Introduction
Pingyangmycin (PYM) has been widely used to treat
head and neck squamous cell carcinoma, the cytotoxic effects of
which depend on its ability to bind to iron and directly damage
DNA, subsequently promoting cell death (1–4). In
addition to inhibiting tumor growth, PYM affects cell apoptosis,
chronic inflammation and angiogenesis (2,5,6). PYM
also exerts a similar mechanism as bleomycin in inducing cell cycle
arrest and cell apoptosis (2). A
previous study suggested that PYM promoted cell apoptosis,
potentially via activating the p53-dependent signaling pathway
(7). PYM, a novel, hydrophilic,
antitumor glycopeptide antibiotic, is produced by
Streptomycetes in the soil of Pingyang county (2). In addition, neochlorogenic acid (NCA),
as one of main components of Cyclocarya paliurus, was
suggested to be involved in suppressing cell apoptosis by
regulating the mitogen-activated protein kinase/AKT signaling
pathway (8). Moreover, NCA, as a
phenolic in pectinase-treated Prunus mume fruit concentrate,
exerts antitumor effects via promoting cell cycle arrest at the S
phase and cell apoptosis via the mitochondrial-dependent apoptotic
pathway, but does not demonstrate cytotoxicity in normal cells
(9). A previous study has also
indicated that NCA may be involved in the anticancer effects of
Leonurus sibiricus extract, which are closely associated
with DNA damage (10). The effects
of phenolic compounds on DNA damage may be attributed to mutagenic
activity, possibly by intercalating with DNA (11).
Topoisomerases function as important ribozymes that
primarily regulate the topological structure of DNA by catalyzing
the formation or disconnection of phosphodiester bonds (12). DNA topoisomerase II α (TOP2A), which
is a target of several anticancer drugs, can be damaged or
suppressed, which hinders the replication and transcription of DNA,
further promoting cell death (13).
The abnormalities in TOP2A caused by anticancer drugs induce DNA
double-strand breaks and lead to cell apoptosis by stabilizing the
TOP2A cleavage complex (14). TOP2A
is necessary for cell replication and is expressed at elevated
levels during cell proliferation (14). In a previous study, compared with
normal cells, cancer cells displayed upregulated expression levels
of topoisomerase, independent of other factors (15,16).
Oral squamous cell carcinoma (OSCC) is the most common type of
malignant oral and maxillofacial tumor (17). Therefore, the present study aimed to
analyze the effects of NCA and PYM on OSCC cells and to investigate
the potential underlying mechanism.
Materials and methods
Cell lines and reagents
HOK (human normal oral keratinocytes), A-253, HSC-4,
CAL-27 and SCC-4 cells lines (The Cell Bank of Type Culture
Collection of the Chinese Academy of Sciences) were cultured in
DMEM (Hyclone; Cytiva) supplemented with 10% FBS (Biowest), 100
U/ml penicillin and 100 µg/ml streptomycin at 37°C with 5%
CO2. PYM (Harbin Pharmaceutical Group Co., Ltd.) was
dissolved in 1 mg/ml PBS to make a stock solution.
Reverse transcription-quantitative PCR
(RT-qPCR)
Cells were washed twice with PBS and total RNA was
extracted using TRIzol® (Invitrogen; Thermo Fisher
Scientific, Inc.). The concentration of RNA was detected using a
spectrophotometer. Total RNA was reverse transcribed into cDNA
using a ReverTra Aceq PCR RT kit at 37°C for 15 min (Toyobo Life
Science). Subsequently, qPCR was performed using the FastStart
Universal SYBR Green Master mix (Sigma-Aldrich; Merck KGaA). The
following primer sequences were used for the qPCR: TOP2A forward,
5′-CATTGAAGACGCTTCGTTATGG-3′ and reverse,
5′-CAGAAGAGAGGGCCAGTTGTG-3′; and β-actin forward,
5′-ATAGCACAGCCTGGATAGCAACGTAC-3′ and reverse,
5′-CACCTTCTACAATGAGCTGCGTGTG-3′. The following thermocycling
conditions were used for the qPCR: Initial denaturation for 1 min
at 95°C; followed by 40 cycles at 95°C for 30 sec and 60°C for 40
sec; and the reaction was then maintained at 72°C for 5 min. TOP2A
mRNA expression levels were quantified using the 2−ΔΔCq
method (18).
Western blotting
Cells were washed with precooled PBS and total
protein was extracted from cells using RIPA lysis buffer
(Invitrogen; Thermo Fisher Scientific, Inc.). Total protein was
quantified using a BCA assay. Proteins (40 µg protein/lane) were
separated via 12% SDS-PAGE and transferred onto PVDF membranes,
which were blocked with 5% skim milk for 2 h at room temperature.
The membranes were incubated with the following primary antibodies
at 4°C overnight: Anti-TOP2A (1:10,000; cat. no. ab52934; Abcam),
anti-CDK1 (1:2,000; cat. no. ab32094; Abcam), anti-Bax (1:1,000;
cat. no. ab32503; Abcam), anti-caspase-3 (1:500; cat. no. ab13847;
Abcam), anti-Bcl-2 (1:1,000; cat. no. ab32124; Abcam), anti-Cyclin
B1 (1:1,000; sc-245; Santa Cruz Biotechnology, Inc.) and anti-GAPDH
(1:5,000; cat. no. ab8245; Abcam) Following the primary antibody
incubation, the membranes were rinsed three times with TBS-0.05%
Tween-20 at 37°C and then incubated with a horseradish
peroxidase-conjugated goat anti-rabbit IgG (1:10,000; cat. no.
ab6721; Abcam) or rabbit anti-mouse IgG (1:10,000; cat. no. ab6728;
Abcam) secondary antibody at 37°C for 2 h. Protein bands were
visualized using an ECL reagent (EMD Millipore). Protein expression
was semi-quantified using ImageJ software version 1.46 (National
Institutes of Health) with GAPDH as the loading control.
Cell transfection
TOP2A overexpression plasmids [pcDNA3.1(+)-TOP2A]
were constructed and packaged by Shanghai GenePharma Co., Ltd.
Cells (1.5×106 cells/well) were transfected with 0.5
µg/µl TOP2A overexpression plasmids or empty plasmids as the
control group using Lipofectamine® 2000 (Invitrogen;
Thermo Fisher Scientific, Inc.) at 37°C for 24 h. Subsequently, the
cells were used for subsequent experiments.
Cell Counting Kit-8 (CCK-8) assay
Cell viability was assessed using a CCK-8 kit (cat.
no. CK04; Dojindo Molecular Technologies, Inc.), according to the
manufacturer's protocol. A total of 8×103 cells/well
were seeded into 96-well plates at 37°C with 5% CO2.
Cells were treated with 10 µg/ml PYM or 20 µM NCA at 37°C for 24 h.
Subsequently, 10 µl CCK-8 solution was added to each well and
incubated at 37°C for 4 h. The absorbance was detected at a
wavelength of 450 nm using a microplate reader (EnSpire;
PerkinElmer, Inc.).
Colony formation assay
Cells were quantified using a blood counting
chamber. Briefly, cells were seeded into a 10 ml petri-dish
(2×102 cells) containing 10 ml DMEM and 10% FBS, and
were cultured for 14 days at 37°C with 5% CO2. When
visible colonies (>50 cells) appeared in the petri-dishes, the
supernatant was discarded, and cells were washed twice with PBS.
Cells were fixed with 5 ml 10% methanol at 4°C for 10 min.
Following 0.1% crystal violet staining (Sigma-Aldrich; Merck KGaA)
at 37°C for 20 min, the number of cell colonies formed was counted
under a light microscope (magnification, ×20; CKX31SF; Olympus
Corporation).
TUNEL staining
Cells were fixed with 4% paraformaldehyde at 37°C
for 20 min and washed twice with PBS. Subsequently, cells were
treated using 0.2% Triton X-100 at 37°C for 5 min and washed twice
with PBS. Cells were incubated with the prepared TUNEL reaction
liquid at 37°C for 1 h. Following incubation, cells were washed
with PBS and stained with 80 µl 20% DAB at 37°C for 10 min. Cells
were subsequently stained with 5 µg/ml hematoxylin at 37°C for 5
sec. Dehydrated transparent neutral gum was used to mount the
sections. The cells were then observed under a fluorescent
microscope (magnification, ×200) in six randomly selected fields of
view to observe TUNEL positive cells.
Cell cycle analysis
The cell cycle distribution was detected using a
Cell Cycle kit (EZCell™ Cell Cycle Analysis kit; BioVision, Inc.).
Cells were seeded (1×105 cells/ml) into 6-well plates.
Following PYM or NCA treatment for 24 h at 37°C, cells were washed
twice with precooled PBS and fixed with precooled 70% ethanol for
24 h at 4°C. Cells were washed twice with PBS and incubated with
400 µl PI (50 µg/ml) and 100 µl RNaseA (100 µg/ml) for 1 h in the
dark at 4°C (BD Biosciences). Cell cycle distribution was analyzed
via flow cytometry (LSR-II; BD Biosciences) and analyzed using
FlowJo version 10 software (FlowJo LLC).
Co-immunoprecipitation (Co-IP)
Cells were collected and washed twice with PBS. Cell
lysate was prepared using 500 µl RIPA lysis buffer (Invitrogen;
Thermo Fisher Scientific, Inc.), centrifuged at 24,148.8 × g for 10
min at 4°C and part of the supernatant was used to perform western
blotting analysis as the input. Then, 1 µg anti-TOP2A (1:10,000;
cat. no. ab52934; Abcam) and anti-rabbit IgG (1:1,000; ab172730;
Abcam; negative control) primary antibodies were incubated with the
remaining supernatant at 4°C overnight. Following the incubation,
10 µl Protein A agarose beads (Cell Signaling Technology, Inc.)
were added to the remaining supernatant to capture antigen-antibody
complexes at 4°C for 4 h. Then, the supernatant was centrifuged for
3 min at 1,509.3 × g at 4°C. Immunoprecipitation was performed by
conducting western blotting according to the aforementioned
protocol.
Statistical analysis
Statistical analysis was performed using GraphPad
Prism 6 software (GraphPad Software, Inc.). Data are presented as
the mean ± SD of ≥3 experimental repeats. Comparisons among
multiple groups were analyzed using one-way ANOVA and a Tukey's
post hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
Expression levels of TOP2A in
different OSCC cell lines
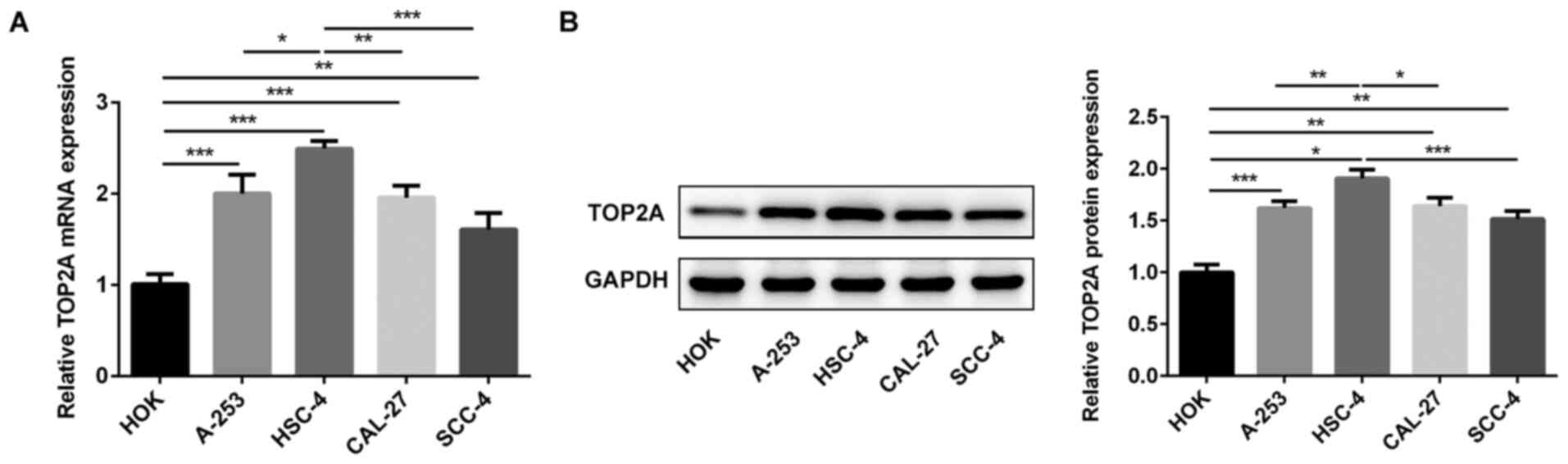
TOP2A expression levels were analyzed in HOK, A-253,
HSC-4, CAL-27 and SCC-4 cells via RT-qPCR and western blotting
(Fig. 1). Compared with HOK cells,
TOP2A expression levels were significantly upregulated in OSCC cell
lines. Moreover, the expression levels of TOP2A in HSC-4 cells were
significantly increased compared with the other OSCC cell lines.
Therefore, HSC-4 cells were used for subsequent experiments.
NCA increases the inhibitory effects
of PYM in OSCC via TOP2A
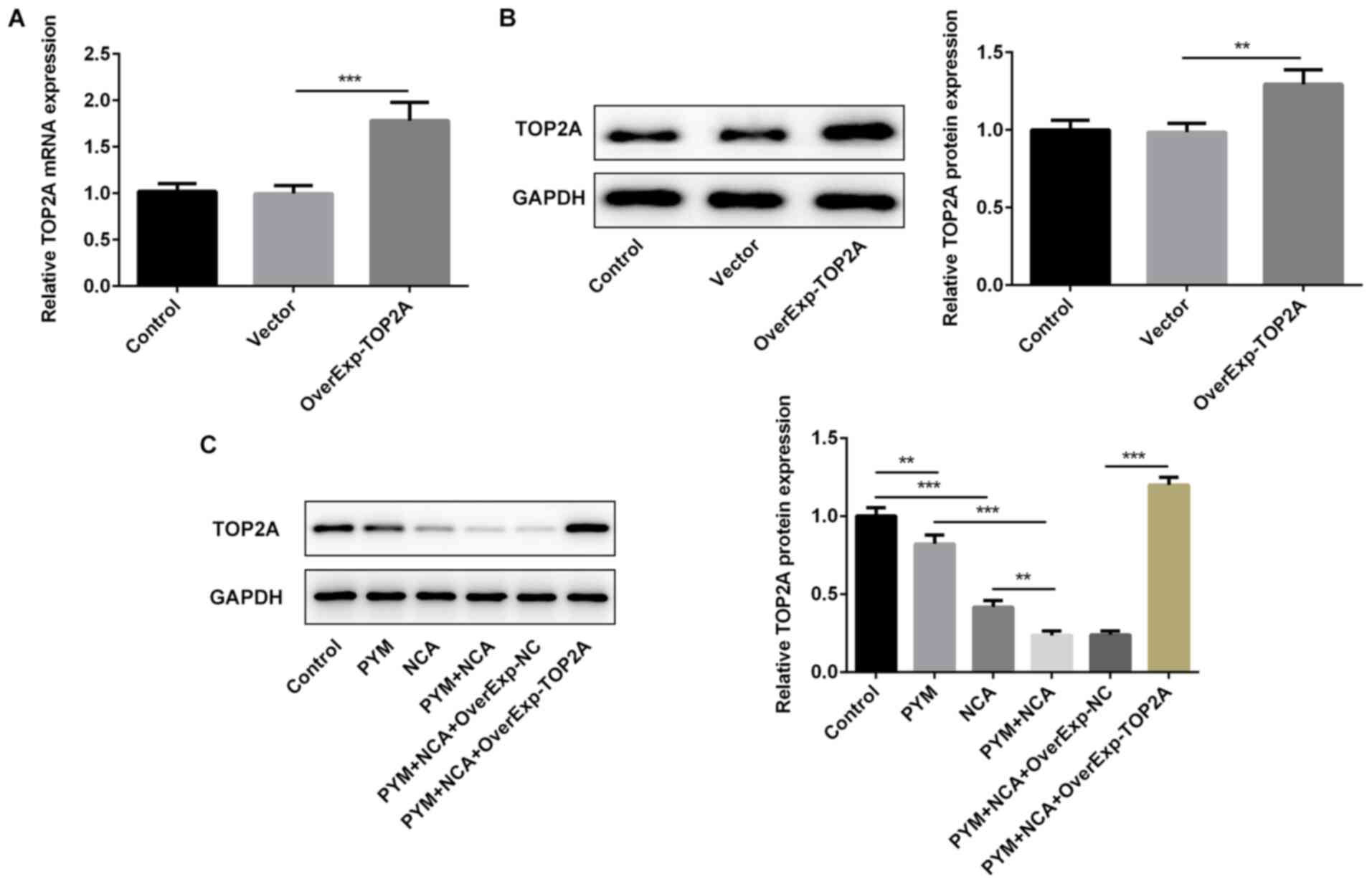
RT-qPCR and western blotting were performed to
assess the transfection efficacy of the TOP2A overexpression
plasmid (Fig. 2A and B). TOP2A
expression levels were significantly downregulated by PYM or NCA
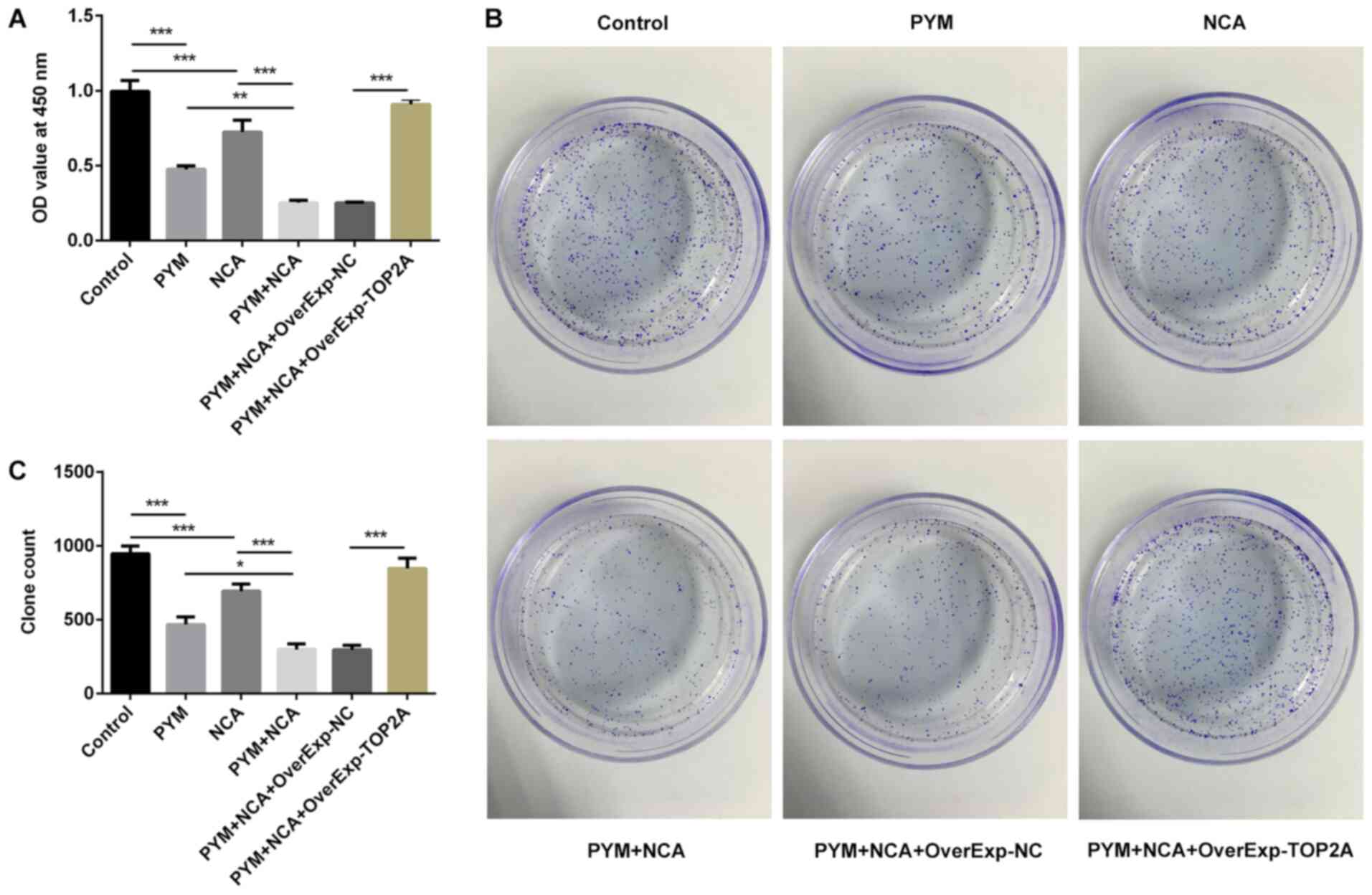
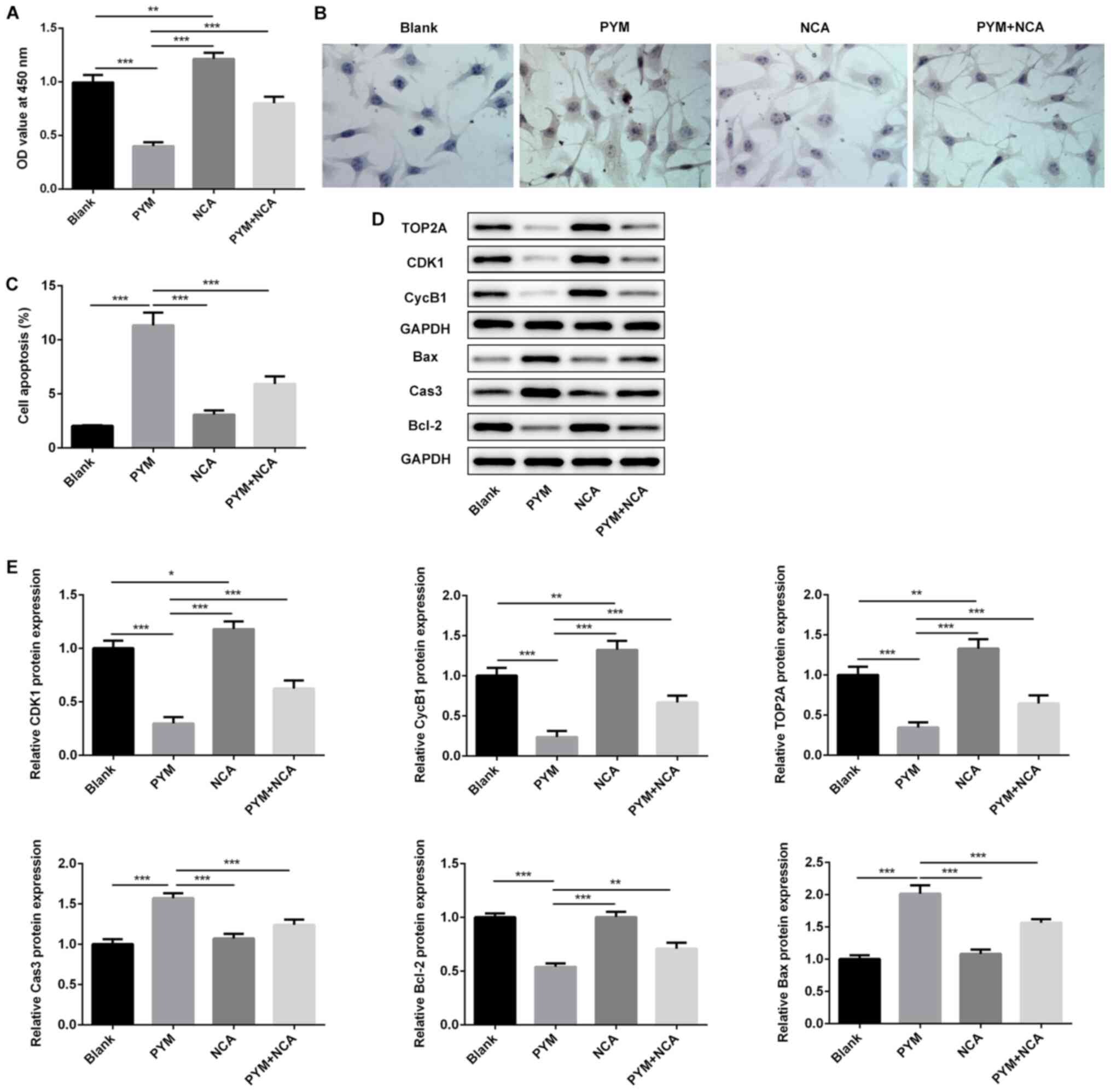
treatment compared with the control group (Fig. 2C). In addition, treatment with PYM
or NCA significantly suppressed HSC-4 cell proliferation, as
determined by conducting CCK-8 and colony formation assays
(Fig. 3A-C). The combination of NCA
and PYM treatment significantly increased the inhibitory effects
induced by either treatment alone on cell proliferation. Moreover,
TOP2A overexpression significantly reversed the suppressive effects
of NCA and PYM on cell proliferation. Therefore, the results
suggested that NCA may enhance the inhibitory effects of PYM on
HSC-4 cell proliferation by regulating TOP2A.
NCA enhances PYM-induced OSCC cell
apoptosis via TOP2A
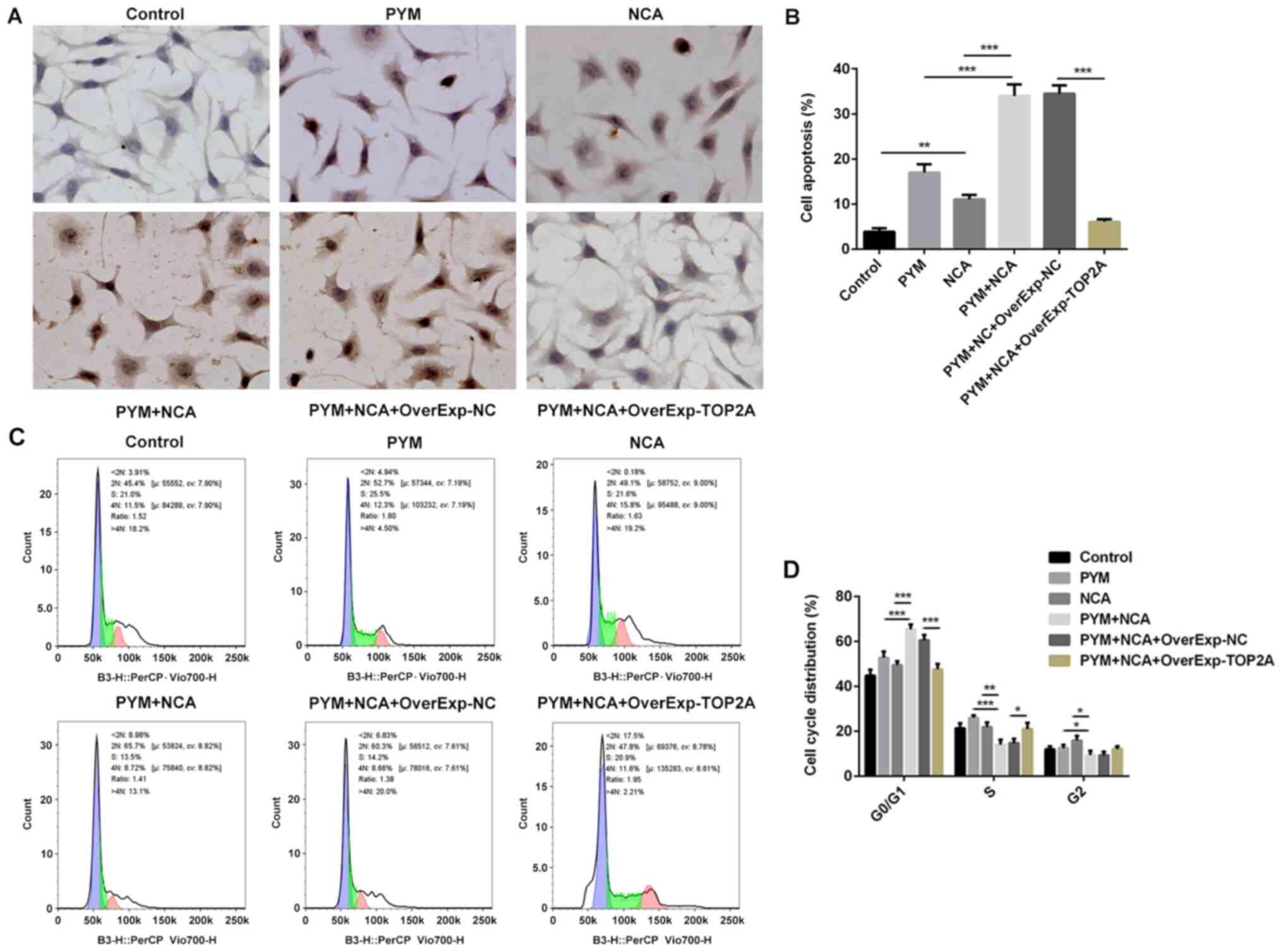
The TUNEL staining results indicated that NCA
enhanced PYM-induced OSCC cell apoptosis (Fig. 4A and B). In addition, the
combination of PYM and NCA treatment significantly increased cell
cycle arrest in the G0/1 and reduced cell
levels in the S phase compared with the PYM or NCA groups (Fig. 4C and D), indicating that the
combination of PYM and NCA treatment may suppress the transition
from G1 to S phase. To analyze the mechanisms underlying
PYM- and NCA-mediated regulation of the cell cycle, the expression
levels of cell cycle-related proteins were detected via western
blotting (Fig. 5A). The results
indicated that the expression levels of CDK1 and cyclin B1 were
significantly downregulated in the PYM and NCA groups compared with
the control group. Moreover, compared with the control group, the
expression levels of the anti-apoptotic protein Bcl-2 were
significantly downregulated, but the expression levels of the
proapoptotic proteins, Bax and caspase-3, were significantly
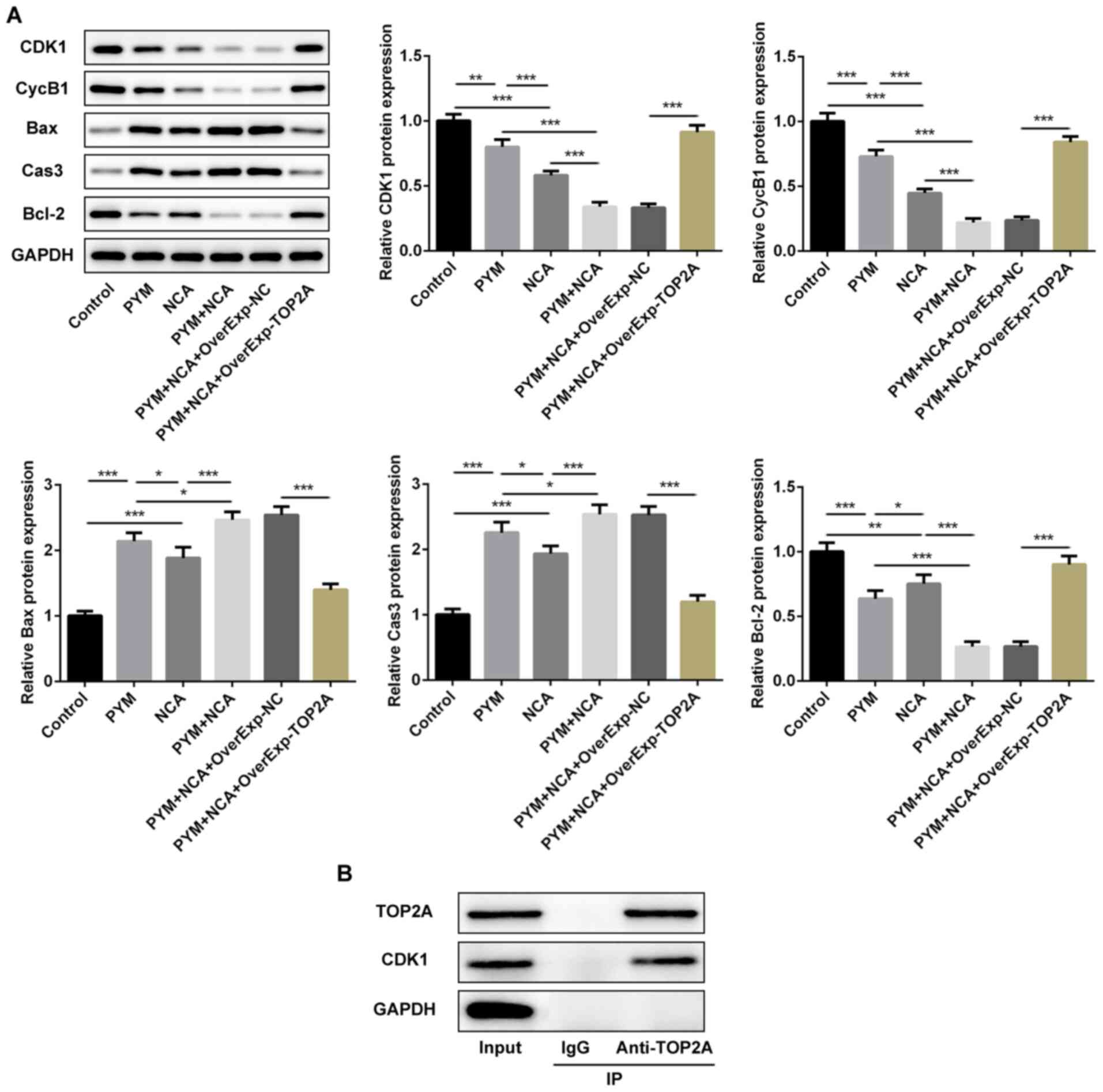
upregulated in the PYM and NCA groups (Fig. 5A). NCA enhanced the effects of PYM
treatment on cell cycle arrest and cell apoptosis, potentially by
regulating CDK1/cyclin B1 expression levels and the
mitochondrial-mediated apoptotic pathway in a TOP2A-dependent
manner. Furthermore, when the supernatant was incubated with an
anti-TOP2A antibody, CDK1 was detected in the immunoprecipitation
complex, which indicated that TOP2A may interact with CDK1
(Fig. 5B).
Effects of NCA on normal oral
epithelial cells
Subsequently, whether NCA exerted protective effects
against PYM-induced oral epithelial cell injury was investigated.
The CCK-8 assay and TUNEL staining results indicated that NCA
markedly decreased the toxicity of PYM in normal oral epithelial
cells (Fig. 6A-C). The results also
demonstrated that in normal cells, the combination of PYM and NCA
significantly reduced cell apoptosis compared with PYM alone
(Fig. 6B and C). Furthermore, NCA
decreased PYM-induced normal cell injury by regulating the cell
cycle and apoptotic pathways (Fig. 6D
and E). In HOK cells, NCA significantly upregulated TOP2A
expression compared with the PYM group (Fig. 6D and E), whereas NCA displayed the
opposite effects in HSC-4 cells (Fig.
2C). Therefore, NCA may regulate TOP2A expression in human
normal oral cells and OSCC cells via different mechanisms.
Discussion
PYM, as one of the main chemotherapy drugs in
preoperative induction chemotherapy for patients with OSCC, has
achieved good efficacy; however, the drug presents with increased
side effects, such as oral mucositis (19,20).
The present study indicated that the combined treatment of PYM and
NCA displayed improved, synergistic effects over the suppression of
HSC-4 cell proliferation. OSCC is the most common malignant type of
oral and maxillofacial tumor worldwide (21,22).
PYM induces a common side effect of chemotherapy, oral mucositis,
which can cause patients oral mucosal pain (23). Therefore, oral care has been
hypothesized to serve an important role in enhancing the wound
healing process during chemotherapy (24). A previous study suggested that drugs
derived from natural products may provide important therapeutic
effects for numerous types of human disease (25).
Honeysuckle is an effective compound that is used in
a number of mouthwashes (26).
Moreover, honeysuckle contains numerous types of organic acid
active ingredients, among which, chlorogenic acid has been
thoroughly studied (27–29). NCA, an isomer of chlorogenic acid,
displays significant anti-inflammatory, antitumor and
immune-promoting effects (30–32).
In the present study, the expression levels of TOP2A were
significantly upregulated in OSCC cells compared with HOK cells.
TOP2A and CDK1 expression levels have been reported to be
upregulated in certain types of cancer, where expression is
negatively associated with patient survival (33–35).
In addition, TOP2A inhibited the incorrect attachment of
microtubules and kinetochores, and was involved in suppressing
tumor cell proliferation (16,36).
Moreover, the TOP2A signaling pathway was also reported to mediate
cell cycle arrest (16). The
present study revealed that NCA promoted PYM-mediated cell cycle
arrest at the G0/1 phase via TOP2A. The checkpoint in
the G1/S phase determines whether the cell proliferates
(37). Under normal conditions,
G1 arrest prevents damaged DNA from replication and
helps to repair the damaged DNA (37). CDK1 serves an important role in
determining mitotic progression (38). CDK1/cyclin complexes phosphorylate
substrates that are involved in triggering centrosome separation,
Golgi dynamics, nuclear envelope breakdown and chromosome
condensation during the G2 phase and early mitosis
(37). Once CDK1 is inactivated
during cell injury, the cells are arrested at the G2
checkpoint to facilitate cell repair (37). However, a previous study has
reported that the absence of CDK1 compensated the CDK2 functions by
driving cells into the S phase (39). The results of the present study
indicated that NCA and PYM treatment may arrest cells in the
G1 phase, potentially by decreasing CDK1 and cyclin B
expression levels. A previous study revealed that downregulated
endogenous lysine acetyltransferase 2A expression levels promoted
G1/S transition, partially via downregulating CDK1
expression in an E2F transcription factor 1-dependent manner
(40). Furthermore, CDK1 has been
reported to be involved in G1 arrest (41). Previously, it was demonstrated that
the promotion of G1/S transition occurred via the
interaction of ZNFX1 antisense RNA 1 with the CDK1/cyclin B complex
(42). The present study indicated
that NCA treatment enhanced the antitumor effects of PYM by
arresting the cell cycle in the G0/1 phase in a
TOP2A-mediated manner. In addition, both NCA and PYM treatment
induced OSCC cell apoptosis, which was partially mediated via
regulating the TOP2A-mediated mitochondrial-dependent apoptotic
pathway. For normal cells, NCA did not inhibit proliferation, but
moderately promoted it, and at the same time reduced PYM-induced
apoptosis. It has been reported that NCA exerts antitumor effects
in a dose-dependent manner in vitro and suppresses tumor
growth in vivo in human gastric carcinoma cells (32). Moreover, NCA reduces ROS production
and suppresses mTOR/PI3K/Akt signaling (32). NCA also possesses strong free
radical scavenging and antioxidant activities (43). Furthermore, NCA has been reported to
activate the nuclear factor erythroid 2-related factor 2 signaling
pathway and induce 5′AMP-activated protein kinase phosphorylation
(44). Research has demonstrated
that Apocynum venetum tea extracts (AVTEs), which contain
NCA, exert antioxidation effects and promote cell survival in 293T
cells, whereas in HepG2 human hepatoma cells, AVTEs display
antitumor effects potentially by inducing cell apoptosis (45). Collectively, the aforementioned
studies and the present study indicated that NCA could display
different effects between normal cells and cancer cells via
different mechanisms. The present study indicated that NCA displays
antioxidant activities by promoting HOK cell survival, as indicated
by the CCK-8 assay and TUNEL staining results. A previous study
indicated that in etomoxir-induced oxidative stress, TOP2A
expression was downregulated (46).
In addition, the dose of NCA used in the present study displayed
different effects in HOK and HSC-4 cells. The Co-IP results
suggested that CDK1 interacted with TOP2A in HSC-4 cells, which
indicated that NCA enhanced the antitumor effects of PYM partly via
regulating the interaction of CDK1 and TOP2A. TOP2A and CDK1
expression levels are upregulated and are associated with low
survival in some types of cancer, such as adrenocortical (47) and hepatocellular carcinoma (48). By constructing a protein-protein
interaction network, a previous study identified a potential
relationship between CDK1 and TOP2A (49–51).
In addition to regulating the cell cycle, CDKs possess a broad
range of biological functions, including an involvement in DNA
damage repair and interactions with other proteins to regulate
tumor growth (52–54). In conclusion, the results of the
present study suggested that NCA may enhance the antitumor effects
of PYM by regulating TOP2A. Moreover, NCA may reduce the toxicity
of PYM in normal oral epithelial cells. Therefore, the use of NCA
may enhance the therapeutic effects of PYM chemotherapy in patients
with OSCC.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
JC and DL made substantial contributions to the
conception and design of the study, acquired, analyzed and
interpreted the data, and drafted and revised the manuscript for
important intellectual content; TZ, WL, SC, GY and XL performed the
experiments and interpreted the data. All authors read and approved
the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Gong JH, Liu XJ, Li Y and Zhen YS:
Pingyangmycin downregulates the expression of EGFR and enhances the
effects of cetuximab on esophageal cancer cells and the xenograft
in athymic mice. Cancer Chemother Pharmacol. 69:1323–1332. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
He Y, Lan Y, Liu Y, Yu H, Han Z, Li X and
Zhang L: Pingyangmycin and bleomycin share the same cytotoxicity
pathway. Molecules. 21:8622016. View Article : Google Scholar
|
|
3
|
Chen J and Stubbe J: Bleomycins: Towards
better therapeutics. Nat Rev Cancer. 5:102–112. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tai KW, Chang YC, Chou LS and Chou MY:
Cytotoxic effect of pingyangmycin on cultured KB cells. Oral Oncol.
34:219–223. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Sun ML, Wang CM and Wen YM: Anticancer
effects of Pingyangmycin-activated carbon nanoparticles against
human oral squamous carcinoma Tca8113 and BcaCD885 cell lines in
vitro. Hua Xi Kou Qiang Yi Xue Za Zhi. 28:257–260. 2010.(In
Chinese). PubMed/NCBI
|
|
6
|
Zhang L, Chen F, Zheng J, Wang H, Qin X
and Pan W: Chitosan-based liposomal thermogels for the controlled
delivery of pingyangmycin: Design, optimization and in vitro and in
vivo studies. Drug Deliv. 25:690–702. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Tu JB, Li QY, Jiang F, Hu XY, Ma RZ, Dong
Q, Zhang H, Pattar P and Li SX: Pingyangmycin stimulates apoptosis
in human hemangioma-derived endothelial cells through activation of
the p53 pathway. Mol Med Rep. 10:301–305. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Ning ZW, Zhai LX, Peng J, Zhao L, Huang T,
Lin CY, Chen WH, Luo Z, Xiao HT and Bian ZX: Simultaneous
UPLC-TQ-MS/MS determination of six active components in rat plasma:
Application in the pharmacokinetic study of Cyclocarya
paliurus leaves. Chin Med. 14:282019. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cho HD, Kim JH, Won YS, Moon KD and Seo
KI: Inhibitory effects of pectinase-treated Prunus mume
fruit concentrate on colorectal cancer proliferation and
angiogenesis of endothelial cells. J Food Sci. 84:3284–3295. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Sitarek P, Kowalczyk T, Santangelo S,
Bialas AJ, Toma M, Wieczfinska J, Sliwinski T and Skala E: The
extract of Leonurus sibiricus transgenic roots with AtPAP1
transcriptional factor induces apoptosis via DNA damage and down
regulation of selected epigenetic factors in human cancer cells.
Neurochem Res. 43:1363–1370. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
de Carvalho MC, Barca FN, Agnez-Lima LP
and de Medeirost SR: Evaluation of mutagenic activity in an extract
of pepper tree stem bark (schinus terebinthifolius raddi). Environ
Mol Mutagen. 42:185–191. 2003. View
Article : Google Scholar : PubMed/NCBI
|
|
12
|
Garcia-Carbonero R and Supko JG: Current
perspectives on the clinical experience, pharmacology, and
continued development of the camptothecins. Clin Cancer Res.
8:641–661. 2002.PubMed/NCBI
|
|
13
|
Chen AY and Liu LF: DNA topoisomerases:
Essential enzymes and lethal targets. Ann Rev Pharmacol Toxicol.
34:191–218. 1994. View Article : Google Scholar
|
|
14
|
Pendleton M, Lindsey RH, Felix CA,
Grimwade D and Osheroff N: Topoisomerase II and leukemia. Ann N Y
Acad Sci. 1310:98–110. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Ni W, Zhang S, Jiang B, Ni R, Xiao M, Lu
C, Liu J, Qu L, Ni H, Zhang W and Zhou P: Identification of
cancer-related gene network in hepatocellular carcinoma by combined
bioinformatic approach and experimental validation. Pathol Res
Pract. 215:1524282019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Liu L, Lin J and He H: Identification of
potential crucial genes associated with the pathogenesis and
prognosis of endometrial cancer. Front Genet. 10:3732019.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Lawal AO, Adisa AO and Effiom OA: A review
of 640 oral squamous cell carcinoma cases in Nigeria. J Clin Exp
Dent. 9:e767–e771. 2017.PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Li Pei, Yang Cheng, GU Xiaoming, et al:
Relationship between the expression of KI-67, VEGF and P16 and the
efficacy of pingyangmycin induced chemotherapy in oral squamous
cell carcinoma. 22:89–92
|
|
20
|
Ding HC: Pingyangmycin in the treatment of
oral squamous cell carcinoma: Effects and side effects. J Applied
Stomatol. 1:3–5. 1998.
|
|
21
|
Siegel R, Ward E, Brawley O and Jemal A:
Cancer statistics, 2011: The impact of eliminating socioeconomic
and racial disparities on premature cancer deaths. CA Cancer J
Clin. 61:212–236. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Peterson DE, Jones JB and Petit RG II:
Randomized, placebo-controlled trial of Saforis for prevention and
treatment of oral mucositis in breast cancer patients receiving
anthracycline-based chemotherapy. Cancer. 109:322–331. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Madan PD, Sequeira PS, Shenoy K and Shetty
J: The effect of three mouthwashes on radiation-induced oral
mucositis in patients with head and neck malignancies: A randomized
control trial. J Cancer Res Ther. 4:3–8. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Iwamoto M, Morikawa T, Narita M, Shibahara
T and Katakura A: Investigation of surgical site infections and
bacteria detected following neck dissection in patients with oral
cancer. Bull Tokyo Dent Coll. 61:1–7. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Li W, Jinyi L, Weiqi F, Xiangjin Z, Liwen
R, Shiwei L, Jinhua W, Tengfei J and Guanhua D:
3-O-acetyl-11-keto-β-boswellic acid exerts anti-tumor effects in
glioblastoma by arresting cell cycle at G2/M phase. J Exp Clin
Cancer Res. 37:1322018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu D, Zhao J, Sun H, He S, Wang J and
Zhang Q: Preparation technology of an antibacterial mouthwash. J
Food Safety Quality Inspection. 10:638–644. 2019.
|
|
27
|
Chaowuttikul C, Palanuvej C and
Ruangrungsi N: Pharmacognostic specification, chlorogenic acid
content, and in vitro antioxidant activities of Lonicera
japonica flowering bud. Pharmacognosy Res. 9:128–132.
2017.PubMed/NCBI
|
|
28
|
Ye LH, Du LJ and Cao J: Fatty acids-based
microemulsion liquid chromatographic determination of multiple
caffeoylquinic acid isomers and caffeic acid in honeysuckle sample.
J Pharm Biomed Anal. 171:22–29. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Chaovanalikit A, Thompson MM and Wrolstad
RE: Characterization and quantification of anthocyanins and
polyphenolics in bluehHoneysuckle (Lonicera caerulea L.). J
Agric Food Chem. 52:848–852. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Zhao Z, Shin HS, Satsu H, Totsuka M and
Shimizu M: 5-caffeoylquinic acid and caffeic acid down-regulate the
oxidative stress- and TNF-alpha-induced secretion of interleukin-8
from Caco-2 cells. J Agric Food Chem. 56:3863–3868. 2008.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Fang W, Ma Y, Wang J, Yang X, Gu Y and Li
Y: In vitro and in vivo antitumor activity of neochlorogenic acid
in human gastric carcinoma cells are complemented with ROS
generation, loss of mitochondrial membrane potential and apoptosis
induction. J BUON. 24:221–226. 2019.PubMed/NCBI
|
|
32
|
De Maria CAB, Moreira Santos MC, José De
Lima Dias U and Marana M: Stabilization of soybean oil with heated
quercetin and 5-caffeoylquinic acid in the presence of ferric ion.
J Agric Food Chem. 48:3935–3938. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hossain MA, Asa TA, Rahman MM, Uddin S,
Moustafa AA, Quinn JMW and Moni MA: Network-based genetic profiling
reveals cellular pathway differences between follicular thyroid
carcinoma and follicular thyroid adenoma. Int J Environ Res Public
Health. 17:13732020. View Article : Google Scholar
|
|
34
|
Pabla S, Conroy JM, Nesline MK, Glenn ST,
Papanicolau-Sengos A, Burgher B, Hagen J, Giamo V, Andreas J, Lenzo
FL, et al: Proliferative potential and resistance to immune
checkpoint blockade in lung cancer patients. J Immunother Cancer.
7:272019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Xue JM, Liu Y, Wan LH and Zhu YX:
Comprehensive analysis of differential gene expression to identify
common gene signatures in multiple cancers. Med Sci Monit.
26:e9199532020. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Coelho PA, Queiroz-Machado J, Carmo AM,
Moutinho-Pereira S, Maiato H and Sunkel CE: Dual role of
topoisomerase II in centromere resolution and aurora B activity.
PLoS Biol. 6:e2072008. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Ben-Shlomo R: Chronodisruption, cell cycle
checkpoints and DNA repair. Indian J Exp Biol. 52:399–403.
2014.PubMed/NCBI
|
|
38
|
Sasaki M, Terabayashi T, Weiss SM and
Ferby I: The tumor suppressor MIG6 controls mitotic progression and
the G2/M DNA damage checkpoint by stabilizing the WEE1 kinase. Cell
Rep. 24:1278–1289. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Satyanarayana A and Kaldis P: Mammalian
cell-cycle regulation: Several Cdks, numerous cyclins and diverse
compensatory mechanisms. Oncogene. 28:2925–2939. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Qiao L, Zhang Q, Zhang W and Chen JJ: The
lysine acetyltransferase GCN5 contributes to human papillomavirus
oncoprotein E7-induced cell proliferation via up-regulating E2F1. J
Cell Mol Med. 22:5333–5345. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fan X and Chen JJ: Role of Cdk1 in DNA
damage-induced G1 checkpoint abrogation by the human papillomavirus
E7 oncogene. Cell Cycle. 13:3249–3259. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Thorenoor N, Faltejskova-Vychytilova P,
Hombach S, Mlcochova J, Kretz M, Svoboda M and Slaby O: Long
non-coding RNA ZFAS1 interacts with CDK1 and is involved in
p53-dependent cell cycle control and apoptosis in colorectal
cancer. Oncotarget. 7:622–637. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Kulisic-Bilusic T, Schnäbele K, Schmöller
I, Dragovic-Uzelac V, Krisko A, Dejanovic B, Milos M and Pifat G:
Antioxidant activity versus cytotoxic and nuclear factor kappa B
regulatory activities on HT-29 cells by natural fruit juices. Eur
Food Res Technol. 228:417–424. 2009. View Article : Google Scholar
|
|
44
|
Gao XH, Zhang SD, Wang LT, Yu L, Zhao XL,
Ni HY, Wang YQ, Wang JD, Shan CH and Fu YJ: Anti-inflammatory
effects of neochlorogenic acid extract from mulberry leaf (Morus
alba L.) against LPS-stimulated inflammatory response through
mediating the AMPK/Nrf2 signaling pathway in A549 cells. Molecules.
25:13852020. View Article : Google Scholar
|
|
45
|
Li C, Huang G, Tan F, Zhou X, Mu J, Zhao X
and Giaouris E: In vitro analysis of antioxidant, anticancer, and
bioactive components of Apocynum venetum tea extracts. J
Food Quality. 2019:1–13. 2019. View Article : Google Scholar
|
|
46
|
Merrill CL, Ni H, Yoon LW, Tirmenstein MA,
Narayanan P, Benavides GR, Easton MJ, Creech DR, Hu CX, McFarland
DC, et al: Etomoxir-induced oxidative stress in HepG2 cells
detected by differential gene expression is confirmed
biochemically. Toxicol Sci. 68:93–101. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Xiao H, Xu D, Chen P, Zeng G, Wang X and
Zhang X: Identification of five genes as a potential biomarker for
predicting progress and prognosis in adrenocortical carcinoma. J
Cancer. 9:4484–4495. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhou Z, Li Y, Hao H, Wang Y, Zhou Z, Wang
Z and Chu X: Screening hub genes as prognostic biomarkers of
hepatocellular carcinoma by bioinformatics analysis. Cell
Transplant. 28 (Suppl 1):S76–S86. 2019. View Article : Google Scholar
|
|
49
|
Zhao ZW, Fan XX, Yang LL, Song JJ, Fang
SJ, Tu JF, Chen MJ, Zheng LY, Wu FZ, Zhang DK, et al: The
identification of a common different gene expression signature in
patients with colorectal cancer. Math Biosci Eng. 16:2942–2958.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Pan Z, Li L, Fang Q, Qian Y, Zhang Y, Zhu
J, Ge M and Huang P: Integrated bioinformatics analysis of master
regulators in anaplastic thyroid carcinoma. Biomed Res Int.
2019:97345762019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zhu X, Wang D, Lin Q, Wu G, Yuan S, Ye F
and Fan Q: Screening key lncRNAs for human rectal adenocarcinoma
based on lncRNA-mRNA functional synergistic network. Cancer Med.
8:3875–3891. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Menon DR, Luo Y, Arcaroli JJ, Liu S,
KrishnanKutty LN, Osborne DG, Li Y, Samson JM, Bagby S, Tan AC, et
al: CDK1 interacts with Sox2 and promotes tumor initiation in human
melanoma. Cancer Res. 78:6561–6574. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Lim S and Kaldis P: Cdks, cyclins and
CKIs: Roles beyond cell cycle regulation. Development.
140:3079–3093. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Gan W, Zhao H, Li T, Liu K and Huang J:
CDK1 interacts with iASPP to regulate colorectal cancer cell
proliferation through p53 pathway. Oncotarget. 8:71618–71629. 2017.
View Article : Google Scholar : PubMed/NCBI
|