Introduction
Oral squamous cell carcinoma (OSCC) is one of the
most common malignancies worldwide. It was estimated that
>50,000 new cases of OSCC were reported in the United States in
2018, which resulted in 10,000 deaths (1). The current therapies used to treat
OSCC include surgery, chemotherapy and radiotherapy (2). Although advances in the diagnosis and
clinical treatment of OSCC have been made, the 5-year survival
rates of patients with OSCC remain relatively low, due to the
aggressive progression and recurrence of the disease (3,4).
Therefore, it is crucial to identify potential biomarkers to
improve the prognosis and therapeutic strategies of OSCC.
MicroRNAs (miRNAs/miRs) are short non-coding RNAs
that regulate the degradation and translational process of mRNAs by
directly binding to the 3′-untranslated region (3′-UTR) of target
genes (5,6). Accumulating evidence has indicated
that the dysregulation of miRNAs contributes to the initiation and
development of various types of human cancer, including pancreatic
cancer (7), gastric cancer
(8) and breast cancer (9). Numerous studies have revealed that
miR-149-3p acts as a tumor suppressor. For example, miR-149-3p was
shown to modulate polo-like kinase 1 (PLK1) levels by competing
with heterogeneous nuclear ribonucleoprotein K in the 3′-UTR of
PLK1 mRNA, which led to the decreased growth of HeLa cells
(10). In addition, miR-149-3p
downregulated S100A4 expression, and suppressed the proliferation
and metastasis of bladder cancer (11). Conversely, a recent study reported
that miR-149-3p attenuated the apoptosis and depletion of
CD8+ T cells, suggesting a potential protective role of
miR-149-3p in CD8+ T cells (12). Moreover, miR-149-3p promoted the
aggressiveness of cancer cells via inhibition of DAB2-interacting
protein in a cell-autonomous and non-autonomous manner (13). However, to the best of our
knowledge, the functional role and mechanisms of miR-149-3p in OSCC
remain unknown.
In the present study, overexpression of miR-149-3p
attenuated cell viability and proliferation, whereas miR-149-3p
knockdown augmented the viability and proliferation of OSCC cells.
The data further demonstrated that miR-149-3p decreased AKT2
expression by directly binding to the 3′-UTR of AKT2 mRNA.
Moreover, AKT2 efficiently restored the inhibited proliferative
capacity of OSCC cells induced by miR-149-3p. Notably,
miR-149-3p-overexpressing OSCC cells exhibited higher sensitivity
to the chemotherapeutic drug 5-fluorouracil (5-Fu). Taken together,
to the best of our knowledge, the present study was the first to
indicate that miR-149-3p inhibited the proliferation of OSCC cells
and improved the efficacy of 5-Fu in OSCC cells by targeting
AKT2.
Materials and methods
Cell culture and reagents
The human OSCC Cal27 and SCC-9 cells were purchased
from American Type Culture Collection. Cal-27 cells were grown in
DMEM (Gibco; Thermo Fisher Scientific, Inc.) with 10% FBS (Gibco;
Thermo Fisher Scientific, Inc.), and SCC-9 cells were cultured in
DMEM/Ham's F-12 medium (Gibco; Thermo Fisher Scientific, Inc.)
supplemented with 400 ng/ml hydrocortisone (Sigma-Aldrich; Merck
KGaA) and 10% FBS, according to the manufacturer's instructions.
The MSK-Leuk1 Cells were maintained in EpiLife™ Medium (Gibco;
Thermo Fisher Scientific, Inc.) supplemented with Human
Keratinocyte Growth Supplement (Gibco; Thermo Fisher Scientific,
Inc.). Cells were maintained in an atmosphere containing 5%
CO2 at 37°C.
The pcDNA3-Myr-HA-Akt2 (plasmid no. 9016; Addgene,
Inc.) was provided as a gift from Professor William Sellers
(Dana-Farber Cancer Institute, USA). 5-Fu was purchased from
Selleck Chemicals and crystal violet was obtained from Sangon
Biotech Co., Ltd.
Transfection
The commercial miR-149-3p mimic (cat. no.
miR10004609), mimic control (cat. no. miR1N0000001), miR-149-3p
inhibitor (cat. no. miR20004609) and inhibitor control (cat. no.
miR2N0000001) were purchased from Guangzhou RiboBio Co., Ltd. Cal27
and SCC-9 cells were grown to ~50% confluence, and then miR-149-3p
mimic (50 nM), mimic control (50 nM), miR-149-3p inhibitor (100 nM)
or inhibitor control (100 nM) were transiently transfected into
OSCC cells using Lipofectamine® RNAiMAX Transfection
Reagent (Invitrogen; Thermo Fisher Scientific, Inc.), according to
the manufacturer's instructions. After 48 h of transfection at
37°C, cells were harvested for subsequent reverse
transcription-quantitative polymerase chain reaction (RT-qPCR)
analysis. For AKT2 overexpression, Cal27 and SCC-9 cells were
plated into 6-well plates, and grown to ~50% confluence. Then, 4 µg
pcDNA3-Myr-HA-Akt2 plasmid or empty pcDNA3 plasmid was transiently
transfected into OSCC cells using Lipofectamine® 3,000
Transfection Reagent (Invitrogen; Thermo Fisher Scientific, Inc.).
After 48 h of transfection at 37°C, cells were harvested for
subsequent RT-qPCR and western blot analysis.
RNA extraction and RT-qPCR
Total RNA was isolated from OSCC cells using
TRIzol® reagent (Invitrogen; Thermo Fisher Scientific,
Inc.). The cDNA was reverse transcribed from RNA at 37°C for 1 h
and 85°C for 5 min using an All-in-One cDNA Synthesis SuperMix
(GeneCopoeia, Inc.). For miRNA quantification, the expression
levels of miR-149-3p were detected using an All-in-one miRNA qPCR
kit (GeneCopoeia, Inc.) according to the manufacturer's
instructions. The following thermocycling conditions for qPCR were
used: Initial denaturation at 95°C for 10 min; and 40 cycles of
denaturation at 95°C for 10 sec, annealing at 60°C for 20 sec, and
extension at 72°C for 15 sec. For AKT2 quantification, the
expression levels of AKT2 were detected with TB Green®
Premix Ex Taq™ II (Tli RNaseH Plus) (Takara Biotechnology Co.,
Ltd.). The following thermocycling conditions for qPCR were used:
Initial denaturation at 95°C for 5 min; and 40 cycles of
denaturation at 95°C for 10 sec, and annealing and extension at
60°C for 45 sec. The following primers were used: miR-149-3p
forward, 5′-ACAGGGAGGGACGGGGG-3′ and reverse,
5′-CAGTGCAGGGTCCGAGGTATT-3′; U6 forward,
5′-CAAATTCGTGAAGCGTTCCATA-3′ and reverse,
5′-AGTGCAGGGTCCGAGGTATTC-3′; AKT2 forward,
5′-TCCGAGGTCGACACAAGGTA-3′ and reverse, 5′-CTGGTCCAGCTCCAGTAAGC-3′;
β-actin forward, 5′-CTCCATCCTGGCCTCGCTGT-3′ and reverse,
5′-GCTGTCACCTTCACCGTTCC-3′. The 2−∆∆Cq method was used
for all qPCR quantification (14).
U6 snRNA and β-actin were used as an endogenous control for
miR-149-3p and AKT2, respectively.
Western blot analysis
OSCC cells were collected and lysed with NP-40 lysis
buffer (Beyotime Institute of Biotechnology). Protein concentration
was quantified using a Pierce Detergent Compatible Bradford Assay
kit (Pierce; Thermo Fisher Scientific, Inc.). Cell lysates (30 µg)
were resolved by SDS-PAGE on 12% gel, and subsequently transferred
to PVDF membranes. After blocking with 5% skimmed milk at room
temperature for 1 h, the membrane was incubated overnight at 4°C
with the following primary antibodies: AKT2 (1:1,000; cat. no.
3063; Cell Signaling Technology, Inc.), cleaved caspase-3 (1:1,000;
cat. no. 9664; Cell Signaling Technology, Inc.), cleaved PARP
(1:1,000; cat. no. 5625; Cell Signaling Technology, Inc.) and
β-actin (1:1,000; cat. no. 8457; Cell Signaling Technology, Inc.).
After incubation with the primary antibody, the membrane was washed
in TBS with 0.05% Tween-20 three times, and then incubated with a
horseradish peroxidase-conjugated goat anti-rabbit antibody
(1:3,000; cat. no. 7074; Cell Signaling Technology, Inc.) at room
temperature for 1 h. The protein bands were developed using
Immobilon™ Western Chemiluminescent HRP Substrate (EMD Millipore).
The images were analyzed by ImageJ software (version no. 1.52a;
National Institutes of Health) using β-actin as the loading
control.
Cell Counting Kit-8 (CCK-8) assay
Cell viability was estimated using the CCK-8 assay
(Dojindo Molecular Technologies, Inc.), according to the
manufacturer's protocol. Briefly, OSCC cells transfected with
miR-149-3p mimic or inhibitor were seeded in 96-well plates
(5×103 cells/well). After 24 h of transfection at 37°C,
the cells were cultured for another 24, 48, 72 or 96 h and then
incubated with 10 µl CCK-8 reagent at 37°C for 2 h. For 5-Fu
treatment, the cells were treated with 10 µM 5-Fu (Selleck
Chemicals) at 37°C for 24 h, and then incubated with 10 µl CCK-8
reagent at 37°C for 2 h. The absorbance of samples at 450 nm was
determined using a microplate reader.
Colony formation assay
OSCC cells transfected with miR-149-3p mimic or
inhibitor were plated in 24-well plates (500 cells/well). Cells
were grown to form colonies for 12 days. Subsequently, the colonies
were stained with crystal violet at room temperature for 20 min,
and colony numbers were calculated using ImageJ (version number
1.52a) software (National Institutes of Health).
Flow cytometry assay
For the dose curve of 5-Fu treatment, OSCC cells
transfected with miR-149-3p mimic or mimic control were treated
with 5, 10 or 20 µM 5-Fu at 37°C for 24 h. For the time curve of
5-Fu treatment, OSCC cells transfected with miR-149-3p mimic or
mimic control were treated with 10 µM 5-Fu at 37°C for 12, 24 or 48
h. Then, cells were harvested and stained Annexin V-FITC and PI at
room temperature for 15 min using the Annexin V-FITC Apoptosis
Detection Kit (Nanjing KeyGen Biotech Co., Ltd.), according to the
manufacturer's instructions. Subsequently, the cells were analyzed
with a flow cytometer (FACSAria™; BD Biosciences) and FlowJo
software (version 10.0.7; Tree Star, Inc.). The cells in Q2
(late-stage apoptosis) and Q3 (early-stage apoptosis) were
considered to be apoptotic cells.
Dual luciferase reporter assay
The target genes of miR-149-3p were predicted using
TargetScan (version 7.2; http://www.targetscan.org/vert_72/) and miRDB (version
6.0; http://www.mirdb.org/index.html). The
mutated (MT) AKT2 3′-UTR fragments containing the putative binding
site of miR-149-3p were generated using Fast MultiSite Mutagenesis
System (cat. no. FM201; TransGen Biotech Co., Ltd.), according to
the manufacturer's protocol. The human wild-type (WT) and MT AKT2
3′-UTR fragments were inserted into the pEZX-FR02 luciferase
reporter vector (GeneCopoeia, Inc.). Cal27 and SCC-9 cells were
seeded in 48-well plates, and grown to ~50% confluence. Then, the
cells were co-transfected with 0.5 µg WT or MT AKT2 3′-UTR and 50
nM miR-149-3p mimic or mimic control using Lipofectamine 3,000
Transfection Reagent. After transfection for 36 h at 37°C, the
luciferase activity was detected using the Dual-Luciferase Reporter
Assay system (Promega Corporation). The firefly luciferase activity
was normalized to the Renilla luciferase activity.
Statistical analysis
All quantitative data are presented as the mean ± SD
for at least three independent experiments. Student's t-test or
one-way ANOVA followed by Tukey's post hoc test were used to assess
the differences between two and more than two groups, respectively.
Pearson correlation analysis was used to analyze the correlation
between the expression levels of miR-149-3p and AKT2 in TCGA head
and neck squamous cell carcinoma dataset (https://www.cancer.gov/about-nci/organization/ccg/research/structural-genomics/tcga)
(15). The statistical analyses
were performed using SPSS software version 22.0 (IBM Corp.).
P<0.05 was considered to indicate a statistically significant
difference.
Results
miR-149-3p inhibits the proliferation
of OSCC cells
To explore the role of miR-149-3p in OSCC
tumorigenesis, the expression levels of miR-149-3p were detected in
OSCC cell lines (Cal27 and SCC-9 cells) and in the non-tumorigenic
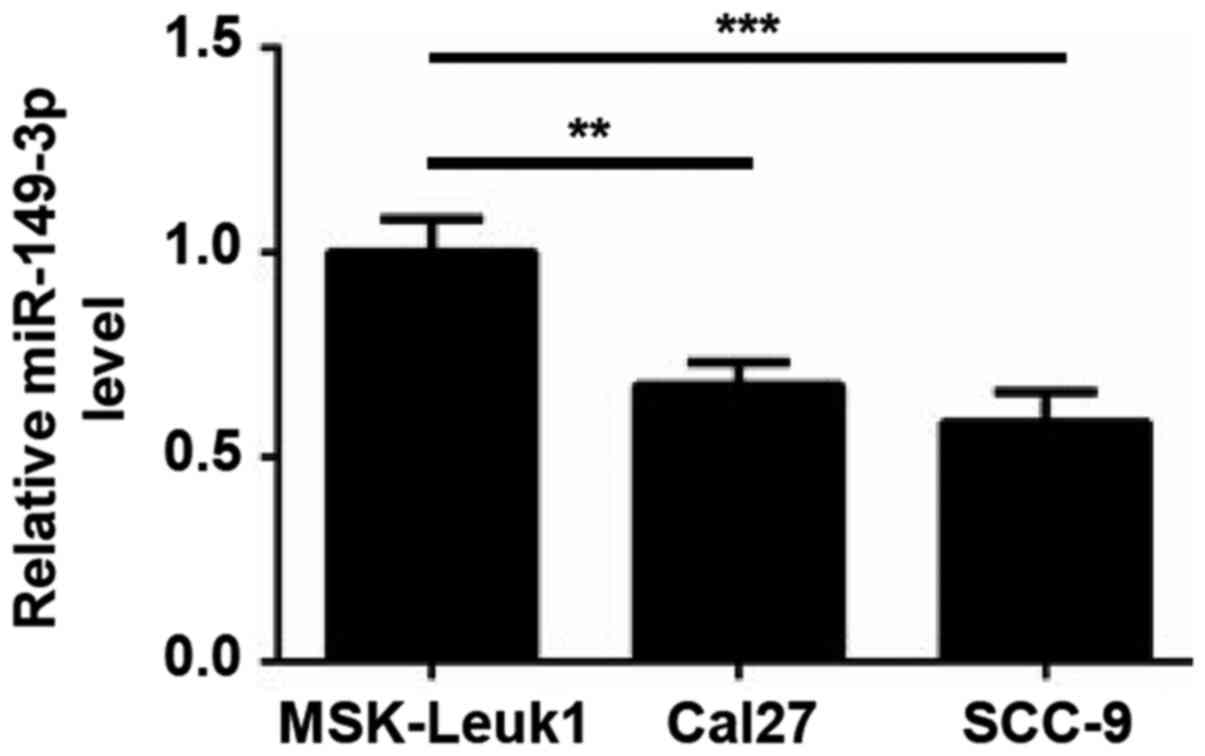
oral epithelial cell line MSK-Leuk1. As shown in Fig. 1, lower expression levels of
miR-149-3p were observed in Cal27 and SCC-9 cells compared with in
MSK-Leuk1 cells, indicating that miR-149-3p may be involved in OSCC
progression. To investigate the functional effect of miR-149-3p on
the proliferation of OSCC cells, Cal27 and SCC-9 cells were
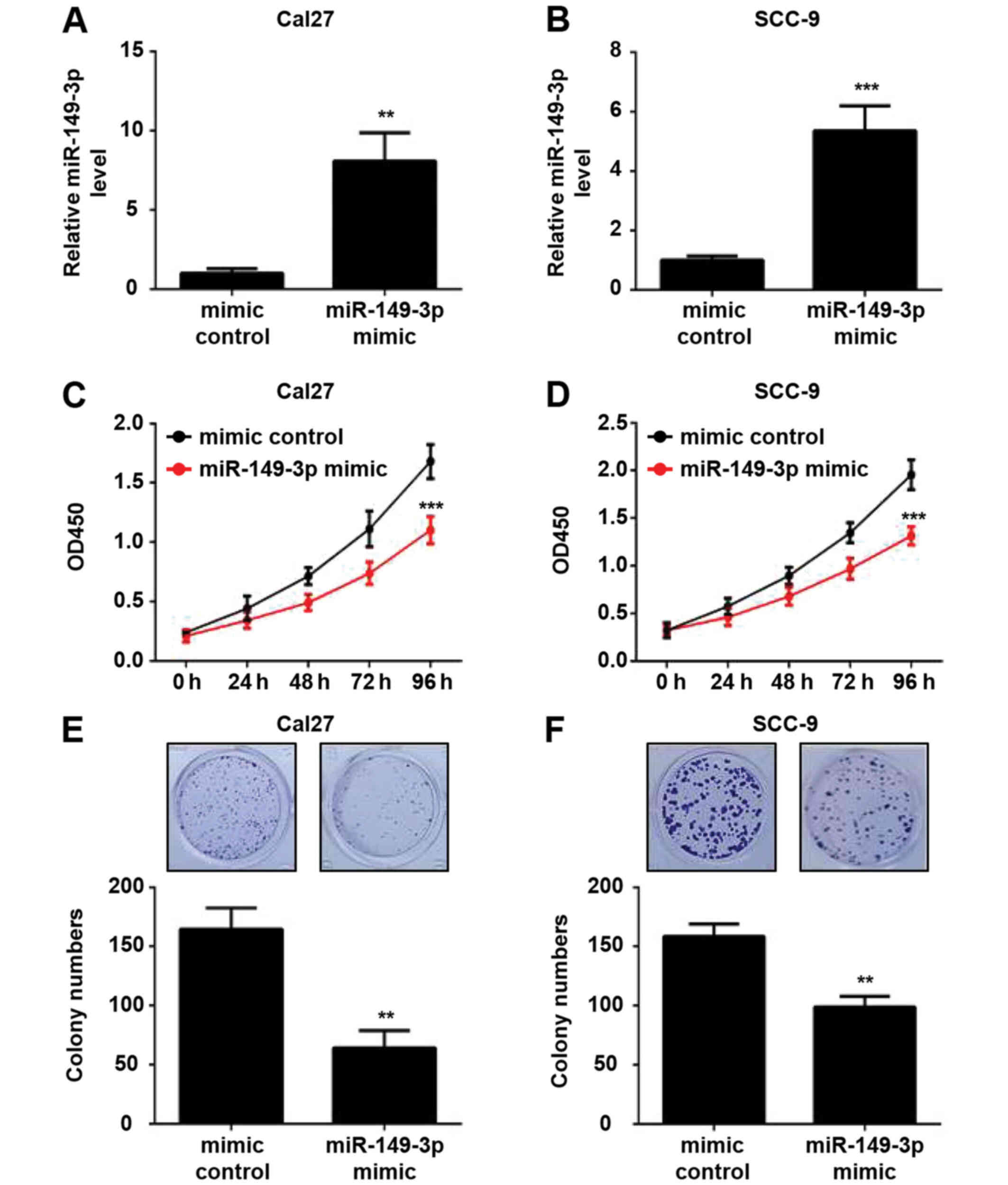
transfected with the miR-149-3p mimic or mimic control.
Post-transfection, the cells were cultured for 48 h and the
expression levels of miR-149-3p were verified by RT-qPCR analysis.
The results revealed an ~8.1- and 5.4-fold increase in the
expression levels of miR-149-3p in miR-149-3p mimic-transfected
Cal27 and SCC-9 cells compared with in mimic control-transfected
cells, respectively (Fig. 2A and
B). CCK-8 assays revealed that miR-149-3p overexpression
significantly suppressed the viability of OSCC cells (Fig. 2C and D). Moreover, the
overexpression of miR-149-3p diminished the colony-forming ability
of OSCC cells (Fig. 2E and F).
These data indicated that miR-149-3p inhibited the proliferation of
OSCC cells.
Knockdown of miR-149-3p promotes the
proliferation of OSCC cells
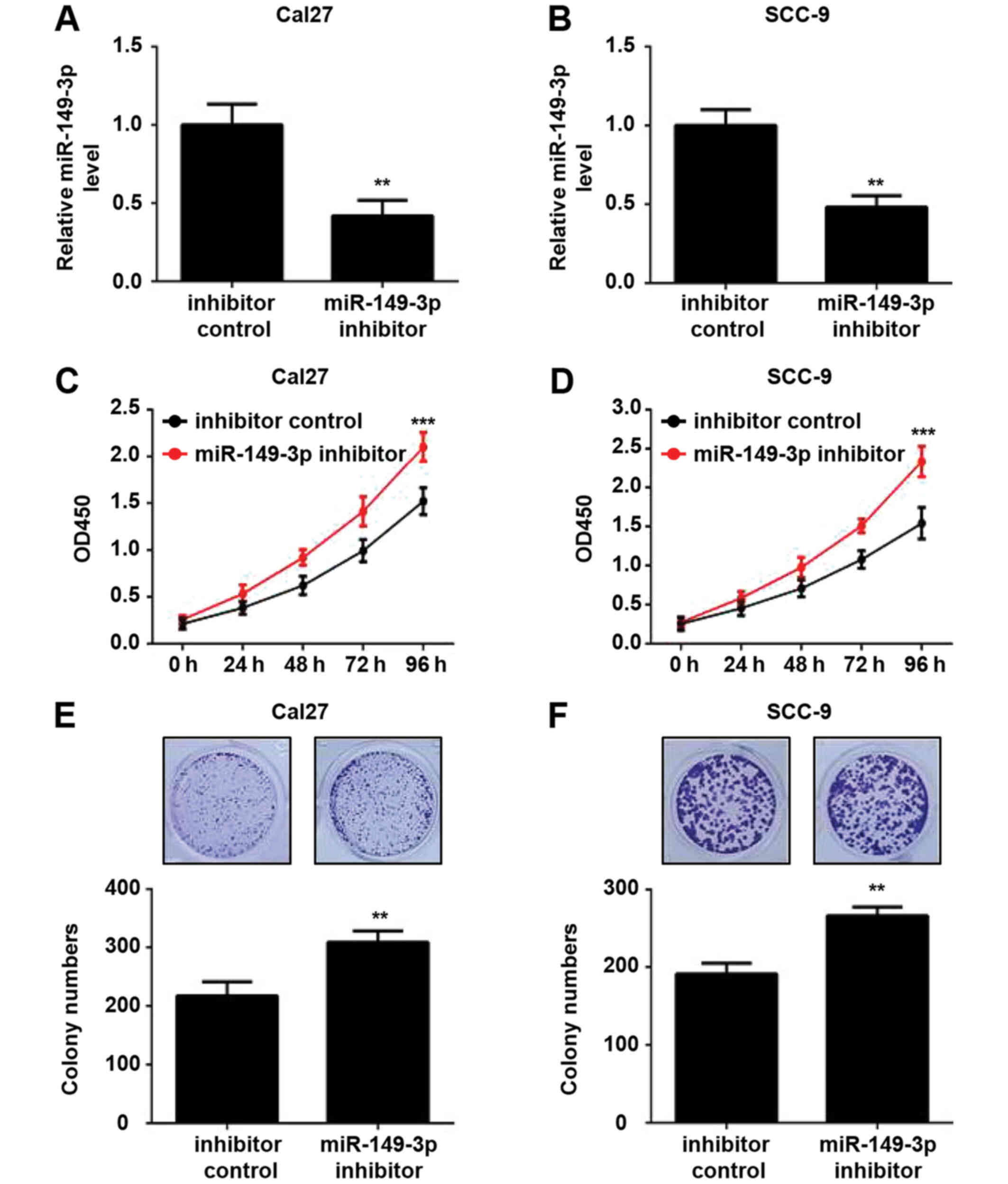
Subsequently, the effects of miR-149-3p knockdown on
the proliferation of OSCC cells were investigated. The expression
levels of miR-149-3p were downregulated in OSCC cells transfected
with the miR-149-3p inhibitor compared with those transfected with
the inhibitor control, as determined by RT-qPCR analysis (Fig. 3A and B). Silencing of miR-149-3p
efficiently augmented the viability of OSCC cells (Fig. 3C and D). Similar results were
observed in the colony formation assay (Fig. 3E and F). These data indicated that
miR-149-3p knockdown promoted the proliferation of OSCC cells.
AKT2 is a direct target of
miR-149-3p
To explore the molecular mechanisms by which
miR-149-3p suppresses the proliferation of OSCC cells, the target
genes of miR-149-3p were analyzed using TargetScan and miRDB
databases; the genes predicted by both analyses were considered
potential target genes of miR-149-3p. According to this strategy, a
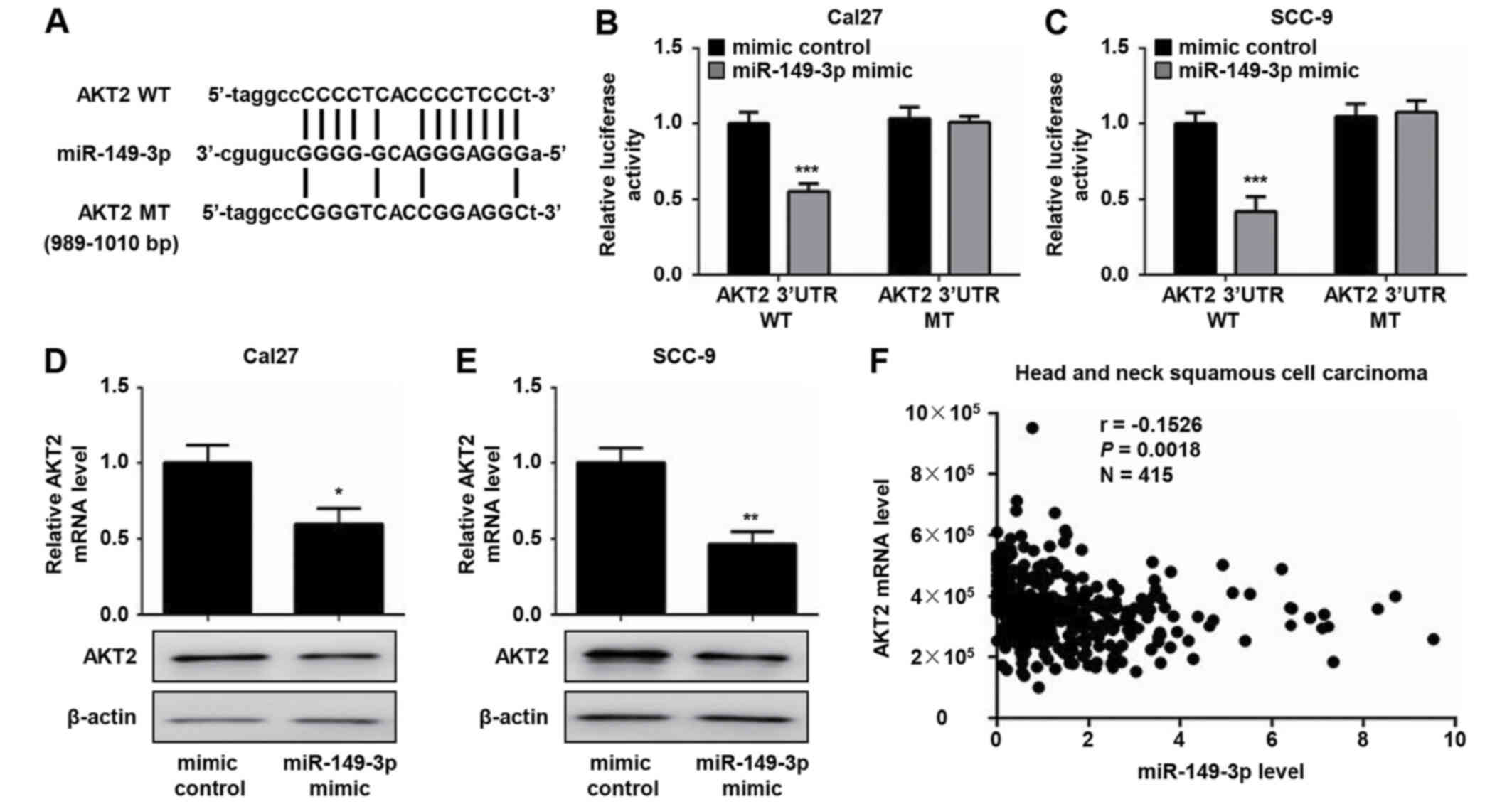
miR-149-3p binding site was identified in the 3′-UTR of AKT2 mRNA
(Fig. 4A). Accumulating evidence
has confirmed that AKT2 is essential for cell survival and
proliferation (16,17). Consequently, the AKT2 gene was
selected for further investigation. To assess whether AKT2 is a
direct target of miR-149-3p, luciferase reporter plasmids that
contain WT or MT 3′-UTR target sequences of AKT2 were constructed.
The dual luciferase reporter assay demonstrated that the miR-149-3p
mimic markedly decreased the luciferase activity of WT AKT2
3′-UTR-transfected Cal27 and SCC-9 cells compared with the mimic
control group (Fig. 4B and C).
Conversely, no effects were noted on the luciferase activity of MT
AKT2 3′-UTR following transfection with the miR-149-3p mimic
(Fig. 4B and C). Moreover, the
overexpression of miR-149-3p resulted in a significant decrease in
the mRNA and protein expression levels of AKT2 in OSCC cells
(Fig. 4D and E). In addition, a
weak negative correlation between AKT2 and miR-149-3p expression
was observed in a TCGA head and neck squamous cell carcinoma
dataset (Fig. 4F). These results
suggested that miR-149-3p could regulate AKT2 expression by
directly binding to the AKT2 3′-UTR.
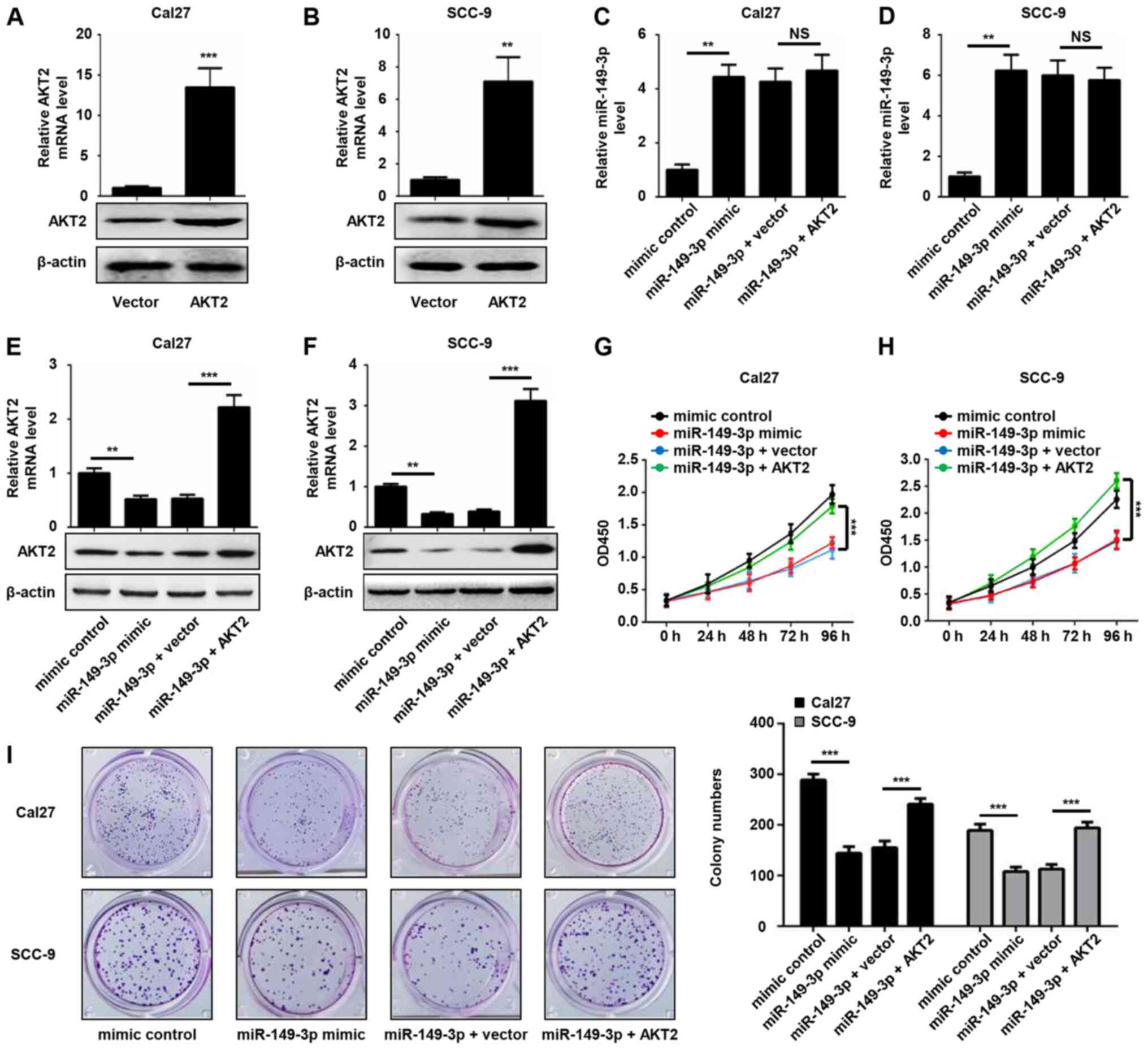
AKT2 restores the inhibitory effect of
miR-149-3p on the proliferation of OSCC cells
To further examine the involvement of AKT2 in
miR-149-3p-mediated cell proliferation, OSCC cells were transfected
with AKT2 plasmid or empty pcDNA3 plasmid, and the expression
levels of AKT2 were validated by RT-qPCR and western blot analysis.
The results revealed that ectopic introduction of AKT2
significantly increased the expression levels of AKT2 in Cal27 and
SCC-9 cells (Fig. 5A and B).
Enforced AKT2 expression did not alter miR-149-3p expression
levels, whereas it rescued the decreased mRNA and protein
expression levels of AKT2 induced by miR-149-3p (Fig. 5C-F). In addition, miR-149-3p-induced
reduced cell viability was significantly attenuated by AKT2
overexpression in OSCC cells (Fig. 5G
and H). A similar phenomenon was revealed by the colony
formation assay (Fig. 5I). These
data indicated that AKT2 was able to reverse the inhibitory effect
of miR-149-3p on the proliferation of OSCC cells.
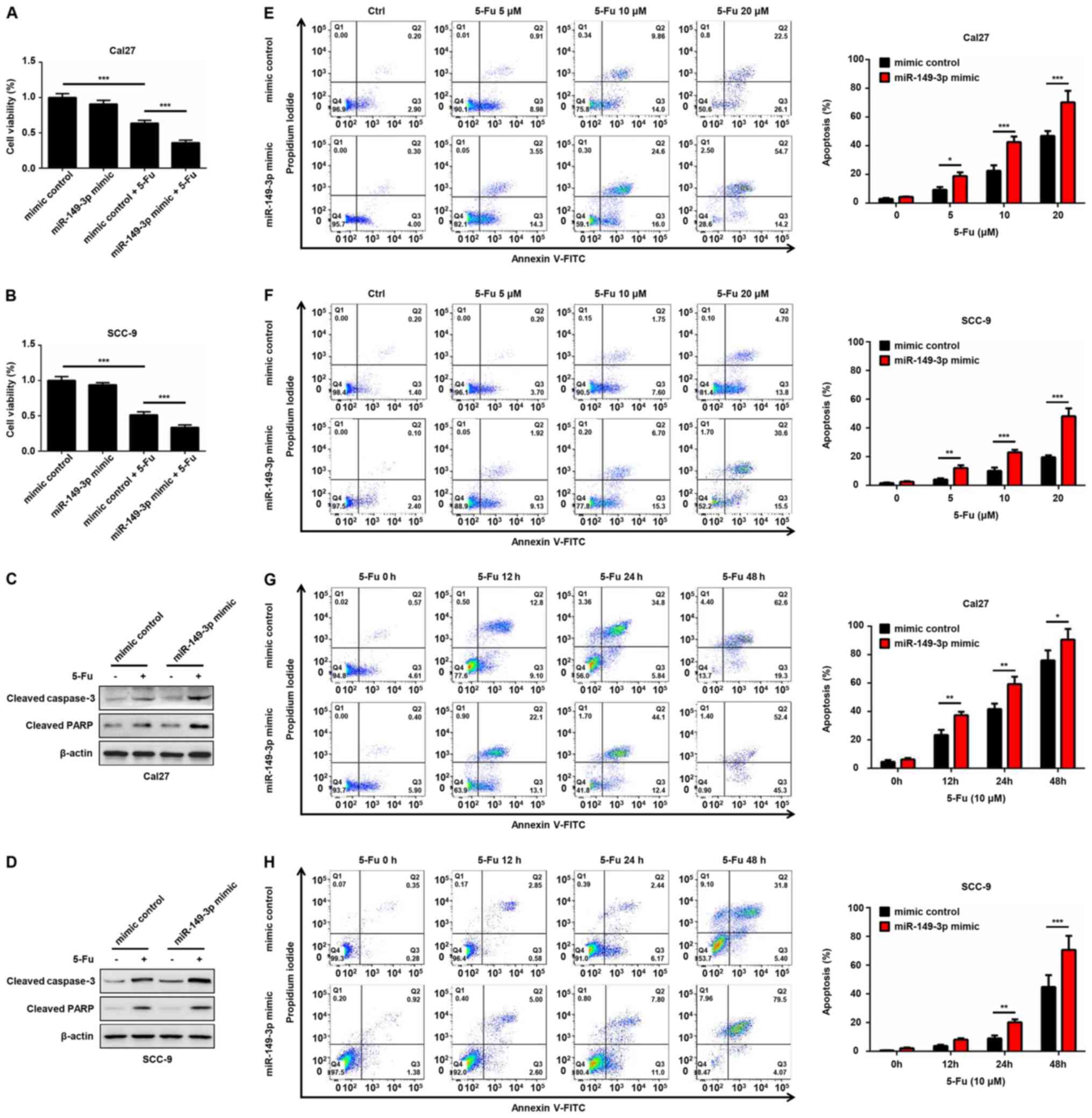
miR-149-3p improves the sensitivity of
OSCC cells to 5-Fu by targeting AKT2
To evaluate whether miR-149-3p contributes to the
anti-tumor effect of chemotherapeutic agents, Cal27 and SCC-9 cells
transfected with miR-149-3p mimic or mimic control were treated
with 5-Fu for 24 h. A significant decrease in cell viability of
5-Fu-treated OSCC cells was noted compared with in the non-treated
group (Fig. 6A and B). Notably,
miR-149-3p mimics enhanced the cytotoxic effects of 5-Fu on OSCC
cells (Fig. 6A and B). Moreover,
the miR-149-3p mimic synergistically increased 5-Fu-induced
elevated expression of cleaved caspase-3 and cleaved PARP (Fig. 6C and D). Flow cytometric analysis
indicated that the number of Annexin V-positive OSCC cells was
markedly increased following 5-Fu treatment in a dose- and
time-dependent manner (Fig. 6E-H).
Notably, the miR-149-3p mimic combined with 5-Fu exhibited an
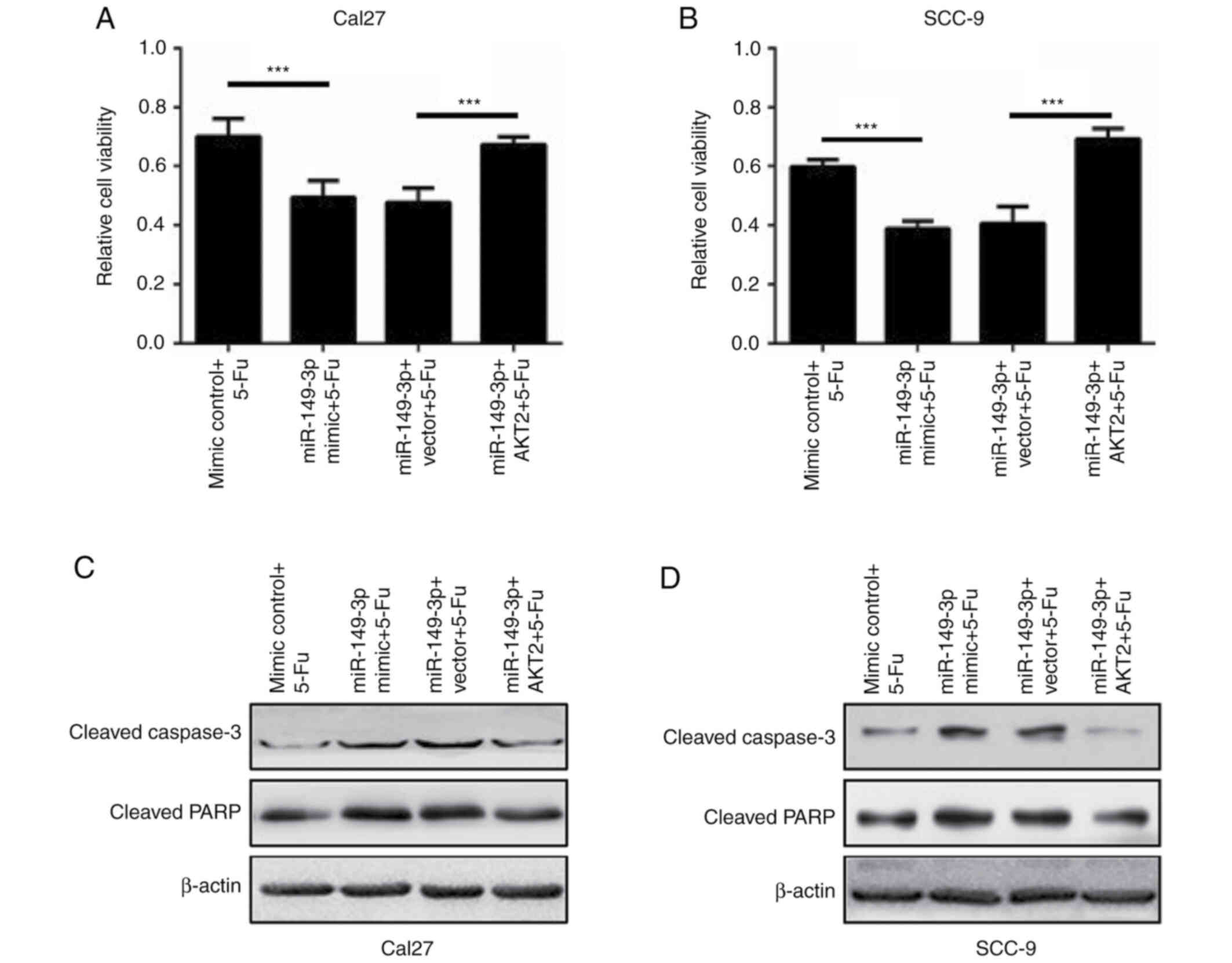
additive effect on the apoptosis of OSCC cells (Fig. 6E-H). Subsequently, the effects of
AKT2 were evaluated on miR-149-3p-modulated cell death following
5-Fu treatment. AKT2 overexpression effectively rescued the reduced
survival of OSCC cells caused by miR-149-3p in the presence of 5-Fu
(Fig. 7A and B). In addition, the
exogenous introduction of AKT2 significantly reversed the activated
caspase-3 and PARP proteins in miR-149-3p-expressing Cal27 and
SCC-9 cells following 5-Fu administration (Fig. 7C and D). These data revealed that
miR-149-3p sensitized OSCC cells to 5-Fu by targeting AKT2.
Discussion
miRNA biogenesis refers to the process of producing
a mature and functional miRNA and includes primary miRNA
transcription and cleavage, and precursor miRNA export and
processing. It has been proposed that miRNA biogenesis is caused
when the guide strand derived from duplex miRNA directly binds and
represses the target mRNA at the translational level, whereas the
passenger strand of miRNA is degraded and displays no function
(18,19). In contrast to this theory,
accumulating evidence has revealed that the passenger strand of
certain precursor miRNAs contributes to the development of several
types of cancer (20,21). A previous study demonstrated that
both miR-149-5p (the guide strand) and miR-149-3p (the passenger
strand) resulted in decreased migratory and invasive capacities by
degrading the mRNA expression of FoxM1 in clear cell renal cell
carcinoma (22). Moreover,
miR-149-5p enhanced chemotherapeutic resistance through
inactivation of the Hippo signaling pathway in ovarian cancer
(23), whereas high levels of
miR-149-3p were associated with the aggressive progression and poor
prognosis of ovarian cancer (24).
These results indicated a similar function between miR-149-3p and
miR-149-5p. However, miR-149-5p and miR-149-3p exhibit diverse
roles in cancer progression. For example, long intergenic
non-protein-coding RNA 472 inhibited tumor growth and metastasis by
sponging and decreasing miR-149-3p expression, leading to the
upregulation of KLLN and activation of p53 signaling in non-small
cell lung cancer (NSCLC) cells, which suggested an oncogenic
function of miR-149-3p (25).
Conversely, miR-149-5p acted as a tumor suppressor in NSCLC
(26,27). In the present study, the data
demonstrated that miR-149-3p overexpression reduced the viability
and colony formation of OSCC cells, whereas knockdown of miR-149-3p
induced the proliferation of OSCC cells. Recent studies have
revealed that lower levels of miR-149-5p were observed in OSCC
tumor tissues compared with in adjacent normal tissues (28), whereas miR-149-5p overexpression
resulted in attenuated proliferation and motility, as well as
increased drug sensitivity, in OSCC cells (29). These data, together with the present
findings, indicated the tumor-suppressive functions of miR-149-5p
and miR-149-3p in OSCC, and suggested that the dual strand of
miR-149 may be a potential biomarker for the generation and
development of OSCC.
The AKT family is an evolutionarily conserved
serine/threonine kinase family that contains three isoforms (AKT1,
AKT2 and AKT3). Emerging evidence has revealed that the
upregulation of AKT2 is essential for tumorigenesis, angiogenesis
and metastasis, and it has been reported to act as an oncogene in
several types of cancer, including hepatocellular carcinoma
(30), ovarian cancer (31) and breast cancer (32). In the present study, the results
demonstrated that the miR-149-3p mimic significantly suppressed the
mRNA and protein expression levels of AKT2. In addition, miR-149-3p
decreased the luciferase activity of WT AKT2 3′-UTR, whereas this
effect was not noted in the MT AKT2 3′-UTR, suggesting that AKT2
was a direct target of miR-149-3p. AKT2 overexpression reversed the
miR-149-3p-mediated inhibitory effect on the proliferation of OSCC
cells. These findings further supported the hypothesis that AKT2
contributes to OSCC progression. In line with the present
observations, AKT2 was previously shown to be upregulated and
activated in OSCC tissues (33),
whereas the reduced expression of AKT2 induced by caffeic acid
phenethyl ester suppressed proliferation and the survival of OSCC
cells (34). Moreover, the
CXCL9/CXCR3 axis promoted invasion and metastasis through
AKT2-mediated epithelial-mesenchymal transition in tongue squamous
cell carcinoma (35). Since AKT2
mRNA has been demonstrated to be regulated by diverse miRNAs, such
as miR-194 (36), miR-625-5p
(37) and miR-92a (38), the crosstalk of these miRNAs that
controls the translational repression of AKT2 requires further
exploration.
5-Fu is one of the first-line chemotherapeutic drugs
used for the treatment of OSCC. Although this drug significantly
improves the survival of patients with OSCC, resistance to 5-Fu
inevitably occurs following the treatment of patients over a long
period of time (39,40). Moreover, the undesired side effects,
including mouth sores, vomiting, nausea and diarrhea, limit the
clinical application of 5-Fu. Increased sensitivity of OSCC cells
to chemotherapeutic drugs is a potential strategy to improve
chemoresistance. In the present study, the results demonstrated
that the miR-149-3p mimic strengthened 5-Fu-induced decreased cell
viability, and increased protein expression levels of cleaved
caspase-3 and cleaved PARP. Caspase-3 is a critical executioner of
apoptosis, and activation of caspase 3 requires proteolytic
processing of its inactive zymogen (total caspase-3) into activated
fragments (cleaved caspase-3). PARP is one of the main cleavage
targets of activated caspase-3, and cleavage of PARP facilitates
cellular disassembly and promotes cell apoptosis (41,42).
As cleaved caspase-3 and cleaved PARP are two representative
markers of apoptosis, the present data further supported that
miR-149-3p contributed to 5-Fu-mediated cell apoptosis. However,
this effect was reversed by AKT2 overexpression, suggesting that
miR-149-3 improved the cytotoxic effects of 5-Fu on OSCC cells by
targeting AKT2. In agreement with these findings, the
methylation-mediated epigenetic silencing of miR-149-3p was
previously discovered to be involved in chemoresistance in breast
cancer cells (43). In addition,
the decreased expression of miR-149-3p was noted in
cisplatin-resistant colon cancer cells (44). Therefore, these observations
indicated that miR-149-3p may be a promising target for overcoming
chemotherapy resistance.
In conclusion, the present study revealed a novel
molecular pathway by which miR-149-3p suppressed the proliferation
of OSCC cells. The mechanism of action involved the negative
regulation of AKT2 mRNA expression. Furthermore, miR-149-3p
enhanced the sensitivity of OSCC cells to 5-Fu. Therefore, these
findings demonstrated an important role of miR-149-3p during OSCC
progression and suggested that the restoration of miR-149-3p
expression may be an effective therapeutic strategy for the
treatment of OSCC.
Acknowledgements
Not applicable.
Funding
This study was supported by the Basic Research
Project of Science and Technology Plan of Shenzhen Bao'an District
(grant nos. 2019JD430 and 2019JD440) and the Medical Scientific
Research Foundation of Guangdong Province (grant no. A2020290).
Availability of data and materials
The datasets used and/or analyzed during the present
study are available from the corresponding author on reasonable
request.
Authors' contributions
QS and WS conceived and designed the study. QS and
HZ performed the experiments and analyzed the data. QS, QL, LC and
DY interpreted the data. QS wrote the manuscript. All authors read
and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2018. CA Cancer J Clin. 68:7–30. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Iamaroon A and Krisanaprakornkit S:
Overexpression and activation of Akt2 protein in oral squamous cell
carcinoma. Oral Oncol. 45:e175–e179. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Leemans CR, Braakhuis BJ and Brakenhoff
RH: The molecular biology of head and neck cancer. Nat Rev Cancer.
11:9–22. 2011. View
Article : Google Scholar : PubMed/NCBI
|
|
4
|
Carvalho AL, Nishimoto IN, Califano JA and
Kowalski LP: Trends in incidence and prognosis for head and neck
cancer in the United States: A site-specific analysis of the SEER
database. Int J Cancer. 114:806–816. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rupaimoole R and Slack FJ: MicroRNA
therapeutics: Towards a new era for the management of cancer and
other diseases. Nat Rev Drug Discov. 16:203–222. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Gebert LFR and MacRae IJ: Regulation of
microRNA function in animals. Nat Rev Mol Cell Biol. 20:21–37.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Nishiwada S, Sho M, Banwait JK, Yamamura
K, Akahori T, Nakamura K, Baba H and Goel A: A microRNA signature
identifies pancreatic ductal adenocarcinoma patients at risk for
lymph node metastases. Gastroenterology. 159:562–574. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhao M, Hou Y, Du YE, Yang L, Qin Y, Peng
M, Liu S, Wan X, Qiao Y, Zeng H, et al: Drosha-independent
miR-6778-5p strengthens gastric cancer stem cell stemness via
regulation of cytosolic one-carbon folate metabolism. Cancer Lett.
478:8–21. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xu Y, Ji K, Wu M, Hao B, Yao KT and Xu Y:
A miRNA-HERC4 pathway promotes breast tumorigenesis by inactivating
tumor suppressor LATS1. Protein Cell. 10:595–605. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Shin CH, Lee H, Kim HR, Choi KH, Joung JG
and Kim HH: Regulation of PLK1 through competition between hnRNPK,
miR-149-3p and miR-193b-5p. Cell Death Differ. 24:1861–1871. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang D, Du G, Xu A, Xi X and Li D:
Expression of miR-149-3p inhibits proliferation, migration, and
invasion of bladder cancer by targeting S100A4. Am J Cancer Res.
7:2209–2219. 2017.PubMed/NCBI
|
|
12
|
Zhang M, Gao D, Shi Y, Wang Y, Joshi R, Yu
Q, Liu D, Alotaibi F, Zhang Y, Wang H, et al: miR-149-3p reverses
CD8+ T-cell exhaustion by reducing inhibitory receptors and
promoting cytokine secretion in breast cancer cells. Open Biol.
9:1900612019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Bellazzo A, Di Minin G, Valentino E,
Sicari D, Torre D, Marchionni L, Serpi F, Stadler MB, Taverna D,
Zuccolotto G, et al: Cell-autonomous and cell non-autonomous
downregulation of tumor suppressor DAB2IP by microRNA-149-3p
promotes aggressiveness of cancer cells. Cell Death Differ.
25:1224–1238. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cancer Genome Atlas Network: Comprehensive
genomic characterization of head and neck squamous cell carcinomas.
Nature. 517:576–582. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Gonzalez E and McGraw TE: The Akt kinases:
Isoform specificity in metabolism and cancer. Cell Cycle.
8:2502–2508. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Hers I, Vincent EE and Tavaré JM: Akt
signalling in health and disease. Cell Signal. 23:1515–1527. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lin S and Gregory RI: MicroRNA biogenesis
pathways in cancer. Nat Rev Cancer. 15:321–333. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Sugawara S, Yamada Y, Arai T, Okato A,
Idichi T, Kato M, Koshizuka K, Ichikawa T and Seki N: Dual strands
of the miR-223 duplex (miR-223-5p and miR-223-3p) inhibit cancer
cell aggressiveness: Targeted genes are involved in bladder cancer
pathogenesis. J Hum Genet. 63:657–668. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Okato A, Arai T, Kojima S, Koshizuka K,
Osako Y, Idichi T, Kurozumi A, Goto Y, Kato M, Naya Y, et al: Dual
strands of pre-miR150 (miR1505p and miR1503p) act as antitumor
miRNAs targeting SPOCK1 in naive and castration-resistant prostate
cancer. Int J Oncol. 51:245–256. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Okato A, Arai T, Yamada Y, Sugawara S,
Koshizuka K, Fujimura L, Kurozumi A, Kato M, Kojima S, Naya Y, et
al: Dual Strands of Pre-miR-149 inhibit cancer cell migration and
invasion through targeting FOXM1 in renal cell carcinoma. Int J Mol
Sci. 18:19692017. View Article : Google Scholar
|
|
23
|
Xu M, Xiao J, Chen M, Yuan L, Li J, Shen H
and Yao S: miR1495p promotes chemotherapeutic resistance in ovarian
cancer via the inactivation of the Hippo signaling pathway. Int J
Oncol. 52:815–827. 2018.PubMed/NCBI
|
|
24
|
Li Y, Liu C, Liao Y, Wang W, Hu B, Lu X
and Cui J: Characterizing the landscape of peritoneal exosomal
microRNAs in patients with ovarian cancer by high-throughput
sequencing. Oncol Lett. 17:539–547. 2019.PubMed/NCBI
|
|
25
|
Zou A, Liu X, Mai Z, Zhang J, Liu Z, Huang
Q, Wu A and Zhou C: LINC00472 Acts as a Tumor Suppressor in NSCLC
through KLLN-Mediated p53-Signaling Pathway via MicroRNA-149-3p and
MicroRNA-4270. Mol Ther Nucleic Acids. 17:563–577. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Liu L, Chen Y, Li Q and Duan P: lncRNA
HNF1A-AS1 modulates non-small cell lung cancer progression by
targeting miR-149-5p/Cdk6. J Cell Biochem. 120:18736–18750. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Li J, Li Y, Wang B, Ma Y and Chen P:
lncRNA-PCAT-1 promotes non-small cell lung cancer progression by
regulating miR-149-5p/LRIG2 axis. J Cell Biochem. Dec 19–2018.(Epub
ahead of print).
|
|
28
|
Lai H, Xu G, Meng H and Zhu H: Association
of SP1 rs1353058818 and STAT3 rs1053004 gene polymorphisms with
human tongue squamous cell carcinoma. Biosci Rep. 23:392019.
|
|
29
|
Luo K, He J, Yu D and Acil Y: MiR-149-5p
regulates cisplatin chemosensitivity, cell growth, and metastasis
of oral squamous cell carcinoma cells by targeting TGFβ2. Int J
Clin Exp Pathol. 12:3728–3739. 2019.PubMed/NCBI
|
|
30
|
Wang Q, Yu WN, Chen X, Peng XD, Jeon SM,
Birnbaum MJ, Guzman G and Hay N: Spontaneous hepatocellular
carcinoma after the combined deletion of Akt isoforms. Cancer Cell.
29:523–535. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Lin X, Shen J, Dan Peng, He X, Xu C, Chen
X, Tanyi JL, Montone K, Fan Y, Huang Q, Zhang L and Zhong X:
RNA-binding protein LIN28B inhibits apoptosis through regulation of
the AKT2/FOXO3A/BIM axis in ovarian cancer cells. Signal Transduct
Target Ther. 3:232018. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Chen X, Ariss MM, Ramakrishnan G, Nogueira
V, Blaha C, Putzbach W, Islam ABMMK, Frolov MV and Hay N:
Cell-autonomous versus systemic akt isoform deletions uncovered new
roles for Akt1 and Akt2 in Breast Cancer. Mol Cell. 80:87–101.e5.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Roy NK, Monisha J, Padmavathi G,
Lalhruaitluanga H, Kumar NS, Singh AK, Bordoloi D, Baruah MN, Ahmed
GN, Longkumar I, et al: Isoform-specific role of akt in oral
squamous cell carcinoma. Biomolecules. 9:2532019. View Article : Google Scholar
|
|
34
|
Kuo YY, Lin HP, Huo C, Su LC, Yang J,
Hsiao PH, Chiang HC, Chung CJ, Wang HD, Chang JY, et al: Caffeic
acid phenethyl ester suppresses proliferation and survival of TW2.6
human oral cancer cells via inhibition of Akt signaling. Int J Mol
Sci. 14:8801–8817. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Li Z, Liu J, Li L, Shao S, Wu J, Bian L
and He Y: Epithelial mesenchymal transition induced by the
CXCL9/CXCR3 axis through AKT activation promotes invasion and
metastasis in tongue squamous cell carcinoma. Oncol Rep.
39:1356–1368. 2018.PubMed/NCBI
|
|
36
|
Wu JC, Chen R, Luo X, Li ZH, Luo SZ and Xu
MY: MicroRNA-194 inactivates hepatic stellate cells and alleviates
liver fibrosis by inhibiting AKT2. World J Gastroenterol.
25:4468–4480. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Qian FH, Deng X, Zhuang QX, Wei B and
Zheng DD: miR6255p suppresses inflammatory responses by targeting
AKT2 in human bronchial epithelial cells. Mol Med Rep.
19:1951–1957. 2019.PubMed/NCBI
|
|
38
|
Yu FY, Xie CQ, Jiang CL, Sun JT, Feng HC,
Li C and Huang XW: MiR-92a inhibits fibroblast-like synoviocyte
proliferation and migration in rheumatoid arthritis by targeting
AKT2. J Biosci. 43:911–919. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Nagata M, Nakayama H, Tanaka T, Yoshida R,
Yoshitake Y, Fukuma D, Kawahara K, Nakagawa Y, Ota K, Hiraki A and
Shinohara M: Overexpression of cIAP2 contributes to 5-FU resistance
and a poor prognosis in oral squamous cell carcinoma. Br J Cancer.
105:1322–1330. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Feng X, Luo Q, Zhang H, Wang H, Chen W,
Meng G and Chen F: The role of NLRP3 inflammasome in 5-fluorouracil
resistance of oral squamous cell carcinoma. J Exp Clin Cancer Res.
36:812017. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Fernandes-Alnemri T, Litwack G and Alnemri
ES: CPP32, a novel human apoptotic protein with homology to
Caenorhabditis elegans cell death protein Ced-3 and mammalian
interleukin-1 beta-converting enzyme. J Biol Chem. 269:30761–30764.
1994.PubMed/NCBI
|
|
42
|
Nicholson DW, Ali A, Thornberry NA,
Vaillancourt JP, Ding CK, Gallant M, Gareau Y, Griffin PR, Labelle
M, Lazebnik YA, et al: Identification and inhibition of the
ICE/CED-3 protease necessary for mammalian apoptosis. Nature.
376:37–43. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
He DX, Gu XT, Li YR, Jiang L, Jin J and Ma
X: Methylation-regulated miR-149 modulates chemoresistance by
targeting GlcNAc N-deacetylase/N-sulfotransferase-1 in human breast
cancer. FEBS J. 281:4718–4730. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Liu Y, Yang C, Zhao Y, Chi Q, Wang Z and
Sun B: Overexpressed methyltransferase-like 1 (METTL1) increased
chemosensitivity of colon cancer cells to cisplatin by regulating
miR-149-3p/S100A4/p53 axis. Aging (Albany NY). 11:12328–12344.
2019. View Article : Google Scholar : PubMed/NCBI
|