Introduction
Osteoarthritis (OA) is the most common form of
arthropathy in elderly individuals worldwide, and is characterized
by local inflammation, articular cartilage damage and degradation
of the cartilage (1). The number of
patients with OA is expected to increase as the population ages,
increasing the psychological and socio-economic burden of patients
(2). Current therapeutic strategies
for OA are aimed at symptomatic control, rather than disease
modification (3); therefore,
extending the existing knowledge of the pathology of OA is required
for the development of effective therapeutic strategies.
OA displays typical heritability, which is why gene
therapy for OA has gained much attention. Clinically, in
vivo gene expression is difficult to control, and the selection
and safety of vectors require confirmation; therefore, OA gene
therapy is currently at the in vitro and in vivo
stages of research (4). Meanwhile,
a growing number of studies have indicated that non-coding RNAs
(ncRNAs) are possible regulators of cell apoptosis (5,6). As a
ncRNA subclass, long ncRNAs (lncRNAs) are >200 nucleotides in
length, and display improved tissue and cell specificity compared
with coding RNAs. lncRNAs are differentially expressed in different
cells, tissues and developmental stages, and are associated with
the occurrence and development of various diseases, including OA
and malignancies (7–10). Moreover, it has been confirmed that
enriched lncRNAs in the cytoplasm typically participate in
post-transcriptional modification by binding to microRNAs
(miRNAs/miRs) or mRNAs (8). For
example, Li et al (11)
reported that Pvt1 oncogene (PVT1) regulated chondrocyte apoptosis
in OA by sponging miR-488-3p. Xiao et al (12) indicated that lncRNA miR4435-2HG,
which regulated chondrocyte cell proliferation and apoptosis, may
be downregulated in OA. Furthermore, it has been reported that
lncRNA small nucleolar RNA host gene 16 (SNHG16) reversed the
effects of miR-15a/16 on the lipopolysaccharide-induced
inflammatory pathway (13). A
recent study reported that SNHG16 may be associated with the
occurrence of OA (14). However,
the biological role of SNHG16 during the development of OA requires
further investigation.
Therefore, the present study aimed to explore the
roles of SNHG16 during the development of OA and the potential
underlying mechanisms. The present study provided a theoretical
basis for OA treatment and a novel perspective for clinical
therapy.
Materials and methods
Cell culture
Previous studies have indicated that CHON-001 cells
treated with interleukin (IL)-1β serve as an in vitro model
of OA; therefore, this model was selected for the present study
(15,16). The human chondrocyte cell line
CHON-001 (American Type Culture Collection) was cultured in
RPMI-1640 medium (Thermo Fisher Scientific, Inc.) supplemented with
10% FBS (Thermo Fisher Scientific, Inc.) and 2 mM glutamine
(Sigma-Aldrich; Merck KGaA) at 37°C at 5% CO2. To induce
the in vitro model of OA, CHON-001 cells were treated with
IL-1β (10 ng/ml) (Sigma-Aldrich; Merck KGaA) for 24 h at room
temperature.
Tissue collection
OA tissues (n=20) and normal joint tissues (n=20)
were obtained from the Second Affiliated Hospital of Inner Mongolia
Medical University (Hohhot, China) between August 2017 and April
2019. OA tissues were obtained from patients with OA. Normal joint
tissues were obtained from patients who had been in a car accident.
Clinical and pathological data of the patients were collected.
Patients were diagnosed with OA according to the American College
of Rheumatology classification criteria for the diagnosis of OA
(17). The present study was
approved by the Institutional Ethical Committee of the Second
Affiliated Hospital of Inner Mongolia Medical University. Written
informed consent was obtained from all participants. Clinical
characteristics of the patients and healthy controls are presented
in Table I.
 | Table I.Clinical characteristics of the
patients and healthy controls. |
Table I.
Clinical characteristics of the
patients and healthy controls.
| Clinical
characteristic | Patients
(n=20) | Healthy controls
(n=20) | P-value |
|---|
| Age (years) | 57.63±7.22 | 57.28±6.55 | 0.8733 |
| Sex
(male/female) | 11/9 | 10/10 | 0.7515 |
| Body mass index
(kg/m2) | 23.86±2.05 | 24.01±1.89 | 0.8112 |
| Family history of
OA (yes/no) | 5/15 | 2/18 | 0.2119 |
Cell transfection
siRNAs targeted against SNHG16 (SNHG16 siRNA1 and
SNHG16 siRNA2; 10 nM) and a negative control siRNA (siRNA-NC) were
purchased from Guangzhou RiboBio Co., Ltd. and transfected into
CHON-001 cells (5×103) using Lipofectamine®
2000 (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Cells were incubated at 37°C for 6 h
before subsequent experiments were performed. Transfection
efficiency was determined using reverse transcription-quantitative
PCR (RT-qPCR). The sequences of the siRNAs were as follows:
Negative control siRNA, 5′-UUCUCCGAACGUGUCACGUTT-3′; SNHG16 siRNA1,
5′-GGAAUGAAGCAACUGAGAUUU−3′; and SNHG16 siRNA2,
5′-GGGTTACGATTGCCCAGAT-3′.
RT-qPCR
Total RNA was extracted from tissues or cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). Total RNA (5–10 µg) was reverse transcribed into cDNA using
the PrimeScript RT reagent kit (Takara Bio, Inc.), according to the
manufacturer's instructions. Subsequently, qPCR was performed using
the SYBR premix Ex Taq II kit (Takara Bio, Inc.). The following
primer pairs were used for qPCR: SNHG16, forward,
5′-CTTCCGCCATGATTGTGAGG-3′, reverse, 5′-AACAGTCCCTCCTTGGTCTC-3′;
β-actin, forward, 5′-AGCGAGCATCCCCCAAAGTT-3′, reverse,
5′-GGGCACGAAGGCTCATCATT-3′; collagen II, forward,
5′-ATGCCACACTCAAGTCCCTCA-3′, reverse, 5′-GTCTCGCCAGTCTCCATGTTG-3′;
MMP13, forward, 5′-CAGAACTTCCCAACCGTATTGAT-3′, reverse,
5′-TGTATTCAAACTGTATGGGTCCG-3′; miR-373-3p, forward,
5′-GGCGGAAGTGCTTCGATTTT-3′, reverse, 5′-GTGCAGGGTCCGAGGTATTC-3′;
and U6, forward, 5′-GCTTCGGCAGCACATATACT-3′ and reverse,
5′-GTGCAGGGTCCGAGGTATTC-3′. The following thermocycling conditions
were used for qPCR: Initial denaturation for 10 min at 95°C; 40
cycles of 95°C for 15 sec and 60°C for 30 sec; and final extension
for 1 min at 60°C. miRNA and mRNA levels were quantified using the
2−ΔΔCq method and normalized to the internal reference
genes β-actin and U6, respectively (18).
Cell Counting Kit-8 (CCK-8) assay
The CCK-8 assay (Beyotime Institute of
Biotechnology) was performed to investigate cell proliferation.
CHON-001 cells (5×103 cells/well) were plated into
96-well plates and treated for 0, 24, 48 or 72 h at room
temperature as follows: 10 ng/ml IL-1β (IL-1β + NC siRNA); 10 ng/ml
IL-1β and subsequently transfected with SNHG16 siRNA1 (IL-1β +
SNHG16 siRNA1). Control cells were left untreated. CCK-8 reagent
(10 µl) was added to each well and incubated for 2 h at 37°C. The
absorbance of each well was measured at a wavelength of 450 nm
using a microplate reader.
Western blotting
Total protein was extracted from OA tissues and
CHON-001 cells using RIPA lysis buffer (Beyotime Institute of
Biotechnology) and quantified using a bicinchoninic acid assay kit
(Thermo Fisher Scientific, Inc.). Proteins (40 µg/lane) were
separated by SDS-PAGE on 10% gels and were transferred onto PVDF
membranes (Thermo Fisher Scientific, Inc.). Subsequently, the
membranes were blocked with 5% skim milk in TBS-Tween (10%) for 1 h
at room temperature. The membranes were then incubated overnight at
4°C with primary antibodies targeted against: Collagen II (1:1,000;
cat. no. ab325034; Abcam), aggrecan (1:1,000; cat. no. ab1816028;
Abcam), MMP13 (1:1,000; cat. no. ab25367; Abcam), p21 (1:1,000;
cat. no. ab14323; Abcam), cleaved caspase-3 (1:1,000; cat. no.
ab43583; Abcam) and β-actin (1:1,000; cat. no. ab06789; Abcam).
Following primary antibody incubation, the membranes were incubated
with a HRP-conjugated secondary antibody (1:5,000; cat. no.
ab20876; Abcam) for 1 h at room temperature. Protein bands were
detected using an ECL kit (Thermo Fisher Scientific, Inc.). Protein
expression was semi-quantified using Image-Pro Plus software
(version 6.0; Media Cybernetics, Inc.) with β-actin as the internal
control.
Target prediction
TargetScan (www.targetscan.org/vert_71/) and microRNA Target
Prediction Database (miRDB) databases (www.mirdb.org)
were used to identify the downstream target genes of SNHG16.
ELISA
The levels of TNF-α in the media of CHON-001 cells
were detected using the TNF-α ELISA kit [cat. no. ab47956, Hangzhou
Multi Sciences (Lianke) Biotech Co., Ltd.], according to the
manufacturer's protocol.
Immunofluorescence
CHON-001 cells were seeded (5×104
cells/well) into 24-well plates and incubated at 37°C for 24 h.
Cells were pre-fixed with 4% paraformaldehyde for 10 min at room
temperature, and fixed in pre-cold methanol at 4°C for another 10
min. Following blocking with 10% BSA (Beyotime Institute of
Biotechnology) at room temperature for 1 h, cells were incubated
overnight at 4°C with anti-Ki67 primary antibody (1:1,000; cat. no.
ab35267; Abcam) and DAPI. Following primary antibody incubation,
cells were incubated with the goat anti-rabbit IgG secondary
antibody (1:5,000; cat. no. ab65734; Abcam) at room temperature for
1 h. Immunofluorescence staining was observed using a CX23
fluorescence microscope (Olympus Corporation) and quantified using
Image-Pro Plus software (version 6.0; Media Cybernetics, Inc.).
Cell apoptosis analysis
CHON-001 cells were seeded (5×104
cells/well) in 6-well plates. Cells were centrifuged at 15 × g for
5 min at 4°C and resuspended in 100 µl binding buffer (BD
Biosciences). Subsequently, 5 µl Annexin V-FITC (BD Biosciences)
and 5 µl propidium (BD Biosciences) iodide were added to the cell
solution for 15 min at 0°C. Early and late apoptotic cells were
analyzed using a FACS flow cytometer (BD Biosciences) and WinMDI
software (version 2.9; Invitrogen; Thermo Fisher Scientific,
Inc.).
Dual-luciferase reporter assay
The partial sequence of the 3′-untranslated region
(UTR) SNHG16 and the 3′-UTR of p21 containing the putative binding
sites of miR-373-3p were synthetized and obtained from Sangon
Biotech Co., Ltd. Subsequently, the sequences were cloned into the
pmirGLO Dual-Luciferase miRNA Target Expression Vector (Promega
Corporation) to construct wild-type (WT) reporter vectors for
SNHG16 and p21. The mutant (MUT) SNHG16 and p21-3′-UTR sequences
containing the putative binding sites of miR-373-3p were created
using the Q5 Site-Directed Mutagenesis kit (New England Biolabs,
Inc.), according to the manufacturer's protocol, and cloned into
pmirGLO vectors to construct MUT reporter vectors for SNHG16 and
p21. The WT and MUT SNHG16 vectors were transfected into CHON-001
cells (5×103 cells/well) with blank (untransfected
cells), 10 nM vector-control (pcDNA-3.1 vector; cat. no. ab06707;
Beyotime Institute of Biotechnology) or 10 nM miR-373-3p mimics
(cat. no. ab35643; Beyotime Institute of Biotechnology) using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.)
according to the manufacturer's instructions. Similarly, the WT and
MUT p21 vectors were transfected into 293T cells with blank,
vector-control or miR-373-3p mimics. At 48 h post-transfection,
cells were collected and the relative luciferase activity was
detected using the Dual-Glo Luciferase assay system (Promega
Corporation), according to the manufacturer's protocol. Firefly
luciferase activity was normalized to Renilla luciferase
activity.
Statistical analysis
Experiments were performed in at least triplicate.
Data are presented as the mean ± SD. Comparisons between two groups
were analyzed using an unpaired Student's t-test. Comparisons among
multiple groups were analyzed using one-way ANOVA followed by
Tukey's post hoc test. Statistical analyses were performed using
GraphPad Prism software (version 7; GraphPad Software, Inc.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
Expression levels of SNHG16 are
upregulated in OA tissues
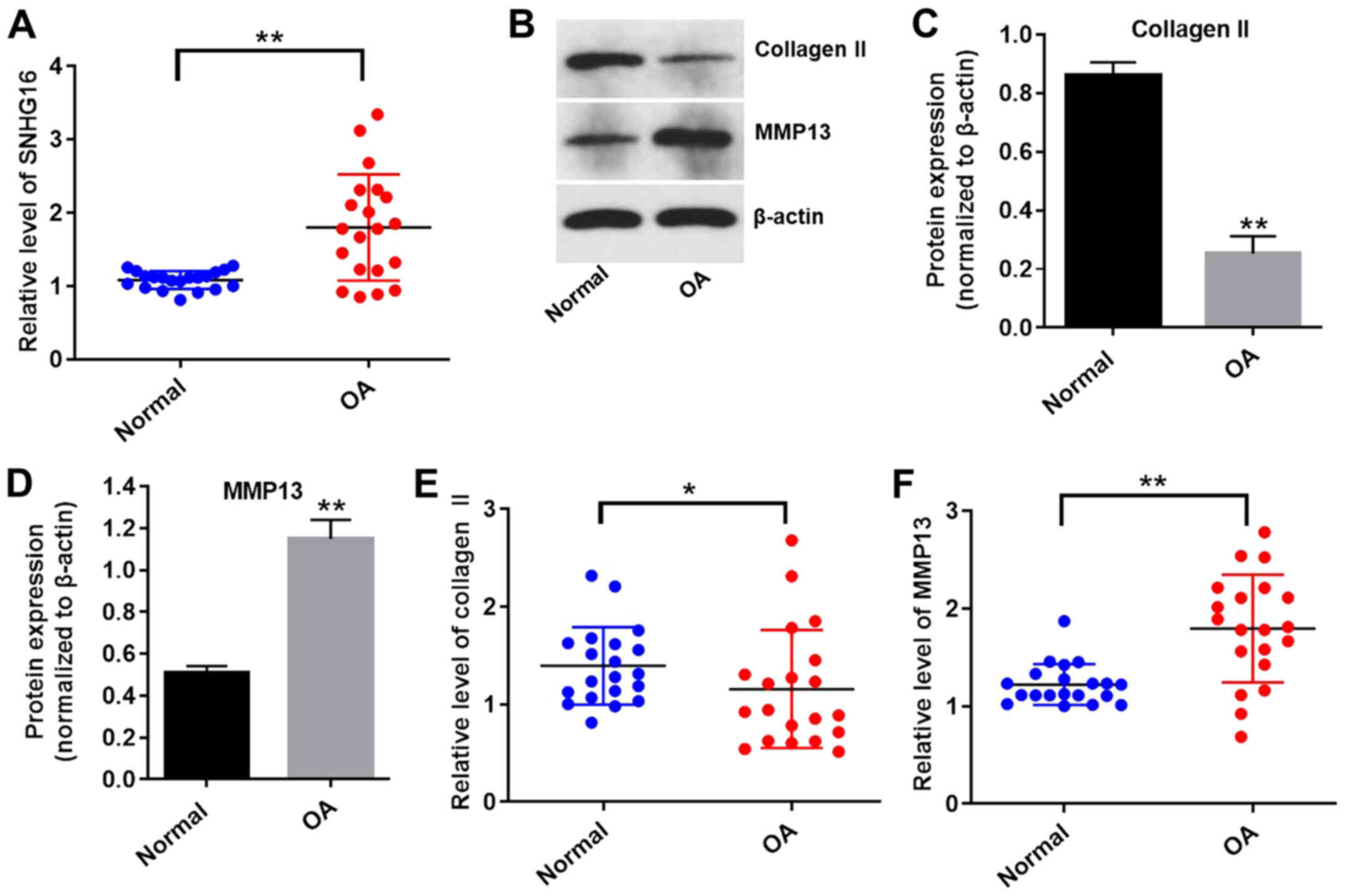
To explore whether the expression levels of lncRNA
SNHG16 were altered in OA, the expression of SNHG16 in OA and
normal tissues was measured using RT-qPCR. The expression levels of
SNHG16 were significantly upregulated in OA tissues compared with
normal tissues (Fig. 1A). The
expression levels of collagen II were significantly reduced, while
the expression levels of MMP13 were significantly upregulated in OA
tissues compared with normal tissues (Fig. 1B-F). These results indicated that
lncRNA SNHG16 was significantly upregulated in OA tissues compared
with normal tissues.
In vitro model of OA was successfully
established
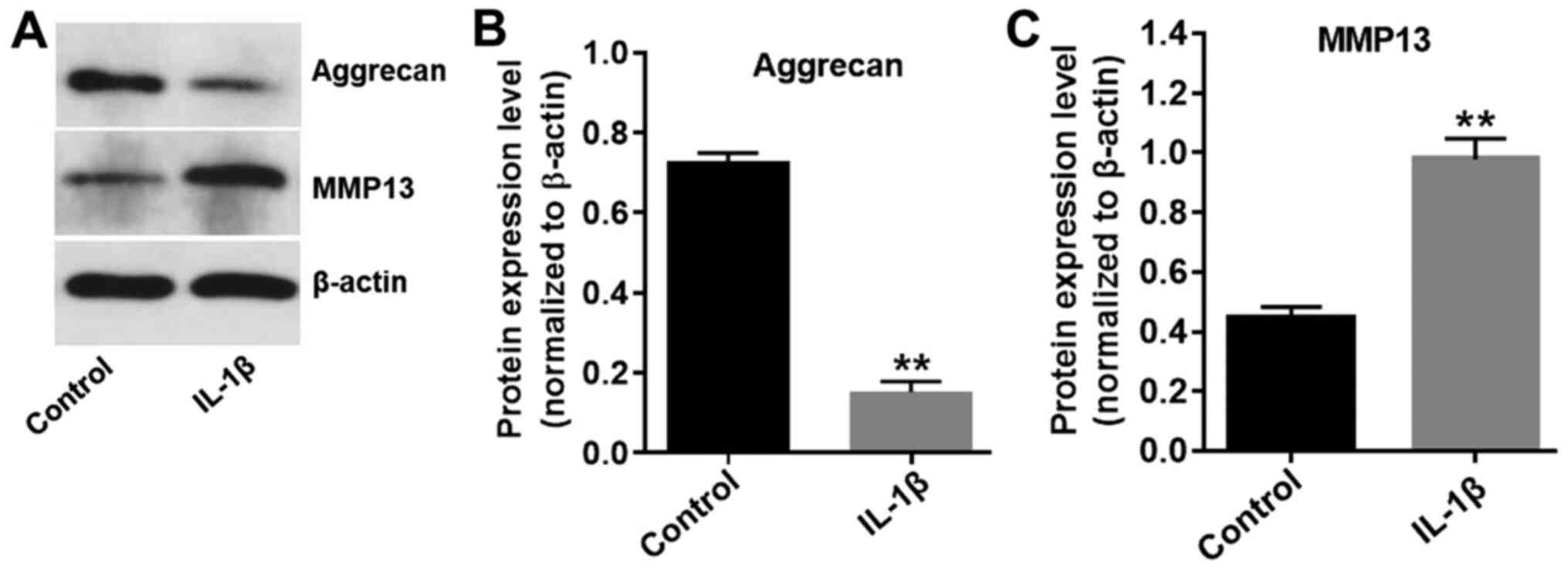
It has previously been demonstrated that IL-1β
contributes to the destruction of articular cartilage (19); therefore, CHON-001 cells were
treated with 10 ng/ml IL-1β for 24 h to develop an in vitro
model of OA. The expression levels of aggrecan in CHON-001 cells
were significantly decreased in the presence of IL-1β (Fig. 2A and B), while IL-1β significantly
increased the expression levels of MMP13 in CHON-001 cells,
compared with the control group (Fig.
2A and C). The results indicated that the in vitro model
of OA was successfully established.
SNHG16 knockdown significantly
inhibits apoptosis and enhances the proliferation of CHON-001
cells
To investigate transfection efficiency, the
expression levels of SNHG16 in CHON-001 cells were measured using
RT-qPCR. SNHG16 siRNA1 significantly decreased the expression
levels of SNHG16 in CHON-001 cells compared with the control group
(Fig. 3A). However, SNHG16 siRNA2
displayed a limited effect on the expression of SNHG16 compared
with the control group (Fig. 3A);
therefore, SNHG16 siRNA1 was used for subsequent experiments. The
CCK-8 assay and Ki-67 staining were performed to assess CHON-001
cell proliferation. CHON-001 cell proliferation was significantly
decreased in the presence of IL-1β compared with the control group,
but co-treatment with SNHG16 siRNA1 partially reversed the effects
of IL-1β on CHON-001 cell proliferation. The SNHG16 siRNA group had
a limited effect on CHON-001 cell proliferation compared with the
control group (Fig. 3B). Similarly,
the proportion of Ki-67-positive CHON-001 cells was significantly
decreased by treatment with IL-1β compared with the control group.
However, SNHG16 knockdown reversed the inhibitory effect of IL-1β
on CHON-001 cell proliferation, as indicated by a significant
increase in the proportion of Ki-67-positive cells (Fig. 3C and D). Flow cytometry was then
performed to investigate CHON-001 cell apoptosis. The results
suggested that IL-1β induced cell apoptosis and co-treatment with
SNHG16 knockdown inhibited the proapoptotic effect of IL-1β on
CHON-001 cells (Fig. 3E and F).
Moreover, IL-1β treatment significantly increased the expression
levels of SNHG16 in CHON-001 cells compared with the control group.
However, the effect of IL-1β on SNHG16 expression was significantly
reversed by SNHG16 knockdown in CHON-001 cells (Fig. 3G). These results suggested that
SNHG16 knockdown inhibited apoptosis and enhanced the proliferation
of CHON-001 cells.
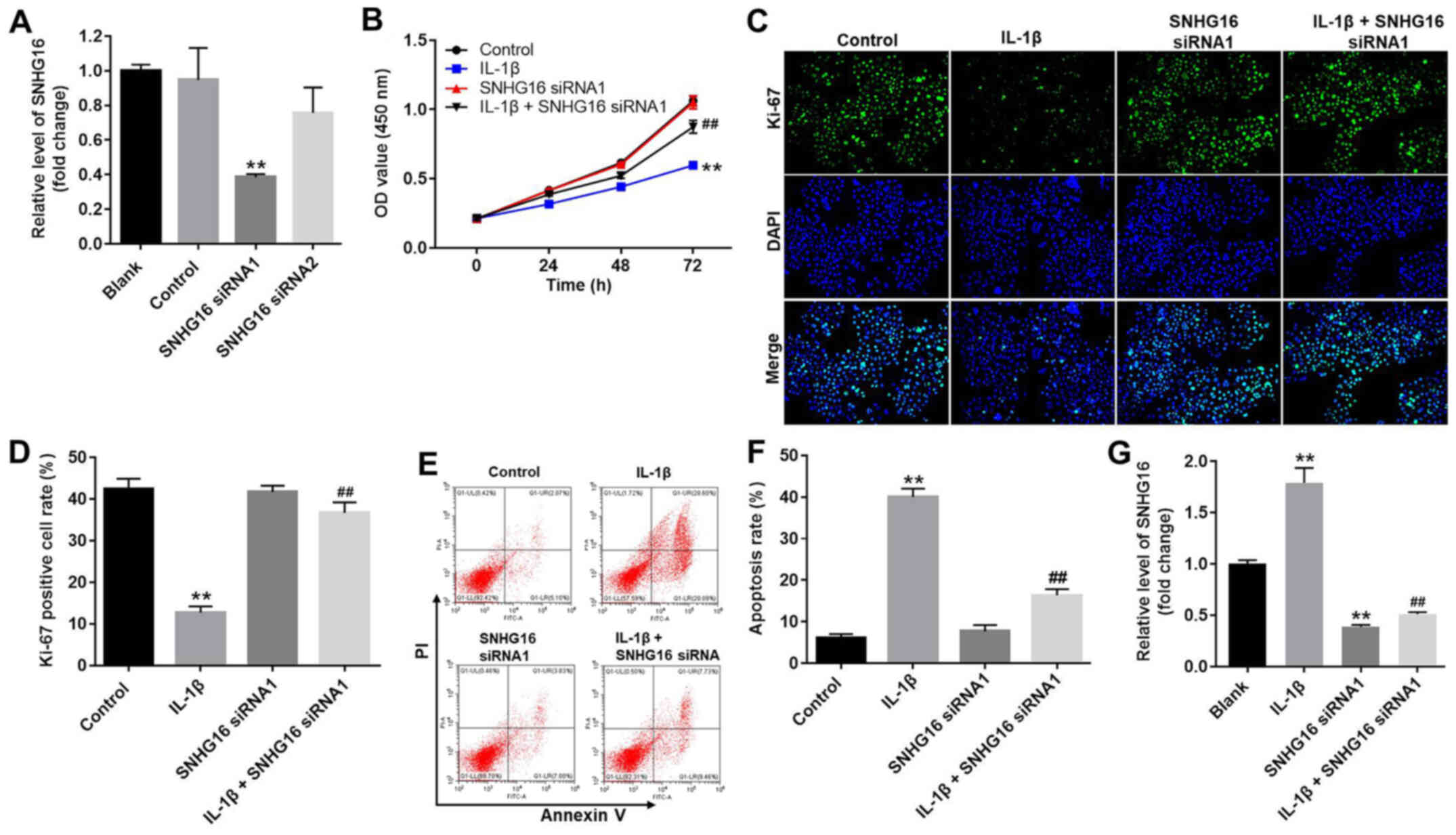 | Figure 3.SNHG16 knockdown inhibits apoptosis
and promotes the proliferation of CHON-001 cells. (A) CHON-001
cells were transfected with SNHG16 siRNA1 or SNHG16 siRNA2.
Transfection efficiency was assessed by RT-qPCR. (B) CHON-001 cells
were treated with IL-1β (10 ng/ml) and/or SNHG16 siRNA1 for 0, 24,
48 or 72 h. The effect of SNHG16 on CHON-001 cell proliferation was
assessed using the Cell Counting Kit-8 assay. CHON-001 cell
proliferation was (C) detected by Ki-67 staining (magnification,
×200) and (D) semi-quantified. CHON-001 cell apoptosis was detected
by Annexin V/PI staining, (E) assessed by flow cytometry and (F)
quantified. (G) Expression levels of SNHG16 in CHON-001 cells were
detected by RT-qPCR. **P<0.01 vs. control group;
##P<0.01 vs. IL-1β group. IL-1β, interleukin-1β; OD,
optical density; PI, propidium iodide; RT-qPCR, reverse
transcription-quantitative PCR; siRNA, small interfering RNA;
SNHG16, small nucleolar RNA host gene 16. |
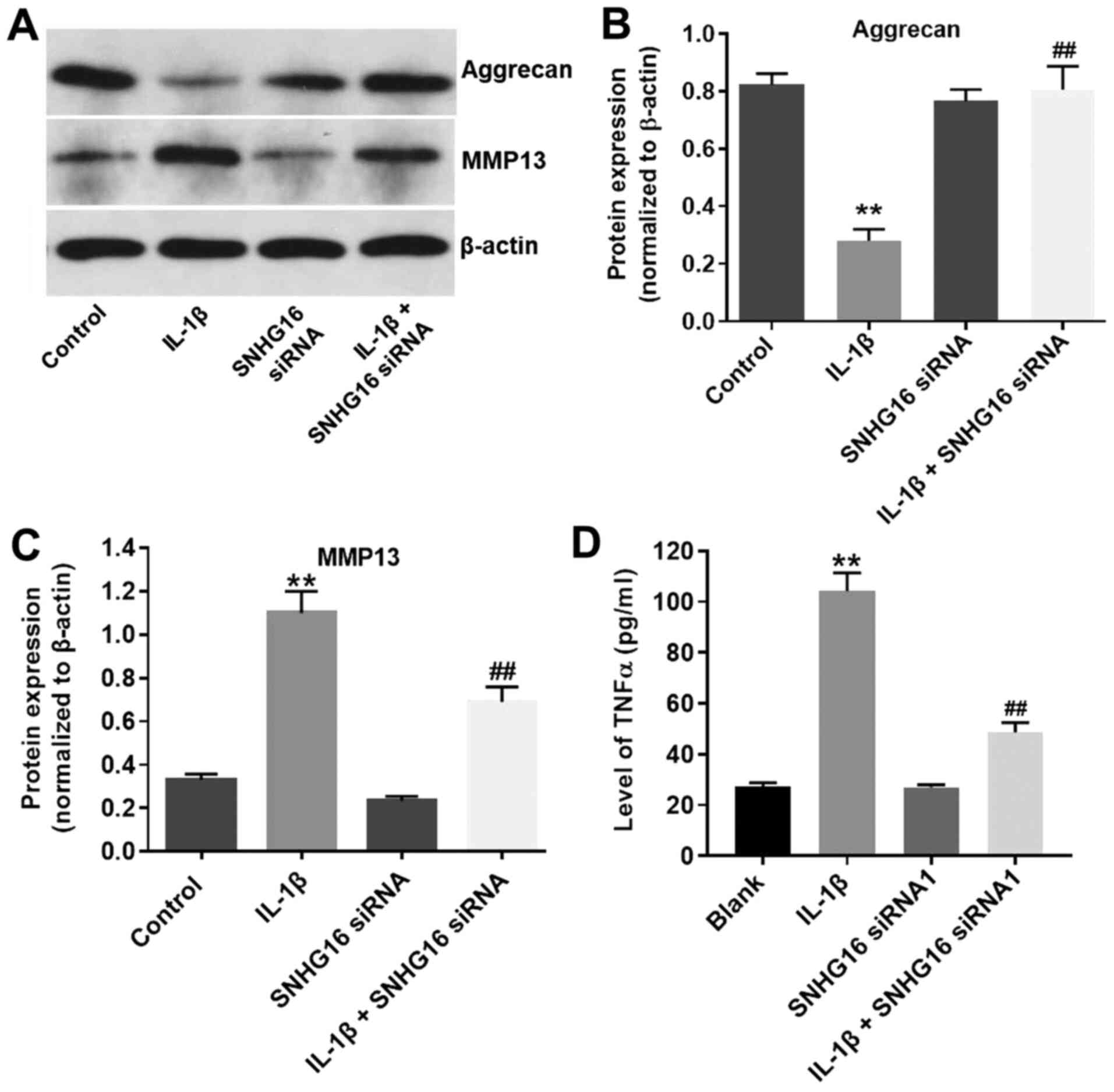
SNHG16 knockdown ameliorates IL-1 β
-induced OA in vitro
To further assess the function of lncRNA SNHG16 in
IL-1β-treated CHON-001 cells, western blotting was performed. IL-1β
significantly decreased the expression of aggrecan in CHON-001
cells compared with the control group, and SNHG16 knockdown
decreased the inhibitory effect of IL-1β on aggrecan expression
(Fig. 4A and B). Furthermore, the
protein expression levels of MMP13 were significantly increased in
the IL-1β group compared with the control group, and SBHG16
knockdown partially reduced the IL-1β-induced effects (Fig. 4A and C). The SNHG16 siRNA1 group
displayed limited effects on the expression levels of aggrecan and
MMP13 compared with the control group (Fig. 4A-C). Additionally, the levels of
TNF-α in the media of CHON-001 cells were significantly increased
by IL-1β treatment compared with the control group, and SNHG16
knockdown partially inhibited IL-1β-mediated effects on TNF-α
(Fig. 4D). The results suggested
that SNHG16 knockdown may ameliorate IL-1β-induced OA in
vitro.
miR-373-3p is the downstream target
gene of SNHG16
To investigate the potential mechanism underlying
the action by which lncRNA SNHG16 ameliorated OA in vitro,
TargetScan and miRDB databases were used to identify the downstream
target genes of SNHG16. SNHG16 displayed a putative miR-373-3p
targeting site (Fig. 5A).
Furthermore, the RT-qPCR results indicated that miR-373-3p mimic
was stably transfected into CHON-001 cells (Fig. 5B). In addition, the luciferase
reporter assay was performed to determine whether miR-373-3p
directly interacted with SNHG16 in CHON-001 cells. The results
indicated that co-transfection of the WT SNHG16 vector with
miR-373-3p mimic significantly decreased luciferase activities
compared with the control group (Fig.
5C). Therefore, the results suggested that SNHG16 bound to
miR-373-3p.
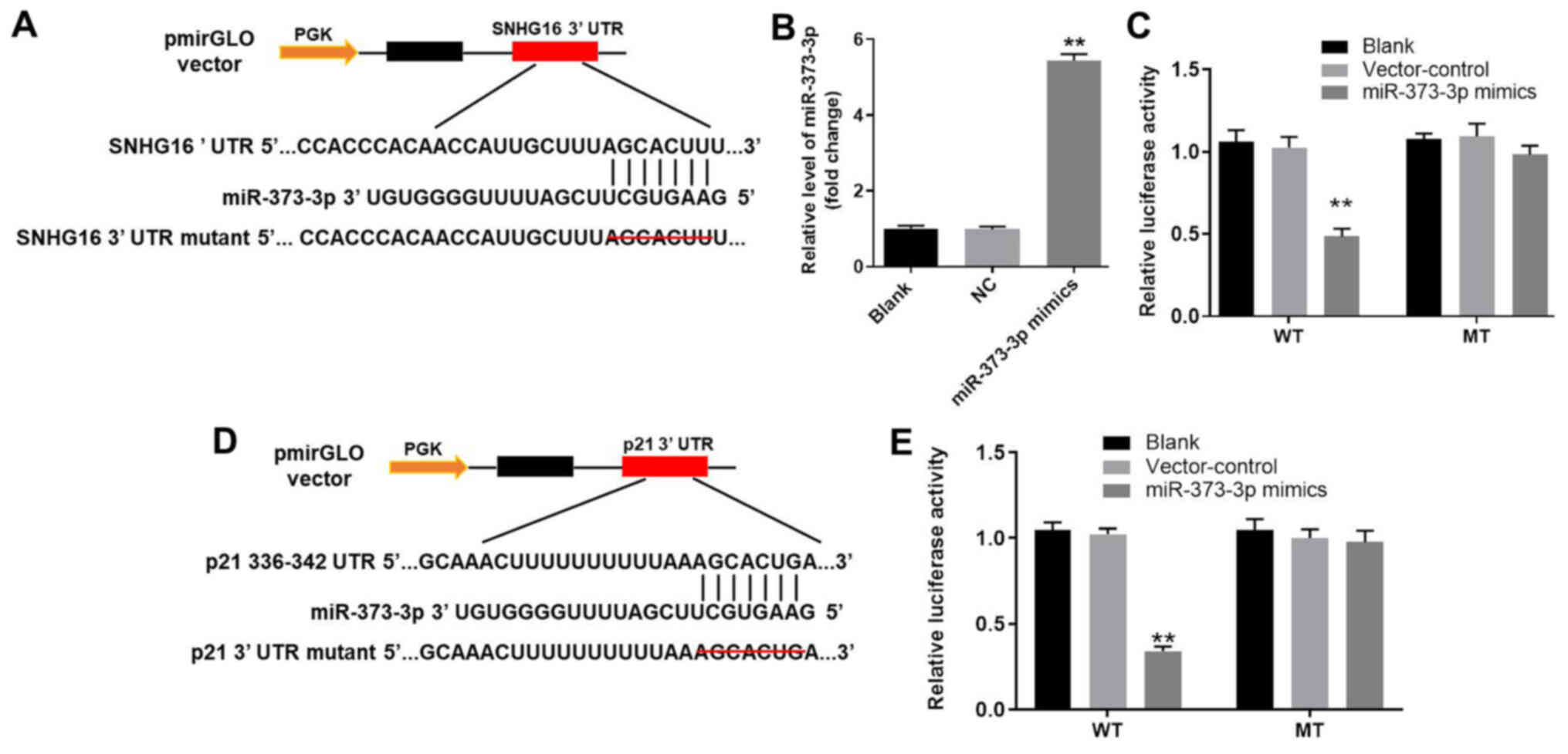 | Figure 5.miR-373-3p is the downstream target
gene of SNHG16 and p21 is the direct target gene of miR-373-3p. (A)
3′-UTR gene structure of SNHG16 at the position of 1,467-1,473
contained the predicted target site of miR-373-3p, with a sequence
of AUCCGCU. (B) miR-373-3p expression in CHON-001 cells was
detected by RT-qPCR. (C) Luciferase activity was measured in
CHON-001 cells following co-transfection with WT/MUT SNHG16 3′-UTR
plasmid and miR-373-3p mimic using a dual luciferase reporter
assay. (D) 3′-UTR gene structure of p21 at the position of 750–756
contained the predicted target site of miR-373-3p, with a sequence
of AACCCUUG. (E) Luciferase activity was measured in CHON-001 cells
following co-transfection with WT/MUT p21 3′-UTR plasmid and
miR-373-3p mimic using a dual luciferase reporter assay.
**P<0.01 vs. the control or blank group. 3′-UTR, 3′-untranslated
region; miR, microRNA; MUT, mutant; NC, negative control; RT-qPCR,
reverse transcription-quantitative PCR; SNHG16, small nucleolar RNA
host gene 16; WT, wild-type. |
p21 is the direct target gene of
miR-373-3p
Subsequently, the TargetScan and miRDB databases
were used to investigate the direct targets of miR-373-3p. The
results suggested that p21 was a potential target of miR-373-3p
(Fig. 5D). In addition, the
luciferase assay demonstrated significantly reduced luciferase
activity in CHON-001 cells post-transfection with WT p21 and
miR-373-3p mimic compared with the control group (Fig. 5E). The results indicated that p21
was the direct target of miR-373-3p.
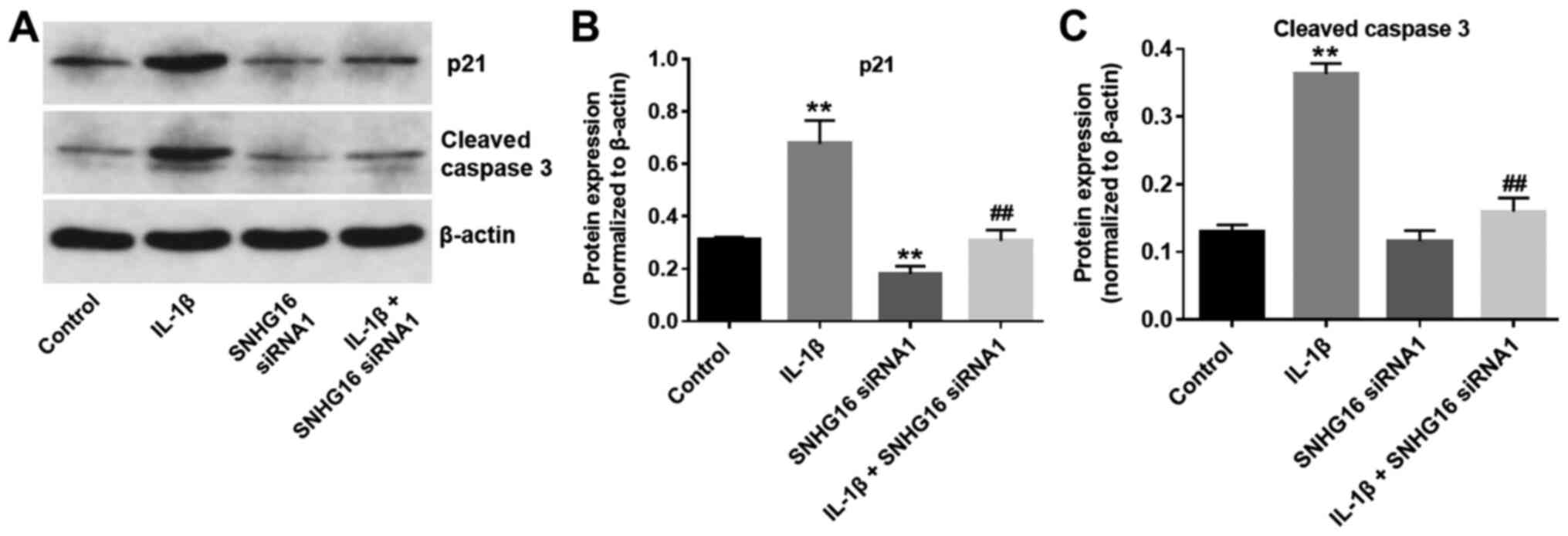
SNHG16 knockdown significantly
decreases the IL-1 β -mediated activation of p21 and cleaved
caspase-3
To further investigate the biological role of SNHG16
during the progression of OA, the expression levels of the cell
cycle regulator, p21, and proapoptotic protein, cleaved caspase-3,
were detected using western blot analysis. The expression levels of
p21 in CHON-001 cells were significantly increased in the IL-1β
group compared with the control group. IL-1β-mediated effects on
p21 expression were significantly rescued by SNHG16 knockdown in
CHON-001 cells (Fig. 6A and B).
Additionally, SNHG16 knockdown significantly reduced p21 expression
levels compared with the control group (Fig. 6A and B). Similarly, IL-1β
significantly increased the expression levels of cleaved caspase-3
compared with the control group. However, SNHG16 knockdown
significantly inhibited the proapoptotic effect of IL-1β on the
expression of cleaved caspase-3 in CHON-001 cells (Fig. 6A and C). The results indicated that
SNHG16 knockdown may decrease the IL-1β-mediated effects on p21 and
cleaved caspase-3 expression.
Discussion
Previous studies have indicated that lncRNAs serve a
critical role during the progression of OA (11,20).
In the present study, it was further suggested that the expression
of lncRNA SNHG16 was significantly upregulated in OA tissues
compared with normal joint tissues. Zhou et al (21) reported that the expression of lncRNA
HOX transcript antisense RNA was significantly upregulated in OA
tissue, which was similar to the results obtained in the present
study. In addition, a recent study reported that the inflammatory
response may have an important role during the progression of OA
(22). Tan et al (23) indicated that the expression levels
of IL-1β were increased in OA tissues. Furthermore, IL-1β promoted
damage in arthritic cartilage and enhanced the expression levels of
matrix-degradation proteins (24).
The aforementioned results indicated that high expression levels of
lncRNA SNHG16 were closely associated with the progression of
OA.
To further explore the biological role of lncRNA
SNHG16 during the development of OA, in vitro experiments
were performed. Collagen II and MMP13 are matrix components of the
cartilage (25,26). Loss of collagen II and upregulation
of MMP13 expression can lead to the development of OA (27–29).
In the present study, CHON-001 cell apoptosis was induced by IL-1β,
whereas SNHG16 knockdown significantly decreased the expression of
cleaved caspase-3, a proapoptotic protein, in IL-1β-induced
CHON-001 cells (30). Lin et
al (31) reported that
luteoloside inhibited IL-1β-induced apoptosis in vitro by
inactivating cleaved caspase-3 (31). The aforementioned results are
consistent with the results of the present study, indicating that
SNHG16 knockdown inhibited CHON-001 cell apoptosis by
downregulating the expression levels of cleaved caspase-3.
Similarly, the expression of collagen II was significantly
decreased and the expression of MMP13 was significantly increased
in OA tissues compared with normal joint tissues, which was
consistent with previous studies (32,33).
Based on the results of the present study, it was suggested that
the in vitro model of OA was successfully established.
Further experiments suggested that SNHG16 knockdown
significantly decreased apoptosis and increased the proliferation
of CHON-001 cells treated with IL-1β. Furthermore, SNHG16 knockdown
significantly decreased the expression levels of aggrecan and
upregulated the expression levels of MMP13 in CHON-001 cells
treated with IL-1β. Similarly, Tu et al (34) suppressed IL-1β-induced cartilage
degradation by decreasing the expression levels of collagen II. A
previous study revealed that the Bushenhuoxue formula attenuated
cartilage degeneration in a mouse model of OA via the transforming
growth factor-β/MMP13 signaling pathway (35). Additionally, the expression levels
of PVT1 were increased in IL-1β-stimulated OA chondrocytes;
therefore, PVT1 knockdown may inhibit the development of OA by
altering inflammatory responses (36). The aforementioned results were
consistent with the results of the present study, indicating that
SNHG16 knockdown may suppress the development of OA in
vitro.
The mechanism underlying the action by which SNHG16
knockdown inhibited the progression of OA in vitro was
investigated. The dual-luciferase reporter assay indicated that
miR-373-3p was the downstream target gene of SNHG16. miRNAs are
highly conserved ncRNAs that display versatile biological functions
(37,38). Lei et al (39) reported that lncRNA SNHG1 inhibited
IL-1β-induced OA by suppressing the miR-16-5p-mediated p38 MAPK and
NF-κB signaling pathway. In addition, lncRNA metastasis associated
lung adenocarcinoma transcript 1 promoted the progression of OA by
regulating the miR-150-5p/AKT3 signaling pathway (40). Meanwhile, miR-373-3p has been
reported to be involved in multiple diseases (41,42).
The present study also investigated the biological effect of
miR-373-3p on IL-1β-treated CHON-001 cell proliferation, suggesting
that it may act as a suppressor during the progression of OA. The
results of the present study further suggested that SNHG16 may
promote the development of OA by sponging miR-373-3p.
It has been reported that miRNAs primarily exert
their function by binding to target genes (43,44).
To explore the potential mechanism underlying the action of
miR-373-3p during the progression of OA, TargetScan and miRDB
databases were used to identify target genes of miR-373-3p. p21, a
cell cycle regulator, was identified as a direct target of
miR-373-3p (45). It has been
reported that p21 may be partially involved in G2 arrest
following cell damage (46). In
addition, p21 deficiencies resulted in patient susceptibility to OA
via STAT3 phosphorylation (47). In
the present study, the luciferase reporter assay further indicated
that p21 was a direct target gene of miR-373-3p. Furthermore, the
results suggested that IL-1β significantly increased the expression
of p21 in CHON-001 cells. Tang et al (48) reported that p21 promoted chondrocyte
apoptosis in OA by sponging miR-451. The results obtained in the
present study suggested that downregulation of p21 may contribute
to the alleviation of IL-1β-induced CHON-001 cell growth
inhibition. Since STAT3 has been reported to be involved in the
progression of OA (49), further
investigation into the effects of SNHG16 on STAT3 signaling should
be conducted. The present study did not include in vivo
experiments due to lack of funds; therefore, future studies should
employ in vivo models to investigate therapeutic targets for
OA.
In conclusion, the present study investigated the
roles of SNHG16 during the development of OA and the underlying
mechanisms. The results indicated that SNHG16 expression was
significantly upregulated in OA tissues compared with normal
tissues, and that knockdown of SNHG16 expression increased the
proliferation and inhibited apoptosis of CHON-001 cells. miR-373-3p
was identified as a downstream target of SNHG16, and p21 was
identified as a target gene of miR-373-3p. The present study
suggested that SNHG16 knockdown inhibited the progression of OA
in vitro, which indicated that SNHG16 may serve as a novel
therapeutic target for OA.
Acknowledgements
The authors would like to thank Dr Zhang (Department
of Surgery, Medical School, Stanford University) for proofreading
the manuscript.
Funding
The present study was supported by Inner Mongolia
Natural Science Foundation [grant no. (2017LH)0309].
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
HF and YY conceived and supervised the study. HF, LD
and YY designed the study. HF and LD performed the experiments and
analyzed the data. All authors reviewed the results and approved
the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Institutional
Ethical Committee of the Second Affiliated Hospital of Inner
Mongolia Medical University. Written informed consent was obtained
from all participants.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Allen KD, Choong PF, Davis AM, Dowsey MM,
Dziedzic KS, Emery C, Hunter DJ, Losina E, Page AE, Roos EM, et al:
Osteoarthritis: Models for appropriate care across the disease
continuum. Best Pract Res Clin Rheumatol. 30:503–535. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Cavalli E, Levinson C, Hertl M, Broguiere
N, Brück O, Mustjoki S, Gerstenberg A, Weber D, Salzmann G,
Steinwachs M, et al: Characterization of polydactyly chondrocytes
and their use in cartilage engineering. Sci Rep. 9:42752019.
View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Taylor N: Nonsurgical management of
osteoarthritis knee pain in the older adult. Clin Geriatr Med.
33:41–51. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Qiong J, Xia Z, Jing L and Haibin W:
Synovial mesenchymal stem cells effectively alleviate
osteoarthritis through promoting the proliferation and
differentiation of meniscus chondrocytes. Eur Rev Med Pharmacol.
24:1645–1655. 2020.
|
|
5
|
Su Y, Wu H, Pavlosky A, Zou LL, Deng X,
Zhang ZX and Jevnikar AM: Regulatory non-coding RNA: New
instruments in the orchestration of cell death. Cell Death Dis.
7:e23332016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zhang Y, Ma L, Wang C, Wang L, Guo Y and
Wang G: Long noncoding RNA LINC00461 induced osteoarthritis
progression by inhibiting miR-30a-5p. Aging (Albany NY).
12:4111–4123. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ai D and Yu F: LncRNA DNM3OS promotes
proliferation and inhibits apoptosis through modulating IGF1
expression by sponging MiR-126 in CHON-001 cells. Diagn Pathol.
14:1062019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shen H, Wang Y, Shi W, Sun G, Hong L and
Zhang Y: LncRNA SNHG5/miR-26a/SOX2 signal axis enhances
proliferation of chondrocyte in osteoarthritis. Acta Biochim
Biophys Sin (Shanghai). 50:191–198. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Qi K, Lin R, Xue C, Liu T, Wang Y, Zhang Y
and Li J: Long non-coding RNA (LncRNA) CAIF is downregulated in
osteoarthritis and inhibits LPS-Induced interleukin 6 (IL-6)
upregulation by downregulation of MiR-1246. Med Sci Monit.
25:8019–8024. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Song J, Ahn C, Chun CH and Jin EJ: A long
non-coding RNA, GAS5, plays a critical role in the regulation of
miR-21 during osteoarthritis. J Orthop Res. 32:1628–1635. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li Y, Li S, Luo Y, Liu Y and Yu N: LncRNA
PVT1 regulates chondrocyte apoptosis in osteoarthritis by acting as
a sponge for miR-488-3p. DNA Cell Biol. 36:571–580. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Xiao Y, Bao Y, Tang L and Wang L: LncRNA
MIR4435-2HG is downregulated in osteoarthritis and regulates
chondrocyte cell proliferation and apoptosis. J Orthop Surg Res.
14:2472019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang W, Lou C, Gao J, Zhang X and Du Y:
LncRNA SNHG16 reverses the effects of miR-15a/16 on LPS-induced
inflammatory pathway. Biomed Pharmacother. 106:1661–1667. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheng W, Hao CY, Zhao S, Zhang LL and Liu
D: SNHG16 promotes the progression of osteoarthritis through
activating microRNA-93-5p/CCND1 axis. Eur Rev Med Pharmacol Sci.
23:9222–9229. 2019.PubMed/NCBI
|
|
15
|
Cheng F, Hu H, Sun K, Yan F and Geng Y:
miR-455-3p enhances chondrocytes apoptosis and inflammation by
targeting COL2A1 in the in vitro osteoarthritis model. Biosci
Biotechnol Biochem. 84:695–702. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wang X, Fan J, Ding X, Sun Y, Cui Z and
Liu W: Tanshinone I inhibits IL-1β-induced apoptosis, inflammation
and extracellular matrix degradation in chondrocytes CHON-001 cells
and attenuates murine osteoarthritis. Drug Des Devel Ther.
13:3559–3568. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Palmieri B, Lodi D and Capone S:
Osteoarthritis and degenerative joint disease: Local treatment
options update. Acta Biomed. 81:94–100. 2010.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li M, Zhao J and Jia L: USP14-mediated
IkBα degradation exacerbates NF-kB activation and IL-1β-stimulated
chondrocyte dedifferentiation. Life Sci. 218:147–152. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Xu J and Xu Y: The lncRNA MEG3
downregulation leads to osteoarthritis progression via miR-16/SMAD7
axis. Cell Biosci. 7:692017. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Zhou W, He X, Chen Z, Fan D, Wang Y, Feng
H, Zhang G, Lu A and Xiao L: LncRNA HOTAIR-mediated Wnt/β-catenin
network modeling to predict and validate therapeutic targets for
cartilage damage. BMC Bioinformatics. 20:4122019. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Paish HL, Baldock TE, Gillespie CS, Del
Carpio Pons A, Mann DA, Deehan DJ, Borthwick LA and Kalson NS:
Chronic, active inflammation in patients with failed total knee
replacements undergoing revision surgery. J Orthop Res.
37:2316–2324. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Tan C, Zhang J, Chen W, Feng F, Yu C, Lu
X, Lin R, Li Z, Huang Y, Zheng L, et al: Inflammatory cytokines via
up-regulation of aquaporins deteriorated the pathogenesis of early
osteoarthritis. PLoS One. 14:e02208462019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Kapoor M, Martel-Pelletier J, Lajeunesse
D, Pelletier JP and Fahmi H: Role of proinflammatory cytokines in
the pathophysiology of osteoarthritis. Nat Rev Rheumatol. 7:33–42.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Zhang Y, Liu S, Guo W, Hao C, Wang M, Li
X, Zhang X, Chen M, Wang Z, Sui X, et al: Coculture of hWJMSCs and
pACs in oriented scaffold enhances hyaline cartilage regeneration
in vitro. Stem Cells Int. 2019:51301522019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang M, Sampson ER, Jin H, Li J, Ke QH, Im
HJ and Chen D: MMP13 is a critical target gene during the
progression of osteoarthritis. Arthritis Res Ther. 15:R52013.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ji B, Ma Y, Wang H, Fang X and Shi P:
Activation of the P38/CREB/MMP13 axis is associated with
osteoarthritis. Drug Des Devel Ther. 13:2195–2204. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zheng W, Feng Z, Lou Y, Chen C, Zhang C,
Tao Z, Li H, Cheng L and Ying X: Silibinin protects against
osteoarthritis through inhibiting the inflammatory response and
cartilage matrix degradation in vitro and in vivo. Oncotarget.
8:99649–99665. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Lorenz J, Seebach E, Hackmayer G, Greth C,
Bauer RJ, Kleinschmidt K, Bettenworth D, Böhm M, Grifka J and
Grässel S: Melanocortin 1 receptor-signaling deficiency results in
an articular cartilage phenotype and accelerates pathogenesis of
surgically induced murine osteoarthritis. PLoS One. 9:e1058582014.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Crowley LC and Waterhouse NJ: Detecting
cleaved caspase-3 in apoptotic cells by flow cytometry. Cold Spring
Harb Protoc. 2016:2016. View Article : Google Scholar
|
|
31
|
Lin J, Chen J, Zhang Z, Xu T, Shao Z, Wang
X, Ding Y, Tian N, Jin H, Sheng S, et al: Luteoloside inhibits
il-1β-induced apoptosis and catabolism in nucleus pulposus cells
and ameliorates intervertebral disk degeneration. Front Pharmacol.
10:8682019. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chakkalakal SA, Heilig J, Baumann U,
Paulsson M and Zaucke F: Impact of arginine to cysteine mutations
in collagen II on protein secretion and cell survival. Int J Mol
Sci. 19:E5412018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Niebler S, Schubert T, Hunziker EB and
Bosserhoff AK: Activating enhancer binding protein 2 epsilon
(AP-2ε)-deficient mice exhibit increased matrix metalloproteinase
13 expression and progressive osteoarthritis development. Arthritis
Res Ther. 17:1192015. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Tu C, Huang X, Xiao Y, Song M, Ma Y, Yan
J, You H and Wu H: Schisandrin A inhibits the IL-1β-induced
inflammation and cartilage degradation via suppression of MAPK and
NF-kB signal pathways in rat chondrocytes. Front Pharmacol.
10:412019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Wang PE, Zhang L, Ying J, Jin X, Luo C, Xu
S, Dong R, Xiao L, Tong P and Jin H: Bushenhuoxue formula
attenuates cartilage degeneration in an osteoarthritic mouse model
through TGF-β/MMP13 signaling. J Transl Med. 16:722018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Zhao Y, Zhao J, Guo X, She J and Liu Y:
Long non-coding RNA PVT1, a molecular sponge for miR-149,
contributes aberrant metabolic dysfunction and inflammation in
IL-1β-simulated osteoarthritic chondrocytes. Biosci Rep.
38:BSR201805762018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Eguchi T and Kuboki T: Cellular
reprogramming using defined factors and MicroRNAs. Stem Cells Int.
2016:75309422016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Tam C, Wong JH, Tsui SK, Zuo T, Chan TF
and Ng TB: LncRNAs with miRNAs in regulation of gastric, liver, and
colorectal cancers: Updates in recent years. Appl Microbiol
Biotechnol. 103:4649–4677. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Lei J, Fu Y, Zhuang Y, Zhang K and Lu D:
LncRNA SNHG1 alleviates IL-1β-induced osteoarthritis by inhibiting
miR-16-5p-mediated p38 MAPK and NF-kB signaling pathways. Biosci
Rep. 39:BSR201915232019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zhang Y, Wang F, Chen G, He R and Yang L:
LncRNA MALAT1 promotes osteoarthritis by modulating miR-150-5p/AKT3
axis. Cell Biosci. 9:542019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Yeon M, Byun J, Kim H, Kim M, Jung HS,
Jeon D, Kim Y and Jeoung D: CAGE binds to beclin1, regulates
autophagic flux and CAGE-derived peptide confers sensitivity to
anti-cancer drugs in non-small cell lung cancer cells. Front Oncol.
8:5992018. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Smith-Vikos T, Liu Z, Parsons C, Gorospe
M, Ferrucci L, Gill TM and Slack FJ: A serum miRNA profile of human
longevity: Findings from the baltimore longitudinal study of aging
(BLSA). Aging (Albany NY). 8:2971–2987. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Adlakha YK and Saini N: Brain microRNAs
and insights into biological functions and therapeutic potential of
brain enriched miRNA-128. Mol Cancer. 13:332014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Ple H, Landry P, Benham A, Coarfa C,
Gunaratne PH and Provost P: The repertoire and features of human
platelet microRNAs. PLoS One. 7:e507462012. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Pozzoli G, Petrucci G, Navarra P, Marei HE
and Cenciarelli C: Aspirin inhibits proliferation and promotes
differentiation of neuroblastoma cells via p21(Waf1) protein
up-regulation and Rb1 pathway modulation. J Cell Mol Med.
23:7078–7087. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Koyano T, Namba M, Kobayashi T, Nakakuni
K, Nakano D, Fukushima M, Nishiyama A and Matsuyama M: The p21
dependent G2 arrest of the cell cycle in epithelial tubular cells
links to the early stage of renal fibrosis. Sci Rep. 9:120592019.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Hayashi S, Fujishiro T, Hashimoto S,
Kanzaki N, Chinzei N, Kihara S, Takayama K, Matsumoto T, Nishida K,
Kurosaka M and Kuroda R: p21 deficiency is susceptible to
osteoarthritis through STAT3 phosphorylation. Arthritis Res Ther.
17:3142015. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Tang L, Ding J, Zhou G and Liu Z:
LncRNAp21 promotes chondrocyte apoptosis in osteoarthritis by
acting as a sponge for miR451. Mol Med Rep. 18:5295–5301.
2018.PubMed/NCBI
|
|
49
|
Xu X, Lv H, Li X, Su H, Zhang X and Yang
J: Danshen attenuates cartilage injuries in osteoarthritis in vivo
and in vitro by activating JAK2/STAT3 and AKT pathways. Exp Anim.
67:127–137. 2018. View Article : Google Scholar : PubMed/NCBI
|