Introduction
Gastric carcinoma (GC) is a malignant tumor
originating from the gastric mucosa (1). The incidence rate of GC is second only
to lung carcinoma and is significantly higher in men than in women
(2). Due to the GC tumor invasion
and metastasis, GC affects the liver, kidney and respiratory
functions and endangers life (3).
The majority of patients with early GC generally have no distinct
symptoms. However, with the growth of tumor, obvious symptoms
appear as the gastric function is affected (4). In clinical practice, surgical
treatment, chemotherapy, traditional Chinese medicine treatment and
other comprehensive therapies are used to ameliorate the symptoms
of patients with GC and prolong the survival (5,6).
Nevertheless, the prognosis of patients with GC in advanced-stage
is still unsatisfactory. Thus, it is important to investigated a
tumor marker and develop a new curative method for GC.
Long non-coding (lnc)RNAs are transcripts >200
nucleotides in length (7).
Increasing evidence has uncovered significant functions of lncRNAs
in various types of cancer (8,9).
Notably, a number of GC-associated lncRNAs such as H19 (10), HOXA11-AS (11) and AK058003 (12) have been reported to modulate the
proliferative, migratory and invasive capacities of GC cells. The
cancer-related lncRNA LIFR-AS1 has been studied in breast (13,14)
and colorectal cancer (15). These
studies have shown that lncRNA LIFR-AS1 is linked to the high
survival rate of breast cancer and can inhibit breast cancer cell
proliferation and migration; in addition, it can regulate the
resistance of colorectal cancer to photodynamic therapy (14,15).
However, the functions of lncRNA LIFR-AS1 in GC remain to be
elucidated.
Micro (mi)RNAs are encoded by endogenous genes with
a length of ~22 nucleotides and possess multiple vital regulating
effects in a variety of cells (16,17).
Accumulating evidence has established that lncRNAs work as miRNAs
sponge to serve functions in the progression of various types of
cancer, including GC (18,19). miR-4698 is situated on chromosome
12, which is linked to metastatic melanoma (20). Notably, Kalhori et al
(21) revealed that miR-4698 can
stop glioblastoma cell proliferation by controlling the PI3K/AKT
pathway (21). Whether miR-4698
takes part in mediating the functions of lncRNA LIFR-AS1 in GC
cells is the focus of the present study.
Microtubule-associated tumor suppressor 1 (MTUS1) is
a tumor suppressor gene at chromosome 8p21.3–22, encoding a
mitochondrial protein and controlling cellular proliferation
(22). Previous studies have
demonstrated that the expression status of MTUS1 is altered in
several types of tumors, such as fibroadenoma and breast cancer
(23), colorectal carcinoma
(24) and bladder cancer (25). Thus far, no study has investigated
whether MTUS1 participates in mediating the functions of lncRNA
LIFR-AS1 in GC cells.
According to the above research, the present study
preliminarily investigated the functions of lncRNA LIFR-AS1 in GC
cell proliferation, migration and invasion and determined the
molecular mechanisms involved. The results from the present study
might offer a new research orientation for GC treatment from the
aspect of lncRNA, miRNA and mRNA axis.
Materials and methods
Collection of samples from patients
with GC
The GC tissues and paired adjacent tissues needed
for the experiments were collected from 41 patients with GC
(Table I). No treatment was
performed on these patients prior to surgery. Patients were
excluded if they had other clinical disorders. The obtained samples
were frozen in liquid nitrogen and stored at −80°C. All patients
personally signed an informed consent. The protocol of the present
study was approved by the Ethics Committee of of Cangzhou People's
Hospital (approval number AF/SC-08/02.0).
 | Table I.Correlation between LRFR-AS1
expression and clinical characteristics. |
Table I.
Correlation between LRFR-AS1
expression and clinical characteristics.
| Variable | N (%) | Relative LRFR-AS1
expression (2−ΔΔCq method) | P-value |
|---|
| Age (year) | 59.2±10.5 | / |
|
| Sex |
|
| 0.2584 |
|
Male | 27 (65.8%) | 0.466±0.185 |
|
|
Female | 14 (34.2%) | 0.541±0.223 |
|
| Histologic
type |
|
| 0.2711 |
|
Adenocarcinoma | 35 (85.4%) | 0.477±0.187 |
|
|
Others | 6 (14.6%) | 0.575±0.264 |
|
| Differentiation
degree |
|
| 0.0383a |
|
Well/Moderately | 18 (43.9%) | 0.564±0.209 |
|
|
Poorly | 23 (56.1%) | 0.435±0.176 |
|
| Depth of
invasion |
|
| 0.1190 |
| T1 and
T2 | 16 (39.0%) | 0.552±0.215 |
|
| T3 and
T4 | 25 (61.0%) | 0.452±0.182 |
|
| Nodal status |
|
| 0.0045b |
|
pN0 | 18 (43.9%) | 0.588±0.184 |
|
|
pN1-3 | 23 (56.1%) | 0.416±0.180 |
|
| TNM stage |
|
| 0.0011b |
| I,
II | 26 (63.4%) | 0.565±0.187 |
|
| III,
IV | 15 (36.6%) | 0.364±0.153 |
|
Cell culture
GC cell lines (MKN45 and AGS) and the gastric
mucosal epithelial cell line (GES-1) were obtained from the Cell
Bank of the Chinese Academy of Sciences. RPMI-1640 medium (Gibco;
Thermo Fisher Scientific, Inc.) containing 10% fetal bovine serum
(FBS; Invitrogen; Thermo Fisher Scientific, Inc.) was used to
culture these cells. The culture conditions were 5% CO2
and 95% air at 37°C.
Cell transfection
The vectors of pcDNA-LIFR-AST and paired pcDNA
negative control (NC), as well as miR-4698 mimics, miR-4698
inhibitors and their corresponding controls (mimics NC and
inhibitors NC) used in this study were synthesized by GenePharma.
The small interfering (si)RNA targeting microtubule-associated
tumor suppressor 1 (MTUS1) was constructed in U6/GFP/Neo vectors
(GenePharma) to silence MTUS1 expression. A scrambled siRNA was
included as NC. The MKN45 and AGS cells were pre-incubated on
6-well plates until they reached ~60% confluence and the
transfection was conducted with Lipofectamine® 3000
(Invitrogen; Thermo Fisher Scientific, Inc.). A total of 50 nM
pcDNA-LIFR-AST or pcDNA NC, 20 nM miR-4698 mimics or mimics NC, 20
nM miR-4698 inhibitors or inhibitors NC and 45 nM si-MTUS1 or si-NC
were used. Cells were harvested 48 h post-transfection for further
studies.
Detection of cell proliferation
When cell transfection was concluded, Cell Counting
Kit-8 (CCK-8) and 5-ethynyl-2′- deoxyuridine (EdU) incorporation
assays (Abcam) were used to assess cell proliferation. Transfected
MKN45 and AGS cells (5×103 cells/well) were cultivated
at 37°C for 1, 2, 3 and 4 days. CCK-8 solution (10 µl) was added
into each well for another 2 h incubation. A microplate reader
(BioTek Instruments, Inc.) was used for examining the absorbance at
450 nm wavelength. For the EdU assay, transfected cells were grown
in 24-well plates at 37°C. The cells were then stained with 40 µM
EdU solution at 37°C for 2 h and then 4% formaldehyde and 10X
Triton X-100 buffer (Sigma-Aldrich; Merck KGaA) were added at room
temperature for 15 min incubation. The reaction mix was added to
fluorescently labelled EdU and was incubated at 37°C for 30 min.
Finally, the EdU positive cells were analyzed by a fluorescence
microscope (Olympus Corporation, magnification, ×100). Images were
captured in five random fields.
Wound healing assay
The transfected MKN45 and AGS cells were cultured in
6-well plates to produce a cell monolayer (100% confluence). The
cell wounds were created using a 100 µl pipette tip. After washing
three times with sterile PBS, cells were further cultured at 37°C
for 24 h and then the width of scratch was observed with an
inverted microscope (Olympus Corporation, magnification, ×100).
Additionally, images were captured at 0 and 24 h after scratching.
ImageJ software (v1.8.0.112, National Institutes of Health) was
used to analyze the images. Images were captured in five random
fields.
Cell invasion
The transfected MKN45 and AGS cells in 200 µl
serum-free RPMI-1640 medium were added to the upper Transwell
chambers (Corning Inc.; 24-well insert, pore size 8 mm). The
Transwell membrane was pre-coated with Matrigel (BD Biosciences).
Meanwhile, the bottom Transwell chambers were filled with 600 µl
RPMI-1640 medium supplemented with 10% FBS. After 24 h incubation
at 37°C, the cells were fixed and subsequently stained with 0.5%
crystal violet solution (Sigma-Aldrich; Merck KGaA) for 15 min at
room temperature. The invasive cells were counted under a light
microscope (Olympus Corporation, magnification, ×100). ImageJ
software (v1.8.0.112, National Institutes of Health) was used to
analyze the images. Images were captured in five random fields.
Luciferase reporter assay
The lncRNA LIFR-AS1 fragment or the 3′-untranslated
region (UTR) of MTUS1 was amplified by performing PCR. Then, the
pmirGLO luciferase vector (Promega Corporation) was used to
establish LIFR-AS1 wild type (WT) and MTUS1-WT. A site-directed
mutagenesis kit (Thermo Fisher Scientific, Inc.) was employed to
generate LIFR-AS1 mutant (MUT) or MTUS1-MUT. All the above
constructed vectors and miR-4698 mimics (500 ng) were
co-transfected into MKN45 and AGS cells using
Lipofectamine® 3000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h transfection, the relative luciferase
activity was assessed using a dual luciferase reporter assay system
(Promega Corporation). Renilla luciferase activity was used
for normalization.
Reverse transcription-quantitative
(RT-q) PCR
Total RNA was isolated from GC tissues and cell
lines (5×106 cells/ml) using TRIzol® reagent
(Thermo Fisher Scientific, Inc.) according to the manufacturer's
protocols. A Reverse Transcription kit (Promega Corporation) was
used to synthesize cDNA according to the manufacturer's protocols.
SYBR® Green PCR Kit (Qiagen, Inc.) was used to observe
lncRNA LIFR-AS1 and MTUS1 expression. All reactions were performed
in a 10 µl reaction volume in triplicate. PCR were with conditions
of 95°C for 50 sec, followed by 40 cycles of 95°C for 12 sec and
55.5°C for 30 sec. For detecting miR-4698 expression, TaqMan
Universal Master Mix II (Takara Biotechnology Co., Ltd.) was
employed for RT-qPCR procedure. β-actin and U6 served as internal
control for standardization of the data. All the data were analyzed
by the 2−ΔΔCq method (26). Primer sequences are listed in
Table II.
 | Table II.Primers used for reverse
transcription-quantitative PCR. |
Table II.
Primers used for reverse
transcription-quantitative PCR.
| Primer name | Sequence |
|---|
| LIFR-AS1 | Forward
5′-GCAAATACTGTGTATTAGTCC-3′ |
|
| Reverse
5′-CCGCTTCCTTGTGAAGAAGGT-3′ |
| miR-4698 | Forward
5′-TGGTACTGATGTGATGGACT-3′ |
|
| Reverse
5′-TCATATCACACAGCACCGAT-3′ |
| MTUS1 | Forward
5′-GAGCTGAGCACTTACAGCAACAA-3′ |
|
| Reverse
5′-TTCAACTGCATTAAGAGCTGTAA-3′ |
| U6 | Forward
5′-CTCGCTTCGGCAGCACA-3′ |
|
| Reverse
5′-AACGCTTCACGAATTTGCGT-3′ |
| β-actin | Forward
5′-CCTGACGCCAACACACTGC-3′ |
|
| Reverse
5′-ATACTCCTGCTTGGTGATCC-3′ |
Western blot assay
Following cell transfection, proteins were extracted
with RIPA buffer with phenylmethanesulfonyl fluoride (Beyotime
Institute of Biotechnology). The protein content was assessed using
the BCA method. Afterwards, total protein (30 µg) was separated via
10% SDS-PAGE (Beyotime Institute of Biotechnology). The separated
proteins were transferred to polyvinylidene fluoride (PVDF)
membranes (EMD Millipore). Following blocking with 5% skimmed milk
powder for 2 h at room temperature, the primary antibodies of MTUS1
(1:1,000, cat. no. sc-393120, Santa Cruz Biotechnology Inc.), MEK
(1:1,000, cat. no. ab32091), phosphorylated (p-)MEK (1:1,000, cat.
no. ab96379), ERK (1:1,000, cat. no. ab184699), p-ERK (1:1,000,
cat. no. ab201015), Cell division cycle-25 (Cdc25)B (1:1,000, cat.
no. ab124819), cyclin-dependent kinase (Cdk)1 (1:1,000, cat. no.
ab133327), p-Cdk1 (1:1,000, cat. no. ab201008) and GAPDH (1:2,000,
cat. no. ab8245; all from Abcam) were incubated with the PVDF
membranes overnight at 4°C. Next, the appropriate secondary
antibodies [1:5,000; goat anti-mouse IgG H&L (HRP) cat. no.
ab205719 or goat anti-rabbit IgG H&L (HRP) cat. no. ab6721,
Abcam] were used to incubate the above membranes for additional 1 h
at room temperature. The protein levels were monitored by enhanced
chemiluminescence reagents (Amersham Biosciences; Cytiva). The
protein bands were quantified using Quantity One software (v4.6.6,
Bio-Rad Laboratories, Inc.).
Bioinformatics analysis
The expression levels of lncRNA LIFR-AS1 in GC
tumors and normal tissues was downloaded from The Cancer Genome
Atlas (TCGA; http://portal.gdc.cancer.gov/). DIANA tools
(http://carolina.imis.athena-innovation.gr/diana_tools/web/index.php)
was used to predict potential binding miRNAs for lncRNA LIFR-AS1.
The potential target gene of miRNA was predicted by TargetScan
software, version 7.2 (http://www.targetscan.org/vert_72/).
Statistical analysis
All data are presented as the mean ± standard
deviation. SPSS 19.0 statistical software (IBM Corp.) was used for
statistical analyses. Relevance between lncRNA LIFR-AS1 and
miR-4698 or MTUS1 was assessed by using Pearson correlation
analysis. Comparisons between different groups were analyzed by
using Student's t-test or one-way ANOVA followed by Tukey's
multiple comparisons test. P<0.05 was considered to indicate a
statistically significant difference.
Results
Downregulation of lncRNA LIFR-AS1 in
GC tissues and cell lines
To investigate the biological role of LIFR-AS1 in
GC, data from normal and tumor samples were obtained from TCGA and
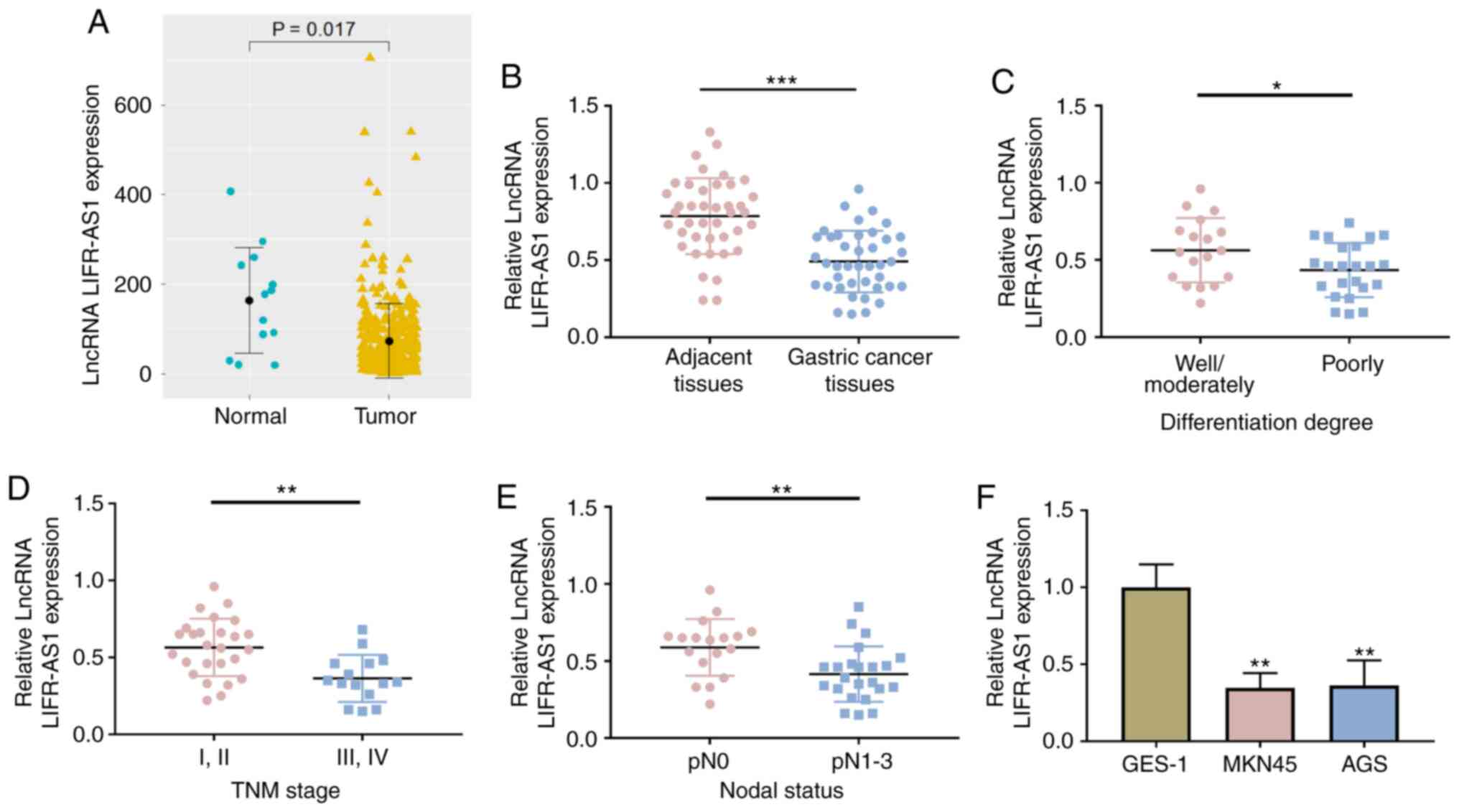
it was found that LIFR-AS1 was significantly downregulated in GC
tissues compared with normal tissues (P=0.017; Fig. 1A). Expression of lncRNA LIFR-AS1 was
examined in 41 GC tissues and the adjacent tissues using RT-qPCR.
The low expression of lncRNA LIFR-AS1 was clearly presented in GC
tissues (P<0.001, Fig. 1B). The
relationship between lncRNA LIFR-AS1 expression and clinical
characteristics is shown in Table
I. In the GC tissues of patients with poorly differentiation
degree, III and IV TNM stage and pN1-3 nodal status, the expression
of lncRNA LIFR-AS1 was significantly reduced (P<0.05, P<0.01;
Fig. 1C-E). Furthermore, the
expression of lncRNA LIFR-AS1 was also explored in GES-1 and GC
cell lines (MKN45 and AGS). It was identified that expression of
lncRNA LIFR-AS1 was lower in MKN45 and AGS cells compared with that
in GES-1 cells (P<0.01; Fig.
1F). These results demonstrated that lncRNA LIFR-AS1 was
downregulated in GC tissues and cells.
Overexpression of lncRNA LIFR-AS1
restrains GC cell proliferation and movement
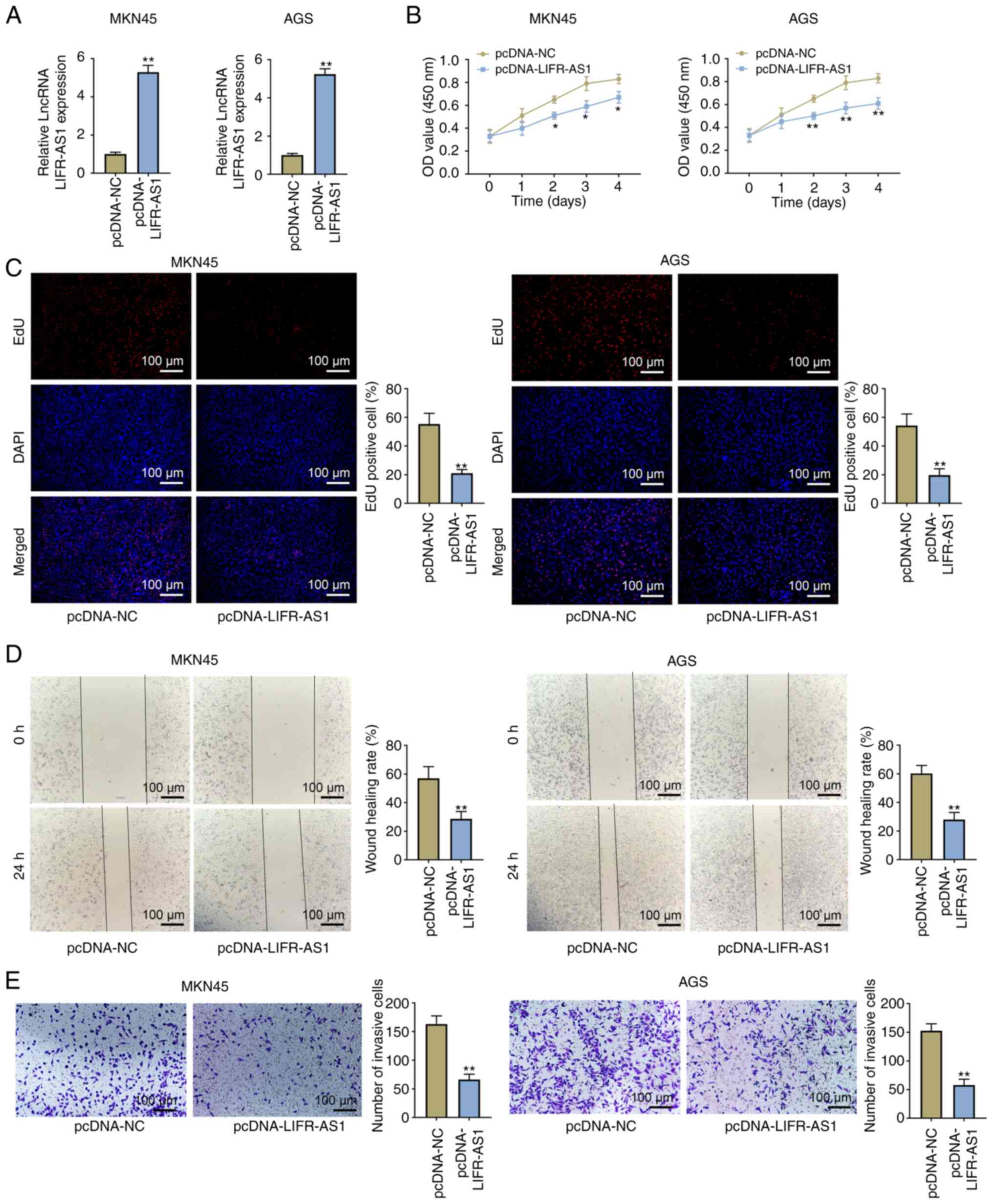
The overexpressed vector of lncRNA LIFR-AS1
(pcDNA-LIFR-AS1) was transfected into MKN45 and AGS cells.
Expression of lncRNA LIFR-AS1 was distinctly upregulated in
pcDNA-LIFR-AS1-transfected MKN45 and AGS cells (P<0.01; Fig. 2A). CCK-8 experimental results
revealed that overexpressed lncRNA LIFR-AS1 reduced cell viability
at days 2, 3 and 4 in MKN45 and AGS cells (P<0.05, P<0.01;
Fig. 2B). In addition, the number
of EdU positive cells was also reduced by overexpressed lncRNA
LIFR-AS1 (P<0.01; Fig. 2C). Cell
migratory and invasive ability was then evaluated by wound and cell
invasion assays. It was noted that overexpressed lncRNA LIFR-AS1
significantly decreased the wound healing areas in MKN45 and AGS
cells (P<0.01; Fig. 2D).
Similary, the capacity of cell invasion was also inhibited by
overexpressed lncRNA LIFR-AS1 (P<0.01; Fig. 2E). All above results suggested that
lncRNA LIFR-AS1 suppressed GC cell proliferation, migration and
invasion.
Negative correlation between lncRNA
LIFR-AS1 and miR-4698
Fig. 3A shows the
predicted binding site of miR-4698 in lncRNA LIFR-AS1. To verify
the predicted sequence binding sites between lncRNA LIFR-AS1 and
miR-4698, luciferase reporter gene assay was performed to further
confirm their association. It was observed that the luciferase
activity was reduced in MKN45 and AGS cells with LIFR-AS1-WT and
miR-4698 mimics co-transfection (P<0.05; Fig. 3B). It was noted that overexpressed
lncRNA LIFR-AS1 decreased miR-4698 expression in MKN45 and AGS
cells (P<0.01; Fig. 3C).
Notably, upregulated miR-4698 expression was found in GC tissues
(P<0.001; Fig. 3D) in addition
to MKN45 and AGS cells (P<0.01; Fig.
3E). A Pearson correlation test revealed a negative correlation
between lncRNA LIFR-AS1 and miR-4698 (P<0.05; Fig. 3F). All these findings demonstrated
the negative correlation between lncRNA LIFR-AS1 and miR-4698 and
that lncRNA LIFR-AS1 worked as a sponge of miR-4698 in GC
cells.
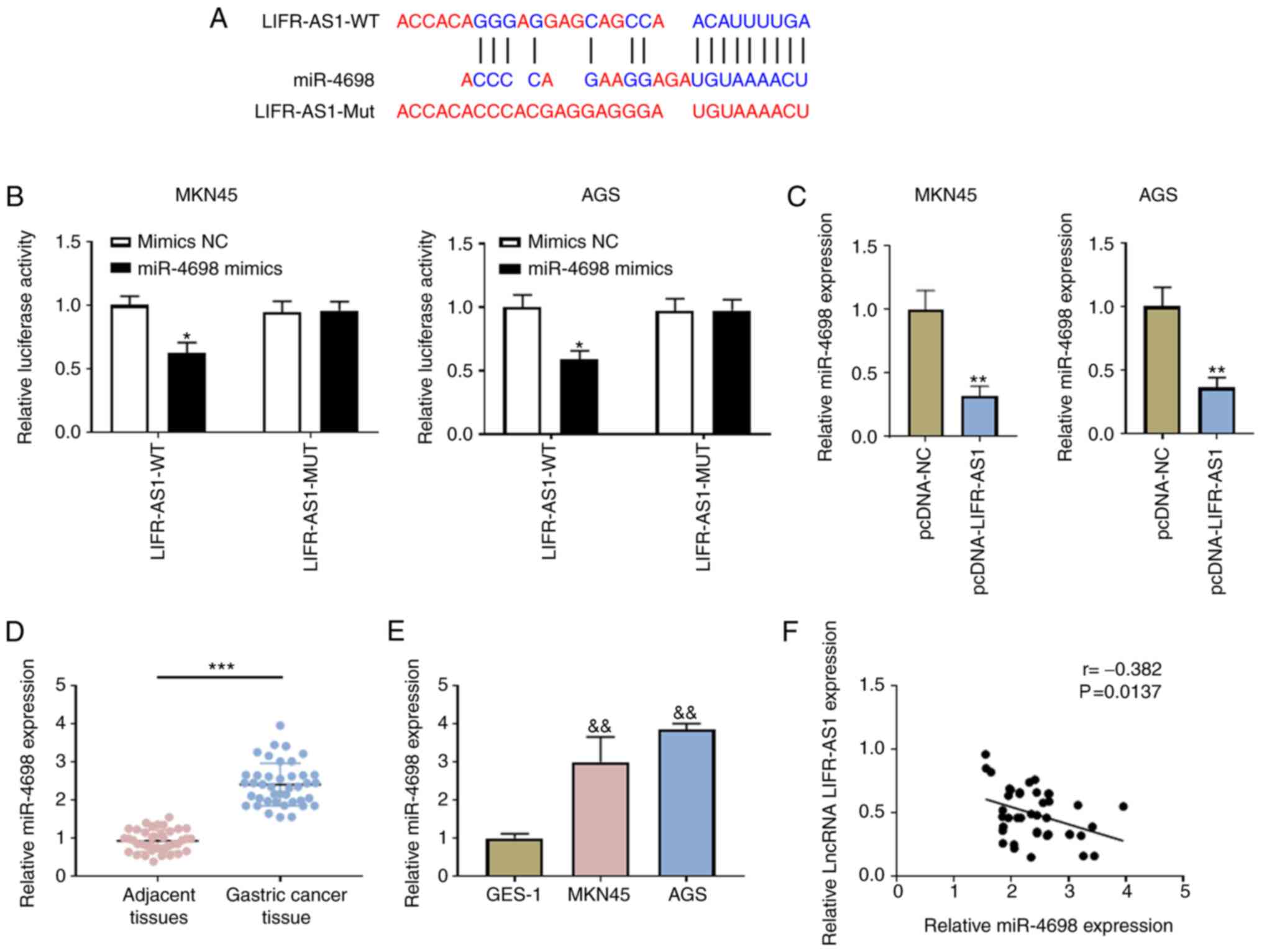 | Figure 3.lncRNA LIFR-AS1 acts as a sponge for
miR-4698. (A) The sequence of binding site between lncRNA LIFR-AS1
and miR-4698 was predicated using the online tool DIANA. (B)
Association between lncRNA LIFR-AS1 and miR-4698 was investigated
using a luciferase reporter system. (C) Expression of miR-4698 was
established following pcDNA-LIFR-AS1 and pcDNA-NC transfection
using RT-qPCR. Expression of miR-4698 in (D) 41 pairs of GC tissues
and matched adjacent tissues and (E) in GES-1 and GC (MKN45 and
AGS) cell lines was evaluated using RT-qPCR. (F) Mutual regulation
between lncRNA LIFR-AS1 and miR-4698 was assessed using Pearson
correlation analysis. *P<0.05 LIFR-AS1-WT+Mimics NC group,
**P<0.01 vs. pcDNA-NC group, &&P<0.01 vs.
GES-1 group, ***P<0.001 vs. adjacent tissues group. lnc., long
non-coding; miR, microRNA; RT-qPCR, reverse
transcription-quantitative PCR; GC, gastric carcinoma; GES-1,
gastric mucosal epithelial cell line; Wt, wild type; Mut, mutant;
NC, negative control. |
MTUS1 is a target of miR-4698 and is
positively associated with lncRNA LIFR-AS1
TargetScan revealed that the 3′-UTR of MTUS1
contained the complementary site for the seed region of miR-4698
(Fig. 4A). Following
co-transfection with miR-4698 mimics and the fragments of MTUS1-WT
or MTUS1-MUT, the change in the luciferase activity was evaluated.
The results demonstrated inhibited luciferase activity in miR-4698
mimics and MTUS1-WT co-transfected cells (P<0.05; Fig. 4B). The overexpressed and suppressed
vectors of miR-4698 (miR-4698 mimics and miR-4698 inhibitors) were
transfected into GC cells and the correlation between miR-4698 and
MTUS1 was monitored. Expression of miR-4698 was clearly enhanced in
miR-4698 mimics-transfected cells (P<0.01), but restricted in
miR-4698 inhibitors-transfected cells (P<0.05; Fig. 4C). Western blotting results revealed
that MTUS1 expression was inhibited by miR-4698 overexpression and
increased by miR-4698 inhibition (P<0.05; Fig. 4D). Fig.
4E and F demonstrate the downregulation of MTUS1 in GC tissues
(P<0.001) and in GC (MKN45 and AGS) cell lines (P<0.01). A
Pearson correlation test further confirmed the negative correlation
between MTUS1 and miR-4698 (Fig.
4G) and the positive correlation between MTUS1 and lncRNA
LIFR-AS1 (Fig. 4H). Together, these
results demonstrated that MTUS1 was a new predicated target gene of
miR-4698 and was also positively regulated by lncRNA LIFR-AS1.
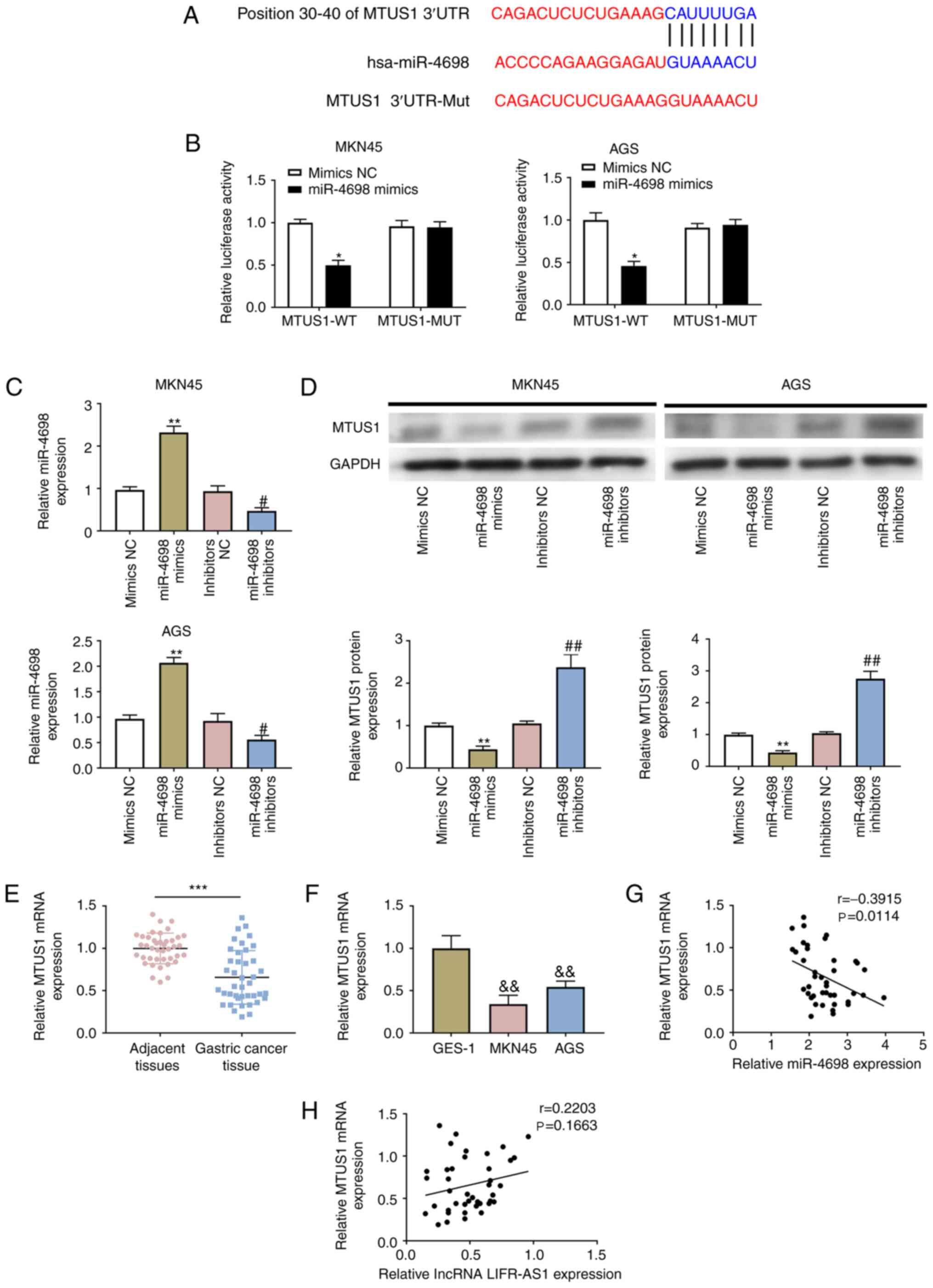 | Figure 4.MTUS1 is a direct target of miR-4698
and is positively regulated by lncRNA LIFR-AS1. (A) The binding
sequence between miR-4698 and MTUS1 was predicted using TargetScan
algorithms. (B) Association between MTUS1 and miR-4698 was
evaluated using a luciferase reporter system. The vectors of
miR-4698 mimics and miR-4698 inhibitors as well as the
corresponding controls (mimics NC and inhibitors NC) were
transfected into GC (MKN45 and AGS) cell lines. (C) The expression
of miR-4698 in above transfected cells was explored using RT-qPCR;
(D) The expression of MTUS1 in above transfected cells was
evaluated using western blotting. Expression of MTUS1 in (E) 41
pairs of GC tissues and matched adjacent tissues and (F) in GES-1
and GC (MKN45 and AGS) cell lines was examined using RT-qPCR.
Interactions between MTUS1 and (G) miR-4698 or (H) lncRNA LIFR-AS1
were investigated using Pearson correlation analysis. *P<0.05
vs. MTUS1-WT+Mimics NC group, **P<0.01 vs. Mimics NC group,
&&P<0.01 vs. GES-1 group, ***P<0.001 vs.
adjacent tissues group, #P<0.05 vs. inhibitors NC
group, ##P<0.01 vs. inhibitors NC group. MTUS1,
microtubule-associated tumor suppressor 1; miR, microRNA; lnc, long
non-coding; NC, negative control; GC, gastric carcinoma; GES-1,
gastric mucosal epithelial cell line; RT-qPCR, reverse
transcription-quantitative PCR. |
MEK/ERK pathway is counteracted by
overexpression of lncRNA LIFR-AS1 via regulation of MTUS1
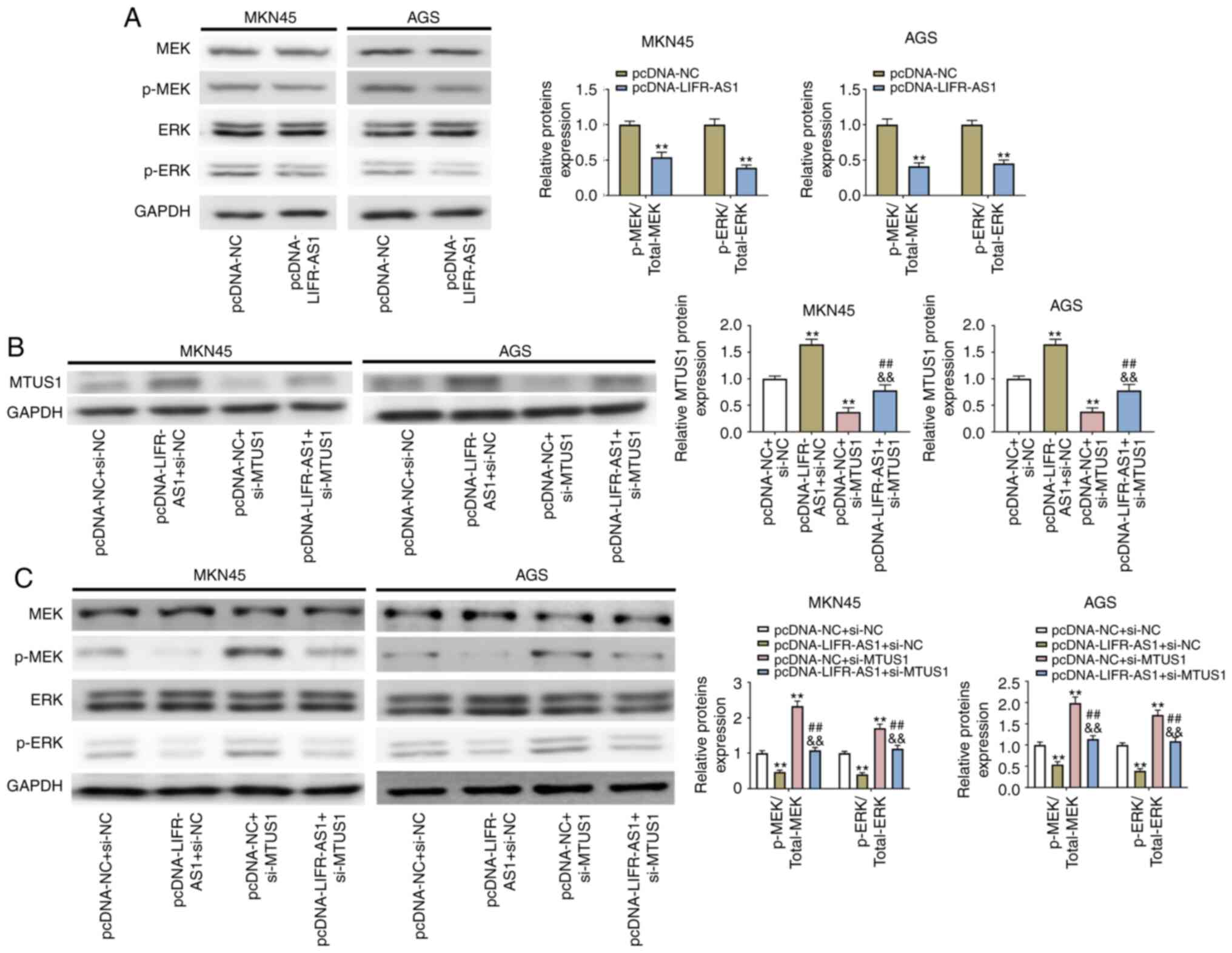
The signaling pathway regulation was assessed when
cells were transfected with the expression vectors of
pcDNA-LIFR-AS1 and si-MTUS1. It was noted that overexpressed lncRNA
LIFR-AS1 inhibited p-MEK and p-ERK expression in GC (MKN45 and AGS)
cell lines (P<0.05; Fig. 5A).
si-MTUS1 vector was transfected into GC cells to inhibit MTUS1
expression and the relationship between lncRNA LIFR-AS1 and MTUS1
explored. It was observed that overexpressed lncRNA LIFR-AS1
upregulated MTUS1 expression in MKN45 and AGS cells (P<0.01;
Fig. 5B). By contrast, silencing of
MTUS1 increased p-MEK and p-ERK expression (P<0.05, Fig. 5C). Following pcDNA-LIFR-AS1 and
si-MTUS1 co-transfection, the suppressed p-MEK and p-ERK expression
in GC (MKN45 and AGS) cell lines was reversed by si-MTUS1
(P<0.01; Fig. 5C). On the basis
of these results, it was hypothesized that the MEK/ERK pathway was
hindered by overexpressed lncRNA LIFR-AS1 modulating MTUS1.
Overexpression of lncRNA LIFR-AS1
inhibits Cdc25B and inactivates Cdk1 signaling
The present study identified that overexpressed
lncRNA LIFR-AS1 inhibited Cdc25B expression in GC (MKN45 and AGS)
cell lines. The tyrosine phosphorylation of Cdk1 was induced by
overexpression of lncRNA LIFR-AS1 in GC (MKN45 and AGS) cell lines
(P<0.05; Fig. 6A). Following
pcDNA-LIFR-AS1 and si-MTUS1 co-transfection, the suppressed Cdc25B
expression and the induced tyrosine phosphorylation of Cdk1 in GC
(MKN45 and AGS) cell lines was reversed by si-MTUS1 (P<0.01;
Fig. 6B). These results
demonstrated that the inhibition of Cdc25B activity by lncRNA
LIFR-AS1 could contribute to Cdk1 tyrosine phosphorylation.
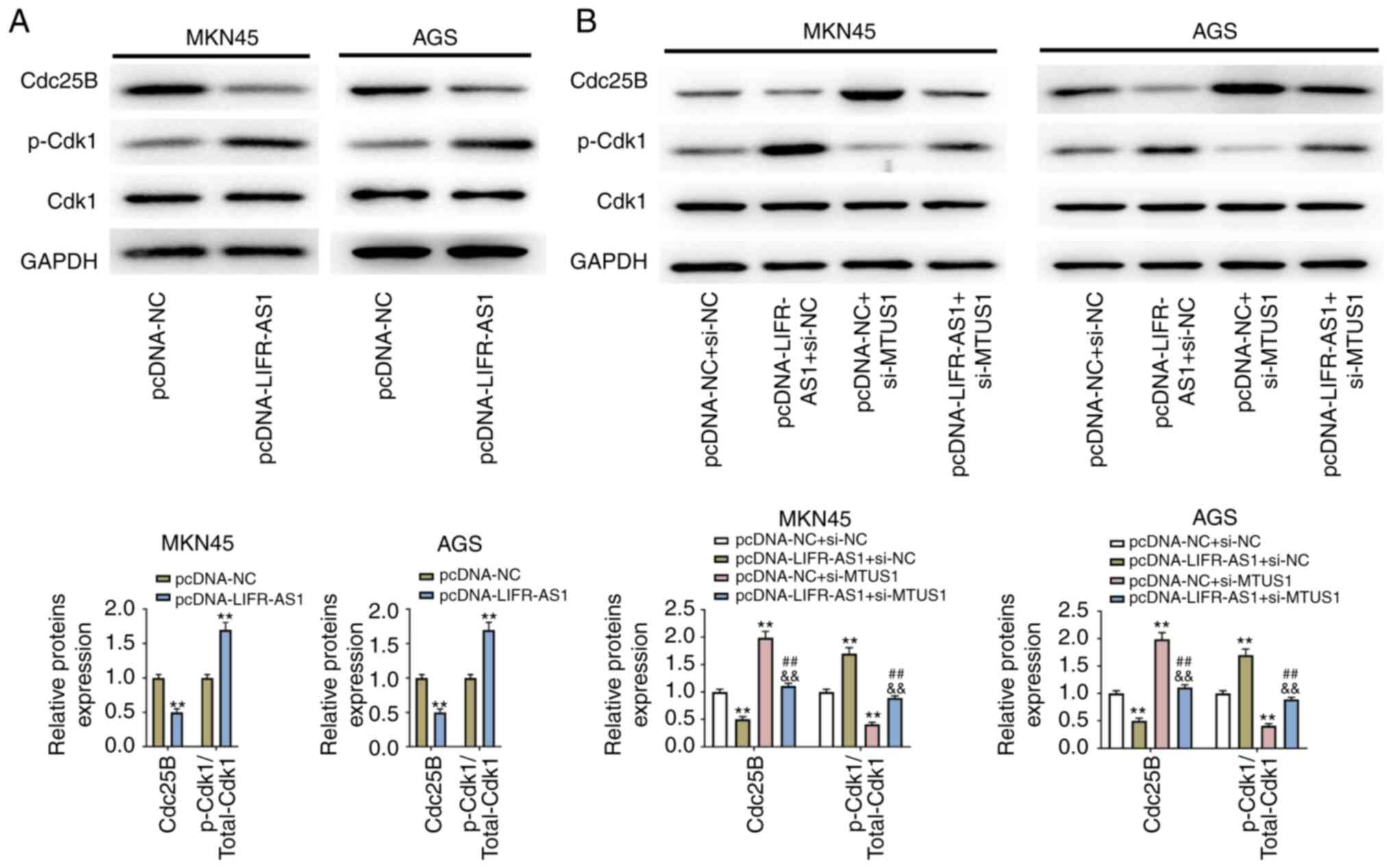 | Figure 6.Overexpression of lncRNA LIFR-AS1
inhibits Cdc25B and inactivates Cdk1 signaling. (A) The vectors of
pcDNA-LIFR-AS1 and pcDNA-NC were transfected into GC (MKN45 and
AGS) cell lines, protein levels of Cdc25B and p-Cdk1 were examined
using western blotting. (B) The vectors of pcDNA-LIFR-AS1, si-MTUS1
and the corresponding controls (pcDNA-NC and si-NC) were
transfected into GC (MKN45 and AGS) cell lines, protein levels of
Cdc25B and p-Cdk1 were examined using western blotting. **P<0.01
vs. pcDNA-NC group, ##P<0.01 vs. pcDNA-LIFR-AS1+si-NC
group, &&P<0.01 vs. pcDNA-NC+si-MTUS1 group.
lnc, long non-coding; Cdc, cell division cycle; Cdk,
cyclin-dependent kinase; NC, negative control; GC, gastric
carcinoma; si, small interfering; MTUS1, microtubule-associated
tumor suppressor 1. |
Discussion
GC has become one of the leading causes of mortality
in individuals and its pathogenesis is still unclear (27). In the treatment of GC priority is
given to surgical operation, whereas the unfavorable prognosis
remains an enormous challenge in clinical practice. Due to their
clinical potential in cancer treatment, lncRNAs have been
extensively studied in GC (28). Wu
et al (29) reported that
lncRNA ZEB2-AS1 is upregulated in gastric cancer and affects cell
proliferation and invasion via miR-143-5p/HIF-1α axis. Xiao et
al (30) found that lncRNA
MALAT1 increases the stemness of gastric cancer cells via enhancing
SOX2 mRNA stability. However, the influence of lncRNA LIFR-AS1 in
GC remains to be elucidated. The present study first investigated
the functions of lncRNA LIFR-AS1 in GC cell proliferation and
movement and uncovered the potential molecular mechanisms.
Decreased expression of lncRNA LIFR-AS1 was identified in GC
tissues and cell lines. Further study revealed that lncRNA LIFR-AS1
inhibited GC cell proliferation, migration and invasion through
sponging miR-4698. In addition, MTUS1 was verified to be a target
gene of miR-4698 and positively regulated by lncRNA LIFR-AS1.
Furthermore, lncRNA LIFR-AS1 blocked the MEK/ERK pathway by
mediating MTUS1.
lncRNA LIFR-AS1 is derived from the antisense
transcription of the LIFR gene located on human chromosome
5p13.1, which serves a crucial role in the pathogenesis of a number
of illnesses (31). A previous
study revealed that lncRNA LIFR-AS1 negatively correlates with
tumor size in uterine fibroids (32). Additionally, Wang et al
(13) found that lncRNA LIFR-AS1 is
downregulated in breast cancer. In accordance with that study, the
present study also observed decreased lncRNA LIFR-AS1 expression in
GC tissues and cell lines. Xu et al (14) explored the effects of lncRNA
LIFR-AS1 on breast cancer cell proliferation and migration and
verify its inhibitory role in breast cancer progression. On the
basis of that research, the functions of lncRNA LIFR-AS1 in GC cell
proliferation, migration and invasion were further investigated.
The present study also demonstrated the suppressed functions of
lncRNA LIFR-AS1 in GC cell proliferation, migration and invasion.
These findings suggested that lncRNA LIFR-AS1 participated in the
control of the GC progression.
Previous studies have demonstrated the vital
regulatory functions of lncRNA-miRNA network in types of human
cancers, such as diabetic pancreatic cancer and colorectal cancer
(33,34). In the field of GC, lncRNA gastric
cancer-related lncRNA1 has been reported to modulate GC cell
proliferation and metastasis by sponging miR-885-3p (35). lncRNA XIST promotes GC cell growth
and invasion by regulating miR-497 (36). miR-4698 is a novel miRNA and its
downregulation is reported in chronic obstructive pulmonary disease
(37). Similarly, a profile of
miRNAs by microarrays in Liu et al (38) demonstrates that miR-4698 expression
is upregulated in gastric cancer stem cells. Considering its
abnormal expression in different diseases, the present study
attempted to uncover the influence of miR-4698 in lncRNA
LIFR-AS1-affected GC cell proliferation, migration and invasion.
Upregulated expression of miR-4698 was observed in GC tissues and
cell lines. In addition, a reciprocal suppression between miR-4698
and lncRNA LIFR-AS1 was identified. Thus, it was concluded that
lncRNA LIFR-AS1 exerted inhibitory functions in GC cell
proliferation, migration and invasion through sponging
miR-4698.
MTUS1 is reported to locate on the reverse strand of
the chromosome 8p22, which can encode angiotensin II AT2
receptor-interacting proteins (39). As a tumor suppressor, MTUS1 has been
investigated in colon and prostate cancer and its aberrant
expression is associated with the development of these cancers
(40,41). The present study also observed the
downregulated expression of MTUS1 in GC tissues and cell lines. In
a previous study, silencing of MTUS1 facilitates GC cell growth and
metastasis (42). The results of
the present study confirmed that MTUS1 was a target gene of
miR-4698. In addition, a positive correlation between MTUS1 and
lncRNA LIFR-AS1 was identified. Given the relationship of lncRNA
LIFR-AS1, miR-4698 and MTUS1, it was hypothesized that MTUS1
probably joined in modulating the functions of lncRNA LIFR-AS1 in
GC.
MEK/ERK is a highly conserved cell signaling
pathway, existing in eukaryotic cells, which regulates diverse cell
behaviors in human cancers (43,44).
The MEK/ERK pathway participates in adjusting the cisplatin
resistance in GC cells (45) and
inactivation of the MEK/ERK pathway can affect the proliferative
ability of GC cells (46). Several
lncRNAs affect cancer cell proliferation and metastasis by
controlling the MEK/ERK pathway (47,48).
However, whether lncRNA LIFR-AS1 regulates the MEK/ERK pathway to
restrain GC cell proliferation and movement remains unreported. The
present study showed that overexpressed lncRNA LIFR-AS1 inhibited
the MEK/ERK pathway. The process was reversed by silencing of
MTUS1. These findings corroborated that the MEK/ERK pathway was
inhibited by overexpression of lncRNA LIFR-AS1 via modulating
MTUS1.
Cdc25 serves an important role in transitions
between cell-cycle phases by dephosphorylating and activating Cdks
(49). ERK-MAP kinases are directly
involved in activating Cdc25 during the G(2)/M transition (50). A previous study reported that
small-molecule Cdc25 inhibitors inhibit Cdc25 enzymatic activities
and induce tyrosine phosphorylation of the Cdc25-targeted Cdks
(51). The present study identified
that lncRNA LIFR-AS1 inhibited Cdc25B activity and contributed to
Cdk1 tyrosine phosphorylation and cell growth inhibition.
The present study had some limitations. First,
animal experiments to investigate the roles of lncRNA LIFR-AS1 in
GC were not performed. Second, the role of lncRNA LIFR-AS1 on tumor
cell apoptosis was unknown. Third, the underlying molecular
mechanism of lncRNA LIFR-AS1 downregulation in GC cells was not
elucidated.
In summary, the present study showed that lncRNA
LIFR-AS1 was downregulated in GC tissues and was associated with
the prognosis of patients with GC. lncRNA LIFR-AS1 inhibited GC
cell proliferation, migration and invasion. Results revealed that
lncRNA LIFR-AS1 interacted with miR-4698 to regulate MTUS1
expression. These results provided a new lncRNA-directed
therapeutics for GC. However, the detailed effects of miR-4698 and
MTUS1 on GC cell behaviors remain to be elucidated.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JC and JZ conceived and designed the study. JZ, XL,
LF, NZ and JY performed the study, collected the data and analyzed
the data. JC wrote the manuscript. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
All patients personally signed an informed consent.
The protocol of the present study was approved by the Ethics
Committee of Cangzhou People's Hospital (approval number
AF/SC-08/02.0).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Zhong Q, Sun Q, Xu GF, Fan XQ, Xu YY, Liu
F, Song SY, Peng CY and Wang L: Differential analysis of lymph node
metastasis in histological mixed-type early gastric carcinoma in
the mucosa and submucosa. World J Gastroenterol. 24:87–95. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Tanahashi T, Yoshida K and Yamaguchi K:
Current status and future perspective of the chemotherapy for
gastric cancer. Nihon Shokakibyo Gakkai Zasshi. 115:500–506.
2018.(In Japanese). PubMed/NCBI
|
|
3
|
Li T, Gao X, Han L, Yu J and Li H:
Identification of hub genes with prognostic values in gastric
cancer by bioinformatics analysis. World J Surg Oncol. 16:1142018.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Dreznik A, Purim O, Idelevich E, Kundel Y,
Sulkes J, Sulkes A and Brenner B: Gastric cancer: Biology and
clinical manifestations in Israel. J Surg Oncol. 105:316–322. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sasaki T: Discussion for gastric cancer
treatment guidelines in Japan. Nihon Rinsho. 61:13–18. 2003.(In
Japanese).
|
|
6
|
Kim H, Kim H, Kim SY, Kim TY, Lee KW, Baek
SK, Kim TY, Ryu MH, Nam BH and Zang DY: Second-line chemotherapy
versus supportive cancertreatment in advanced gastric cancer: A
meta-analysis. Ann Oncol. 24:2850–2854. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Cong P, Zhang G, Chen Z, Chen Y and Zhang
L: lncRNApred: Classification of long non-coding RNAs and
protein-coding transcripts by the ensemble algorithm with a new
hybrid feature. PLoS One. 11:e01545672016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Cao MX, Jiang YP, Tang YL and Liang XH:
The crosstalk between lncRNA and microRNA in cancer metastasis:
Orchestrating the epithelial-mesenchymal plasticity. Oncotarget.
8:12472–12483. 2015. View Article : Google Scholar
|
|
9
|
Chen D, Sun Q, Cheng X, Zhang L, Song W,
Zhou D, Lin J and Wang W: Genome-wide analysis of long noncoding
RNA (lncRNA) expression in colorectal cancer tissues from patients
with liver metastasis. Cancer Med. 5:1629–1639. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Li H, Yu B, Li J, Su L, Yan M, Zhu Z and
Liu B: Overexpression of lncRNA H19 enhances carcinogenesis and
metastasis of gastric cancer. Oncotarget. 5:2318–2329. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sun M, Nie F, Wang Y, Zhang Z, Hou J, He
D, Xie M, Xu L, De W, Wang Z and Wang J: lncRNA HOXA11-AS promotes
proliferation and invasion of gastric cancer by scaffolding the
chromatin modification factors PRC2, LSD1 and DNMT1. Cancer Res.
76:6299–6310. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Wang Y, Liu X, Zhang H, Sun L, Zhou Y, Jin
H, Zhang H, Zhang H, Liu J, Guo H, et al: Hypoxia-inducible
lncRNA-AK058003 promotes gastric cancer metastasis by targeting
γ-synuclein. Neoplasia. 16:1094–1106. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Wang L, Li J, Zhao H, Hu J, Ping Y, Li F,
Lan Y, Xu C, Xiao Y and Li X: Identifying the crosstalk of
dysfunctional pathways mediated by lncRNAs in breast cancer
subtypes. Mol Biosyst. 12:711–720. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xu F, Li H and Hu C: LIFR-AS1 modulates
Sufu to inhibit cell proliferation and migration by miR-197-3p in
breast cancer. Biosci Rep. 39:BSR201805512019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Liu K, Yao H, Wen Y, Zhao H, Zhou N, Lei S
and Xiong L: Functional role of a long non-coding RNA
LIFR-AS1/miR-29a/TNFAIP3 axis in colorectal cancer resistance to
pohotodynamic therapy. Biochim Biophys Acta Mol Basis Dis.
1864:2871–2880. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yanaihara N and Harris C: MicroRNA
involvement in human cancers. Clin Chem. 59:1811–1812. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Matamala N, Vargas MT, González-Cámpora R,
Miñambres R, Arias JI, Menéndez P, Andrés-León E, Gómez-López G,
Yanowsky K, Calvete-Candenas J, et al: Tumor MicroRNA expression
profiling identifies circulating MicroRNAs for Early breast cancer
detection. Clin Chem. 61:1098–1096. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yu Y, Nangia-Makker P, Farhana L and
Majumdar A: A novel mechanism of lncRNA and miRNA interaction:
CCAT2 regulates miR-145 expression by suppressing its maturation
process in colon cancer cells. Mol Cancer. 16:1552017. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hao NB, He YF, Li XQ, Wang K and Wang RL:
The role of miRNA and lncRNA in gastric cancer. Oncotarget.
8:81572–81582. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Mikkelsen LH, Andersen MK, Andreasen S,
Larsen AC, Tan Q, Toft PB, Wadt K and Heegaard S: Global microRNA
profiling of metastatic conjunctival melanoma. Melanoma Res.
29:465–473. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kalhori MR, Arefian E, Fallah Atanaki F,
Kavousi K and Soleimani M: miR-548× and miR-4698 controlled cell
proliferation by affecting the PI3K/AKT signaling pathway in
glioblastoma cell lines. Sci Rep. 10:15582020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Seibold S, Rudroff C, Weber M, Galle J,
Wanner C and Marx M: Identification of a new tumor suppressor gene
located at chromosome 8p21.3-22. FASEB J. 17:1180–1182. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kara M, Kaplan M, Bozgeyik I, Ozcan O,
Celik OI, Bozgeyik E and Yumrutas O: MTUS1 tumor suppressor and its
miRNA regulators in fibroadenoma and breast cancer. Gene.
587:173–177. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Ozcan O, Kara M, Yumrutas O, Bozgeyik E,
Bozgeyik I and Celik OI: MTUS1 and its targeting miRNAs in
colorectal carcinoma: Significant associations. Tumour Biol.
37:6637–6645. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Xiao J, Chen JX, Zhu YP, Zhou LY, Shu QA
and Chen LW: Reduced expression of MTUS1 mRNA is correlated with
poor prognosis in bladder cancer. Oncol Lett. 4:113–118. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Livak K and Schmittgen T: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Van Cutsem E, Sagaert X, Topal B,
Haustermans K and Prenen H: Gastric cancer. Lancet. 388:2654–2664.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Zhang G, Li S, Lu J, Ge Y, Wang Q, Ma G,
Zhao Q, Wu D, Gong W, Du M, et al: lncRNA MT1JP functions as a
ceRNA in regulating FBXW7 through competitively binding to
miR-92a-3p in gastric cancer. Mol Cancer. 17:872018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wu F, Gao H, Liu K, Gao B, Ren H, Li Z and
Liu F: The lncRNA ZEB2-AS1 is upregulated in gastric cancer and
affects cell proliferation and invasion via miR-143-5p/HIF-1α axis.
Onco Targets Ther. 12:657–667. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Xiao Y, Pan J, Geng Q and Wang G: lncRNA
MALAT1 increases the stemness of gastric cancer cells via enhancing
SOX2 mRNA stability. FEBS Open Bio. 9:1212–1222. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Frey HA, Stout MJ, Pearson LN, Tuuli MG,
Cahill AG, Strauss JF III, Gomez LM, Parry S, Allsworth JE and
Macones GA: Genetic variation associated with preterm birth in
African-American women. Am J Obstet Gynecol. 215:235.e1–e8. 2016.
View Article : Google Scholar
|
|
32
|
Aissani B, Zhang K and Wiener H: Genetic
determinants of uterine fibroid size in the multiethnic NIEHS
uterine fibroid study. Int J Mol Epidemiol Genet. 6:9–19.
2015.PubMed/NCBI
|
|
33
|
Yao K, Wang Q, Jia J and Zhao H: A
competing endogenous RNA network identifies novel mRNA, miRNA and
lncRNA markers for the prognosis of diabetic pancreatic cancer.
Tumor Biol. 39:10104283177078822017. View Article : Google Scholar
|
|
34
|
Matboli M, Shafei A and Ali M, El-Din
Ahmed TS, Naser M, Abdel-Rahman T, Anber N and Ali M: Role of
extracellular lncRNA-SNHG14/miRNA-3940-5p/NAP12 mRNA in colorectal
cancer. Arch Physiol Biochem. 1–7. 2019.doi:
10.1080/13813455.2019.1650070 (Epub ahead of print). View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Lin Z, Zhou Z, Guo H, He Y, Pang X, Zhang
X, Liu Y, Ao X, Li P and Wang J: Long noncoding RNA gastric
cancer-related lncRNA1 mediates gastric malignancy through
miRNA-885-3p and cyclin-dependent kinase 4. Cell Death Dis.
9:6072018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Ma L, Zhou Y, Luo X, Gao H, Deng X and
Jiang Y: Long non-coding RNA XIST promotes cell growth and invasion
through regulating miR-497/MACC1 axis in gastric cancer.
Oncotarget. 8:4125–4135. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Shen Z, Tang W, Guo J and Sun S:
miR-483-5p plays a protective role in chronic obstructive pulmonary
disease. Int J Mol Med. 40:193–200. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Liu J, Ma L, Wang Z, Wang L, Liu C, Chen R
and Zhang J: MicroRNA expression profile of gastric cancer stem
cells in the MKN-45 cancer cell line. Acta Biochim Biophys Sin
(Shanghai). 46:92–99. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Di Benedetto M, Bièche I, Deshayes F,
Vacher S, Nouet S, Collura V, Seitz I, Louis S, Pineau P,
Amsellem-Ouazana D, et al: Structural organization and expression
of human MTUS1, a candidate 8p22 tumor suppressor gene encoding a
family of angiotensin II AT2 receptor-interacting proteins, ATIP.
Gene. 380:127–136. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Zuern C, Heimrich J, Kaufmann R, Richter
KK, Settmacher U, Wanner C, Galle J and Seibold S: Down-regulation
of MTUS1 in human colon tumors. Oncol Rep. 23:183–189.
2010.PubMed/NCBI
|
|
41
|
Louis SN, Chow L, Rezmann L, Krezel MA,
Catt KJ, Tikellis C, Frauman AG and Louis WJ: Expression and
function of ATIP/MTUS1 in human prostate cancer cell lines.
Prostate. 70:1563–1574. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Li X, Liu H, Yu T, Dong Z, Tang L and Sun
X: Loss of MTUS1 in gastric cancer promotes tumor growth and
metastasis. Neoplasma. 61:128–135. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhang X, Liu G, Ding L, Jiang T, Shao S,
Gao Y and Lu Y: HOXA3 promotes tumor growth of human colon cancer
through activating EGFR/Ras/Raf/MEK/ERK signaling pathway. J Cell
Biochem. 119:2864–2874. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Hu G, Zhang J, Xu F, Deng H, Zhang W, Kang
S and Liang W: SLP-2 inhibits cisplatin-induced apoptosis through
MEK/ERK signaling and mitochondrial apoptosis pathway in cervical
cancer cells. Cancer Sci. 109:1357–1368. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Fu X, Feng J, Zeng D, Ding Y, Yu C and
Yang B: PAK4 confers cisplatin resistance in gastric cancer cells
via PI3K/Akt- and MEK/ERK-dependent pathways. Biosci Rep.
34:e000942014. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Lin Z, Zhang C, Zhang M, Xu D, Fang Y,
Zhou Z, Chen X, Qin N and Zhang X: Targeting Cadherin-17
Inactivates Ras/Raf/MEK/ERK signaling and inhibits cell
proliferation in gastric cancer. PLoS One. 9:e852962014. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Zhuang R, Zhang X, Lu D, Wang J, Zhuo J,
Wei X, Ling Q, Xie H, Zheng S and Xu X: lncRNA DRHC inhibits
proliferation and invasion in hepatocellular carcinoma via
c-Myb-regulated MEK/ERK signaling. Mol Carcinog. 58:366–375. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Wang Y, Gu J, Lin X, Yan W, Yang W and Wu
G: lncRNA BANCR promotes EMT in PTC via the Raf/MEK/ERK signaling
pathway. Oncol Lett. 15:5865–5870. 2018.PubMed/NCBI
|
|
49
|
Sur S and Agrawal DK: Phosphatases and
kinases regulating CDC25 activity in the cell cycle: Clinical
implications of CDC25 overexpression and potential treatment
strategies. Mol Cell Biochem. 416:33–46. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Wang R, He G, Nelman-Gonzalez M, Ashorn
CL, Gallick GE, Stukenberg PT, Kirschner MW and Kuang J: Regulation
of Cdc25C by ERK-MAP kinases during the G2/M transition. Cell.
128:1119–1132. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Peyregne VP, Kar S, Ham SW, Wang M, Wang Z
and Carr BI: Novel hydroxyl naphthoquinones with potent Cdc25
antagonizing and growth inhibitory properties. Mol Cancer Ther.
4:595–602. 2005. View Article : Google Scholar : PubMed/NCBI
|