Introduction
Hepatolithiasis is a common disease that poses a
serious health threat to the Chinese population, with an incidence
rate of 2–25% (1). The underlying
pathology of hepatolithiasis involves chronic proliferative
cholangitis of the mucus-producing major intrahepatic bile duct;
this represents the key lesion associated with this condition
(2,3). The proliferative glands and biliary
epithelium produce high amounts of mucin to initiate the process of
stone formation and development, which comprise calcium salts and
lipids (4). This is an important
pathological mechanism that can lead to chronic proliferative
cholangitis and stone formation (5). Bacterial infection within the biliary
system is closely related to the development of cholelithiasis, in
which Gram-negative bacteria, including Escherichia coli and
Klebsiella, produce lipopolysaccharides (LPS), a common
substance in infectious bile and gallstone cores (6,7). A
total of 22 mucin genes have been identified to date (8). Of these, mucin 5AC (MUC5AC) has been
identified as the most important mucin secreted by the bile duct
(9,10). This protein is expressed at high
levels in patients with primary hepatolithiasis and is an important
nuclear-promoting factor (11).
Previous studies from our research group reported that under normal
conditions, MUC5AC is only expressed at low levels in the human
intrahepatic bile duct (12).
However, the expression of MUC5AC is known to significantly
increase following LPS treatment, and there is a positive
association between MUC5AC expression and LPS concentration
(13). At present, studies on the
regulatory mechanisms underlying the overexpression of MUC5AC in
response to a variety of factors have primarily focused on the
airway epithelium and nasal mucosa (10). The regulation of MUC5AC expression
is a result of the enhanced transcription of the MUC5AC gene;
however, little is known about such mechanisms in the biliary
system (14). Therefore, it is
important to elucidate the specific mechanisms underlying mucin
synthesis in the bile duct epithelium to develop strategies to
inhibit the oversecretion of bile duct mucus. This would create a
new therapeutic target for hepatolithiasis and thus help to prevent
and treat this disease.
The transcription initiation site of the MUC5AC gene
is located 48 base pairs (bp) upstream of the translation
initiation codon (15). There is a
sequence ~1,000 bp upstream of the initiation codon that features a
dense distribution of transcription factor binding sites; these
binding sites constitute the primary area that regulates the
expression of the MUC5AC gene (15). This region regulates MUC5AC in a
manner that shows differential expression in different tissues and
cells; this occurs via the combined action of different
transcription factors (9).
Specificity protein 1 (Sp1), a member of the SP family, is a
ubiquitously expressed transcription factor (7). Sp1 contains a zinc finger domain at
the C terminal, which can specifically recognize the GC box element
in DNA sequences and participate in the transcriptional regulation
of various genes, such as vascular endothelial growth factor (VEGF)
and human epidermal growth factor receptor 2 (HER-2) (16,17).
Studies have demonstrated that Sp1 is the primary regulator of
MUC5AC expression, and Sp1 expression can directly activate the
promoter of MUC5AC, induce MUC5AC gene transcription and promote
the high expression of MUC5AC in mucin (15). Electrophoretic mobility shift assays
revealed that following activation, Sp1 protein translocates into
the nucleus and binds to the regulatory element corresponding to
the MUC5AC gene promoter, which is the main targeting element for
MUC5AC transcription (16). Other
previous studies have demonstrated that even in the same tissue,
Sp1 expression induced by various stimuli is not consistent at the
binding site of control elements in the MUC5AC promoter (15,18). A
previous study reported that the Sp1 binding site within the 324–64
bp sequence of the MUC5AC 5′-upstream region is an important
regulatory element for neutrophil elastase-induced MUC5AC gene
expression (19). Another study
investigating cigarette smoking-induced secretion of MUC5AC from
epithelial cells in the airway revealed that smoking induced Sp1
protein expression, phosphorylation, translocation to the nucleus
and binding to the MUC5AC promoter-3724/-3224 bp corresponding
sequences, indicating that smoking can induce high expression of
MUC5AC; while the rest of the original cis-element exerted no such
function (18). The present study
focused on a regulatory role for Sp1 in transcription and the
binding site of Sp1 and the MUC5AC promoter in LPS-induced
overexpression of MUC5AC in the bile duct epithelium. This
represents the primary focus of the current study.
Further studies on the regulation of Sp1 expression
found that in LPS-treated RAW264.7 macrophages, microRNA
(miRNA/miR)-130b can directly interact with the 3′-untranslated
region (3′-UTR) of the Sp1 gene and inhibit the expression of Sp1
(20), suggesting that the
expression of Sp1 is regulated by miR-130b. miRNAs are a class of
small non-coding s RNA molecules (21,22)
that can specifically bind to the 3′-UTR of the target mRNA by
complementary pairing, which blocks gene expression by degrading or
inhibiting the target gene mRNA (23,24).
Abnormal miRNA expression can lead to the disturbance of the
corresponding regulatory network and represents an important factor
underlying the occurrence of diseases (25,26).
Therefore, determining whether the regulation of Sp1 expression is
mediated by miR-130b during the overexpression of MUC5AC in the
LPS-induced bile duct epithelium formed the second aim of the
current study.
Materials and methods
Ethics
All animal studies were approved by the local ethics
committee of the Affiliated Shengjing Hospital of China Medical
University (approval no. 2017PS231K). All animals were maintained
under 12-h light/dark cycles and had free access to food and water
at 25°C and normal atmospheric pressure. All handling procedures
aimed to minimize suffering and conformed to the standards of the
Ethics Committee of Shengjing Hospital.
Cell lines and cell culture
Human intrahepatic biliary epithelial cells
(HIBEpiCs) were purchased from ScienCell Research Laboratories,
Inc., and cultivated in epithelial cell medium containing 500 ml
basal epithelium medium (ScienCell Research Laboratories, Inc.), 5
ml epithelial cell growth supplement (ScienCell Research
Laboratories, Inc.), 10 ml fetal bovine serum (ScienCell Research
Laboratories, Inc.) and 5 ml penicillin/streptomycin solution
(ScienCell Research Laboratories, Inc.) in an incubator at 37°C
with 5% CO2. The cell medium was refreshed every 1–2
days, and passaged at a ratio of 1:2 or 1:3 every 3 to 5 days.
Cell pretreatment and administration
of LPS and inhibitors
To investigate the effect of LPS exposure, HIBEpiCs
were treated with different concentrations of LPS (1, 10 and 100
µg/ml and a negative control, which was without LPS exposure) for
24 h once cells have reached 60–70% confluency. Mithramycin A (MA;
10 µg/ml; Sigma-Aldrich; Merck KGaA), an Sp1 inhibitor, was used
for inhibitory experiments. HIBEpiCs were incubated with 1 µg/ml MA
and LPS for 24 h at 37°C.
Western blotting
Protein expression was measured by western blotting,
as described previously (3,6), with some modifications. The
precipitate protein was lysed in lysis buffer (Beyotime Institute
of Biotechnology), and protein concentrations were measured using a
Bradford protein assay kit (Bio-Rad Laboratories, Inc.). Equal
amounts of protein (15 µg) were separated via SDS-PAGE with an 8%
gel and then transferred onto polyvinylidene difluoride membranes.
Membranes were subsequently blocked with 5% nonfat milk in TBS with
0.1% Tween-20 (TBS-T) at room temperature for 1.5 h to eliminate
non-specific binding. Membranes were then incubated overnight with
primary antibodies against Sp1 (1:1,000; cat. no. ab124804; Abcam)
and GADPH (1:3,000; cat. no. 60004-1-Ig; ProteinTech Group, Inc.)
at 4°C. Subsequently, membranes were washed three times in TBS-T
and incubated with secondary antibodies 2 h at room temperature
(horseradish peroxidase-conjugated anti-rabbit immunoglobulin G;
1:3,000; cat. nos. WB0177 and A23210; Beijing Zhongshan Golden
Bridge Biotechnology Co., Ltd.). Immunoreactive bands were then
visualized using an enhanced chemiluminescent kit (EMD Millipore)
according to the manufacturer's instructions. Band densities were
quantified using Gel-Pro Analyzer densitometry software (version
6.3; Media Cybernetics, Inc.
Reverse transcription-quantitative PCR
(RT-qPCR)
Total RNA was extracted from HIBEpiCs cell cultures
using TRIzol® (Invitrogen; Thermo Fisher Scientific,
Inc.). RNA concentration was determined by spectrophotometry.
RT-qPCR was performed using the PrimeScript RT reagent kit
according to the manufacturer's instructions with gDNA Eraser
(Takara Bio, Inc.) and SYBR Green Premix Ex Taq (Takara Bio, Inc.).
RT-qPCR was performed as follows: 10 min at 95°C, followed by 35
cycles of 15 sec at 95°C and 40 sec at 55°C. The following primer
pairs were used for the qPCR: MUC5AC, forward,
5′-AGCCGGCAACCTACTACTCG-3′ and reverse,
5′-AAGTGGTCATAGGCTTCGTGC-3′; Sp1 forward,
5′-TGGCAGCAGCAGTACCAATGGC-3′ and reverse,
5′-CCAGGTAGTCCTGTCAGAACTT-3′; miR-130b-3p
CAGUGCAAUGAUGAAAGGGCAU-polyA; GADPH forward,
5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse, 5′-GCCATCACGCCACAGTTTC-3′
and U6, forward, 5′-GGAACGATACAGAGAAGATTAGC-3′ and reverse,
5′-TGGAACGCTTCACGAATTTGCG-3′. Relative expression levels were
calculated using the 2−ΔΔCq method (6) following normalization to the
expression of GAPDH or U6.
Immunohistochemistry (IHC)
IHC was performed to detect the expression of MUC5AC
in bile duct tissue obtained from clinical specimens. Tissues were
obtained from patients who underwent partial hepatectomy due to
intrahepatic bile duct stones in the General Surgery Department of
Shengjing Hospital between September 2017 and November 2018; these
tissues acted as an experimental group. Normal adjacent tissue,
obtained from hepatic hemangioma resection surgery, were selected
as the control group. The sample collecting procedure was approved
by the local ethics committee of the Affiliated Shengjing Hospital
of China Medical University (approval no. 2017PS231K). All patients
provided written informed consent. The bile duct tissue was fixed
overnight in 4% paraformaldehyde at 4°C, sectioned into 4-µm
slices. To block endogenous peroxidase activity, each section was
incubated with 0.3% H2O2 at room temperature
for 10 min. After blocking in 5% bovine serum albumin (Beijing
ZSGB-BIO Technology, Ltd.) for 20 min at room temperature, the
sections were incubated with primary MUC5AC antibody (1:1,000; cat.
no. ab3649; Abcam) overnight at 4°C. The next day, sections were
washed three times using PBS, then incubated with biotin-conjugated
anti-mouse IgG (1:100; cat. no. BM2004; Wuhan Boster Biological
Technology, Ltd.) secondary antibody for 20 min at room
temperature. Lastly, the sections were incubated with
HRP-streptavidin (1:1,000; cat. no. BIR701-3; Beijing Borsi
Technology Co., Ltd.) at room temperature for 10 min, then
visualized using a DAB peroxidase kit (1:20; cat. no. AR1000; Wuhan
Boster Biological Technology, Ltd.) at room temperature for 1 min,
then counterstained with hematoxylin at room temperature for 3 min.
The stained slides were visualized under light microscope
(magnification, ×200 and ×400; Eclipse Ci-L; Nikon Corporation) and
the images were analyzed using ImageJ 1.51K (National Institutes of
Health). Quantitative analysis was completed based on the
percentage of positive cells with MUC5AC staining (0–25%, low;
25–50%, medium; >50%, high). The details of clinical specimens
were shown in Table I.
 | Table I.The details of clinical specimens. |
Table I.
The details of clinical specimens.
| Variable | Control group
n=8 | Patients with
hepatoliths n=10 |
|---|
| Age, years | 52±16 | 61±24 |
| Male (n, %) | 5 (62.5%) | 6 (60%) |
| Female (n, %) | 3 (37.5%) | 4 (40%) |
Immunofluorescence (IF)
Tissue IF
Fluorescent co-staining experiments were performed
to verify the association between MUC5AC expression (cat. no.
ab3649; Abcam) and Sp1 expression (cat. no. ab124804; Abcam) in
bile duct tissue. IF was performed as the aforementioned IHC
procedure. Tissues were incubated with fluorescent secondary
antibody (1:100; cat. no. FITC-10835; ProteinTech Group, Inc.).
Subsequently, sections were incubated with DAPI for 5 min to stain
cell nuclei. The stained slides were visualized under fluorescence
microscope (magnification, ×200 and ×400; Eclipse Ci-L; Nikon
Corporation) and the images were analyzed using ImageJ 1.51K
(National Institutes of Health). Quantitative analysis was
completed based on the percentage of positive cells with MUC5AC and
Sp1 staining (0–25%, low; 25–50%, medium; >50%, high).
Cell IF
HIBEpiCs pretreated with 100 µg/ml of LPS for 24 h
were used as the experimental group for cell IF analysis. Variation
in MUC5AC and Sp1 expression were then compared between
experimental and negative control groups. Following LPS treatment,
cultured HIBEpiCs were fixed with −20°C methanol for 10 min, washed
in PBS three times for 5 min, then blocked with 1% BSA (cat. no
A8020; Beijing Solarbio Science & Technology Co., Ltd.) in PBS
for 30 min at room temperature. Following washing in PBS three
times for 5 min, the sections were then simultaneously incubated
with MUC5AC (cat. no. ab3649) and Sp1 (cat. no. ab124804) primary
antibodies at 1:100 dilution (Abcam) overnight at 4°C. Sections
were then washed in PBS three times for 5 min and incubated in
fluorescent secondary antibody (goat anti-rat; cat. no. SA00009-1
and goat anti-rabbit; cat. no. FITC-10835; both 1:200; ProteinTech
Group, Inc.) for 1 h at room temperature. Nuclei were then stained
with DAPI for 5 min at room temperature.
Enzyme-linked immunosorbent assay
(ELISA)
ELISA was used to detect the concentration of MUC5AC
in HIBEpiCs cell supernatants, which was produced by collecting and
centrifuging cells in an ultrafiltration tube (EMD Millipore) at
2,000 × g 5 min at room temperature. Subsequently, 250 µl
concentrated cell supernatant solution was obtained from the
control, LPS exposure, Sp1 shRNA transfection and miR130b mimics
intervention groups and assayed using the human MUC5AC ELISA assay
kit (cat. no. CSB-E10109h; Cusabio Technology, LLC) to determine
MU5AC concentration.
Cell transfection
Short hairpin RNA (shRNA) plasmids were for the
specific inhibition of Sp1 and miR-130b mimic and inhibitor were
purchased from Shanghai GeneChem Co., Ltd. A total of 8 µg shRNA
plasmids and miR-130b mimic were transfected into HIBEpiCs using
Lipofiter Liposomal transfection reagent (Hanbio Biotechnology Co.,
Ltd.) for 48 h before experimentation. Transfection efficiency of
shRNA efficiency was measured by RT-qPCR and western blotting. The
sequences of the shRNAs and miRs used are as follows: Sp1
overexpression plasmids (cat, no. 51288; Genechem Co., Ltd.),
5′-AGAAGGAGAGCAAAACCAGC-3′; Sp1 suppression shRNA (cat, no. 4090;
Genechem Co., Ltd.), 5′-CCTGGTGCAAACCAACAGATT-3′; miR-130b mimic
(cat, no. 14159; Genechem Co., Ltd.), 5′-ATGCCCTTTCATCATTGCACTG-3′
and miR-130b inhibitor (cat. no. 18930; Genechem Co., Ltd.),
5′-ATGCCCTTTCATCATTGCACTG-3′.
Luciferase reporter gene assay
Luciferase reporter gene assays were performed to
verify the direct binding effect and exact binding position between
miR-130b and Sp1 3′-UTR sequences. The binding site was predicted
using an online tool (http://www.targetscan.org). Plasmids (Genechem Co.,
Ltd.) for miR-130b, the Sp1 3′-UTR and mutant Sp1 3′-UTR (3′UTR-mu
were constructed (Table II).
miR-130b was then co-transfected by Nanofectin transfection reagent
(PAA Laboratories GmbH; GE Healthcare) with Sp1 3′-UTR and Sp1
3′UTR-mu into HIBEpiCs. Luciferase activities were then detected by
a Dual-Luciferase Reporter Assay system (Promega Corporation) 48 h
after transfection. The data were quantified by normalizing to
Renilla luciferase activity.
 | Table II.Primers used for the luciferase
reporter gene assay. |
Table II.
Primers used for the luciferase
reporter gene assay.
| Primer | Sequence |
|---|
| miR-130b-F |
5′-TGTGGAAAGGACGCGGGATCGCCCCAGCCAGCCTGCATTC-3′ |
| miR-130b-R |
5′-CAGCGGTTTAAACTTAAGCTAAAAAACACTTACCCTCTGG-3′ |
| Sp1 3′UTR-F |
5′-GAGGAGTTGTGTTTGTGGAC-3′ |
| Sp1 3′UTR-R |
5′-GACGATAGTCATGCCCCGCG-3′ |
| Sp1 3′UTR-mut-F |
5′-GAGGAGTTGTGTTTGTGGAC-3′ |
| Sp1 3′UTR-mut R |
5′-GACGATAGTCATGCCCCGCG-3′ |
Chromatin immunoprecipitation (ChIP)
assay
Human HIBEpiCs were fixed with 1% formaldehyde for
15 min at room temperature and quenched with 0.125 M glycine.
Chromatin was then isolated using lysis buffer (Active Motif,
Inc.). Lysates were sonicated using the EpiShear™ Probe Sonicator
(Active Motif, Inc.) with an EpiShear™ Cooled Sonication Platform
(Active Motif, Inc.), and the DNA was sheared to an average length
of 300–500 bp. Genomic DNA was then prepared by treating aliquots
of chromatin with RNase, proteinase K and heating for
decrosslinking overnight at 65°C, followed by purification using
the QIAquick PCR Purification kit (Qiagen China Co., Ltd.). The
resultant DNA was then quantified on a NanoDrop™ spectrophotometer
(Thermo Fisher Scientific, Inc.). Extrapolation to the original
chromatin volume allowed quantitation of the total chromatin yield.
Genomic DNA regions of interest were then isolated using antibodies
against Sp1 (1:150; cat, no. ab231778; Abcam). Complexes were
eluted from the beads with SDS buffer and subjected to RNase and
proteinase K treatment (10 mg/ml) at 45°C. Crosslinks were reversed
by overnight incubation with 5 M NaCl at 65°C, and Chip DNA was
purified using a QIAquick PCR Purification kit (Qiagen China Co.,
Ltd.). qPCR reactions were performed in triplicate using TB Green™
Premix Ex Taq™ (Takara Bio, Inc.) on a CFX Connect™ Real-Time PCR
system (Bio-Rad Laboratories, Inc.). Two positive and two negative
control sites were tested, plus three test sites in one gene of
interest. The resultant signals were normalized for primer
efficiency by performing qPCR for each primer pair using input DNA
(pooled unprecipitated genomic DNA from each cell sample). Primer
sequences for chromatin immunoprecipitation-quantitative PCR were
shown in Table III.
 | Table III.Primer sequences for chromatin
immunoprecipitation- quantitative PCR. |
Table III.
Primer sequences for chromatin
immunoprecipitation- quantitative PCR.
| Binding site | Primer
sequence |
|---|
| Sp1 binding | F:
5′-CACTCCTGCCACATGTGAAG-3′ |
| site A | R:
5′-GTTCTGAGCACTTGCTTCCA-3′ |
| Sp1 binding | F:
5′-TCAGGAGACAGAAGCAGG-3′ |
| site B | R:
5′-TGAGGAGTGAGTGAGCCAAG-3′ |
| Sp1 binding | F:
5′-TCGGAAACTGGGCTCTATCC-3′ |
| site C | R:
5′-CCCTCAGCAGCCTCTGAGGA-3′ |
Rat lithogene model
A total of 30 6-week-old male Sprague-Dawley rats
(body weight 220±15 g) were purchased from Beijing Dingguo
Changsheng Biotechnology Co., Ltd. A total of 18 Sprague-Dawley
rats were randomly divided into two groups (n=9 for control group
and n=9 for experiment group): Pseudo-operation control group and
experimental group. The rats were first anesthetized with
pentobarbital sodium (intraperitoneal injection, 30 mg/kg),
underwent a laparotomy and a polyethylene (PE) tube was inserted
into their common bile duct and fixed. Rats were then injected with
the appropriate drugs (5 mg/ml LPS for the experimental group and
distilled water for the control group) via the PE tubes over a
period of 3 days. The pseudo-operation control group was injected
with distilled water (100 µl) and the experimental group was
injected with LPS (5 mg/kg), at a total volume of 100 µl. The PE
tubes were closed for 12 h after drug injection and then re-opened.
The common bile duct and intrahepatic bile duct were harvested 15
days after surgery. The rats were anesthetized with pentobarbital
sodium (intraperitoneal injection, 60 mg/kg) and then sacrificed by
cervical dislocation while bile duct samples were obtained. The
expression levels of MUC5AC, Sp1 and miR-130b were evaluated by
western blotting, RT-qPCR and IHC. No rats died during the modeling
period.
Rat biochemical analysis
Blood samples were taken from each group of rats and
analyzed for serum total bilirubin, ALT and AST levels using a
biochemical analyzer (cat. no. 98-11084-01; cat. no. 98-24010-US;
cat. no. 98-24016-US; Catalyst One; IDEXX).
Statistical analysis
Data are presented as the mean ± SD. All figures
were created using SPSS software (version 20.0; SPSS Inc.). Data
were analyzed using one-way ANOVA and Tukey's post hoc test for
multiple group comparisons. Pearson algorithm for bivariate
correlation analysis was used in positive distribution data.
Pearson of Spearman algorithm for bivariate correlation analysis
was used in nonpositive distribution data. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression levels of miR-130b, Sp1 and
MUC5AC differed between intrahepatic cholangiolithiasis tissue and
normal tissue
Bacterial infection of the biliary tract produces
LPS, which then induces increased expression of MUC5AC (2,3,6). The
aim of the present study was to identify the underlying mechanism
of this through a series of experiments. First, an IHC assay was
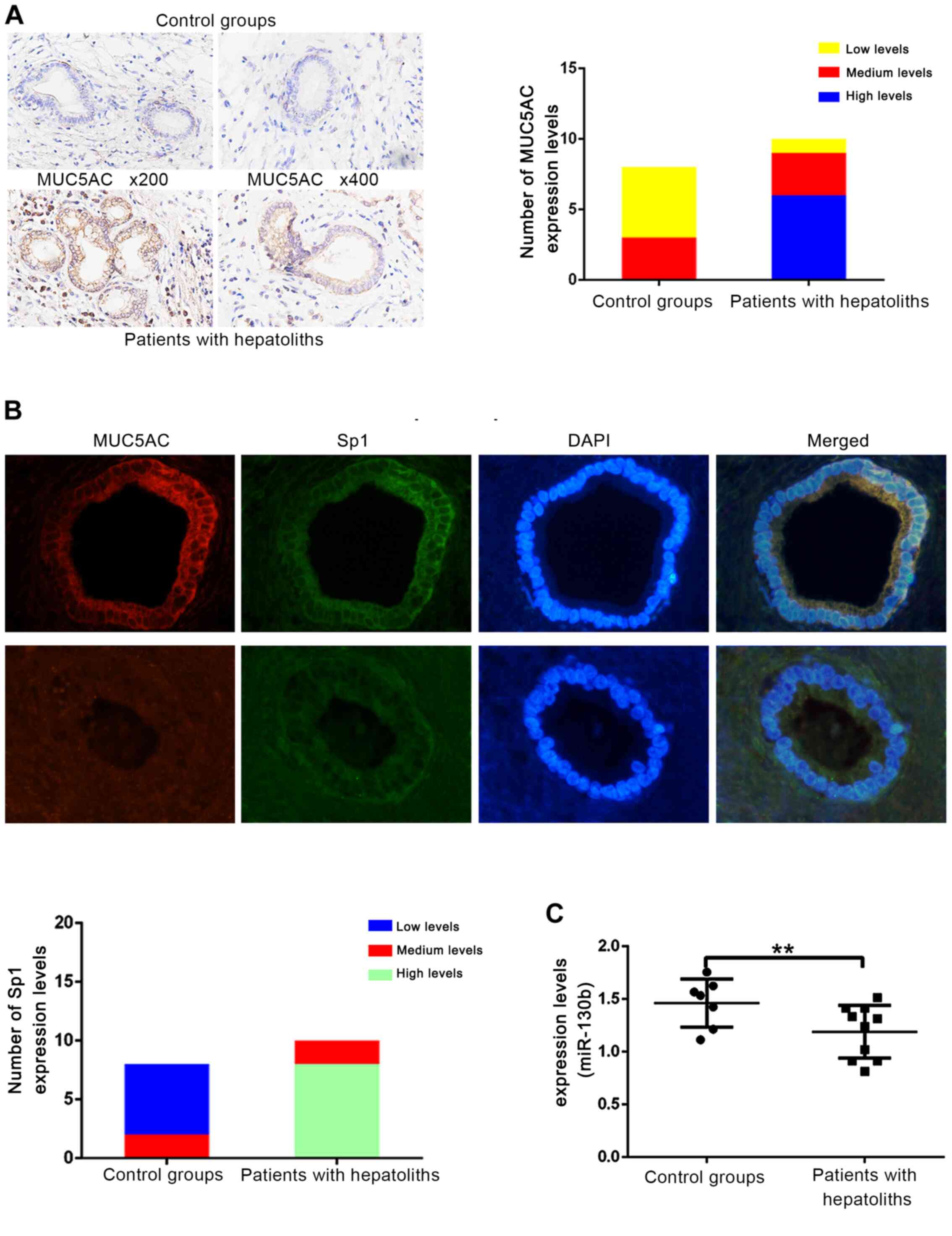
performed to stain and compare the expression of MUC5AC in clinical
paraffin-embedded sections from an intrahepatic cholangiolithiasis
group and a control group. Based on the IHC results, MUC5AC
staining was markedly higher in the intrahepatic cholangiolithiasis
group compared with the control group (Fig. 1A). The regulatory function of Sp1 on
MUC5AC expression and the effect of miR-130b on Sp1 expression, has
been described briefly for diseases in other body systems (15,18,19);
however, their effects in biliary tract diseases is unknown.
Therefore, MUC5AC and Sp1 were stained in clinical paraffin
sections using an IF assay. Sp1 expression was higher in the
intrahepatic cholangiolithiasis compared with the control group
(Fig. 1B). The expression of
miR-130b was significantly lower (Fig.
1C) in the cholangiolithiasis group compared with the control
group. These findings suggested that miR-130b and Sp1 expressional
changes may be associated with MUC5AC changes and the occurrence of
intrahepetic gallstones.
LPS induces the expression of MUC5AC,
Sp1 and miR-130b in HIBEpiCs in a concentration-dependent
manner
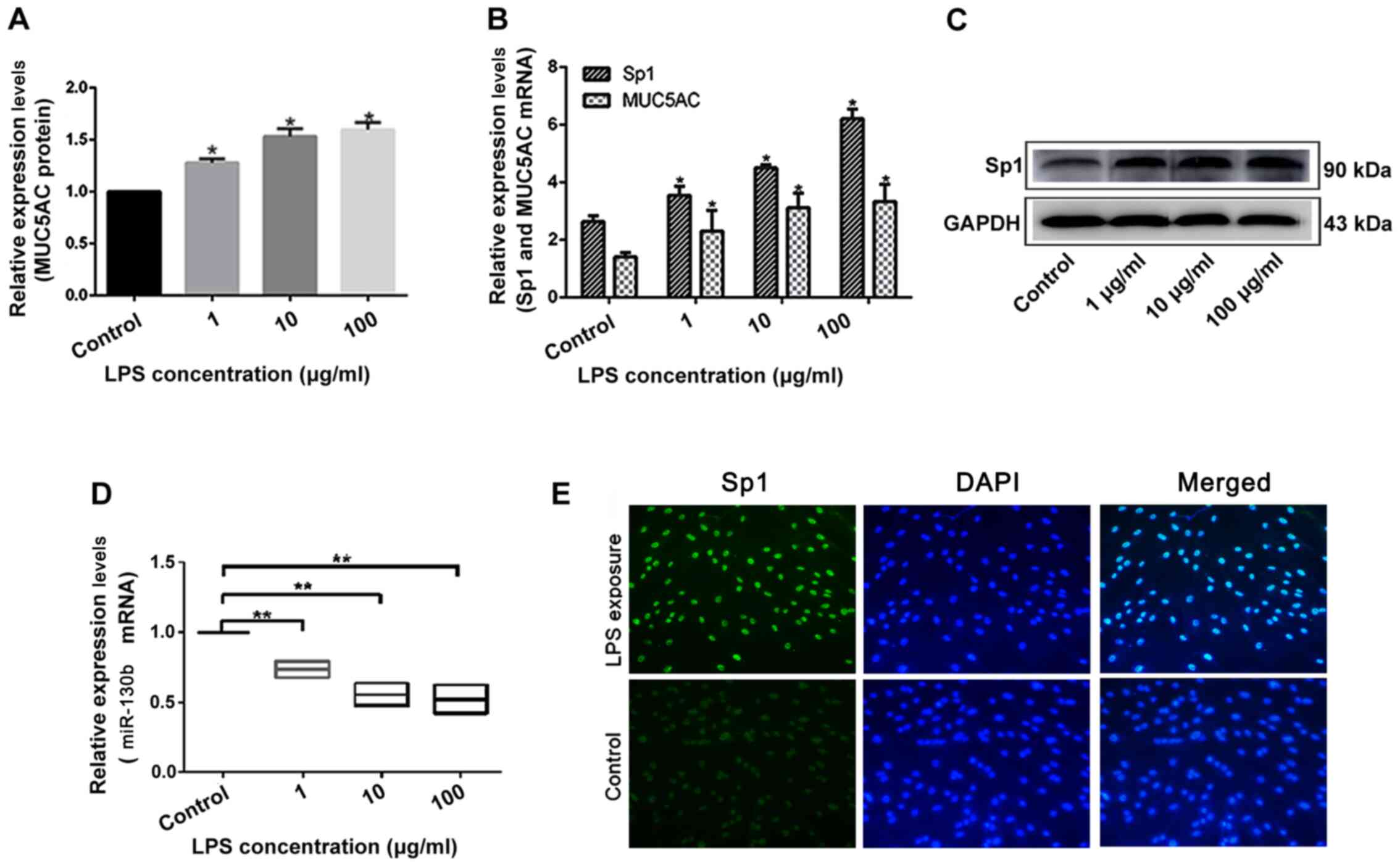
HIBEpiCs were incubated with of 1, 10 and 100 µg/ml
LPS for 24 h. Using ELISA and RT-qPCR, it was found that the
secretion of MUC5AC in the cell supernatant (Fig. 2A), and the expression of MUC5AC mRNA
(Fig. 2B) significantly increased
after LPS intervention in a concentration-dependent manner.
Similarly, Sp1 mRNA (Fig. 2B) and
protein expression levels (Fig. 2C)
increased in HIBEpiCs following LPS interference in a
concentration-dependent manner. Meanwhile, miR-130b levels
significantly decreased upon increasing LPS concentration (Fig. 2D). The Sp1 DNA-binding effect in
LPS-induced cell nuclei was also higher compared with the control
group (Fig. 2E). Thus, changes in
MUC5AC expression in the intrahepatic cholangiolithiasis biliary
tissue after LPS treatment was consistent with changes in Sp1
expression and contrasted the changes observed in miR-130b
expression. These findings showed that the miR-130b-Sp1-MUC5AC
pathway may play a role in LPS-induced upregulation of MUC5AC in
the bile duct epithelium.
Changes in the expression of miR-130b,
Sp1 and MUC5AC in an intrahepatic bile duct stone animal model
To further investigate the miR-130b-Sp1-MUC5AC
signaling pathway, an intrahepatic bile duct stone animal model was
established (Fig. 3A). After 15
days of modeling (Fig. 3B),
compared with normal rats, the serum total bilirubin and leukocyte
indices of the rats in the experiment group significantly
increased, which indicated that the biliary tract of the model rats
presented inflammation and obstruction caused by pigment stones
(Fig. 3C). Bile samples from the
model rats were collected after 15 days of modeling. Compared with
bile of rats in control group, bile from the rats in experiment
group were turbid and formed yellow crystals (Fig. 3D).
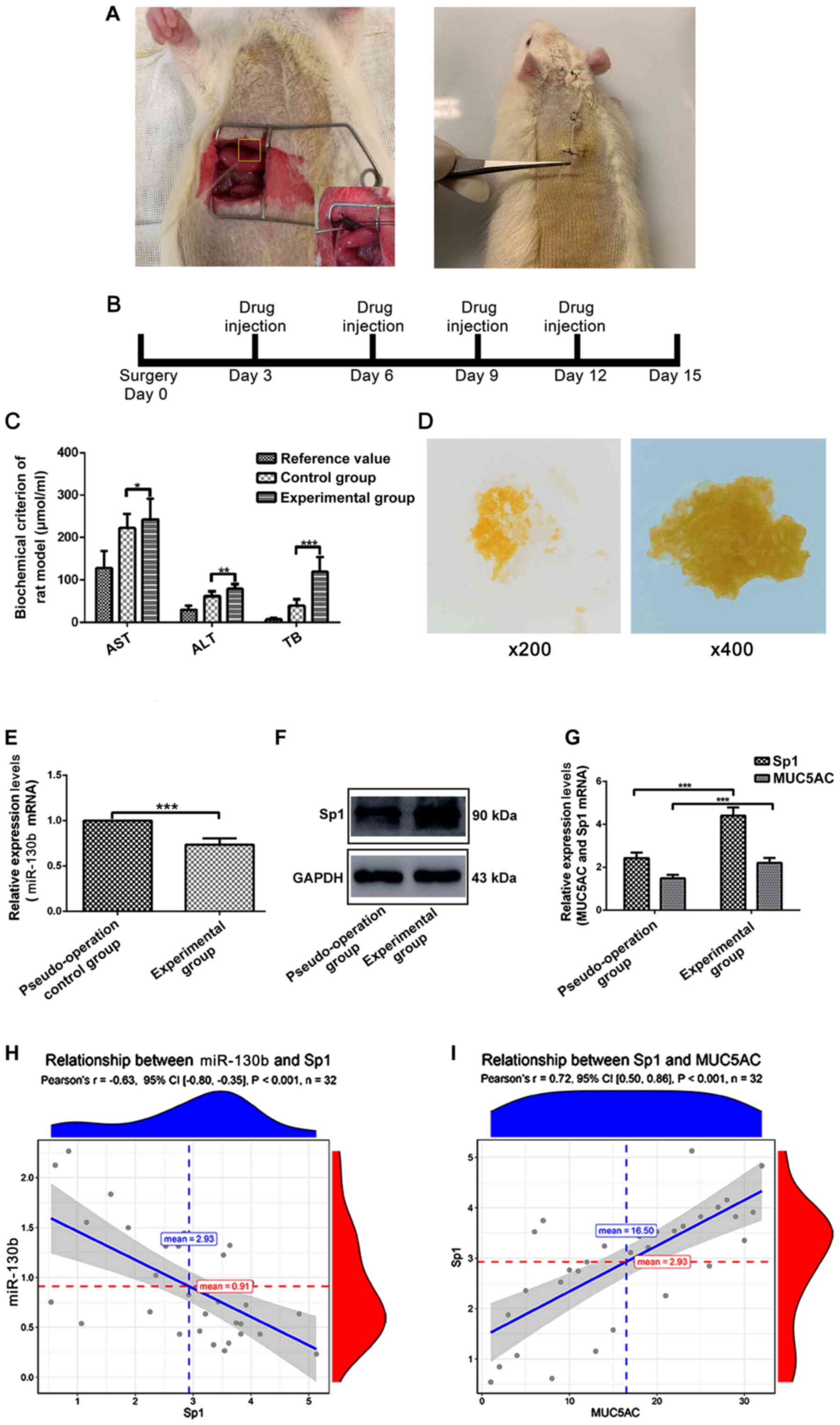 | Figure 3.MUC5AC, Sp1 and miR-130b levels in an
intrahepatic bile duct stone animal model. (A) Sprague-Dawley rats
underwent laparotomy and a PE tube was inserted into their common
bile duct and fixed. The PE tube was placed from the back of the
neck through a subcutaneous tunnel and fixed. (B) Days of each drug
injection. (C) Rat serum levels of AST, ALT and TB were measured.
(D) Images of rat bile smears after modeling. (E) Western blotting
was performed to measure Sp1 expression in bile duct tissues from
different groups. (F) RT-qPCR assay to detect expression of
miR-130b in bile duct tissues across different groups. (G) RT-qPCR
assay to detect expression levels of Sp1 and MUC5AC in bile duct
tissues from different groups. (H) Correlation analysis of Sp1 and
miR-130b mRNA expression levels. (I) Correlation analysis of MUC5AC
and Sp1 mRNA expression levels. *P<0.05, **P<0.01 and
***P<0.0001. AST, aspartate transaminase; ALT, alanine
aminotransferase; TB, total bilirubin; MUC5AC, mucin 5AC; Sp1,
specificity protein 1; miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; PE, polyethylene. |
RT-qPCR results indicated that the expression of
miR-130b was significantly lower in the experimental group compared
with the pseudo-operation group (Fig.
3E). Western blotting and RT-qPCR further indicated that
compared with the pseudo-operation group, Sp1 protein and mRNA
expression in the experimental group were higher (Fig. 3F and G). RT-qPCR results indicated
that the mRNA expression of MUC5AC significantly increased in the
experimental group compared with the pseudo-operation group
(Fig. 3G). Collectively, this
series of in vivo experiments suggested that upregulation of
MUC5AC expression in intrahepatic bile duct stone tissue was due to
expressional changes of miR-130b and Sp1.
Correlation analysis of miR-130b, Sp1 and MUC5AC
mRNA expression levels is shown in Fig.
3H and I. The results indicated that miR-130b and Sp1 mRNA
expression levels in bile duct samples showed a significantly
negative correlation, whereas mRNA expression levels of Sp1 and
MUC5AC showed a significant positive correlation.
Sp1 directly regulates MUC5AC
expression by binding with its promoter sequence
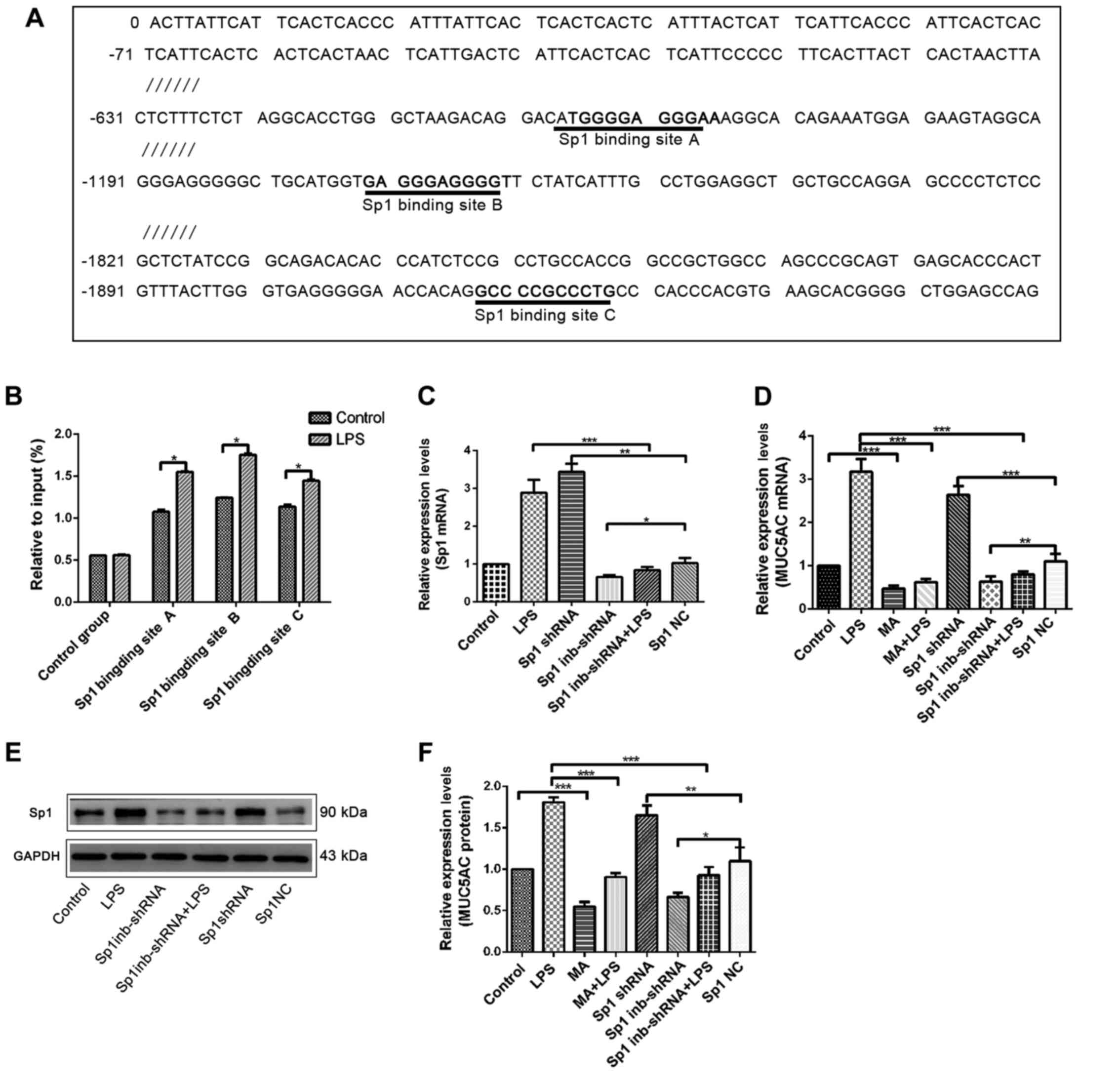
To verify the direct binding of Sp1 to the MUC5AC
promoter sequence and identify the corresponding binding sites, the
possible binding sites of Sp1 to the MUC5AC promoter sequences were
predicted using online tools. The predictions indicated that there
are three possible binding sites, located at positions 665–675,
1209–1219 and 1918–1928 of the MUC5AC promoter sequence (Fig. 4A). Three ChIP-qPCR primers were
designed according to these three binding sites (Table II). Cells in the experimental (10
µg/ml LPS treatment for 24 h) and control groups were then
evaluated using a ChIP-qPCR assay. All three binding sites played a
role in Sp1 binding. The levels Sp1 binding at the three binding
sites were significantly higher in the experimental group compared
with the control group (Fig. 4B).
This indicated that these three binding sites play a role in
regulating the effect of Sp1 on MUC5AC expression and that LPS
induction significantly increased the levels of Sp1 binding.
To provide further evidence for the regulatory
effect of Sp1 on MUC5AC expression, shRNA was used to overexpress
Sp1 or suppress Sp1 expression, or cells were pre-treated with MA.
RT-qPCR, western blotting and ELISA were then performed to detect
the changes in Sp1 and MUC5AC levels. Experimental results showed
that Sp1 and MUC5AC mRNA in the overexpression group was
significantly higher compared with the negative control group. A
marked decline in the Sp1 suppressed group compared with the
negative control group was also observed (Fig. 4C and D). The western blotting
results showed that, Sp1 protein expression was significantly
increased in the Sp1 overexpression group, but significantly lower
in the Sp1 suppressed-expression group (Fig. 4E). ELISA results showed that
compared with their respective controls, MUC5AC levels in the Sp1
overexpression group significantly increased, while MUC5AC levels
in the MA and Sp1 suppressed-expression group significantly
decreased (Fig. 4F).
These results indicated that the effect of
LPS-induced MUC5AC upregulation in the bile duct epithelium was
regulated by Sp1. Increase in Sp1 expression can promote this
effect and decreased Sp1 expression can reverse this.
miR-130b directly inhibits Sp1
expression by binding with its 3′-UTR sequence
Next, the potential binding sites of miR-130b and
the Sp1 3′-UTR were predicted using online tools (http://www.targetscan.org/vert_72/). A binding
site was found for miR-130b at position 720–726 in the Sp1 3′-UTR
sequence (Fig. 5A). Next,
luciferase vectors containing full length miR-130b and Sp1 3′-UTR
regions were constructed (Fig. 5A),
and these luciferase vectors were co-transfected into HIBEpiCs for
luciferase reporter gene assays. The results indicated that,
compared with the negative control group, the fluorescence
intensity of the 3′UTR+mir group significantly decreased (Fig. 5B), thus showing that miR-130b binds
directly to the Sp1 3′-UTR sequences. A point mutation vector for
miR-130b was constructed (Fig. 5A),
and this was co-transfected with the Sp1 3′-UTR fragment into
HIBEpiCs. It was found that the fluorescence intensity of
3′UTR-nu+miR group relatively increased compared with 3′UTR+mir
group (Fig. 5C).
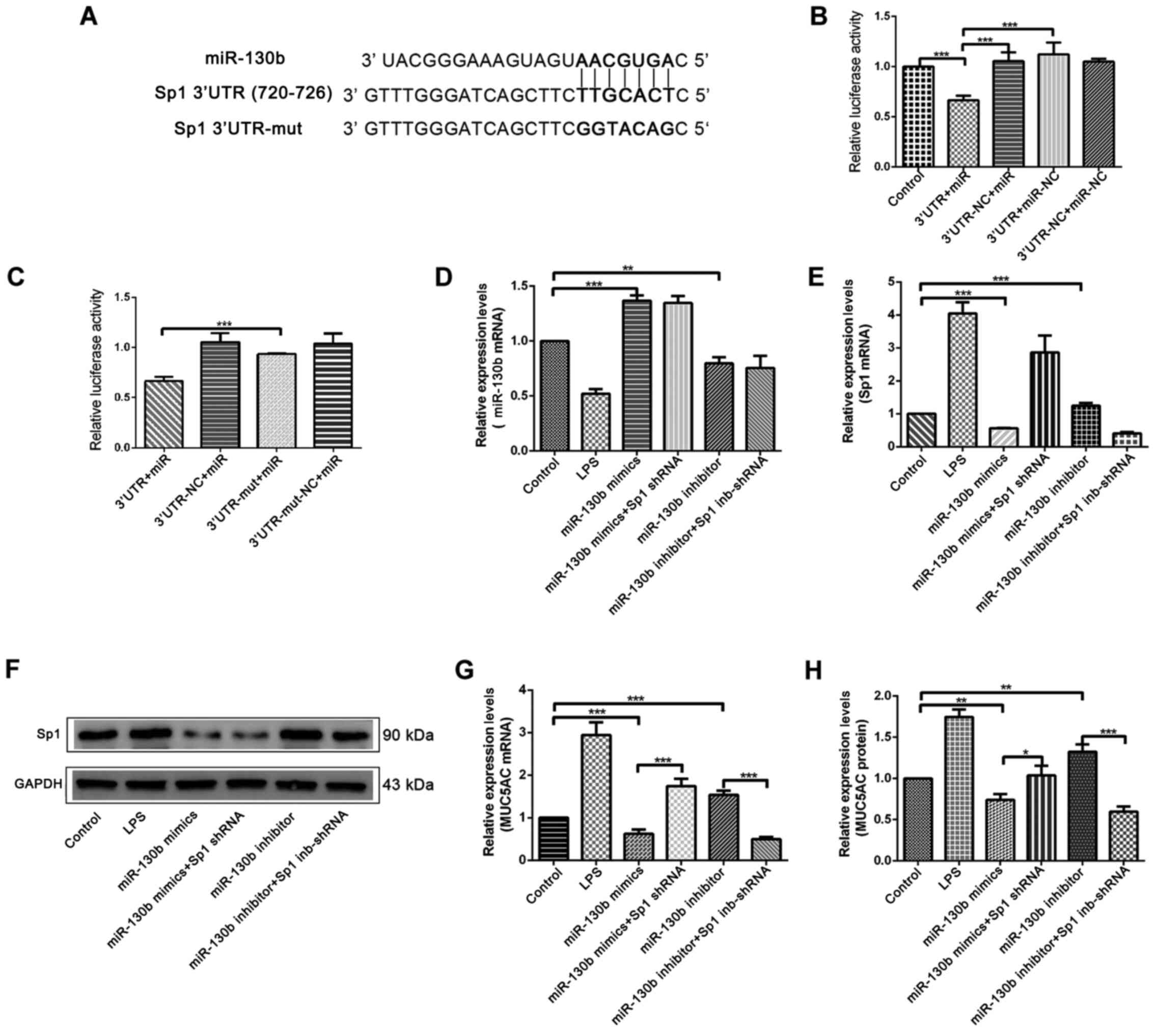 | Figure 5.Investigation of the direct binding
position between miR-130b and the 3′-UTR sequence of Sp1 and
further verification of the regulatory effect of miR-130b on MUC5AC
and Sp1 expression. (A) Online tools were used to predict the
binding position of miR-130b to the 3′-UTR sequence of Sp1, and the
corresponding luciferase primer for a natural plasmid and a mutated
plasmid were established. (B and C) Luciferase gene detection for
miR-130b and the Sp1 3′-UTR sequence. HIBEpiCs were transfected
with miR-130b overexpression mimics or miR-130b inhibitors for 24
h. (D) RT-qPCR detected changes in miR-130b expression in HIBEpiCs.
(E) RT-qPCR detected changes in of Sp1 mRNA expression in HIBEpiCs.
(F) Western blotting detected changes Sp1 protein expression in
HIBEpiCs. (G) RT-qPCR detected changes in of MUC5AC mRNA expression
in HIBEpiCs. (H) ELISA detected changes in MUC5AC levels in HIBEpiC
supernatant. *P<0.05, **P<0.01 and ***P<0.0001. MUC5AC,
mucin 5AC; Sp1, specificity protein 1; miR, microRNA; HIBEpiCs,
human intrahepatic biliary epithelial cells; RT-qPCR, reverse
transcription-quantitative PCR; 3′-UTR, 3′-untranslated region;
LPS, lipopolysaccharide; NC, negative control; mut, mutant; shRNA,
short hairpin RNA; inb, inhibitor. |
To verify the suppressive effect of miR-130b on the
expression of Sp1, a miR-130b overexpression mimic and an
inhibitor-expression plasmid were transfected into HIBEpiCs for a
functional recovery assay, and the expression levels of miR-130b,
Sp1 and MUC5AC were detected by western blotting, RT-qPCR and
ELISA. RT-qPCR results for miR-130b detection showed that, compared
with the control group, the expression of miR-130b significantly
increased in the miR-130b mimics group, while the expression
significantly decreased in the inhibitor group (Fig. 5D). RT-qPCR and western blotting
results further showed that the mRNA and protein expression of Sp1
decreased in the miR-130b mimics group, and increased in the
miR-130b inhibitor group, when compared with the control group
(Fig. 5E and F). ELISA and RT-qPCR
results further showed that compared with the control group, the
mRNA and protein expression of MUC5AC in the miR-130b mimics group
was significantly lower, but significantly higher in the miR-130b
inhibitor group. In addition, the increase of MUC5AC induced by Sp1
shRNA can be suppressed by miR-130b mimics (Fig. 5G and H).
These results indicated that LPS-induced MUC5AC
upregulation was achieved by increasing Sp1 expression and Sp1
expression was directly regulated by miR-130b. Hence, the
miR-130b-Sp1-MUC5AC signaling pathway may play a role in the
development of intrahepatic bile stones.
Discussion
Intrahepatic cholelithiasis is a common disease in
the biliary system among the Chinese population (1). At present, the surgical and clinical
treatment method of intrahepatic cholelithiasis is relatively
limited in hepatectomy or choledochoscopic lithotripsy, however,
these method were accompanied with numerous complications such as
intrahepatic insufficiency and recurrence of calculus after
treatment (27). Therefore, it is
of importance to study the detailed mechanism of the disease and
develop new treatment methods. Previous studies showed that LPS
produced by bacterial infection of bile duct could increase MUC5AC
secretion in the bile duct epithelium, which then promotes bile
duct stone formation (3,6). However, to the best of our knowledge,
the mechanism of MUC5AC overexpression following bacterial
infection has not been elucidated. The present study showed that
MUC5AC and Sp1 expression in the bile duct tissue of patients with
hepatolithiasis was higher compared with the control group, while
the expression of miR-130b was relatively low. In addition,
LPS-treated bile duct epithelial cells and in vivo
experiments of Sprague-Dawley rats showed similar results.
Therefore, it was concluded there may be a mutual regulation
between miR_130b, Sp1 and MUC5AC secretion. ChIP and luciferase
reporter assays confirmed that miR-130b has one binding site in the
3′-UTR region of Sp1, which may regulate Sp1 expression by binding
to this site. Sp1 contains three binding sites in the promoter
sequence of MUC5AC, which may regulate MUC5AC secretion when Sp1
binding with these three sites (15,18).
In cell transfection experiments, it was found that the expression
of Sp1 and MUC5AC decreased when miR-130b was overexpressed, while
inhibition of miR-130b expression could promote the secretion of
Sp-1 and MUC5AC. Similarly, in the case Sp1 overexpression, MUC5AC
secretion increased, while inhibition of Sp1 expression resulted in
decreased MUC5AC expression. This further confirmed the regulatory
effects of miR-130b to Sp1 and Sp1 to MUC5AC. Therefore, the
present study concluded that miR-130b and Sp1 may play a role in
MUC5AC overexpression in the intrahepatic bile duct epithelium
caused by bacterial infection. Utilizing the regulatory effect of
miR-130b and Sp1 to promote or inhibit miR-130b and Sp1 expression
may provide a new strategy and method for the treatment of
hepatolithiasis. Of note, MUC5AC overexpression is not the only
factor responsible for intrahepatic bile duct stone formation.
Although influencing miR-130b and Sp1 expression can partly
regulate MUC5AC secretion, it may not completely avoid stone
formation. Therefore, how to effectively use miR-130b, Sp1 and
MUC5AC as a signal transduction pathway for the treatment of
hepatolithiasis still needs more further experimental research.
Acknowledgements
Not applicable.
Funding
This study was funded by the Outstanding Scientific
Fund of Shengjing Hospital (grant no. A250) and the National
Natural Science Foundation of China (grant no. A376).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
SW and JK designed the study. XW, ZZ, CY and JH
performed the experiments. XW, YF and YT contributed to data
collection, statistical analysis, data interpretation and
manuscript preparation. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
The study was approved by the local ethics committee
of the Affiliated Shengjing Hospital of China Medical University
(approval no. 2017PS231K).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kim HJ, Kim JS, Joo MK, Lee BJ, Kim JH,
Yeon JE, Park JJ, Byun KS and Bak YT: Hepatolithiasis and
intrahepatic cholangiocarcinoma: A review. World J Gastroenterol.
21:13418–13431. 2015. View Article : Google Scholar
|
|
2
|
Kim HJ, Kim SH, Chae GB, Lee SJ and Kang
CD: Increased expression of mucin 5AC mRNA and decreased expression
of epidermal growth-factor receptor mRNA in gallstone patients.
Tohoku J Exp Med. 214:139–144. 2008. View Article : Google Scholar
|
|
3
|
Wu SD, Yu H and Sun JM: Bacteriological
and electron microscopic examination of primary intrahepatic
stones. Hepatobiliary Pancreat Dis Int. 5:228–231. 2006.
|
|
4
|
Yoo KS, Choi HS, Jun DW, Lee HL, Lee OY,
Yoon BC, Lee KG, Paik SS, Kim YS and Lee J: MUC Expression in
gallbladder epithelial tissues in cholesterol-associated
gallbladder disease. Gut Liver. 10:851–858. 2016. View Article : Google Scholar
|
|
5
|
Chuang SC, His E and Lee KT: Mucin genes
in gallstone disease. Clin Chim Acta. 413:1466–1471. 2012.
View Article : Google Scholar
|
|
6
|
Li M, Tian Y, Wu S, Yu H and Li Y: LPS
stimulates MUC5AC expression in human biliary epithelial cells:
Whether there exists a possible pathway of PKC/NADPH/ROS? Mol Cell
Biochem. 385:87–93. 2014. View Article : Google Scholar
|
|
7
|
Tan NY and Khachigian LM: Sp1
phosphorylation and its regulation of gene transcription. Mol Cell
Biol. 29:2483–2488. 2009. View Article : Google Scholar
|
|
8
|
Baños-Lara Mdel R, Piao B and
Guerrero-Plata A: Differential mucin expression by respiratory
syncytial virus and human metapneumovirus infection in human
epithelial cells. Mediators Inflamm. 2015:3472922015.
|
|
9
|
Zen Y, Harada K, Sasaki M, Tsuneyama K,
Katayanagi K, Yamamoto Y and Nakanuma Y: Lipopolysaccharide induces
overexpression of MUC2 and MUC5AC in cultured biliary epithelial
cells: Possible key phenomenon of hepatolithiasis. Am J Pathol.
161:1475–1484. 2002. View Article : Google Scholar
|
|
10
|
Vilkin A, Nudelman I, Morgenstern S,
Geller A, Dayan YB, Levi Z, Rodionov G, Hardy B, Konikoff F, Gobbic
D and Niv Y: Gallbladder inflammation is associated with increase
in mucin expression and pigmented stone formation. Dig Dis Sci.
52:1613–1620. 2007. View Article : Google Scholar
|
|
11
|
Corfield AP, Myerscough N, Longman R,
Sylvester P, Arul S and Pignatelli M: Mucins and mucosal protection
in the gastrointestinal tract: New prospects for mucins in the
pathology of gastrointestinal disease. Gut. 47:589–594. 2000.
View Article : Google Scholar
|
|
12
|
Ma J, Rubin BK and Voynow JA: Mucins,
mucus and goblet cells. Chest. 154:169–176. 2017. View Article : Google Scholar
|
|
13
|
Ali MS and Pearson JP: Upper airway mucin
gene expression: A review. Laryngoscope. 117:932–938. 2010.
View Article : Google Scholar
|
|
14
|
Rose MC and Voynow JA: Respiratory tract
mucin genes and mucin glycoproteins in health and disease. Physiol
Rev. 86:245–278. 2006. View Article : Google Scholar
|
|
15
|
Hewson CA, Edbrooke MR and Johnston SL:
PMA induces the MUC5AC respiratory mucin in human bronchial
epithelial cells, via PKC, EGF/TGF-alpha, Ras/Raf, MEK, ERK and
Sp-1-dependent mechanisms. J Mol Biol. 344:683–695. 2004.
View Article : Google Scholar
|
|
16
|
Huang C and Xie K: Crosstalk of Sp1 and
Stat3 signaling in pancreatic cancer pathogenesis. Cytokine Growth
Factor Rev. 23:25–35. 2012. View Article : Google Scholar
|
|
17
|
Jiang W, Jin Z, Zhou F, Cui J and Wang L
and Wang L: High co-expression of Sp1 and HER-2 is correlated with
poor prognosis of gastric cancer patients. Surg Oncol. 24:220–225.
2015. View Article : Google Scholar
|
|
18
|
Di YP, Zhao J and Harper R: Cigarette
smoke induces MUC5AC protein expression through the activation of
Sp1. J Biol Chem. 287:27948–27958. 2012. View Article : Google Scholar
|
|
19
|
Oyanagi T, Takizawa T, Aizawa A, Solongo
O, Yagi H, Nishida Y, Koyama H, Saitoh A and Arakawa H: Suppression
of MUC5AC expression in human bronchial epithelial cells by
interferon-γ. Allergol Int. 66:75–82. 2016. View Article : Google Scholar
|
|
20
|
Aslam F, Palumbo L, Augenlicht LH and
Velcich A: The Sp family of transcription factors in the regulation
of the human and mouse MUC2 gene promoters. Cancer Res. 61:570–576.
2001.
|
|
21
|
Luke B and David E: Airway mucus and
asthma: The role of MUC5AC and MUC5B. J Clin Med. 6:1122017.
View Article : Google Scholar
|
|
22
|
Zheng H, Dong X, Liu N, Xia W, Zhou L and
Chen X, Yang Z and Chen X: Regulation and mechanism of mouse
miR-130a/b in metabolism-related inflammation. Int J Biochem Cell
Biol. 74:72–83. 2016. View Article : Google Scholar
|
|
23
|
Li L, Gao F, Jiang Y, Yu L, Zhou Y, Zheng
H, Tong W, Yang S, Xia T, Qu Z and Tong G: Cellular miR-130b
inhibits replication of porcine reproductive and respiratory
syndrome virus in vitro and in vivo. Sci Rep. 5:170102015.
View Article : Google Scholar
|
|
24
|
Kaur K, Bhatia H and Datta M: MicroRNAs in
hepatic pathophysiology in diabetes. World J Diabetes. 2:158–163.
2011. View Article : Google Scholar
|
|
25
|
Contreras J and Rao DS: MicroRNAs in
inflammation and immune responses. Leukemia. 26:404–413. 2012.
View Article : Google Scholar
|
|
26
|
Kim C, Lee H, Cho YM, Kwon OJ, Kim W and
Lee EK: TNFalpha-induced miR-130 resulted in adipocyte dysfunction
during obesity-related inflammation. FEBS Lett. 587:3853–3858.
2013. View Article : Google Scholar
|
|
27
|
Tazuma S, Unno M, Igarashi Y, Inui K,
Uchiyama K, Kai M, Tsuyuguchi T, Maguchi H, Mori T, Yamaguchi K, et
al: Evidence-based clinical practice guidelines for cholelithiasis
2016. J Gastroenterol. 52:276–300. 2017. View Article : Google Scholar
|