Introduction
Allergic rhinitis (AR) is a chronic inflammatory
disorder of the nasal mucosa, which affects 10–30% of the worldwide
population (1). There are two types
of AR: Seasonal and perennial (2).
Both types of AR typically manifest with pruritus, sneezing,
rhinorrhea and nasal congestion (3). Inherited genetics and environmental
exposure contribute to the development and progression of AR, which
frequently leads to asthma (4).
Considering the comprehensive pathogenesis, AR is often
misdiagnosed and patients develop a resistance to drug treatments.
Therefore, there is an urgent need to determine the molecular
mechanisms underlying the progression and development of AR.
Circular RNAs (circRNAs) are a novel class of
non-coding RNAs characterized by covalently closed-loop structures
(5). Several studies have indicated
that circRNA dysregulation is involved in inflammatory-related
diseases, including AR. For example, Liu and Cao (6) reported that circDdx17 downregulated
microRNA (miR/miRNA)-17 expression and alleviated AR symptoms and
pathological conditions. Zhu et al (7) indicated that circHIPK3 overexpression
accelerated T helper cell (Th)2 differentiation of
ovalbumin-induced CD4+ T cells and aggravated the nasal
symptoms of AR mice. However, the exact function of circRNA
arrestin domain-containing 3 (circARRDC3) in the progression of AR
remains unknown.
Granulocyte-macrophage colony-stimulating factor
(GM-CSF) and eotaxin are pro-inflammatory cytokines in allergic
airway inflammation that are synthesized and released by airway
epithelial cells, infiltrating leukocytes and fibroblasts in
response to allergens and inflammatory mediators (8). Mucus hypersecretion, particularly
mucin 5AC (MUC5AC) expression, is a common feature of allergic
airway disorders (9). Accumulating
studies have indicated that the inflammatory factors GM-CSF,
eotaxin and MUC5AC are indicators of AR (8,10).
Therefore, the aim of the present study was to evaluate the
relationship between circARRDC3 and GM-CSF, eotaxin or MUC5AC.
miRNAs are a type of short non-coding RNAs with a
length of ~22 nucleotides (11,12),
which have been implicated in the pathogenesis of AR. For instance,
Teng et al (13)
demonstrated that miR-143 suppressed IL-13-induced inflammatory
cytokine production by targeting IL-13 receptor α1 in AR. Liu et
al (14) reported that miR-487b
ameliorated AR via inactivation of the IL-33/sulfotransferase
signaling pathway. Chen et al (15) indicated that miR-21 was associated
with antenatal immunoglobulin E production and development of AR.
Furthermore, miR-375 has been reported to play an inhibitory role
in the development of allergic inflammation (16). Nevertheless, the biological role of
miR-375 in AR needs to be further elucidated. IL-13, a typical Th2
cytokine, is involved in mucus hypersecretion and the release of
inflammatory mediators by airway epithelial cells (17). A growing number of studies focused
on preventing or reducing allergic airway reactions by targeting
IL-13. For instance, Lv et al (18) indicated that IL-37 attenuated
IL-4/IL-13-induced eotaxin production and lung eosinophilia in
murine allergic asthma. Ashraf et al (19) reported that oxyresveratrol could
improve allergic asthma by downregulating the expression levels of
IL-4, IL-5 and IL-13. Since IL-13 is closely related to the
pathogenesis of allergic diseases, the IL-13 induction model has
been widely used in AR research.
The aim of the present study was to investigate the
role of circARRDC3 in IL-13-induced nasal epithelial cells (NECs).
The results indicated that circARRDC3 contributed to IL-13-induced
inflammatory cytokine and mucus production in NECs via the
miR-375/krueppel-like factor 4 (KLF4) axis.
Materials and methods
Tissue collection
Nasal mucosa samples from the inferior turbinate
mucosa were obtained from 20 healthy subjects (13 males and 7
females; age, 32.1±16.0) and 20 patients with AR. (males and 9
females; age, 36.4±13.8). Samples were stored at −80°C until
further use. The present study was approved by The Ethics Committee
of The Shanghai Ninth People's Hospital (Shanghai, China). Written
informed consent was obtained from all patients.
Cell culture
Under local anesthesia, inferior turbinate nasal
epithelial cells (NECs) were obtained using the nasal fragment
method (20). Cells were cultured
in bronchial epithelial cell growth medium (BEGM; Lonza Group,
Ltd.) with 10% FBS (Gibco; Thermo Fisher Scientific, Inc.) in a
humidified 5% CO2 incubator. The NECs were passaged when
a 70–80% fused monolayer was present. Cells from passage 2 were
used for subsequent experiments.
Cell transfection
Small interfering RNA (siRNA) targeting circARRDC3
(si-circARRDC3) and negative control (si-NC), miR-375 mimic and
negative control (NC mimic), and miR-375 inhibitor and negative
control (NC inhibitor) were purchased from Shanghai GenePharma Co.,
Ltd. circARRDC3 or KLF4 cDNA was amplified and subcloned into
pcDNA3.1 vectors (Shanghai GenePharma Co., Ltd.) to overexpress
circARRDC3 or KLF4. pcDNA3.1 empty vector (pcDNA3.1) as a
control.
All plasmids (0.5 µg) and oligonucleotides (50 nM)
were transfected into NECs cells (6×105 cells/well)
using Lipofectamine™ 2000 (Thermo Fisher Scientific, Inc.). After
transfection for 48 h, the cells were used for subsequent assays.
The sequences of the oligonucleotides used for transfection were as
follows: i) si-circARRDC3, 5′-CAGACCTCATCACTCAGAA-3′; ii) siNC,
5′-CCGAAGGCCGTGTCCCGAG-3′; iii) miR-375 mimic,
5′-UUUGUUCGUUCGGCUCGCGUGA-3′; iv) NC mimic,
5′-UUUGUACUACACAAAAGUACUG-3′; v) miR-375 inhibitor,
5′-UCACGCGAGCCGAACGAACAAA-3′; and vi) NC inhibitor,
5′-CAGUACUUUUGUGUAGUACAAA-3′.
IL-13 stimulation of NECs
Following lentivirus infection, NECs
(0.75×105/well) were stimulated with 50 ng/ml IL-13
(R&D Systems, Inc.) for 14 days at 37°C in BEGM without
hydrocortisone. The media was replaced twice per week. Cell
supernatant and pellets were collected centrifuged for 3 min at 4°C
for reverse transcription-quantitative PCR (RT-qPCR).
MTT assay
Transfected cells were seeded into 96-well culture
plates (2×104 cells/well). A total of ~20 µl MTT
solution was added to each well, and the cell culture plates were
incubated at 37°C for 4 h. DMSO (150 µl) was added following
supernatant removal. The absorbance was determined at a wavelength
of 490 nm using a microplate reader (Bio-Rad Laboratories,
Inc.).
Luciferase reporter assay
The Starbase (version 2.0; http://starbase.sysu.edu.cn/starbase2/) and TargetScan
(Release 7.2; http://www.targetscan.org/vert_72/) online tools were
used to predict the downstream targets of circARRDC3 and miR-375,
respectively (21). Wild-type and
mutant circARRDC3 or KLF4 sequences were subcloned into the pmirGLO
vector (Promega Corporation). Subsequently, the vectors were
co-transfected with miR-375 mimic, NC mimic, miR-375 inhibitor or
NC inhibitor into 293T cells (2×105 cells/well) using
Lipofectamine® 2000 (Thermo Fisher Scientific, Inc.).
Following 48 h of transfection, a dual-luciferase reporter system
(Promega Corporation) was used to measure luciferase activity.
Firefly luciferase activity was normalized to Renilla
luciferase.
RT-qPCR
Total RNA was extracted from NECs or nasal mucosal
specimens using TRIzol® reagent (Invitrogen; Thermo
Fisher Scientific, Inc.). RT was performed using the PrimeScript
Reagent kit (Takara Bio, Inc.) at 37°C for 30 min. qPCR was
performed using the SYBR-Green PCR Master Mix kit (Takara Bio,
Inc.) on the ABI Prism 7300 Sequence Detection system (Applied
Biosystems; Thermo Fisher Scientific, Inc.). The thermocycling
conditions consisted of an initial denaturation at 95°C for 15 sec,
followed by 40 cycles of denaturation at 94°C for 30 sec, annealing
at 60°C for 20 sec, and extension at 72°C for 40 sec. GAPDH and U6
were used as internal controls. Gene expression was analyzed using
the 2−∆∆Cq method (22).
The following primer pairs were used for the qPCR: circARRDC3
forward, 5′-TCCTATTGCTCTTCCTTGTGG-3′ and reverse,
5′-AAAAAGTCAGGGCAGCAGAG-3′; miR-375 forward,
5′-AAACCGGACCTGAGCGTTTT-3′ and reverse, 5′-CCGAACGAACAAAACGCTCA-3′;
KLF4 forward, 5′-TCTCCCACATGAAGCGACTT-3′ and reverse,
5′-ATGGGTCAGCGAATTGGAGA-3′; GAPDH forward,
5′-TCGTGGAAGGACTCATGACC-3′ and reverse, 5′-ATGATGTTCTGGAGAGCCCC-3′;
and U6 forward, 5′-CTCGCTTCGGCAGCACATATACTA-3′ and reverse,
5′-ACGAATTTGCGTGTCATCCTTGCG-3′; GM-CSF forward,
5′-ATGTGGCTGCAGAGCCTGCTGC-3′ and reverse, 5′-CTCCCAGCAGTCAAAGGG-3′;
eotaxin forward, 5′-CCCCTTCAGCGACTAGAGAG-3′ and reverse,
5′-TCTTGGGGTCGGCACAGAT-3′; MUC5AC forward, 5′-TGATCATCCAGCAGGGCT-3′
and reverse, 5′-CCGAGCTCAGAGGACATATGGG-3′.
Western blot analysis
NECs were lysed with pre-cooled RIPA lysis buffer
(Beyotime Institute of Biotechnology) and quantified using a BCA
Protein Assay kit (Beyotime Institute of Biotechnology). A total of
20 µg protein/lane was added into each well of a vertical
electrophoresis tank and separated by SDS-PAGE on 10% gels.
Subsequently, the proteins were transferred to PVDF membranes.
After blocking with 5% skim-milk for 1 h at room temperature, the
membranes were incubated with primary antibodies specific for KLF4
(1:1,000; Novous; cat. no. IMG-6081A) and GAPDH antibody (1:500;
Santa Cruz Biotechnology, Inc.; cat. no. sc-47724) at 4°C
overnight. The membranes were washed three times with 0.1%
TBS-Tween-20 (each time 5 min), then incubated with goat
anti-rabbit/mouse HRP-conjugated secondary antibodies (1:5,000;
Abcam; cat. nos. ab205718 and ab205719) at room temperature for 1
h. Protein bands were visualized using the Enhanced
Chemiluminescence Plus kit (Thermo Fisher Scientific, Inc.),
according to the manufacturer's instructions. Protein bands were
quantified using Quantity One software (version 4.6.6; Bio-Rad
Laboratories, Inc.).
TUNEL assay
The TUNEL assay was performed using an In Situ Cell
Death Detection kit (Roche Diagnostics GmbH) according to the
manufacturer's instructions. After rising twice by PBS, NECs cells
fixed by 4% formaldehyde for 25 min at 4°C. The cells were
permeabilized by 0.2% Triton X-100 for 5 min. The cells were then
equilibrated with 100 µl Equilibration buffer for 10 min at room
temperature. Subsequently, cells were labeled with 50 µl terminal
deoxynucleotidyl transferase (TdT) reaction cocktail for 45 min at
37°C, followed by treatment with Click-iT reaction cocktail. The
nucleus was stained with hematoxylin or methyl green at room
temperature for 1 h. A fluorescence microscope (magnification,
×100; Olympus Corporation) was utilized to observe TUNEL-positive
cells in at least 5 fields of view.
Statistical analysis
Statistical analyses were performed using SPSS 23.0
software (IBM Corp.). Data are presented as the mean ± SD from at
least three independent experiments. Student's t-test or one-way
ANOVA followed by Tukey's post hoc test were used for comparisons
between groups. Correlation analysis was carried out using
Pearson's correlation coefficient. P<0.05 was considered to
indicate a statistically significant difference.
Results
circARRDC3 regulates IL-13-induced
inflammatory cytokines, mucus production, cell viability and
apoptosis of NECs
To determine the effects of circARRDC3 on AR
development, si-circARRDC3 or pcDNA3.1-circARRDC3 were transfected
into NECs to inhibit or overexpress circARRDC3 (Fig. 1A). As shown in Fig. 1B-D, knockdown of circARRDC3
downregulated the levels of GM-CSF, eotaxin and MUC5AC, whereas
circARRDC3 overexpression upregulated the levels of GM-CSF, eotaxin
and MUC5AC. In addition, circARRDC3 interference significantly
promoted the viability of IL-13-treated NECs, whereas circARRDC3
overexpression significantly repressed cell viability (Fig. 1E). Furthermore, knockdown of
circARRDC3 significantly suppressed the apoptosis of IL-13-treated
NECs, and the addition of circARRDC3 significantly promoted cell
apoptosis (Fig. 1F). Collectively,
the data indicated that circARRDC3 may contribute to AR
development.
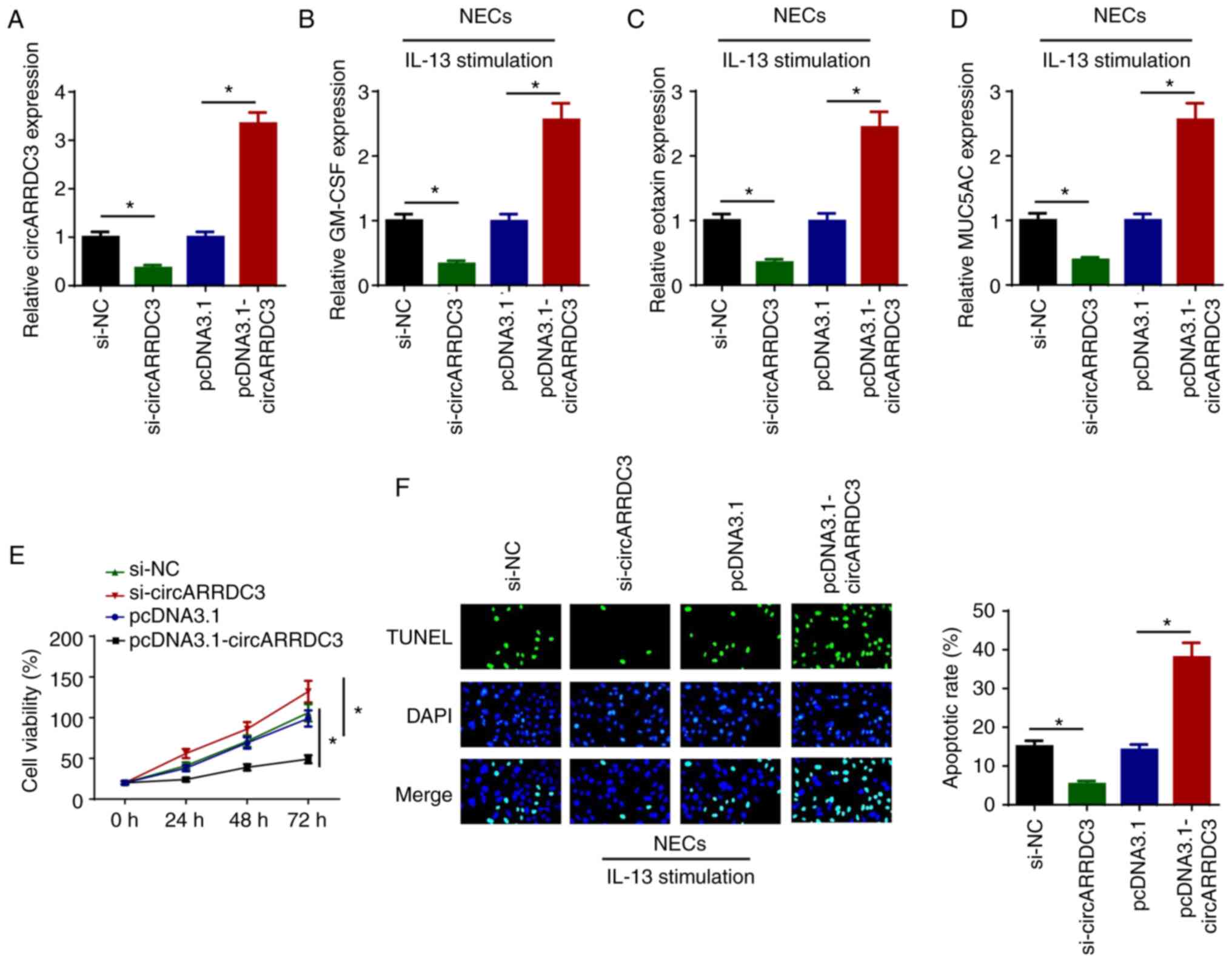 | Figure 1.circARRDC3 regulates IL-13-induced
inflammatory cytokines, mucus production, cell viability and
apoptosis of NECs. (A) circARRDC3 expression levels were detected
by RT-qPCR in NECs transfected with si-NC, si-circARRDC3, pcDNA3.1
and pcDNA3.1-circARRDC3. The levels of (B) GM-CSF, (C) eotaxin and
(D) MUC5AC were detected by RT-qPCR in IL-13-induced NECs
transfected with si-NC, si-circARRDC3, pcDNA3.1 and
pcDNA3.1-circARRDC3. (E) Cell viability was examined by an MTT
assay in IL-13-induced NECs transfected with si-NC, si-circARRDC3,
pcDNA3.1 and pcDNA3.1-circARRDC3. (F) Cell apoptosis was examined
using a TUNEL assay in IL-13-induced NECs transfected with si-NC,
si-circARRDC3, pcDNA3.1 and pcDNA3.1-circARRDC3. Magnification,
×100. *P<0.05. ARRDC3, arrestin domain-containing 3; circ-,
circular RNA; NEC, nasal epithelial cell; RT-qPCR, reverse
transcription-quantitative PCR; si-, small interfering RNA; NC,
negative control; GM-CSF, granulocyte-macrophage colony-stimulating
factor; MUC5AC, mucin 5AC. |
miR-375 interacts with circARRDC3
Based on bioinformatics analysis (StarBase 2.0),
miR-375 was predicted to be a downstream target of circARRDC3
(Fig. 2A). RT-qPCR indicated that
the expression of miR-375 was upregulated in NECs transfected with
miR-375 mimic and was downregulated in NECs transfected with
miR-375 inhibitor (Fig. 2B). As
shown in Fig. 2C, transfection with
the miR-375 mimic decreased and miR-375 inhibitor increased the
luciferase activity of circARRDC3-WT, while no significant
difference was observed in the circARRDC3-Mut group. To determine
whether miR-375 was critical for circARRDC3-mediated AR
development, si-NC, si-circARRDC3 and si-circARRDC3 + miR-375
inhibitor were transfected into IL-13-treated NECs. RT-qPCR
revealed that knockdown of circARRDC3 significantly upregulated
miR-375 expression, which was reversed following miR-375 inhibitor
transfection (Fig. 2D). Moreover,
transfection with the miR-375 inhibitor reversed the inhibitory
effects of circARRDC3 knockdown on the levels of GM-CSF, eotaxin
and MUC5AC in IL-13-induced NECs (Fig.
2E-G). Further results indicated that circARRDC3 knockdown
promoted the viability of IL-13-induced NECs, whereas the effects
were reversed by miR-375 inhibition (Fig. 2H). Furthermore, the depletion of
circARRDC3 suppressed the apoptosis of IL-13-induced NECs, which
was abolished following miR-375 inhibitor transfection (Fig. 2I). In summary, these results
revealed that circARRDC3 regulated AR progression by targeting
miR-375 in IL-13-induced NECs.
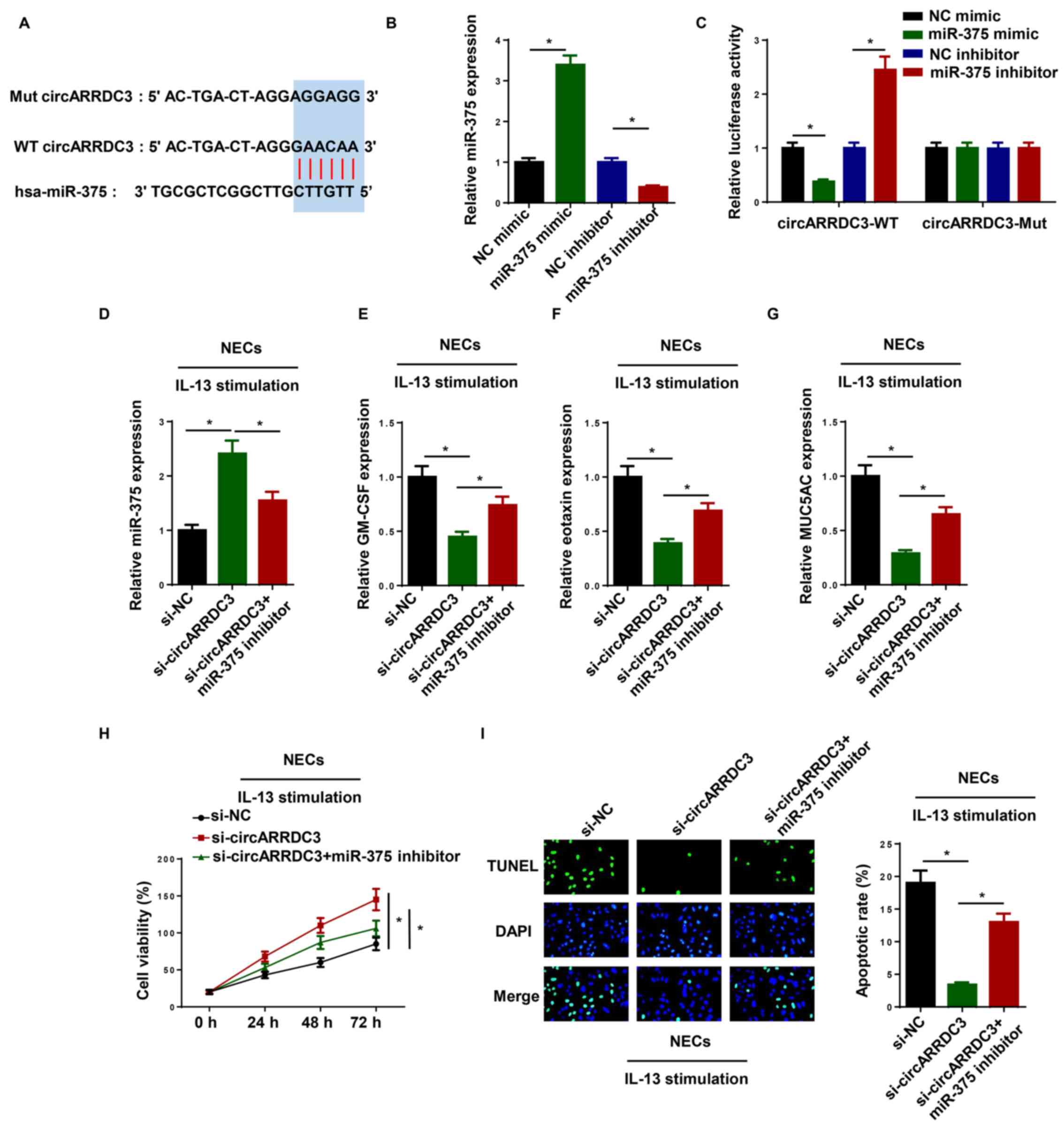 | Figure 2.miR-375 interacts with circARRDC3.
(A) Bioinformatics prediction of putative binding site at
circARRDC3 by miR-375. (B) RT-qPCR was performed to determine the
expression of miR-375 in NECs transfected with miR-375 mimic or
miR-375 inhibitor. (C) Dual luciferase reporter assay showed
luciferase activity in WT circARRDC3-containing NECs transfected
with NC mimic, miR-375 mimic, NC inhibitor and miR-375 inhibitor.
(D) miR-375 expression levels were detected by RT-qPCR in
IL-13-induced NECs transfected with si-NC, si-circARRDC3, and
si-circARRDC3 + miR-375 inhibitor. The levels of (E) GM-CSF, (F)
eotaxin (G) and MUC5AC were detected by RT-qPCR in IL-13-induced
NECs transfected with si-NC, si-circARRDC3, and si-circARRDC3 +
miR-375 inhibitor. (H) Cell viability was examined by an MTT assay
in IL-13-induced NECs transfected with si-NC, si-circARRDC3, and
si-circARRDC3 + miR-375 inhibitor. (I) Cell apoptosis was examined
using a TUNEL assay in IL-13-induced NECs transfected with si-NC,
si-circARRDC3, si-circARRDC3 + miR-375 inhibitor. Magnification,
×100. *P<0.05. ARRDC3, arrestin domain-containing 3; circ-,
circular RNA; NEC, nasal epithelial cell; RT-qPCR, reverse
transcription-quantitative PCR; si-, small interfering RNA; NC,
negative control; GM-CSF, granulocyte-macrophage colony-stimulating
factor; MUC5AC, mucin 5AC; miR, microRNA; WT, wild-type; Mut,
mutant. |
Overexpression of KLF4 reverses the
miR-375-mediated inhibitory effects on AR development
To investigate the potential effects of miR-375 in
NECs, bioinformatics tools (TargetScan) were used to predict the
possible targets. It was found that the 3′-UTR of KLF4 had direct
binding sites for miR-375 (Fig.
3A). In addition, the luciferase reporter assay indicated that
transfection with the miR-375 mimic decreased and the miR-375
inhibitor increased the luciferase activity of KLF4-WT, while there
were no changes observed in the KLF4-Mut group (Fig. 3B). Moreover, RT-qPCR and western
blotting revealed that miR-375 upregulation decreased KLF4
expression and miR-375 downregulation increased KLF4 expression
(Fig. 3C and D). Collectively, the
results suggested that miR-375 could directly interact with KLF4
and significantly inhibit its expression.
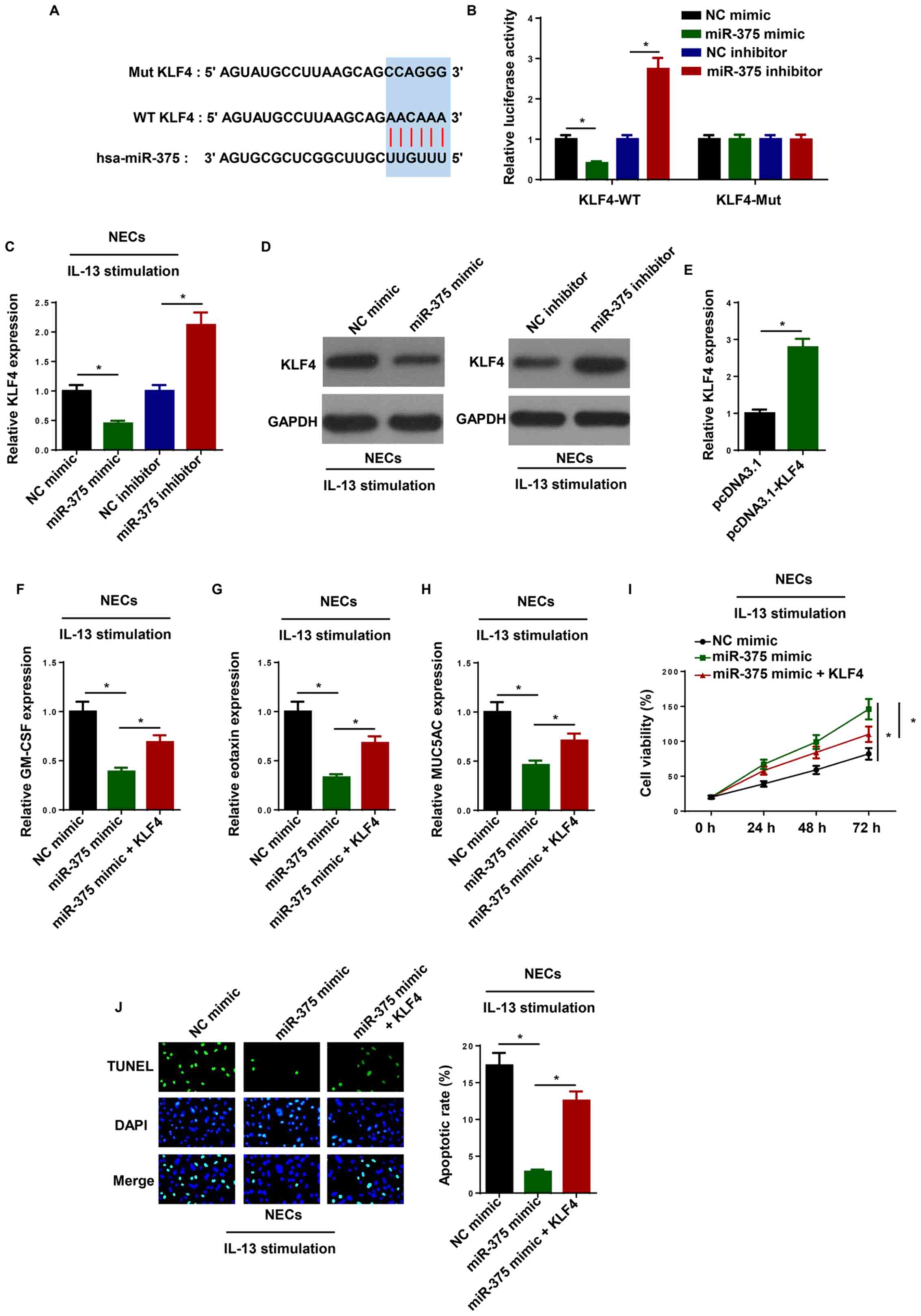 | Figure 3.Overexpression of KLF4 reverses
miR-375-mediated inhibitory effects on AR development. (A)
Bioinformatics prediction of putative binding site at 3’-UTR of
KLF4 by miR-375. (B) Dual luciferase reporter assay showed
luciferase activity in WT KLF4-containing NECs transfected with NC
mimic, miR-375 mimic, NC inhibitor and miR-375 inhibitor. The
levels of KLF4 were detected by (C) RT-qPCR and (D) western
blotting in IL-13-induced NECs transfected with NC mimic, miR-375
mimic, NC inhibitor and miR-375 inhibitor. (E) RT-qPCR was
performed to determine KLF4 expression in NECs transfected with
pcDNA3.1 and pcDNA3.1-KLF4. The levels of (F) GM-CSF, (G) eotaxin
and (H) MUC5AC were detected by RT-qPCR in IL-13-induced NECs
transfected with NC mimic, miR-375 mimic, and miR-375 mimic + KLF4.
(I) Cell viability was examined by an MTT assay in IL-13-induced
NECs transfected with NC mimic, miR-375 mimic, and miR-375 mimic +
KLF4. (J) Cell apoptosis was examined using a TUNEL assay in
IL-13-induced NECs transfected with NC mimic, miR-375 mimic, and
miR-375 mimic + KLF4. Magnification, ×100. *P<0.05. NEC, nasal
epithelial cell; RT-qPCR, reverse transcription-quantitative PCR;
NC, negative control; GM-CSF, granulocyte-macrophage
colony-stimulating factor; MUC5AC, mucin 5AC; miR, microRNA; WT,
wild-type; Mut, mutant; KLF4, krueppel-like factor 4; UTR,
untranslated region. |
To further assess the roles of KLF4 and miR-375 in
AR, NC mimic, miR-375 mimic, and miR-375 mimic + KLF4 were
transfected into IL-13-induced NECs. RT-qPCR showed the KLF4
expression was significantly increased in NECs transfected with
pcDNA3.1-KLF4 (Fig. 3E). As shown
in Fig. 3F-H, KLF4 overexpression
reversed the suppressive effects of miR-375 mimic transfection on
the expression levels of GM-CSF, eotaxin and MUC5AC. In addition,
the upregulation of miR-375 promoted cell viability in
IL-13-induced NECs, while these effects were abolished by KLF4
overexpression (Fig. 3I).
Furthermore, miR-375 mimic inhibited apoptosis of IL-13-induced
NECs, which was partially reversed following KLF4 overexpression
(Fig. 3J). The results indicated
that miR-375 expression negatively regulated KLF4 expression by
directly interacting with each other in IL-13-induced NECs.
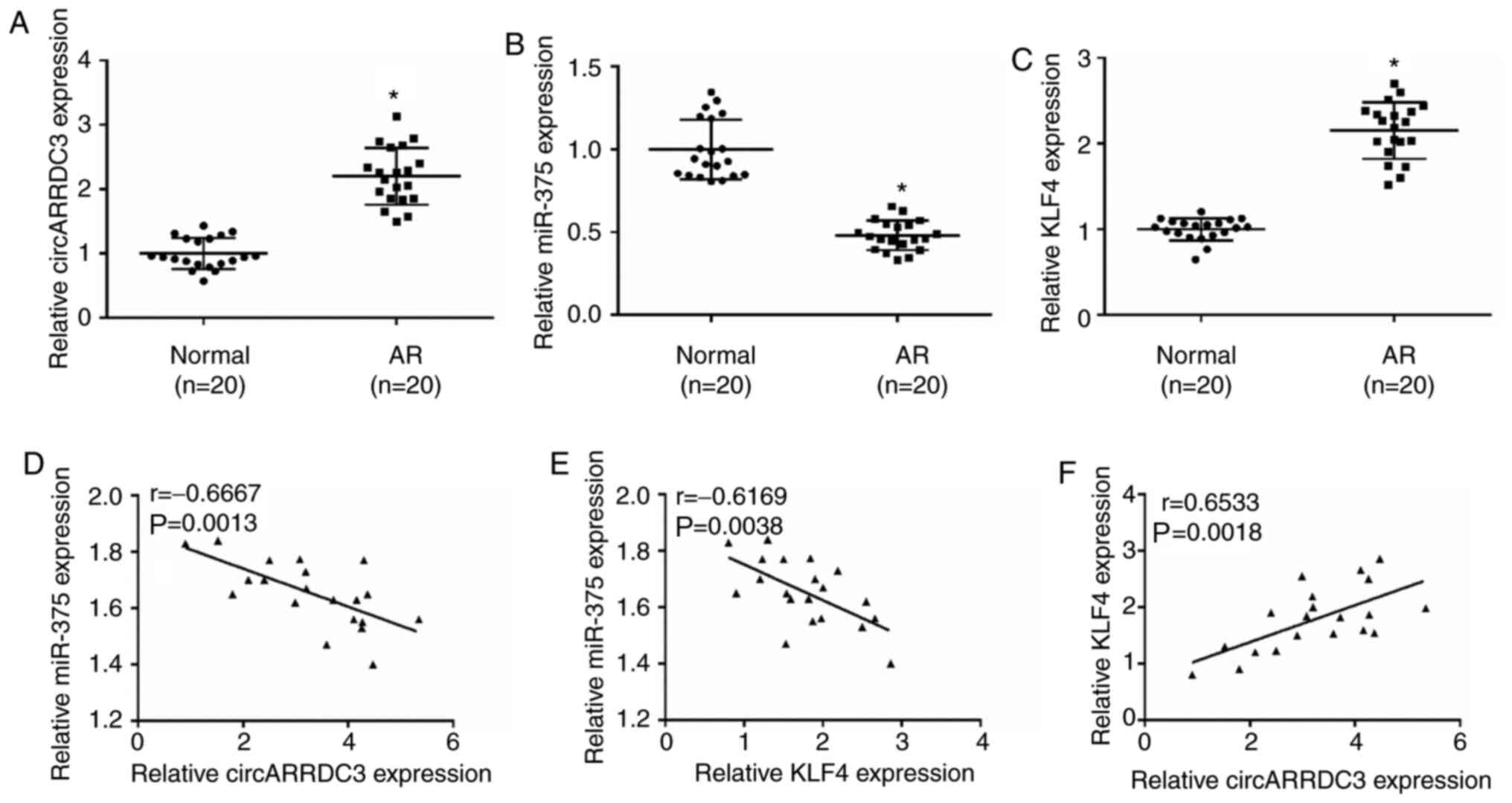
circARRDC3, miR-375 and KLF4 are
dysregulated in AR tissues
RT-qPCR showed that circARRDC3 and KLF4 were
increased, and miR-375 was decreased in AR tissues (Fig. 4A-C). Furthermore, miR-375 expression
was negatively correlated with circARRDC3 or KLF4 expression.
Whereas, circARRDC3 expression was positively correlated with KLF4
expression in AR tissues (Fig.
4D-F). To summarize, circARRDC3, miR-375 and KLF4 were
dysregulated in the clinical nasal mucosa of patients with AR.
Discussion
Increasing evidence has indicated that circRNAs are
involved in inflammatory activities of several diseases (23,24).
For example, Qin et al (25)
indicated that circRNA-9119 suppressed polyriboinosinic
polyribocytidylic acid-induced inflammation in Leydig and Sertoli
cells via toll-like receptor 3 and antiviral innate immune response
receptor RIG-I signalling pathways. Yang et al (26) reported that circ_0000950 promoted
neuronal apoptosis and increased inflammatory cytokine levels in
Alzheimer's disease. To the best of our knowledge, no published
studies have investigated the association between circARRDC3 and
AR. The present study observed that circARRDC3 knockdown
significantly suppressed levels of GM-CSF, eotaxin and MUC5AC, and
promoted cell viability in IL-13-induced NECs, suggesting that
circARRDC3 promoted AR development.
Recent studies have indicated that circRNAs could
serve as miRNA sponges to suppress miRNA expression, and thereby
suppress miRNA function in inflammatory disease. For example, Ye
et al (27) reported that
dysregulation of circRNA_103516 participated in the molecular
mechanism of inflammatory bowel disease by regulating miR-19b-1-5p.
Zhang et al (28)
demonstrated that circARF3 acted as an endogenous miR-103 sponge to
inhibit mitophagy-mediated adipose inflammation both in
vitro and in vivo. Xue et al (29) indicated that circ_0000638 suppressed
neodymium oxide-induced bronchial epithelial cell inflammation via
the miR-498-5p/NF-κB axis. The present study found that miR-375 is
a downstream target of circARRDC3. Knockdown of circARRDC3
suppressed inflammatory cytokines, mucus production and cell
apoptosis in IL-13-induced NECs, which was reversed following
miR-375 inhibitor transfection. In addition, the effects of
circARRDC3 knockdown on promoting the viability of IL-13-induced
NECs were abolished by decreasing miR-375 expression. These results
indicated that circARRDC3 regulated the development of AR via
targeting miR-375.
Our previous study showed that miR-375 was
significantly downregulated in AR mice and significantly
ameliorated AR via the JAK2/STAT3 pathway (30). Considering the comprehensive
mechanisms underlying AR progression, we further investigated
several AR-related genes, including KLF4. KLF4 is a transcription
factor required for epithelial-mesenchymal transition and
establishes the skin barrier (31,32).
Moreover, Kurotaki et al (33) revealed that KLF4 is essential for
murine monocyte differentiation. Using bioinformatics analysis, Liu
et al (34) found that KLF4
was significantly upregulated in patients with seasonal allergic
rhinitis. Mao et al (35)
found a direct association between miR-375 and KLF4, which prompted
us to assess the assosciation between miR-375 and KLF4 and their
potential effects in AR progression and development. The present
study found that miR-375 could directly bind to KLF4 in AR tissues.
In addition, KLF4 overexpression abolished the suppressive effects
of miR-375 mimic on GM-CSF, eotaxin and MUC5AC expression levels
and apotosis in IL-13-induced NECs. Moreover, KLF4 overexpression
reversed the miR-375 mimic-induced effects on cell viability. To
the best of our knowledge, the present study was the first to
demonstrate that circARRDC3 promoted IL-13-induced inflammatory
cytokine and mucus production in NECs via the miR-375/KLF4 axis. In
addition, miR-375 was negatively correlated with circARRDC3 and
KLF4 expression in AR tissues, whereas circARRDC3 expression was
positively correlated with KLF4 expression.
In conclusion, the present study demonstrated that
circARRDC3 promoted AR progression by regulating the miR-375/KLF4
axis, providing a potential therapeutic target for the treatment of
AR. This study primarily focused on the role of circARRDC3 in
IL-13-induced NECs in vitro. Therefore, the function of
circARRDC3 in the primary tissues of patients with AR and in animal
experiments will be investigated in the future.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author upon reasonable
request.
Authors' contributions
TW and PW conceived and designed the study. DC, ZX,
and LY contributed to the acquisition, analysis, and interpretation
of data. TW and PW drafted and revised the manuscript. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
The present study was approved by the ethics
committee of the Shanghai Ninth People's Hospital (Shanghai,
China). Written informed consent was obtained from all
patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Ozdoganoglu T and Songu M: The burden of
allergic rhinitis and asthma. Ther Adv Respir Dis. 6:11–23. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bauchau V and Durham SR: Epidemiological
characterization of the intermittent and persistent types of
allergic rhinitis. Allergy. 60:350–353. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leynadier F, Mees K, Arendt C and Pinelli
ME: Efficacy and safety of levocetirizine in seasonal allergic
rhinitis. Acta Otorhinolaryngol Belg. 55:305–312. 2001.PubMed/NCBI
|
|
4
|
Wang DY: Risk factors of allergic
rhinitis: Genetic or environmental? Ther Clin Risk Manag.
1:115–123. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Liu J and Cao Z: Protective Effect of
Circular RNA (CircRNA) Ddx17 in Ovalbumin (OVA)-Induced Allergic
Rhinitis (AR) Mice. Med Sci Monit. 26:e9190832020.PubMed/NCBI
|
|
7
|
Zhu X, Wang X, Wang Y and Zhao Y: The
regulatory network among CircHIPK3, LncGAS5, and miR-495 promotes
Th2 differentiation in allergic rhinitis. Cell Death Dis.
11:2162020. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Yue L, Yin X, Hao F, Dong J, Ren X, Xu O
and Shan C: Long noncoding RNA Linc00632 inhibits
interleukin-13-induced inflammatory cytokine and mucus production
in nasal epithelial cells. J Innate Immun. 12:116–128. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Shin IS, Lee MY, Jeon WY, Shin NR, Seo CS
and Ha H: EBM84 attenuates airway inflammation and mucus
hypersecretion in an ovalbumin-induced murine model of asthma. Int
J Mol Med. 31:982–988. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Wang L, Lv Q, Song X, Jiang K and Zhang J:
ADRB2 suppresses IL-13-induced allergic rhinitis inflammatory
cytokine regulated by miR-15a-5p. Hum Cell. 32:306–315. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Carleton M, Cleary MA and Linsley PS:
MicroRNAs and cell cycle regulation. Cell Cycle. 6:2127–2132. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Chen S, Liu Y, Wang Y and Xue Z: LncRNA
CCAT1 promotes colorectal cancer yumorigenesis via a
miR-181b-5p/TUSC3 axis. OncoTargets Ther. 12:9215–9225. 2019.
View Article : Google Scholar
|
|
13
|
Teng Y, Zhang R, Liu C, Zhou L, Wang H,
Zhuang W, Huang Y and Hong Z: miR-143 inhibits
interleukin-13-induced inflammatory cytokine and mucus production
in nasal epithelial cells from allergic rhinitis patients by
targeting IL13Rα1. Biochem Biophys Res Commun. 457:58–64. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Liu HC, Liao Y and Liu CQ: miR-487b
mitigates allergic rhinitis through inhibition of the IL-33/ST2
signaling pathway. Eur Rev Med Pharmacol Sci. 22:8076–8083.
2018.PubMed/NCBI
|
|
15
|
Chen RF, Huang HC, Ou CY, Hsu TY, Chuang
H, Chang JC, Wang L, Kuo HC and Yang KD: MicroRNA-21 expression in
neonatal blood associated with antenatal immunoglobulin E
production and development of allergic rhinitis. Clin Exp Allergy.
40:1482–1490. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Dissanayake E and Inoue Y: MicroRNAs in
allergic disease. Curr Allergy Asthma Rep. 16:672016. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Ingram JL and Kraft M: IL-13 in asthma and
allergic disease: Asthma phenotypes and targeted therapies. J
Allergy Clin Immunol. 130:829–842; quiz 843–844. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lv J, Xiong Y, Li W, Cui X, Cheng X, Leng
Q and He R: IL-37 inhibits IL-4/IL-13-induced CCL11 production and
lung eosinophilia in murine allergic asthma. Allergy. 73:1642–1652.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Ashraf MI, Shahzad M and Shabbir A:
Oxyresveratrol ameliorates allergic airway inflammation via
attenuation of IL-4, IL-5, and IL-13 expression levels. Cytokine.
76:375–381. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhu X, Wang X, Wang Y and Zhao Y: Exosomal
long non-coding RNA GAS5 suppresses Th1 differentiation and
promotes Th2 differentiation via downregulating EZH2 and T-bet in
allergic rhinitis. Mol Immunol. 118:30–39. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Agarwal V, Bell GW, Nam JW and Bartel DP:
Predicting effective microRNA target sites in mammalian mRNAs.
eLife. 4:42015. View Article : Google Scholar
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hwang JY, Kim EB, Ka H and Lee CK:
Identification of the porcine XIST gene and its differential CpG
methylation status in male and female pig cells. PLoS One.
8:e736772013. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shenoda BB, Tian Y, Alexander GM,
Aradillas-Lopez E, Schwartzman RJ and Ajit SK: miR-34a-mediated
regulation of XIST in female cells under inflammation. J Pain Res.
11:935–945. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Qin L, Lin J and Xie X: CircRNA-9119
suppresses poly I:C induced inflammation in Leydig and Sertoli
cells via TLR3 and RIG-I signal pathways. Mol Med. 25:282019.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Yang H, Wang H, Shang H, Chen X, Yang S,
Qu Y, Ding J and Li X: Circular RNA circ_0000950 promotes neuron
apoptosis, suppresses neurite outgrowth and elevates inflammatory
cytokines levels via directly sponging miR-103 in Alzheimer's
disease. Cell Cycle. 18:2197–2214. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Ye YL, Yin J, Hu T, Zhang LP, Wu LY and
Pang Z: Increased circulating circular RNA_103516 is a novel
biomarker for inflammatory bowel disease in adult patients. World J
Gastroenterol. 25:6273–6288. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang Z, Zhang T, Feng R, Huang H, Xia T
and Sun C: circARF3 alleviates mitophagy-mediated inflammation by
targeting miR-103/TRAF3 in mouse adipose tissue. Mol Ther Nucleic
Acids. 14:192–203. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Xue H, Yu F, Zhang X, Liu L and Huang L:
circ_0000638 inhibits neodymium oxide-induced bronchial epithelial
cell inflammation through the miR-498-5p/NF-κB axis. Ecotoxicol
Environ Saf. 195:1104552020. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Wang T, Chen D, Wang P, Xu Z and Li Y:
miR-375 prevents nasal mucosa cells from apoptosis and ameliorates
allergic rhinitis via inhibiting JAK2/STAT3 pathway. Biomed
Pharmacother. 103:621–627. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Segre JA, Bauer C and Fuchs E: Klf4 is a
transcription factor required for establishing the barrier function
of the skin. Nat Genet. 22:356–360. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Tiwari N, Meyer-Schaller N, Arnold P,
Antoniadis H, Pachkov M, van Nimwegen E and Christofori G: Klf4 is
a transcriptional regulator of genes critical for EMT, including
Jnk1 (Mapk8). PLoS One. 8:e573292013. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Kurotaki D, Osato N, Nishiyama A, Yamamoto
M, Ban T, Sato H, Nakabayashi J, Umehara M, Miyake N, Matsumoto N,
et al: Essential role of the IRF8-KLF4 transcription factor cascade
in murine monocyte differentiation. Blood. 121:1839–1849. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Liu Y, Shi J and Chen X: Identification of
novel targets for seasonal allergic rhinitis during and outside the
pollen season by microarray analysis. Acta Otolaryngol.
135:1330–1336. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Mao Q, Quan T, Luo B, Guo X, Liu L and
Zheng Q: MiR-375 targets KLF4 and impacts the proliferation of
colorectal carcinoma. Tumour Biol. 37:463–471. 2016. View Article : Google Scholar : PubMed/NCBI
|