Introduction
Age-associated osteoporosis can be referred to as a
systemic impairment of bone formation and enhancement of bone
marrow fat accumulation (1,2). Bone mesenchymal stem cells (BMSCs) can
differentiate into osteoblasts or adipocytes and promote bone
regeneration (3–5). Tissue regeneration and repair via the
differentiation of BMSCs has been a popular topic in regenerative
medicine. At the molecular level, the interactions between hormones
and transcription factors control mesenchymal stem cell
differentiation into osteocytes. The major transcription factors
that play a key role in BMSCs differentiation into osteocytes
include runt-related transcription factor 2 (RUNX2) and osterix
(OSX) (6). Besides, previous
studies have used several indicators to detect osteogenic
differentiation of BMSCs, such as alkaline phosphatase (ALP),
osteocalcin (OCN) secretion, RUNX2, OSX, bone sialoprotein (BSP)
and osteopontin (OPN) (7,8).
Although BMSCs gradually lose their capacity to
differentiate into osteoblasts, they tend to differentiate into
adipocytes in the aging process (9–11). As
a result, the ability to form bone declines with age. This
association between bone formation and aging is the main cause of
the high incidence of age-associated osteoporosis (12,13).
Osteoporosis symptoms usually result in fragile bones, thus causing
the elderly to experience mobility problems (14). More troubling is that the elderly
tend to be more vulnerable to suffer from bone fracture from
accidental falls, which increases the financial and emotional
burden of family members (15–17).
Thus, the treatment of age-associated osteoporosis is of great
concern in society. This public health issue has attracted a great
deal of attention among medical staff as well as researchers. The
purpose of the present study was to gain further insights into the
mechanism of bone formation and the occurrence of age-associated
osteoporosis using cellular and molecular methods.
Proprotein convertase subtilisin/kexin (PCSK)
enzymes are responsible for activating a wide variety of hormones
and protein precursors, ranging from growth factors to
extracellular pathogens (18).
These enzymes cleave and convert their immature target proproteins
into an active functional form. One of the important members
belonging to this enzyme family is PCSK5, which plays an essential
role in a number of cell activities. As described in a
bioinformatics analysis, different Notch signaling components, such
as PCSK5, are differential expression factors in
osteogenic-differentiation progression (19). In another study, silencing PCSK5 in
mice led to severe malformations, bone morphogenic defects and
early embryonic lethality, with the absence of growth
differentiating factor 11 (20).
Based on these previous data, it was speculated that PCSK5 could be
a putative therapeutic factor for age-associated osteoporosis.
However, no study has comprehensively demonstrated the role of
PCSK5, thus the present study was designed to understand how PCSK5
exerts its function in age-associated osteoporosis using BMSCs.
Previous studies have explored the role of
miR-338-3p (microRNA-338-3p) in osteoclast formation. A study
investigated the expression profile of miR-338-3p in BMSC
differentiation, which found that the expression level of
miR-338-3p declined as the osteoblast underwent differentiation
(21). Thus, miR-338-3p has the
potential to participate in the suppression of osteoclast formation
and the acceleration of age-associated osteoporosis. Moreover,
another study reported that miR-338-3p overexpression in osteoclast
precursor cells restricted osteoclast formation (22). Similarly, a study concerning the
function of miR-338-3p in osteoclast differentiation and activation
demonstrated that miR-338-3p was markedly downregulated during this
process (23). In the present
study, the mechanism of miR-338-3p in osteoclast formation and the
role of the PCSK5 gene in age-associated osteoporosis was
explored.
The aim of the present study paper was to study the
axis of miR-338-3p targeting PCSK5 in bone formation by exploring
their roles in osteoblastic differentiation. The findings of the
present study will help to explore some novel therapeutic methods
for age-associated osteoporosis.
Materials and methods
Bioinformatics analysis
The GEO profile (GSE35959) (24) was downloaded from the National
Center for Biotechnology Information (https://www.ncbi.nlm.nih.gov/). The dataset was used
to screen the downregulated differentially expressed genes (DEGs)
with P<0.05 and log2-fold-change (log2FC)
<-2. Metascape (http://metascape.org/gp/index.html#/main/step1) was
subsequently used to enrich the biological processes of the top 100
downregulated DEGs. Next, TargetScan Human v7.1 (http://www.targetscan.org/vert_71/), starBase
v2.0 (http://starbase.sysu.edu.cn/index.php) and miRDB
(http://mirdb.org/) were applied to predict the miRNAs
that bind to the PCSK5 3′ untranslated region (UTR). The
overlapping miRNAs were identified using Venny 2.1 (https://bioinfogp.cnb.csic.es/tools/venny/).
Hematoxylin and eosin staining
(H&E)
H&E staining was employed to detect the
morphology of the distal tibia in rats. A total of 18 male rats,
six in each group, were divided into young group (3 months; 260±10
g), middle-aged group (12 months; 360±10 g) and old group (18
months; 450±10 g). The Sprague-Dawley rats were purchased from
Nanjing Junke Bioengineering (Jiangsu, China). Rats were kept on a
12-h light/dark cycle at 20–25°C with 60% relative humidity under
specific pathogen-free conditions, and fed a non-purified diet with
free access to water ad libitum. All animal procedures were
approved by the Animal Ethics Committee of Affiliated Hospital of
Jianghan University (approval no. JH-20191014-23; Jiangan,
China).
Rats were euthanized by CO2 asphyxiation
(fill rate of 20% of the chamber volume/min with CO2),
and the tibia was harvested after the rats had no vital signs, such
as lack of breath and faded eye color. The tibia of the rats was
subsequently fixed in 10% formalin solution for 2 days at room
temperature and decalcified slowly with EDTA. Next, the tissue was
dehydrated at room temperature in gradient concentrations of
ethanol (70, 80, 90, 95 and 100%) every 2–5 min, embedded in
paraffin and then sliced to 5-µm thick sections. The tissue
sections were subsequently stained with hematoxylin for 5 min and
treated with 1% hydrochloric acid ethanol solution for 5 sec at
room temperature. After that, a 1% ammonia solution was used to
reverse the blue, 1% eosin solution was used to stain the cytoplasm
for 3 min, and the sections were finally dehydrated until
translucent at room temperature. Finally, the images were sealed
with neutral resin and analyzed with IPP6.0 image analysis software
(Media Cybernetics, Inc.).
Cell culture and differentiation
BMSCs were obtained from the femur and tibia of rats
at the age of 3 (young), 12 (middle-aged) and 18- (old-aged)
months. The procedure was carried out as follows: i) firstly, the
medullary cavity was thoroughly flushed the using an α-modified
Eagle's medium (αMEM; Invitrogen; Thermo Fisher Scientific, Inc.)
without ascorbic acid to collect the cells. The cells were then
allowed attach in complete αMEM with 10% FBS, 1% penicillin and
streptomycin (all from Gibco; Thermo Fisher Scientific, Inc.) at
37°C, 5% CO2 for 72 h. Following this step, BMSCs were
finally obtained by replacing the fresh culture medium. For the
induction of osteogenic differentiation, BMSCs were cultured for 16
days in osteogenesis induction αMEM medium with 10% FBS, 300 ng/ml
bone morphogenetic protein 2 (Gibco; Thermo Fisher Scientific,
Inc.), 50 µg/ml ascorbic acid (Gibco; Thermo Fisher Scientific,
Inc.) and 5 mM β-glycerolphosphate (Gibco; Thermo Fisher
Scientific, Inc.).
Cell transfection
Small interference (si)RNA si-PCSK5, miR-338-3p
mimic, miR-338-3p inhibitor and their non-targeting sequence
negative controls (NC) were designed by Genewiz, Inc. The
corresponding sequences are listed in Table SI. The transfection was performed
in middle-aged rat-derived BMSCs at the point of ~70% cell
confluence using Lipofectamine® 3000 reagent
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. The densities of BMSCs seeded into 6-,
12-, 24- and 96-well plates were 2.5×106,
1×106, 5×105 and 5×104 cells per
ml, respectively. The transfection concentration of si-PCSK5 was
2.5 µM, while that of miR-338-3p mimic, miR-338-3p inhibitor and
negative control was 50 nM. The untransfected BMSCs were set as the
CON group. After transfection for 48 h, the transfection efficiency
was detected using reverse transcription-quantitative (RT-q)PCR and
the follow-up experiments were carried out.
RNA isolation, cDNA synthesis and
RT-qPCR
A complete RNA isolation was performed using the
Cell Total RNA Isolation kit (ForeGene). Using RT Easy™ II (Fore
Gene), according to the manufacturer's protocol, cDNA synthesis was
performed. Following which, mRNA expression and miRNA expression
levels were measured by RT-qPCR using the Bio-Rad CFX96 Touch
system (Bio-Rad Laboratories, Inc.) and iTaq™ Universal SYBR GREEN
Supermix (Bio-Rad Laboratories, Inc.). The RT-qPCR thermocycling
conditions were: 50°C for 2 min, at 95°C for 30 sec, and then 40
cycles of 15 sec at 95°C and 30 sec at 60°C. Two reference genes U6
and GAPDH were used to quantify miRNA and mRNA, respectively. To
calculate mRNA expression, the 2−ΔΔCq (25) method was employed. The sequences of
primers are listed in Table I.
 | Table I.Primer sequences for reverse
transcription-quantitative PCR. |
Table I.
Primer sequences for reverse
transcription-quantitative PCR.
| Gene | Primer sequences
(5′→3′) |
|---|
| miR-338-3p | Forward:
TCCAGCATCAGTGATTTTGTTG |
|
| Reverse:
GTGCAGGGTCCGAGGT |
| U6 | Forward:
GCTTCGGCAGCACATATA |
|
| Reverse:
CGCTTCACGAATTTGCGT |
| PCSK5 | Forward:
CAACACACATCCTTGCCAGTC |
|
| Reverse:
ATGTTCTTCCCCGTGTAGCC |
| OSX | Forward:
CCTCCTCAGCTCACCTTCTC |
|
| Reverse:
GTTGGGAGCCCAAATAGAAA |
| RUNX2 | Forward:
GACCAGTCTTACCCCTCCTACC |
|
| Reverse:
CTGCCTGGCTCTTCTTACTGAG |
| OPN | Forward:
CAGTTGTCCCCACAGTAGACAC |
|
| Reverse:
GTGATGTCCTCGTCTGTAGCATC |
| BSP | Forward:
GAATGGCCTGTGCTTTCTCAA |
|
| Reverse:
TCGGATGAGTCACTACTGCCC |
| GAPDH | Forward:
GCTGGTCATCAACGGGAAA |
|
| Reverse:
CGCCAGTAGACTCCACGACAT |
Western blotting
For the collection of protein samples, the cells
were washed with cold PBS. The cells were then lysed with radio
immunoprecipitation assay lysis buffer (Roche Diagnostics GmBH)
containing 10 mM Tris, pH 7.5, 150 mM NaCl, 0.1% SDS, 1.0% Triton
X-100, 1% deoxycholate, 5 mM EDTA, 1 mM sodium orthovanadate, 1 mM
phenylmethylsulfonyl fluoride and protease inhibitor cocktail on
ice for 15 min. The cells were then transferred into pre-cooled 1.5
ml centrifuge tubes. After obtaining the supernatant (protein
samples), their concentrations were measured using a BCA Protein
Assay Reagent kit (Pierce; Thermo Fisher Scientific, Inc.) Next, 30
µg protein sample was added into the wells of an 8% SDS-PAGE gel
for electrophoresis at 80 V for 20 min and later 100 V for 1 h. The
PVDF membrane was subsequently used in the present experiment, and
the transfer current was set at 200 mA. Following this step, the
protein membrane was added to the prepared PBS solution and rinsed
twice. The PBS solution, which contained 5% skimmed milk powder,
was then added for a 60-min blocking at room temperature. After
washing with PBS, anti-PCSK5 (1:500; cat. no. 16470-1-AP) and
β-actin (1:1,000; cat. no. 20536-1-AP) (both from Proteintech
Group, Inc.) were added and incubated overnight at 4°C.
HRP-conjugated Affinipure goat anti-rabbit IgG (H+L) (1:5,000; cat.
no. SA00001-2; Proteintech, Group, Inc.) was added to the membrane
for incubation at room temperature for 1 h. After the secondary
antibody incubation, the product was washed three times with PBS.
The immunoreactive bands were visualized using the ECL-PLUS kit
(Cytiva) according to the manufacturer's manual. The results and
the images were analyzed with the Gel image analysis system and the
Gel-Pro Analyzer 3.1 (Media Cybernetics, Inc.), respectively. The
gray value of the bands was analyzed by Image J2× software (Rawak
Software Inc.). In this procedure, β-actin was used as the
reference gene.
ALP activity and osteocalcin secretion
detection
BMSCs (1×105 cells/well) were seeded onto
a 24-well plate with an osteogenesis induction αMEM for 48 h. Using
the colorimetric ALP Activity Assay kit (Abcam), the cells were
subjected to lysis to detect ALP activity. More specifically, BMSCs
were harvested, washed with cold PBS, resuspended in 50 µl Assay
Buffer, and then homogenized with a Dounce homogenizer (Thermo
Fisher Scientific, Inc.) on ice. Subsequently, the samples were
centrifuged at 4°C at 13,000 × g for 15 min in a cold
microcentrifuge. This step was performed to remove any insoluble
materials. Following that, the supernatant was collected and
transferred into a new tube. Next, 20 µl STOP Solution was added to
sample background control wells to terminate ALP activity in these
samples. Next, 50 µl pNPP solution (5 mM) was added to each well
containing sample and background sample controls. After that, 10 µl
ALP enzyme solution was added to each pNPP standard well for an
incubation period of 60 min at 25°C in the dark. The reaction was
halted by adding 20 µl STOP Solution. The optical absorbance was
finally measured at 450 nm using a microplate reader (Thermo Fisher
Scientific, Inc.).
In order to detect osteocalcin secretion, BMSCs
(treated with 0.25% trypsin) were first mixed with PBS and then
transferred into single-cell suspension repeatedly. The cells were
later crushed completely by repeated freeze-thaw at −20°C. After
ensuring cell suspension was centrifuged for 20 min at 1,667 × g
and 4°C, the supernatant was collected. The osteocalcin secretion
was later detected with the Osteocalcin ELISA Detection kit (cat.
no. AC-11F1; Immunodiagnostic Systems, Ltd.).
Alizarin Red S staining
Alizarin Red S staining was performed to evaluate
mineralization deposition in BMSCs, which had been cultured in a
6-well plate with osteogenesis induction medium for 21 days. The
cells were incubated with 1 ml 10% formaldehyde for 10 min at room
temperature. After which, the cells were washed twice with PBS.
Then, 1 ml 2% Alizarin Red S solution (pH was adjusted to ~4.5 with
0.5% ammonium hydroxide) was added to the cells for 15 min cellular
staining at 25°C. Following the staining, the stained cells were
thoroughly washed with double-distilled water (ddH2O).
The stained cells in each well were subsequently captured by an
optical microscope (Olympus Corporation) at ×200 magnification.
Following which, the positively stained area (red) was observed in
Image Pro Plus 6.0 (Media Cybernetics, Inc.).
Dual-luciferase reporter assay
The binding site of miR-338-3p to PCSK5 3′UTR was
‘AUGCUGG’, which was also the sequence of PCSK5 wild type (PCSK5
WT). To generate PCSK5 mutant type (PCSK5 MUT), ‘AUGCUGG’ was
replaced by ‘UACGACC’. Next, the pmiR-GLO reporter vector with
PCSK5 WT (100 ng) or MUT (100 ng) was co-transfected with
miR-338-3p mimic (50 nM) or mimic NC (50 nM) using Lipofectamine
3000 reagent. The pmiR-GLO reporter vector, miR-338-3p mimic and
mimic NC were all purchased from Promega Corporation. BMSCs were
subsequently seeded in a 96-well plate for an incubation period of
48 h after the co-transfection. The relative luciferase activity,
which could be a strong reference for the association between
miR-338-3p and its target gene PCSK5, was measured using a
dual-luciferase reporter assay system (Promega Corporation).
Renilla luciferase acted as an internal control to normalize the
luciferase activity.
RNA pull-down
The pull-down assay with biotinylated miRNA was
performed according to the methodology used in a previous study
(26). BMSCs were seeded into a
6-well plate. Following which, 100 nM biotinylated miR-338-3p mimic
(Bio-miR-338-3p) or biotinylated negative control (Bio-NC)
(Guangzhou RiboBio Co., Ltd.) was transfected into BMSCs. After the
48-h transfection, the cells were washed with PBS and then lysed
using a specific lysis buffer containing 20 mM Tris, pH 7.5, 200 mM
NaCl, 2.5 mM MgCl2, 0.05% Igepal, 60 U/ml Superase-In
(cat. no. AM2694; Invitrogen; Thermo Fisher Scientific, Inc.), 1 mM
DTT (cat. no. 3483-12-3; EMD Millipore), and protease inhibitors.
Streptavidin magnetic beads 100 µl (Invitrogen; Thermo Fisher
Scientific, Inc.) were subsequently used to pull down the
biotin-coupled RNA complex after incubating the samples for 4 h.
Next, the beads were attracted by a magnetic grate (Invitrogen;
Thermo Fisher Scientific, Inc.). The abundance of PCSK5 was
eventually evaluated using RT-qPCR.
Statistical analysis
Three independent experiments were performed, and
quantitative data were presented as the mean ± standard deviation
(SD). Statistical significance was determined by employing
Student's unpaired t-tests or one-way ANOVA with Dunnett's multiple
comparisons test or Tukey's multiple comparisons test. P<0.05
was considered to indicate a statistically significant
difference.
Results
PCSK5 and miR-338-3p are key
regulators of BMSCs
The 222 downregulated DEGs were first screened out
from GSE35959 with P<0.05 and log2FC <-2. The top
100 downregulated DEGs were subsequently uploaded to Metascape for
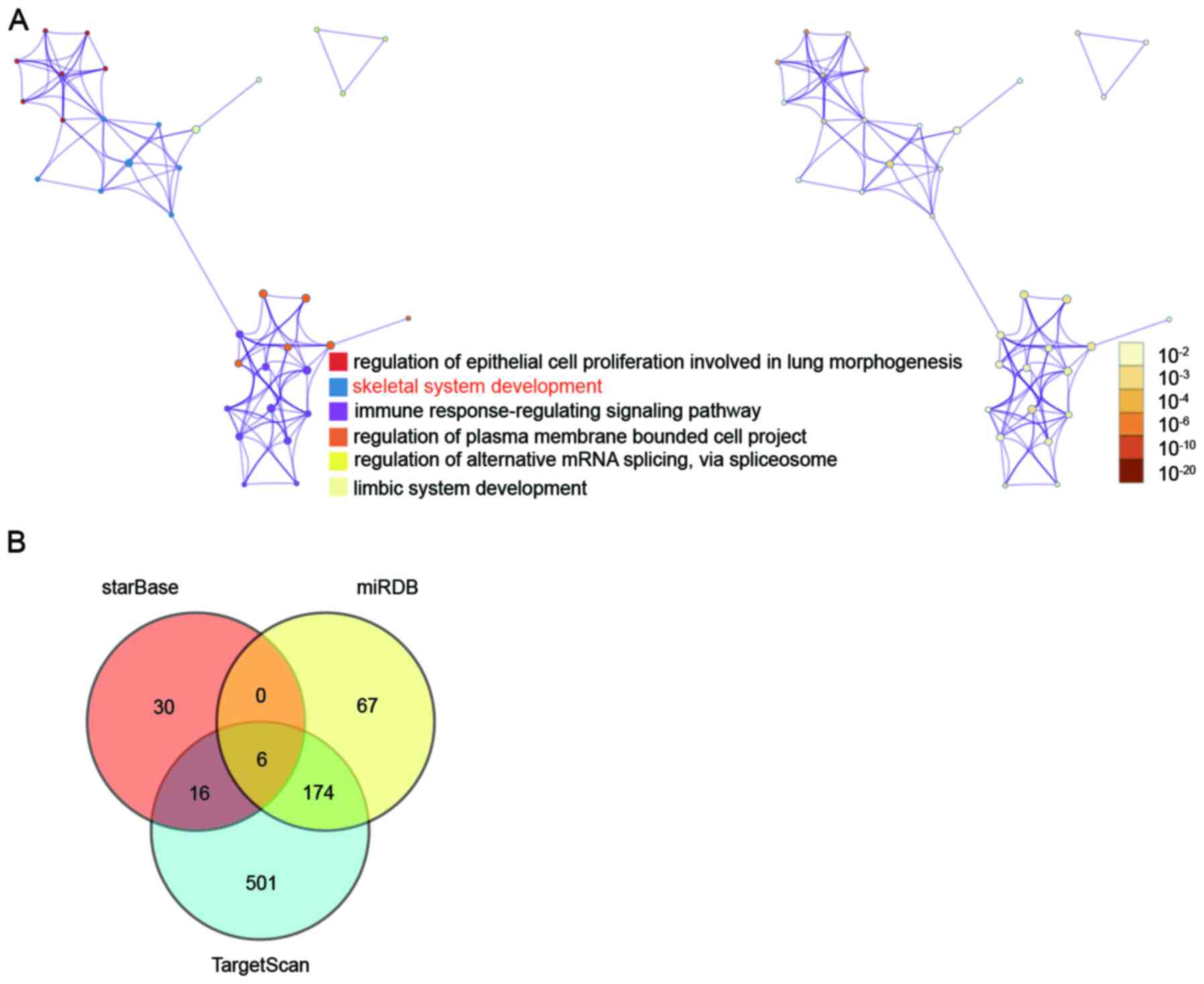
enrichment analysis. As shown in Fig.
1A, the skeletal system development was identified as the key
biological process involving eight genes: FGFR2, NFIB, PCSK5,
RPL38, SFRP2, COL14A1, WASF2 and ANKRD11. In one study, PCSK5 was
reported to be responsible for bone development (27). Besides, mice lacking PCSK5 exhibit
bone morphogenic defects (20).
PCSK5 was, therefore, identified as the gene of interest to be
investigated in the present study. To identify the key miRNAs that
could target PCSK5, starBase, miRDB and TargetScan were applied.
Finally, six overlapping miRNAs were found by Venny 2.1.0 (Fig. 1B); they included hsa-miR-29a-3p,
hsa-miR-29b-3p, hsa-miR-29c-3p, hsa-miR-338-3p, hsa-miR-577 and
hsa-miR-664b-3p. Based on a previous study, miR-338-3p required
further investigation due to its downregulation during osteoblast
differentiation (21).
PCSK5 contributes to osteoblastic
differentiation of BMSCs
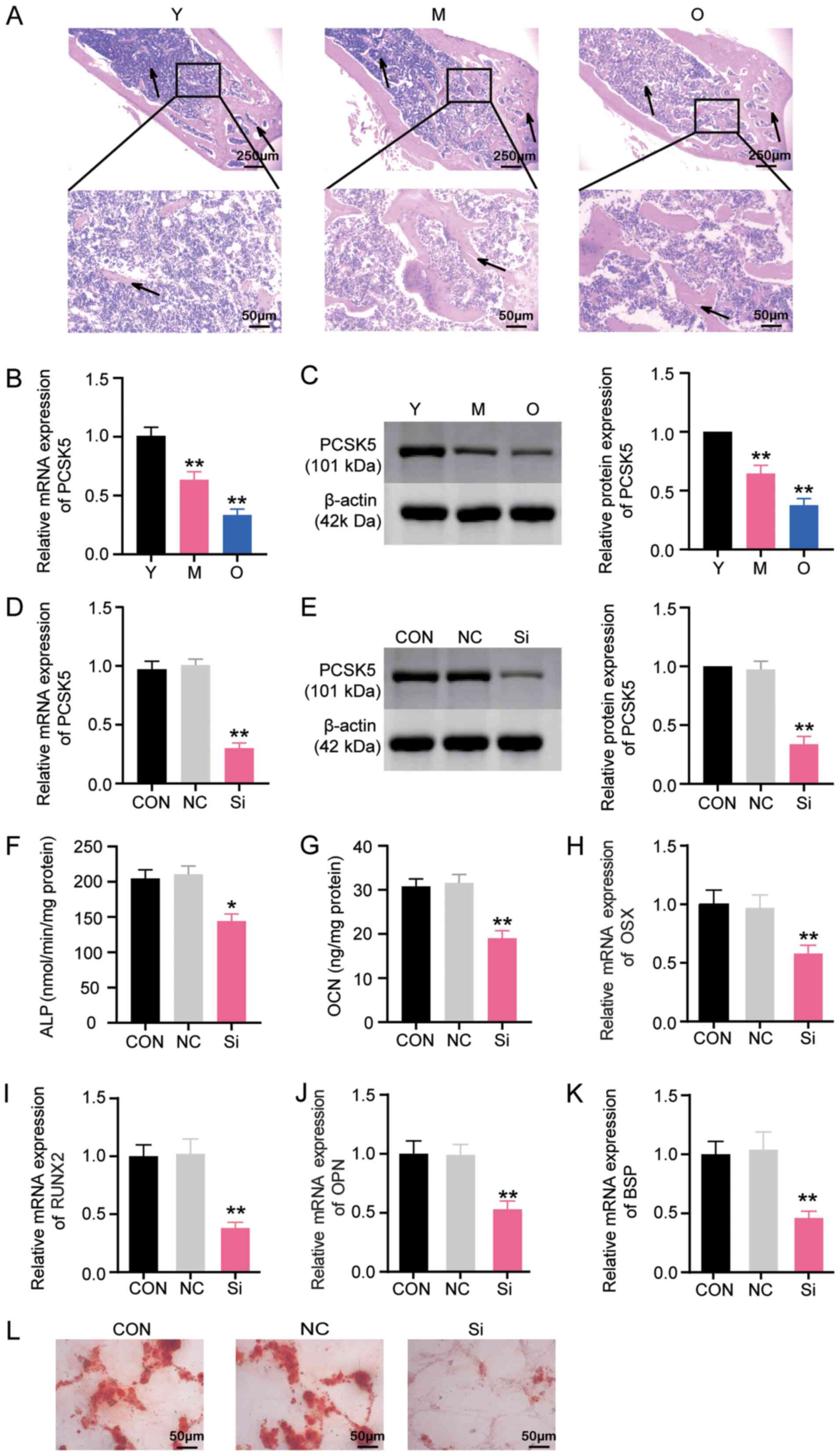
H&E staining was first used to examine the
morphology of the proximal tibial of 3-month-old young rats (Y),
12-month-old middle-aged rats (M) and 18-month-old aged rats (O).
Findings revealed that with aging, obvious bone defects existed in
the metaphysis of the proximal tibial. More specifically, the
osteoid content decreased, and the fracture and separation of the
trabecular meshwork appeared (Fig.
2A). Next, mRNA and protein expression of PCSK5 in BMSCs
harvested from rats of different age stages was measured, including
3-month-old young rats (Y), 12-month-old middle-aged rats (M) and
18-month-old aged rats (O). It was observed that both mRNA and
protein expression of PCSK5 declined as the age of the rats
increased (Fig. 2B and C). More
specifically, the mRNA expression of PCSK5 in the M group was 60%
of the Y group, while the O group was only 30% of the Y group
(Fig. 2B). The protein expression
of PCSK5 in the M group was 70% of the Y group, whereas the O group
was only 40% of the Y group (Fig.
2C). To obtain an improved understanding of the role of PCSK5
in age-associated osteoporosis progression, si-PCSK5 was
synthesized to knock down PCSK5, and was transfected into BMSCs
derived from middle-aged rats. The results of RT-qPCR showed that
si-PCSK5 could notably decrease the mRNA expression of PCSK5 by
~70% compared with the control group (Fig. 2D). Western blotting results also
showed that the protein expression of PCSK5 was reduced by si-PCSK5
by ~65% compared with the control group (Fig. 2E). These results demonstrated that
si-PCSK5 could successfully decrease the expression of PCSK5 at
both mRNA and protein levels. Therefore, si-PCSK5 was employed in
the subsequent experiments.
 | Figure 2.PCSK5 contributes to osteoblastic
differentiation of BMSCs. (A) Representative images of the proximal
tibial metaphysis from rats of the Y, M and O groups detected by
staining with hematoxylin and eosin. Magnifications, ×40 and ×200.
n=6. (B) Expression of PCSK5 mRNA in BMSCs from rats in Y, M and O
groups using RT-qPCR. n=6. (C) Expression of PCSK5 protein in BMSCs
of rats in Y, M and O groups using western blot assay. n=6. (D)
Inhibitory efficiency of si-PCSK5 in BMSCs by RT-qPCR. (E)
Inhibitory efficiency of si-PCSK5 in BMSCs using western blot
analysis. (F) Analysis of ALP activity after the transfection of
si-PCSK5 or NC into BMSCs induced for osteoblastic differentiation
for 48 h. (G) OCN secretion detection after the transfection of
si-PCSK5 or NC into BMSCs induced for differentiation for 48 h.
mRNA expression detection of (H) OSX, (I) RUNX2, (J) OPN and (K)
BSP after the transfection of si-PCSK5 or NC into BMSCs induced for
differentiation for 48 h. (L) Representative images of Alizarin Red
S staining of BMSCs after osteoblastic differentiation for 21 days.
Magnification, ×200. Data are presented as the mean ± SD.
Statistical significance was determined using one-way ANOVA. n=3.
*P<0.05 and **P<0.001 vs. control group. BMSCs, bone
mesenchymal stem cells; PCSK5, proprotein convertase
subtilisin/kexin type 5; Y, 3-month-old young rats; M, 12-month-old
middle-aged rats; O, 18-month-old aged rats; OPN, osteopontin;
RT-qPCR:, reverse transcription-quantitative PCR; si-PCSK5, small
interfering RNA targeting PCSK5; NC, negative control siRNA; ALP,
alkaline phosphatase; OCN, osteocalcin secretion; OSX, osterix;
RUNX2, runt-related gene 2; OPN, osteopontin; BSP, bone
sialoprotein; CON, normal control. |
To comprehensively unravel the role of PCSK5 in
osteoblastic differentiation, si-PCSK5 or its NC was transfected
into BMSCs for 48 h, which were induced to differentiate into
osteoblasts. Since OCN, ALP, OSX, RUNX2, OPN and BSP are popular
markers for osteoblastic differentiation, these markers were
detected to investigate the process of osteoblastic
differentiation. After testing for ALP activity and osteocalcin
secretion, it was found that si-PCSK5 restrained ALP activity by
30% compared with the control group (Fig. 2F) and that it suppressed osteocalcin
secretion by ~35% compared with the control group (Fig. 2G). The mRNA expression levels of
OSX, RUNX2, OPN and BSP (Fig. 2H-K)
were also examined. The findings indicated that the mRNA expression
levels of OSX and OPN decreased by ~40%, whereas the mRNA
expression levels of RUNX2 and BSP decreased by ~60% (Fig. 2H-K). Furthermore, Alizarin Red S
staining was performed after a 21-day induction of osteoblastic
differentiation. The images showed that si-PCSK5 suppressed
mineralized nodule formation in BMSCs (Fig. 2L). Taken together, the results
demonstrated that PCSK5 contributes to the osteoblastic
differentiation of BMSCs.
PCSK5 is a target of miR-338-3p
To explore the mechanism of PCSK5 in the regulation
of age-associated osteoporosis, the targets upstream of PCSK5 were
investigated. The literature has established that a number of genes
are mediated by various miRNAs. Hence, TargetScan was used to
predict the potential miRNAs that could bind to PCSK5 3′UTR. Venny
2.1 analyses revealed that miR-338-3p could target PCSK5 (Fig. 1B). To confirm this, RT-qPCR was used
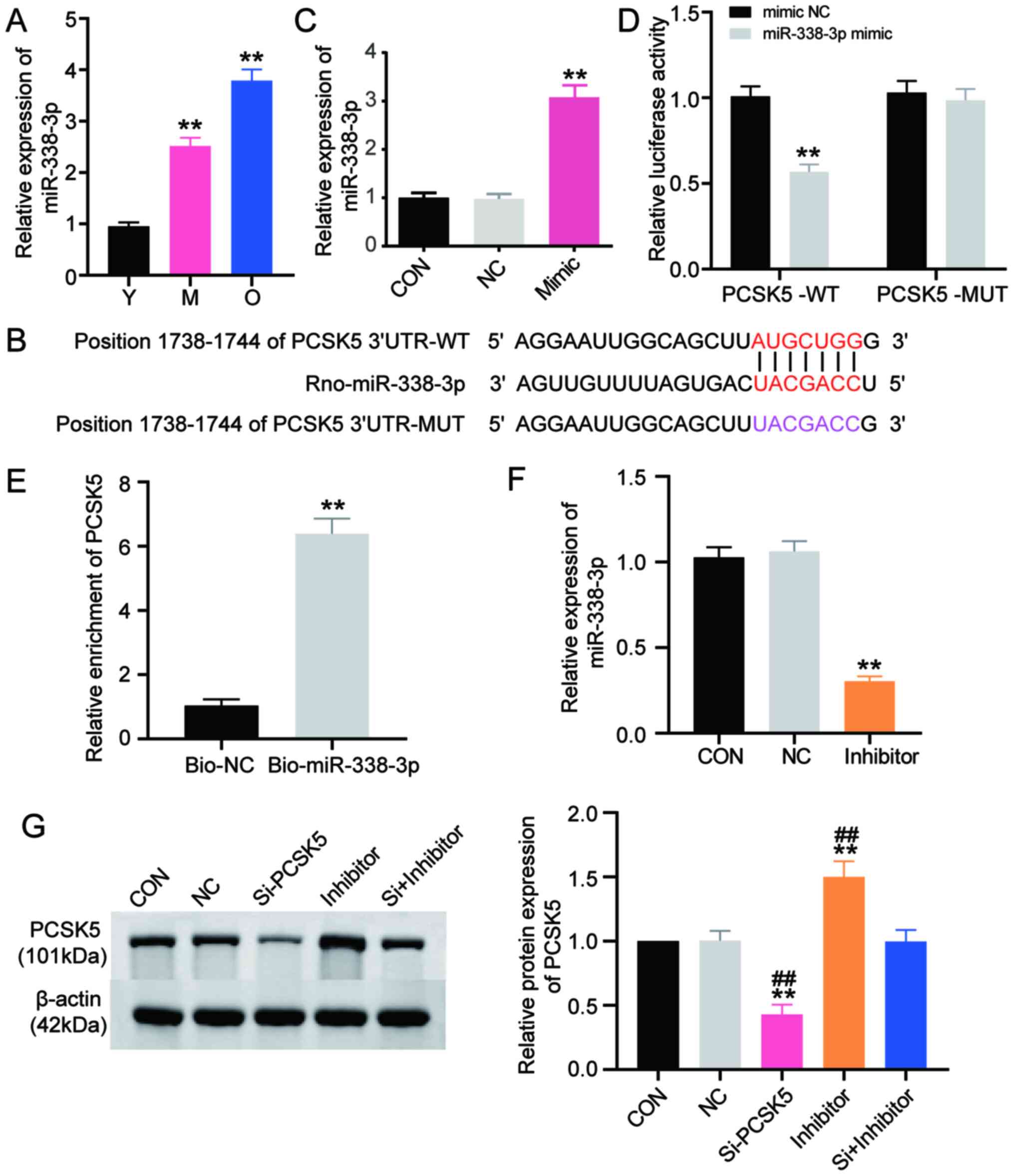
to examine the mRNA expression of miR-338-3p in BMSCs from the Y, M
and O groups of rats. The results showed that the expression of
miR-338-3p in the M group was upregulated 2.5 times as much as the
Y group (Fig. 3A). However, the
expression of miR-338-3p in the O group was found to be
significantly upregulated 4 times as much as the Y group. This
outcome was completely opposite to that of PCSK5 (Fig. 3A). Next, a dual-luciferase reporter
assay kit was utilized to verify whether PCSK5 was the exact target
gene of miR-338-3p. The wild-type (PCSK5-WT) and mutant type
(PCSK5-MUT) of PCSK5 were first generated (Fig. 3B) and then co-transfected with
miR-338-3p mimic or mimic NC into BMSCs. Following transfection
with miR-338-3p mimic, RT-qPCR demonstrated that the expression
level of miR-338-3p increased by ~3 times compared with the mimic
NC group (Fig. 3C). Following
co-transfection, as illustrated in Fig.
3D, the group of PCSK5-WT plus miR-338-3p mimic suppressed
luciferase activity by 40% compared with the group of PCSK5-WT plus
mimic NC. Luciferase activity was not influenced in the mutant
group, meaning miR-338-3p could bind to PCSK5 3′UTR; this was
further confirmed by the RNA pull-down assay. As depicted in
Fig. 3E, the expression of PCSK5
was dramatically enriched in the Bio-miR-338-3p group by >6
times compared with the Bio-NC group. This result suggested that
miR-338-3p could target PCSK5. Subsequently, miR-338-3p inhibitor
was applied to silence miR-338-3p. The inhibitory efficiency
detected using RT-qPCR showed that miR-338-3p inhibitor
successfully decreased the expression of miR-338-3p to a level that
was 30% of the control group (Fig.
3F). The expression of PCSK5 protein was detected by western
blotting, following transfection with si-PCSK5, miR-338-3p
inhibitor or si-PCSK5 plus miR-338-3p inhibitor in BMSCs. The
results indicated that the expression of PCSK5 protein was
suppressed by si-PCSK5 by 60%, while it was enhanced by the
miR-338-3p inhibitor by 50% compared with the control group
(Fig. 3G). The expression of PCSK5
protein, which was suppressed by si-PCSK5, could even be reversed
by the miR-338-3p inhibitor. These results revealed that the
expression of PCSK5 protein could be increased if miR-338-3p was
silenced. Given that miR-338-3p could suppress PCSK5 on the protein
level, miR-338-3p could target PCSK5.
 | Figure 3.PCSK5 is a target of miR-338-3p. (A)
Expression of miR-338-3p in BMSCs from rats in groups Y, M and O by
RT-qPCR. (B) PCSK5-WT or PCSK5-MUT without or with the binding
sequence of miR-338-3p. (C) Transfection efficiency of miR-338-3p
mimic in BMSCs was detected by RT-qPCR. (D) The association between
miR-338-3p and PCSK5 was detected using dual-luciferase reporter
assay. (E) The target association of miR-338-3p with PCSK5 was
detected with RNA pull-down assay system. (F) Inhibitory efficiency
of miR-338-3p inhibitor in BMSCs by RT-qPCR. (G) Expression of
PCSK5 protein was measured using western blotting when miR-338-3p
was silenced by the miR-338-3p inhibitor. Data are presented as the
mean ± SD. Statistical significance was determined with unpaired
Student's t-test or one-way ANOVA. n=3. **P<0.001 vs. control
group; ##P<0.001 vs. si+inhibitor group. PCSK5,
proprotein convertase subtilisin/kexin type 5; miR, microRNA;
RT-qPCR, reverse transcription-quantitative PCR; Y, 3-month-old
young rats; M, 12-month-old middle-aged rats; O, 18-month-old aged
rats; BMSCs, bone mesenchymal stem cells; Bio-NC, biotinylated
negative control; bio-miR-338-3p, biotinylated miR-338-3p mimic;
CON, normal control; NC, negative control; si-PCSK5, small
interfering RNA targeting PCSK5; inhibitor, miR-338-3p inhibitor;
UTR, untranslated region; WT, wild-type; MUT, mutant. |
miR-338-3p inhibits osteoblastic
differentiation of BMSCs
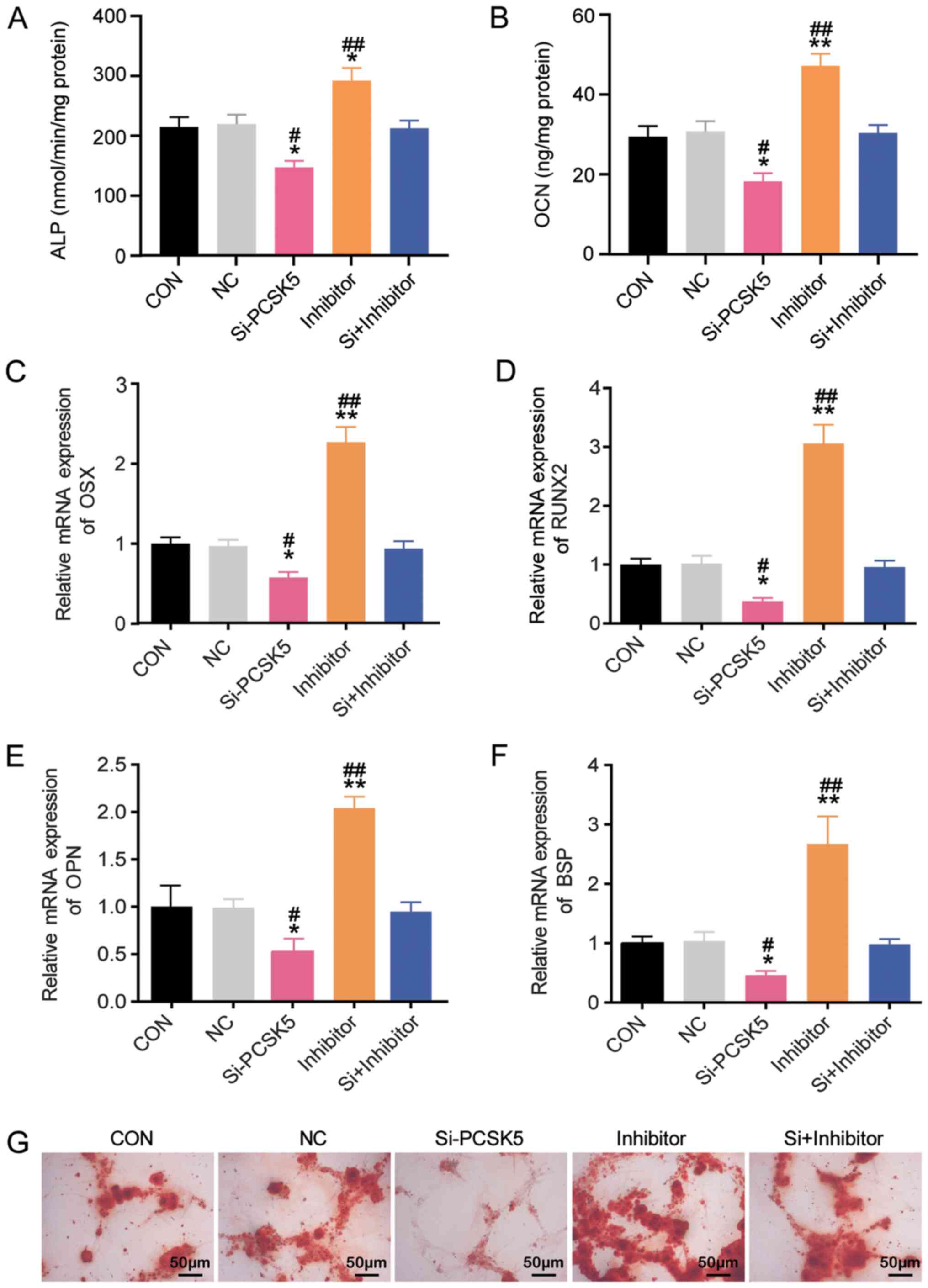
Given that PCSK5 is a potential target of
miR-338-3p, whether or not miR-338-3p exerted the opposite effect
to PCSK5 on osteoblastic differentiation of BMSCs was verified. To
confirm this, BMSCs were transfected with si-PCSK5, miR-338-3p
inhibitor, si-PCSK5 plus miR-338-3p inhibitor, and NC. Similar to
the exploration of PCSK5, ALP activity, OCN secretion, OSX, RUNX2,
OPN and BSP mRNA expression and Alizarin Red S staining were
conducted in BMSCs. As shown in Fig.
4A, the miR-338-3p inhibitor increased ALP activity by 40%
compared to the control group, and it compromised the inhibitory
effect of si-PCSK5 on ALP activity. Compared with the control
group, the secretion of OCN was enhanced by 60% when miR-338-3p was
inhibited (Fig. 4B). Also, the
braking effect of si-PCSK5 on osteocalcin secretion was restored by
the miR-338-3p inhibitor (Fig. 4B).
Compared with the control group, miR-338-3p inhibitor increased the
mRNA expression of OSX, RUNX2, OPN and BSP by ~2.3, 3, 2 and 2.5
times, and it weakened the inhibition of si-PCSK5 on OSX, RUNX2,
BSP and OPN mRNA expression (Fig.
4C-F). Alizarin Red S staining showed that mineralized nodule
formation was facilitated in BMSCs with the miR-338-3p inhibitor
(Fig. 4G) and that the miR-338-3p
inhibitor could reverse the effect of si-PCSK5. Overall, these
results demonstrated that miR-338-3p inhibited osteoblastic
differentiation of BMSCs, thereby playing an opposite role of PCSK5
in age-associated osteoporosis.
 | Figure 4.miR-338-3p inhibits osteoblastic
differentiation of BMSCs by targeting PCSK5. (A) ALP activity was
evaluated after the transfection of si-PCSK5, miR-338-3p inhibitor
or si-PCSK5 plus miR-338-3p inhibitor in BMSCs. (B) OCN secretion
was detected after transfection with si-PCSK5, miR-338-3p inhibitor
or si-PCSK5 plus miR-338-3p inhibitor in BMSCs. (C) OSX mRNA
expression was measured after transfection with si-PCSK5,
miR-338-3p inhibitor or si-PCSK5 plus miR-338-3p inhibitor in
BMSCs. (D) RUNX2 mRNA expression was evaluated after transfection
with si-PCSK5, miR-338-3p inhibitor or si-PCSK5 plus miR-338-3p
inhibitor in BMSCs. (E) OPN mRNA expression was measured after
transfection with si-PCSK5, miR-338-3p inhibitor or si-PCSK5 plus
miR-338-3p inhibitor in BMSCs. (F) BSP mRNA expression was measured
after transfection with si-PCSK5, miR-338-3p inhibitor or si-PCSK5
plus miR-338-3p inhibitor in BMSCs. (G) Alizarin Red S staining was
measured after transfection with si-PCSK5, miR-338-3p inhibitor or
si-PCSK5 plus miR-338-3p inhibitor in BMSCs. Magnification, ×200.
Data are presented as the mean ± SD. Statistical significance was
determined using one-way ANOVA. n=3. *P<0.05 and **P<0.001
vs. control group; #P<0.05 and
##P<0.001 vs. si+inhibitor group. PCSK5, proprotein
convertase subtilisin/kexin type 5; BMSCs, bone mesenchymal stem
cells; miR, microRNA; OSX, osterix; ALP, alkaline phosphatase; OCN,
osteocalcin; OSX, osterix; RUNX2, Runt-related gene 2; OPN,
osteopontin; BSP, bone sialoprotein; CON, normal control; NC,
negative control; inhibitor, miR-338-3p inhibitor; si-PCSK5, small
interfering RNA targeting PCSK5. |
Discussion
In the present study, the regulatory mechanism of
miR-338-3p and its target gene PCSK5 on osteogenic differentiation
was investigated in vitro using BMSCs. The aim was to gain
more insights into the effects of miR-338-3p and its target gene
PCSK5 on the osteogenesis process, as well as understanding their
roles in age-associated osteoporosis. The experimental results
showed that miR-338-3p inhibited osteogenic differentiation, while
its target gene PCSK5 accelerated osteogenic differentiation.
Therefore, miR-338-3p may be a potential regulator that inhibits
osteogenesis and promotes age-associated osteoporosis, even though
its target gene PCSK5 had the opposite effect. A previous study
reported that PCSK5 was localized in bone tissues and expressed in
osteoblasts and osteocytes (28).
The detection of mRNA and protein expression of PCSK5 in BMSCs in
the present study at different stages of the aging process
confirmed that PCSK5 had an inverse relationship with age. Thus,
PCSK5 was suspected to be involved in the regulation of bone
formation. A previous study suggested that PCSK5 was a putative
regulator in skeletal development (27). Similarly, the present identified
PCSK5 as an essential regulatory factor in bone formation by
contributing to osteoblastic differentiation. As osteogenesis is a
process that highly involves osteoblastic differentiation (29), the promotional effect of PCSK5 on
osteoblastic differentiation reflected its positive role in
osteogenesis. Bone deterioration is even caused by a decrease in
osteogenesis function (30). Based
on this, it is proposed that PCSK5 is crucial in protecting against
age-associated osteoporosis.
In addition, the role of miR-338-3p in osteoblastic
differentiation was illustrated in the present study. To be more
specific, it was verified that miR-338-3p could target PCSK5. The
findings revealed that miR-338-3p showed an opposite trend of
expression in BMSCs with age and exerted negative functions on
PCSK5, thereby limiting bone formation. Several studies also
reported that an increasing number of miRNAs could regulate BMSC
differentiation (31–33). In the present study, increased
expression of miR-338-3p was observed in BMSCs of older rat
samples, which indicated a counter-productive role of miR-338-3p in
osteogenesis. Furthermore, it was demonstrated that the enrichment
of miR-338-3p resulted in the reduction of osteoblastic
differentiation in BMSCs. The present results were consistent with
an in vitro analysis that showed that miR-338-3p inhibited
the expression of osteoblastic differentiation markers, such as
OSX, which suppressed osteoblastic differentiation (21). Playing an inhibitory role in
osteoblastic differentiation, miR-338-3p could also be an inducer
of bone loss and age-associated osteoporosis.
According to previous studies, several factors
contribute to promote bone formation and suppress bone resorption
(34–36). In the present study, experiments
were not designed that were relevant to investigate the crosstalk
among miR-338-3p or PCSK5. Future research may focus on this
process and explain the underlying mechanism of miR-338-3p or PCSK5
in osteogenesis as well as age-associated osteoporosis.
In summary, the experiments carried out in the
present study demonstrated that PCSK5 targeted by miR-338-3p could
restrict the process of age-associated osteoporosis by contributing
to osteogenesis. Thus, PCSK5 and its upstream target miR-338-3p
could be valuable biomarkers for the prevention and healing of
age-associated osteoporosis.
Supplementary Material
Supporting Data
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JT conceived and designed the study. MZ acquired the
data. XL performed the data analysis. GR interpreted the data. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
All animal procedures were approved by the Animal
Ethics Committee of Affiliated Hospital of Jianghan University
(approval no. JH-20191014-23; Jiangan, China).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Kiernan J, Hu S, Grynpas MD, Davies JE and
Stanford WL: Systemic Mesenchymal Stromal Cell Transplantation
Prevents Functional Bone Loss in a Mouse Model of Age-Related
Osteoporosis. Stem Cells Transl Med. 5:683–693. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Compston JE, McClung MR and Leslie WD:
Osteoporosis. Lancet. 393:364–376. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Prockop DJ: Marrow stromal cells as stem
cells for nonhematopoietic tissues. Science. 276:71–74. 1997.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Chen Q, Shou P, Zheng C, Jiang M, Cao G,
Yang Q, Cao J, Xie N, Velletri T, Zhang X, et al: Fate decision of
mesenchymal stem cells: Adipocytes or osteoblasts? Cell Death
Differ. 23:1128–1139. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Tae JY, Lee H, Lee H, Ko Y and Park JB:
Osteogenic potential of cell spheroids composed of varying ratios
of gingiva-derived and bone marrow stem cells using concave
microwells. Exp Ther Med. 16:2287–2294. 2018.PubMed/NCBI
|
|
6
|
Almalki SG and Agrawal DK: Key
transcription factors in the differentiation of mesenchymal stem
cells. Differentiation. 92:41–51. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
He S, Yang S, Zhang Y, Li X, Gao D, Zhong
Y, Cao L, Ma H, Liu Y, Li G, et al: LncRNA ODIR1 inhibits
osteogenic differentiation of hUC-MSCs through the
FBXO25/H2BK120ub/H3K4me3/OSX axis. Cell Death Dis. 10:9472019.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yin Q, Wang J, Fu Q, Gu S and Rui Y:
CircRUNX2 through has-miR-203 regulates RUNX2 to prevent
osteoporosis. J Cell Mol Med. 22:6112–6121. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li CJ, Cheng P, Liang MK, Chen YS, Lu Q,
Wang JY, Xia ZY, Zhou HD, Cao X, Xie H, et al: MicroRNA-188
regulates age-related switch between osteoblast and adipocyte
differentiation. J Clin Invest. 125:1509–1522. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Moerman EJ, Teng K, Lipschitz DA and
Lecka-Czernik B: Aging activates adipogenic and suppresses
osteogenic programs in mesenchymal marrow stroma/stem cells: The
role of PPAR-gamma2 transcription factor and TGF-beta/BMP signaling
pathways. Aging Cell. 3:379–389. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Wu J, Zhang W, Ran Q, Xiang Y, Zhong JF,
Li SC and Li Z: The differentiation balance of bone marrow
mesenchymal stem cells is crucial to hematopoiesis. Stem Cells Int.
2018:15401482018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yu B and Wang CY: Osteoporosis: The result
of an ‘aged’ bone microenvironment. Trends Mol Med. 22:641–644.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Khosla S and Hofbauer LC: Osteoporosis
treatment: Recent developments and ongoing challenges. Lancet
Diabetes Endocrinol. 5:898–907. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Cheung CL, Xiao SM and Kung AW: Genetic
epidemiology of age-related osteoporosis and its clinical
applications. Nat Rev Rheumatol. 6:507–517. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Carey JJ, Lewiecki EM, Blank RD, Shuhart
CR and Buehring B: Treatment of low bone density or osteoporosis to
prevent fractures in men and women. Ann Intern Med. 167:9012017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lorentzon M and Cummings SR: Osteoporosis:
The evolution of a diagnosis. J Intern Med. 277:650–661. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Sànchez-Riera L, Carnahan E, Vos T,
Veerman L, Norman R, Lim SS, Hoy D, Smith E, Wilson N, Nolla JM, et
al: The global burden attributable to low bone mineral density. Ann
Rheum Dis. 73:1635–1645. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Turpeinen H, Ortutay Z and Pesu M:
Genetics of the first seven proprotein convertase enzymes in health
and disease. Curr Genomics. 14:453–467. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Osathanon T, Manokawinchoke J, Sa-Ard-Iam
N, Mahanonda R, Pavasant P and Suwanwela J: Jagged1 promotes
mineralization in human bone-derived cells. Arch Oral Biol.
99:134–140. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
McPherron AC, Lawler AM and Lee SJ:
Regulation of anterior/posterior patterning of the axial skeleton
by growth/differentiation factor 11. Nat Genet. 22:260–264. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu H, Sun Q, Wan C, Li L, Zhang L and
Chen Z: MicroRNA-338-3p regulates osteogenic differentiation of
mouse bone marrow stromal stem cells by targeting Runx2 and Fgfr2.
J Cell Physiol. 229:1494–1502. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Zhang XH, Geng GL, Su B, Liang CP, Wang F
and Bao JC: MicroRNA-338-3p inhibits glucocorticoid-induced
osteoclast formation through RANKL targeting. Genet Mol Res.
15:gmr.15037674. 2016.
|
|
23
|
Niu D, Gong Z, Sun X, Yuan J, Zheng T,
Wang X, Fan X, Mao Y, Liu X, Tang B, et al: miR-338-3p regulates
osteoclastogenesis via targeting IKKβ gene. In Vitro Cell Dev Biol
Anim. 55:243–251. 2019. View Article : Google Scholar
|
|
24
|
Benisch P, Schilling T, Klein-Hitpass L,
Frey SP, Seefried L, Raaijmakers N, Krug M, Regensburger M, Zeck S,
Schinke T, et al: The transcriptional profile of mesenchymal stem
cell populations in primary osteoporosis is distinct and shows
overexpression of osteogenic inhibitors. PLoS One. 7:e451422012.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Y, Zheng F, Xiao X, Xie F, Tao D, Huang
C, Liu D, Wang M, Wang L, Zeng F, et al: CircHIPK3 sponges miR-558
to suppress heparanase expression in bladder cancer cells. EMBO
Rep. 18:1646–1659. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Szumska D, Pieles G, Essalmani R, Bilski
M, Mesnard D, Kaur K, Franklyn A, El Omari K, Jefferis J, Bentham
J, et al: VACTERL/caudal regression/Currarino syndrome-like
malformations in mice with mutation in the proprotein convertase
Pcsk5. Genes Dev. 22:1465–1477. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Hoac B, Susan-Resiga D, Essalmani R,
Marcinkiweicz E, Seidah NG and McKee MD: Osteopontin as a novel
substrate for the proprotein convertase 5/6 (PCSK5) in bone. Bone.
107:45–55. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Marie PJ: Targeting integrins to promote
bone formation and repair. Nat Rev Endocrinol. 9:288–295. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eastell R, O'Neill TW, Hofbauer LC,
Langdahl B, Reid IR, Gold DT and Cummings SR: Postmenopausal
osteoporosis. Nat Rev Dis Primers. 2:160692016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Long H, Sun B, Cheng L, Zhao S, Zhu Y,
Zhao R and Zhu J: miR-139-5p represses BMSC osteogenesis via
targeting Wnt/β-catenin signaling pathway. DNA Cell Biol.
36:715–724. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Zhang S, Liu Y, Zheng Z, Zeng X, Liu D,
Wang C and Ting K: MicroRNA-223 suppresses osteoblast
differentiation by inhibiting DHRS3. Cell Physiol Biochem.
47:667–679. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Fu L, Liu H and Lei W: MiR-596 inhibits
osteoblastic differentiation and cell proliferation by targeting
Smad3 in steroid-induced osteonecrosis of femoral head. J Orthop
Surg Res. 15:1732020. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhou S, Qian B, Wang L, Zhang C, Hogan MV
and Li H: Altered bone-regulating myokine expression in skeletal
muscle Of Duchenne muscular dystrophy mouse models. Muscle Nerve.
58:573–582. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Bartold M, Gronthos S, Haynes D and
Ivanovski S: Mesenchymal stem cells and biologic factors leading to
bone formation. J Clin Periodontol. 46 (Suppl 21):12–32. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Alcorta-Sevillano N, Macías I, Rodríguez
CI and Infante A: Crucial role of Lamin A/C in the migration and
differentiation of MSCs in bone. Cells. 9:E13302020. View Article : Google Scholar : PubMed/NCBI
|