. 352:169–175. 2016. View Article : Google Scholar
|
|
6
|
Tanabe S, Ishido K, Matsumoto T, Kosaka T,
Oda I, Suzuki H, Fujisaki J, Ono H, Kawata N, Oyama T, et al:
Long-term outcomes of endoscopic submucosal dissection for early
gastric cancer: A multicenter collaborative study. Gastric Cancer.
20 (Suppl 1):45–52. 2017. View Article : Google Scholar
|
|
7
|
Bang YJ, Van Cutsem E, Feyereislova A,
Chung HC, Shen L, Sawaki A, Lordick F, Ohtsu A, Omuro Y, Satoh T,
et al ToGA Trial Investigators, : Trastuzumab in combination with
chemotherapy versus chemotherapy alone for treatment of
HER2-positive advanced gastric or gastro-oesophageal junction
cancer (ToGA): A phase 3, open-label, randomised controlled trial.
Lancet. 376:687–697. 2010. View Article : Google Scholar
|
|
8
|
Kang YK, Satoh T, Ryu MH, Chao Y, Kato K,
Chung HC, Chen JS, Muro K, Kang WK, Yoshikawa T, et al: Nivolumab
(ONO-4538/BMS-936558) as salvage treatment after second or
later-line chemotherapy for advanced gastric or gastro-esophageal
junction cancer (AGC): a double-blinded, randomized, phase III
trial. J Clin Oncol. 35 (Suppl 4):2. 2017. View Article : Google Scholar
|
|
9
|
Xu X, Qian LJ, Su XY, He KF, Jin KT, Gu
LH, Feng JG, Li GL, Zhou Q, Xu ZZ, et al: Establishment and
characterization of GCSR1, a multi-drug resistant signet ring cell
gastric cancer cell line. Int J Oncol. 46:2479–2487. 2015.
View Article : Google Scholar
|
|
10
|
Cheng J, Kapranov P, Drenkow J, Dike S,
Brubaker S, Patel S, Long J, Stern D, Tammana H, Helt G, et al:
Transcriptional maps of 10 human chromosomes at 5-nucleotide
resolution. Science. 308:1149–1154. 2005. View Article : Google Scholar
|
|
11
|
Birney E, Stamatoyannopoulos JA, Dutta A,
Guigó R, Gingeras TR, Margulies EH, Weng Z, Snyder M, Dermitzakis
ET, Thurman RE, et al Children's Hospital Oakland Research
Institute, : Identification and analysis of functional elements in
1% of the human genome by the ENCODE pilot project. Nature.
447:799–816. 2007. View Article : Google Scholar
|
|
12
|
Carninci P, Kasukawa T, Katayama S, Gough
J, Frith MC, Maeda N, Oyama R, Ravasi T, Lenhard B, Wells C, et al
Consortium F; RIKEN Genome Exploration Research Group and Genome
Science Group, : (Genome Network Project Core Group) The
transcriptional landscape of the mammalian genome. Science.
309:1559–1563. 2005. View Article : Google Scholar
|
|
13
|
Geisler S and Coller J: RNA in unexpected
places: Long non-coding RNA functions in diverse cellular contexts.
Nat Rev Mol Cell Biol. 14:699–712. 2013. View Article : Google Scholar
|
|
14
|
Hirakata S and Siomi MC: piRNA biogenesis
in the germline: From transcription of piRNA genomic sources to
piRNA maturation. Biochim Biophys Acta. 1859:82–92. 2016.
View Article : Google Scholar
|
|
15
|
Jacquier A: The complex eukaryotic
transcriptome: Unexpected pervasive transcription and novel small
RNAs. Nat Rev Genet. 10:833–844. 2009. View
Article : Google Scholar
|
|
16
|
Kim VN, Han J and Siomi MC: Biogenesis of
small RNAs in animals. Nat Rev Mol Cell Biol. 10:126–139. 2009.
View Article : Google Scholar
|
|
17
|
Clark MB, Choudhary A, Smith MA, Taft RJ
and Mattick JS, Taft RJ and Mattick JS: The dark matter rises: The
expanding world of regulatory RNAs. Essays Biochem. 54:1–16. 2013.
View Article : Google Scholar
|
|
18
|
Li H, Hong J and Wijayakulathilaka WS:
Long non-coding RNA SNHG4 promotes cervical cancer progression
through regulating c-Met via targeting miR-148a-3p. Cell Cycle.
18:3313–3324. 2019. View Article : Google Scholar
|
|
19
|
Chen X, Chen Z, Yu S, Nie F, Yan S, Ma P,
Chen Q, Wei C, Fu H, Xu T, et al: Long noncoding RNA LINC01234
functions as a competing endogenous RNA to regulate CBFB expression
by sponging miR-204-5p in gastric cancer. Clin Cancer Res.
24:2002–2014. 2018. View Article : Google Scholar
|
|
20
|
In H, Solsky I, Palis B, Langdon-Embry M,
Ajani J and Sano T: Validation of the 8th edition of the AJCC TNM
staging system for gastric cancer using the national cancer
database. Ann Surg Oncol. 24:3683–3691. 2017. View Article : Google Scholar
|
|
21
|
Nagy Á, Lánczky A, Menyhárt O and Győrffy
B: Validation of miRNA prognostic power in hepatocellular carcinoma
using expression data of independent datasets. Sci Rep. 8:92272018.
View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar
|
|
23
|
Guarino M: Epithelial-mesenchymal
transition and tumour invasion. Int J Biochem Cell Biol.
39:2153–2160. 2007. View Article : Google Scholar
|
|
24
|
Romeo E, Caserta CA, Rumio C and Marcucci
F: The vicious cross-talk between tumor cells with an EMT phenotype
and cells of the immune system. Cells. 8:82019. View Article : Google Scholar
|
|
25
|
Thiery JP: Epithelial-mesenchymal
transitions in development and pathologies. Curr Opin Cell Biol.
15:740–746. 2003. View Article : Google Scholar
|
|
26
|
Wheelock MJ, Shintani Y, Maeda M, Fukumoto
Y and Johnson KR: Cadherin switching. J Cell Sci. 121:727–735.
2008. View Article : Google Scholar
|
|
27
|
Nishizuka M, Komada R and Imagawa M:
Knockdown of RhoE expression enhances TGF-β-induced EMT
(epithelial-to-mesenchymal transition) in cervical cancer HeLa
cells. Int J Mol Sci. 20:202019. View Article : Google Scholar
|
|
28
|
Zhou J, Cheng H, Wang Z, Chen H, Suo C,
Zhang H, Zhang J, Yang Y, Geng L, Gu M, et al: Bortezomib
attenuates renal interstitial fibrosis in kidney transplantation
via regulating the EMT induced by TNF-α-Smurf1-Akt-mTOR-P70S6K
pathway. J Cell Mol Med. 23:5390–5402. 2019. View Article : Google Scholar
|
|
29
|
Xu R, Feng F, Yu X, Liu Z and Lao L:
LncRNA SNHG4 promotes tumour growth by sponging miR-224-3p and
predicts poor survival and recurrence in human osteosarcoma. Cell
Prolif. 51:e125152018. View Article : Google Scholar
|
|
30
|
Tang Y, Wu L, Zhao M, Zhao G, Mao S, Wang
L, Liu S and Wang X: LncRNA SNHG4 promotes the proliferation,
migration, invasiveness, and epithelial-mesenchymal transition of
lung cancer cells by regulating miR-98-5p. Biochem Cell Biol.
97:767–776. 2019. View Article : Google Scholar
|
|
31
|
Bian Z, Jin L, Zhang J, Yin Y, Quan C, Hu
Y, Feng Y, Liu H, Fei B, Mao Y, et al: LncRNA-UCA1 enhances cell
proliferation and 5-fluorouracil resistance in colorectal cancer by
inhibiting miR-204-5p. Sci Rep. 6:238922016. View Article : Google Scholar
|
|
32
|
Yin Y, Zhang B, Wang W, Fei B, Quan C,
Zhang J, Song M, Bian Z, Wang Q, Ni S, et al: miR-204-5p inhibits
proliferation and invasion and enhances chemotherapeutic
sensitivity of colorectal cancer cells by downregulating RAB22A.
Clin Cancer Res. 20:6187–6199. 2014. View Article : Google Scholar
|
|
33
|
Liu L, Wang J, Li X, Ma J, Shi C, Zhu H,
Xi Q, Zhang J, Zhao X and Gu M: MiR-204-5p suppresses cell
proliferation by inhibiting IGFBP5 in papillary thyroid carcinoma.
Biochem Biophys Res Commun. 457:621–626. 2015. View Article : Google Scholar
|