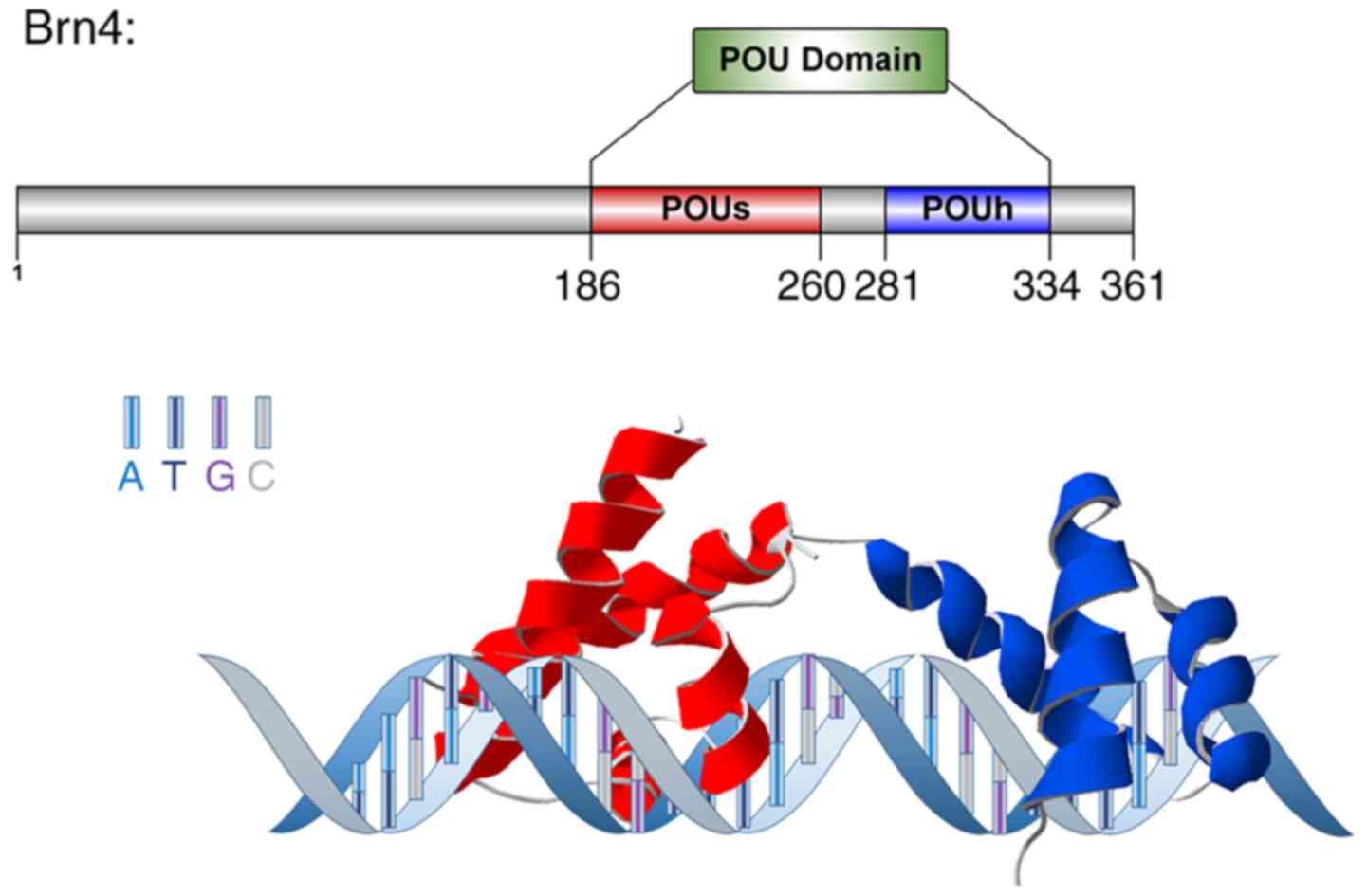
The transcription factor brain (Brn)4 is a member of
the POU protein family. In 1984, Parslow et al (1) first identified a highly conserved
sequence ~70 base pairs upstream of the transcription initiation
site of the immunoglobulin heavy and light chain genes, which was
considered to be the area involved in the transcriptional
regulation of gene expression. Subsequently, DNA elements with
similar functions and highly conserved sequences were found at the
origin of adenoviral DNA replication and at the promoters of
nucleolar small RNA and Histone 2B genes. Given the similarity of
the 150–160 amino acids in the sequences in the mammalian proteins,
pituitary-specific positive transcription factor 1, Oct1, 2 and
nematode Uncoordinated-86, the homologous sequence was named the
POU domain (an acronym of the transcription factors) (2).
The POU domain is a binding site for DNA binding
proteins. It comprises two subdomains linked by a 15–56 amino acid
sequence junction region: i) The N-terminus, which contains the
POU-specific domain (POUS) comprising 76–78 amino acids,
and ii) the C-terminus, which comprises the POU homology domain
(POUH) composed of 60 amino acids (3). Proteins with a POU domain are named
POU proteins, and serve as transcription factors that have
important roles in embryonic development and cell fate-directed
processes by binding to specific nucleotide sequences and
regulating the transcription of downstream target genes (4). According to the degree of homology of
the POU domain, the POU gene can be divided into six classes, which
encode the POU1-6 protein families, respectively, each of which
contains different members (5).
The POU3 family contains four members, Tst-1 [also
known as POU domain, class 3, transcription factor 1 (POU3F1),
Oct6, Test1, and SCIP], Brn1 (also known as POU3F3 and Oct8), Brn2
(also known as POU3F2 and Oct7) and Brn4 (also known as POU3F4,
Oct9, deafness 3 and deafness X-linked 2). Tst-1 was originally
identified in rat testes and glial cells, as well as in the
developing brain and skin (5), and
it is an important regulator of neurogenesis and epidermal
differentiation (6). It has a dual
regulatory function, not only does it activate neural lineage genes
(such as Sox2), but it also represses neural-inhibitory signals
(such as Wnt and bone morphogenetic protein), thus inhibiting cell
proliferation (7,8). The degradation of POU3F1, caused by
numerous factors such as DNA damage, starvation, and oxidative
stress, was reported to induce neural tube defects (9). Brn1 is involved in the development of
the central nervous system, and its loss-of-function mutations were
identified to cause developmental delays, intellectual disability
and impairments in language skills (10). It was also found to be expressed in
the developing kidney (11). In
previous studies, Brn1 knockout mice were discovered to have
neurological defects, including locomotor and auditory impairments,
but also a thicker ascending loop of the limb of Henle and
decreased numbers of nephrons, thus resulting in renal failure and
perinatal death (12,13). Brn2 has been reported to serve an
important role in the development of the central nervous system; it
has been associated with bipolar disorder (14), and it promotes neurogenesis by
regulating the expression levels of neurotrophin 3 (15). Brn2 knockout mice were also revealed
to suffer from cognitive impairment and decreased numbers of
newborn neurons were found in the dentate gyrus of the hippocampus
(16). Another previous study
suggested that Brn2 may promote human gastric carcinoma cell
proliferation, migration and invasion by binding to the promoter of
tumor-associated NADH oxidase (17). It was also shown to serve a role in
the development of hepatocellular carcinoma through the long
non-coding RNA brain cytoplasmic RNA 1/microRNA-490-3p/Brn2 axis
(18). Brn4, also known as POU3F4,
Oct9, deafness 3 and deafness X-linked 2, was named after it was
initially discovered in brain tissue (19). The following sections of the present
review aimed to concisely summarize the research progress on Brn4
regarding its chromosomal location and species homology, protein
molecular structure and tissue distribution, and related biological
processes.
Brn4 is a single copy gene located on the human
chromosome X q21.1, spanning 3,867 nucleotides. Two
polydeoxyadenine nucleotides in the POU3 family gene sequence are
considered to be formed by the reverse transcription of mRNA and
re-entry into chromosomal sequences via genetic recombination
(20). In the absence of introns,
Brn4 mRNA leaves the nucleus and enters the cytoplasm without
undergoing post-transcriptional splicing, thus resulting in rapid
gene expression (20). Brn4 was
formed before the human and rodent gene duplication that occurred
during evolution (5); therefore, it
has high homology between humans and mice, and investigations using
rodent models can provide valuable references for studies of
clinical diseases. In addition, a previous study of a lactose
operon reporter gene identified the presence of ≥2 positive
cis-regulatory elements of 6 kb in length upstream of the
promoter sequence in Brn4 (21),
indicating that Brn4 may be highly expressed at certain stages of
development and may be important in histogenesis.
The transcription factor Brn4 is a DNA-binding
protein that recognizes a specific octamer motif (ATGCAAAT)
(4). Brn4 comprises 361 amino acids
and has a molecular weight of ~39.43 kDa. Both the POUS
and POUH domains of Brn4 contain a helix-turn-helix
motif, which can form a hairpin structure that binds DNA sequences
and exerts biological effects such as transcriptional activation
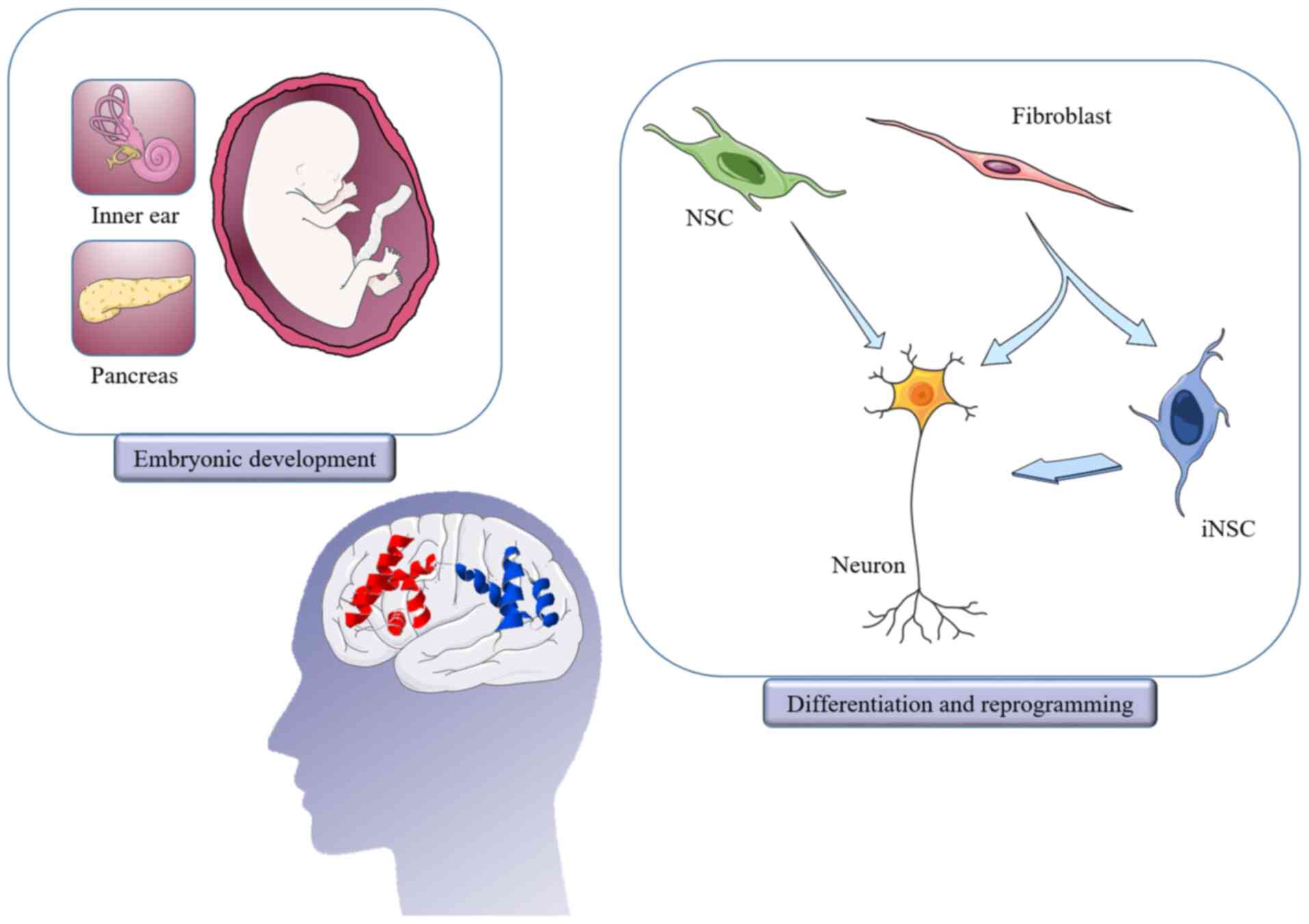
(Fig. 1) (4). During development in mammals, the POU3
gene family has been reported to be widely expressed in
ectoderm-derived tissue, such as the nervous system and auditory
vesicles (22). In addition, Brn4
has been detected in the embryonic pancreas and adult glucagon
cells (23,24). A previous study investigating the
role of the POU3 gene during development in Xenopus laevis
found that Brn4 was expressed in the embryonic neural tube, brain,
developmental ear vesicles, distal renal tubules and connective
tubules (11). RNA sequencing
results of the genome-wide analysis of 272 mouse cell and tissue
types revealed the widespread expression of Brn4 during nervous
system development, including within the neural crest,
neuroectoderm, hypothalamus, pituitary, hippocampus and inner ear
(5). However, in contrast to the
extensive distribution in embryonic development, in adult animals,
Brn4 expression was reported to be limited to a small part of the
forebrain, including the supraoptic nucleus and paraventricular
nucleus of the thalamus.
The inner ear is part of the anatomical structure of
the ear and is located in the cavity of the sacral rock (25). Through a series of complicated
canals, it forms a structure called the labyrinth, which consists
of the bony and membranous labyrinths (25). The bony labyrinth contains the
vestibular system, which is associated with balance, and the
cochlea, which is associated with hearing (26). The cochlea converts sound into nerve
impulses that transmit external signals to the central nervous
system through the auditory nerve, thus resulting in hearing
(26). Inner ear development is a
complex process involving a series of morphological changes caused
by the interaction between ectodermal epithelial cells and
mesenchymal cells (27).
Although Brn4 gene mutations have been widely
confirmed to cause severe inner ear dysplasia, its specific
molecular mechanism remains unclear. Moreover, how to effectively
prevent the occurrence and development of this genetic disease and
how to correct the abnormal cochlear structure caused by the
mutation of Brn4 require further research.
The pancreas originates from the dorsal and ventral
buds of the endoderm, which are later referred to as the dorsal and
ventral lobes; these develop into the ‘tail’ and ‘head’ of the
adult pancreas, respectively (48).
The pancreas functions as both an exocrine and endocrine gland. The
endocrine function involves the pancreatic islets, which lack a
fixed form and are found scattered throughout the pancreas
(49). The most representative
cells in the islets are glucagon-secreting α cells (15–20% of islet
cells) and insulin-secreting β cells (65–80% of islet cells)
(50). The balance of insulin and
glucagon is crucial for the maintenance of blood glucose
homeostasis (50).
Islet α cells are the first endocrine cells formed
in the developing pancreas on embryonic day 9.5 in mice, and Brn4
was revealed to promote their differentiation (Fig. 2) (51–53).
Islet α cells first express the proglucagon gene, and the produced
proglucagon is further processed into glucagon, which increases
blood glucose levels (53).
Previous studies identified that the proximal G1 promoter element
of the proglucagon gene, an AT-rich DNA sequence, was the binding
site for Brn4 (24). Upregulated
expression levels of Brn4 have been detected in islet cell lines
producing glucagon and rat pancreatic islet cells, thus indicating
its importance in pancreatic development. Heller et al
(23) reported that Brn4, expressed
in the E10 pancreas (Fig. 2), was
the only confirmed islet α cell-specific transcription factor.
However, Brn4 deletion did not generate abnormalities in pancreatic
bud formation, pancreatic islet α cell numbers or physiological
function in mice. Therefore, the synthesis and secretion of
glucagon appears to depend on the participation of numerous
factors, which may overlap in function.
Clinically, diabetes is typically treated with oral
medication or insulin injection (54). The complications of diabetes have
long been a prominent cause of human disabilities such as
musculoskeletal abnormalities and microangiopathy (54,55).
One previous study discovered that the differentiated islet cells
from human marrow mesenchymal cells highly expressed pancreatic
transcription factors, such as Brn4, in induced islet-like cell
aggregates, suggesting that Brn4 may be targeted to induce the
regeneration of islet cells to treat clinical diseases including
diabetes, pancreatic cancer and abnormal glucose metabolism
(56).
Self-proliferating neural stem cells (NSCs) are
present throughout life in mammalian brains. These cells are mainly
distributed in the subventricular zone and the hippocampal
subgranular zone, and they can differentiate into neurons and glial
cells (57). Since Brn4 was found
to mediate striatal neuron precursor differentiation in 1999
(58), its role in neural
regeneration has attracted significant attention (Fig. 2). Nuclear receptor-related 1
(Nurr1), a midbrain dopaminergic (DA) neuron-specific transcription
factor, was reported to promote the differentiation of NSCs into DA
neurons, increase the levels of dopamine in the striatum and
ameliorate behavior defects in rats with Parkinson's disease. The
expression of Brn4 also promoted the Nurr1-induced significant
increase in the viability and maturity of DA neurons (59). Brn4 was also discovered to induce
the expression of glial cell line-derived neurotrophic factor,
which contributed to the survival and maturation of DA neurons,
together with its receptors, GDNF family receptor α-1 and Ret
(60). In addition, following
denervation surgery, the expression levels of Brn4 in the
hippocampus of rats were found to be upregulated, and reached a
peak at 14 days post-injury (61).
Functional experiments have indicated that Brn4 not only promotes
the differentiation of NSCs into neurons but also enhances the
maturation of neurons (61,62). This change in Brn4 may be due in
part to the activation of the PI3K/AKT signaling pathway caused by
upregulated expression levels of insulin-like growth factor
(63). In addition, the paired box
protein Pax 6/barrier-to-autointegration factor complex was
demonstrated to drive neurogenesis by directly activating the
expression of transcription factors, such as Brn4, during
neurodevelopment (64).
Investigations to determine target genes of Brn4 using RNA
sequencing have revealed that, alongside the expression of Brn4,
the expression levels of genes involved in neuronal development and
maturation were upregulated, whereas those of genes associated with
maintaining the pluripotency of NSCs were downregulated.
Furthermore, C-terminal-binding protein 2 and Notch2 were suggested
to be direct targets of Brn4 (65,66).
Fetal brain tissue can be used for cell replacement
therapy in neurodegenerative diseases, such as Parkinson's disease,
but it is limited by donor deficiency and cannot be standardized
(67). Fortunately, the induced
differentiation into specific cell groups after the expansion of
NSCs in vitro can solve this problem. For example, in both
the midbrain and the hippocampus, Brn4 promoted the differentiation
of NSCs into neurons (Fig. 2) and
the maturation of new neurons (58–62,68).
Therefore, Brn4 may provide a target for novel drugs in the
treatment of neurodegenerative diseases.
The co-overexpression of Brn2, Sox2 and forkhead box
G2 was identified to be able to reprogram human or mouse
fibroblasts and astrocytes into neural precursor cells, which are
also called induced DA precursors, owing to their DA
differentiation ability (73–75).
Under spontaneous neuronal differentiation condition, these induced
neural precursor cells were observed to differentiate into
glutamatergic and GABAergic neuronal subtypes (73). In previous studies, Brn2 combined
with guanine nucleotide-binding protein subunit β-like protein and
myelin transcription factor 1-like protein was used to reprogram
somatic cells into functional neurons (76–79). A
previous study suggested that Brn2 itself may be sufficient to
achieve the aforementioned somatic cell reprogramming effect, and
that changes in cell culture conditions determine the direction of
induction (80).
Brn4, in combination with Sox2, KLF4 and c-Myc is
called BSKM; BSKM was found to successfully induce fibroblast
reprogramming into iNSCs (Fig. 2).
These cells, after being transplanted into the stem cell pool in
the adult mouse brain, exhibited true pluripotency in terms of
proliferation and differentiation into neurons and glial cells
(81,82). The reprogramming efficiency was
further improved by the addition of E47 or transcription factor 3
(83). Oct4 and Brn4 were
discovered to bind to Sox2 and form a dimer in a similar manner by
cooperativity by sequencing, a method to determine the
cooperativity parameters of transcription factors (84). Although both OKSM and BSKM can
reprogram somatic cells into iNSCs (85), they exhibit several differences; for
example, owing to the subtle differences between Oct4 and Brn4 in
the POUS domain (86),
BSKM was observed to directly transform somatic cells into iNSCs
without requiring the state of transient iPSCs induced by OKSM
(Fig. 2) (81,86).
Thus, BSKM may have lower tumorgenicity and an enhanced clinical
practical value.
Moreover, reprogramming human or mouse fibroblasts
or astrocytes into functional neurons or neuronal precursor cells
by Brn4 is not uncommon (87–89),
and these studies suggested that neurons induced by Brn4 in
vitro could be used as cell resources for substitutive therapy
to treat nerve injury or neurodegenerative diseases. However,
numerous issues require further investigations and research, such
as whether induced neurons can completely replace the damaged
cells; whether these cells have the long-term characteristics of
nerve cells in producing and conducting excitation; and how to
accurately intervene to modulate the direction, cycle and speed of
cell differentiation.
With further medical advancements, the survival rate
of patients with various genetic, chronic and nervous system
diseases has increased significantly (90). However, improving patient comfort
and wellbeing is currently the ultimate goal of medical care
(90,91). As aforementioned, Brn4 was
discovered to promote the survival and synaptic growth of SGNs in
the inner ear (28) and maintain
proper intra-cochlear lymphatic potential by regulating connexins
(22). However, to the best of our
knowledge, its downstream target genes and the specific molecular
mechanisms leading to structural abnormalities of the inner ear
remain unclear. It was hypothesized that Brn4 may not only promote
the growth of neurons, but it may also directly affect the signal
transduction between cells and serve an important role in the
osteogenic differentiation of the inner ear. Brn4−/−
mice were reported to exhibit vertical nodding and an abnormal gait
(30), thereby suggesting that Brn4
might affect the sense of balance and voluntary movement
function.
Although Brn4 was once considered a specific
transcription factor of islet α cells that upregulates the
expression of the proglucagon gene, the pancreatic development of
Brn4−/− mice is not significantly altered, and Brn4
expression levels were also upregulated in insulin-like cells with
the function of decreasing blood glucose in diabetic model mice
(23,24,51,52,56).
However, to the best of our knowledge, the mechanism via which Brn4
maintains blood glucose stability and the quantitative balance
among different islet cell types has not been reported. It can be
suggested that Brn4 may provide an adequate energy supply for
insulin synthesis in islet β cells by promoting glucagon secretion
in islet α cells. Accordingly, the feedback regulation of both
hormones and blood glucose levels would ultimately maintain Brn4
homeostasis.
Brn4 was identified to not only promote the
differentiation of NSCs and the maturation of newborn neurons
(59–61,63–66),
but also induced somatic cells to transform into neural lineage
cells (87–89). However, the majority of the previous
studies have focused on the number rather than the function of
neurons following Brn4-induced differentiation. Further in-depth
studies are required to determine whether the neurons induced by
Brn4 have the long-term function of generating and conducting
excitation; whether they produce effective synaptic connections
with surrounding cells after being transplanted into the injured
site; and how to improve the conversion ratio of neurons.
The microenvironment affects cell fate transitions;
therefore, Brn4-mediated downstream gene transcription may provide
a suitable microenvironment for neurogenesis, and the expression
changes observed in numerous neural lineage-related genes following
Brn4 treatment support this hypothesis (65). Previously, Brn4 was revealed to be a
driving factor in neuroendocrine differentiation, thus promoting
the occurrence and development of castration-resistant prostate
cancer, together with Brn2. It was reported to be used as both a
predictive marker of advanced castration-resistant prostate cancer
and as a novel target to address the issue of resistance to
enzalutamide treatment (92).
Due to its specificity of distribution and its high
degree of homology across species, Brn4 is clearly important in
development and biological functions including promoting the
differentiation of NSCs into neurons and inducing reprogramming of
somatic cells (Fig. 2). However, to
the best of our knowledge, multiple aspects of the regulatory
mechanism of its downstream biological processes remain unknown.
Elucidation of the links between Brn4 and clinical diseases will
require further investigations and experimental data. The present
review aimed to concisely summarize the reported structure and
biological properties of Brn4, with the aim of providing novel
ideas for research in related fields.
Not applicable.
The present review was funded by grants from The
National Natural Science Foundation of China (grant no. 31171038),
Jiangsu Natural Science Foundation (grant no. BK2011385), Jiangsu
‘333’ program funding (grant no. BRA2016450) and The Application
Research Project of Nantong City (grant no. MS12017015-3).
Not applicable.
YW collected the data and drafted the manuscript.
XuZ and JW assisted with manuscript preparation. GJ and XiZ revised
the final manuscript. XiZ conceived and designed the idea for this
paper. All authors read and approved the final manuscript.
Not applicable.
Not applicable.
The authors declare that they have no competing
interests.
|
1
|
Parslow TG, Blair DL, Murphy WJ and
Granner DK: Structure of the 5′ ends of immunoglobulin genes: A
novel conserved sequence. Proc Natl Acad Sci USA. 81:2650–2654.
1984. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Herr W, Sturm RA, Clerc RG, Corcoran LM,
Baltimore D, Sharp PA, Ingraham HA, Rosenfeld MG, Finney M, Ruvkun
G, et al: The POU domain: A large conserved region in the mammalian
pit-1, oct-1, 2, and Caenorhabditis elegans unc-86 gene products.
Genes Dev. 2:1513–1516. 1988. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Liu L, Li Y, Wang Y, Zhao P, Wei S, Li Z,
Chang H and He H: Biochemical characterization and functional
analysis of the POU transcription factor POU-M2 of Bombyx mori. Int
J Biol Macromol. 86:701–708. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Tang X and Engstrom Y: Regulation of
immune and tissue homeostasis by Drosophila POU factors. Insect
Biochem Mol Biol. 109:24–30. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Malik V, Zimmer D and Jauch R: Diversity
among POU transcription factors in chromatin recognition and cell
fate reprogramming. Cell Mol Life Sci. 75:1587–1612. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Fionda C, Di Bona D, Kosta A, Stabile H,
Santoni A and Cippitelli M: The POU-domain transcription factor
Oct-6/POU3F1 as a regulator of cellular response to genotoxic
stress. Cancers (Basel). 11:8102019. View Article : Google Scholar
|
|
7
|
Barral A, Rollan I, Sanchez-Iranzo H,
Jawaid W, Badia-Careaga C, Menchero S, Gomez MJ, Torroja C,
Sanchez-Cabo F, Göttgens B, et al: Nanog regulates Pou3f1
expression at the exit from pluripotency during gastrulation. Biol
Open. 8:bio0463672019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Song L, Sun N, Peng G, Chen J, Han JD and
Jing N: Genome-wide ChIP-seq and RNA-seq analyses of Pou3f1 during
mouse pluripotent stem cell neural fate commitment. Genom Data.
5:375–377. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li G, Jiapaer Z, Weng R, Hui Y, Jia W, Xi
J, Wang G, Zhu S, Zhang X, Feng D, et al: Dysregulation of the
SIRT1/OCT6 axis contributes to environmental stress-induced neural
induction defects. Stem Cell Reports. 8:1270–1286. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Snijders Blok L, Kleefstra T, Venselaar H,
Maas S, Kroes HY, Lachmeijer AM, van Gassen KL, Firth HV, Tomkins
S, Bodek S, et al: De novo variants disturbing the transactivation
capacity of POU3F3 cause a characteristic neurodevelopmental
disorder. Am J Hum Genet. 105:403–412. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Cosse-Etchepare C, Gervi I, Buisson I,
Formery L, Schubert M, Riou JF, Umbhauer M and Le Bouffant R: Pou3f
transcription factor expression during embryonic development
highlights distinct pou3f3 and pou3f4 localization in the Xenopus
laevis kidney. Int J Dev Biol. 62:325–333. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Kumar S, Rathkolb B, Kemter E, Sabrautzki
S, Michel D, Adler T, Becker L, Beckers J, Busch DH, Garrett L, et
al: Generation and standardized, systemic phenotypic analysis of
Pou3f3L423P mutant mice. PLoS One. 11:e01504722016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Rieger A, Kemter E, Kumar S, Popper B,
Aigner B, Wolf E, Wanke R and Blutke A: Missense mutation of POU
domain class 3 transcription factor 3 in Pou3f3L423P mice causes
reduced nephron number and impaired development of the thick
ascending limb of the loop of henle. PLoS One. 11:e01589772016.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Chen C, Meng Q, Xia Y, Ding C, Wang L, Dai
R, Cheng L, Gunaratne P, Gibbs RA, Min S, et al: The transcription
factor POU3F2 regulates a gene coexpression network in brain tissue
from patients with psychiatric disorders. Sci Transl Med.
10:eaat81782018. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lin YJ, Hsin IL, Sun HS, Lin S, Lai YL,
Chen HY, Chen TY, Chen YP, Shen YT and Wu HM: NTF3 is a novel
target gene of the transcription factor POU3F2 and is required for
neuronal differentiation. Mol Neurobiol. 55:8403–8413. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Hashizume K, Yamanaka M and Ueda S: POU3F2
participates in cognitive function and adult hippocampal
neurogenesis via mammalian-characteristic amino acid repeats. Genes
Brain Behav. 17:118–125. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Chen HY, Lee YH, Chen HY, Yeh CA, Chueh PJ
and Lin YM: Capsaicin inhibited aggressive phenotypes through
downregulation of tumor-associated NADH Oxidase (tNOX) by POU
domain transcription factor POU3F2. Molecules. 21:7332016.
View Article : Google Scholar
|
|
18
|
Ding S, Jin Y, Hao Q, Kang Y and Ma R:
LncRNA BCYRN1/miR-490-3p/POU3F2, served as a ceRNA network, is
connected with worse survival rate of hepatocellular carcinoma
patients and promotes tumor cell growth and metastasis. Cancer Cell
Int. 20:62020. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Serrano-Saiz E, Leyva-Diaz E, De La Cruz E
and Hobert O: BRN3-type POU homeobox genes maintain the identity of
mature postmitotic neurons in nematodes and mice. Curr Biol.
28:2813–2823.e2. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Hara Y, Rovescalli AC, Kim Y and Nirenberg
M: Structure and evolution of four POU domain genes expressed in
mouse brain. Proc Natl Acad Sci USA. 89:3280–3284. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Heydemann A, Nguyen LC and Crenshaw EB
III: Regulatory regions from the Brn4 promoter direct LACZ
expression to the developing forebrain and neural tube. Brain Res
Dev Brain Res. 128:83–90. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Phippard D, Heydemann A, Lechner M, Lu L,
Lee D, Kyin T and Crenshaw EB III: Changes in the subcellular
localization of the Brn4 gene product precede mesenchymal
remodeling of the otic capsule. Hear Res. 120:77–85. 1998.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Heller RS, Stoffers DA, Liu A, Schedl A,
Crenshaw EB III, Madsen OD and Serup P: The role of Brn4/Pou3f4 and
Pax6 in forming the pancreatic glucagon cell identity. Dev Biol.
268:123–134. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Hussain MA, Lee J, Miller CP and Habener
JF: POU domain transcription factor brain 4 confers pancreatic
alpha-cell-specific expression of the proglucagon gene through
interaction with a novel proximal promoter G1 element. Mol Cell
Biol. 17:7186–7194. 1997. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Lim R and Brichta AM: Anatomical and
physiological development of the human inner ear. Hear Res.
338:9–21. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Kanzaki S: Gene delivery into the inner
ear and its clinical implications for hearing and balance.
Molecules. 23:25072018. View Article : Google Scholar
|
|
27
|
Roccio M and Edge AS: Inner ear organoids:
New tools to understand neurosensory cell development, degeneration
and regeneration. Development. 146:dev1771882019. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Brooks PM, Rose KP, MacRae ML, Rangoussis
KM, Gurjar M, Hertzano R and Coate TM: Pou3f4-expressing otic
mesenchyme cells promote spiral ganglion neuron survival in the
postnatal mouse cochlea. J Comp Neurol. 528:1967–1985. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Kidokoro Y, Karasawa K, Minowa O, Sugitani
Y, Noda T, Ikeda K and Kamiya K: Deficiency of transcription factor
Brn4 disrupts cochlear gap junction plaques in a model of DFN3
non-syndromic deafness. PLoS One. 9:e1082162014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Phippard D, Lu L, Lee D, Saunders JC and
Crenshaw EB III: Targeted mutagenesis of the POU-domain gene
Brn4/Pou3f4 causes developmental defects in the inner ear. J
Neurosci. 19:5980–5989. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
de Kok YJ, van der Maarel SM,
Bitner-Glindzicz M, Huber I, Monaco AP, Malcolm S, Pembrey ME,
Ropers HH and Cremers FP: Association between X-linked mixed
deafness and mutations in the POU domain gene POU3F4. Science.
267:685–688. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ocak E, Duman D and Tekin M: Genetic
causes of inner ear anomalies: A review from the Turkish study
group for inner ear anomalies. Balkan Med J. 36:206–211. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
DeSmidt AA, Zou B, Grati M, Yan D, Mittal
R, Yao Q, Richmond MT, Denyer S, Liu XZ and Lu Z: Zebrafish model
for nonsyndromic X-linked sensorineural deafness, DFNX1. Anat Rec
(Hoboken). 303:544–555. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Yizhar-Barnea O, Valensisi C, Jayavelu ND,
Kishore K, Andrus C, Koffler-Brill T, Ushakov K, Perl K, Noy Y,
Bhonker Y, et al: DNA methylation dynamics during embryonic
development and postnatal maturation of the mouse auditory sensory
epithelium. Sci Rep. 8:173482018. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Gong WX, Gong RZ and Zhao B: HRCT and MRI
findings in X-linked non-syndromic deafness patients with a POU3F4
mutation. Int J Pediatr Otorhinolaryngol. 78:1756–1762. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Corvino V, Apisa P, Malesci R, Laria C,
Auletta G and Franzé A: X-linked sensorineural hearing loss: A
literature review. Curr Genomics. 19:327–338. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Barashkov NA, Klarov LA, Teryutin FM,
Solovyev AV, Pshennikova VG, Konnikova EE, Romanov GP, Tobokhov AV,
Morozov IV, Bondar AA, et al: A novel pathogenic variant
c.975G>A (p.Trp325*) in the POU3F4 gene in Yakut family (Eastern
Siberia, Russia) with the X-linked deafness-2 (DFNX2). Int J
Pediatr Otorhinolaryngol. 104:94–97. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Giannantonio S, Agolini E, Scorpecci A,
Anzivino R, Bellacchio E, Cocciadiferro D, Novelli A, Digilio MC
and Marsella P: Genetic identification and molecular modeling
characterization of a novel POU3F4 variant in two Italian deaf
brothers. Int J Pediatr Otorhinolaryngol. 129:1097902020.
View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Ozyilmaz B, Mercan GC, Kirbiyik O, Özdemir
TR, Özkara S, Kaya ÖÖ, Kutbay YB, Erdoğan KM, Güvenç MS and Koç A:
First-line molecular genetic evaluation of autosomal recessive
non-syndromic hearing loss. Turk Arch Otorhinolaryngol. 57:140–148.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Jang JH, Oh J, Han JH, Park HR, Kim BJ,
Lee S, Kim MY, Lee S, Oh DY, Choung YH and Choi BY: Identification
of a novel frameshift variant of POU3F4 and genetic counseling of
Korean incomplete partition type III subjects based on detailed
genotypes. Genet Test Mol Biomarkers. 23:423–427. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Han JJ, Nguyen PD, Oh DY, Han JH, Kim AR,
Kim MY, Park HR, Tran LH, Dung NH, Koo JW, et al: Elucidation of
the unique mutation spectrum of severe hearing loss in a Vietnamese
pediatric population. Sci Rep. 9:16042019. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Su Y, Gao X, Huang SS, Mao JN, Huang BQ,
Zhao JD, Kang DY, Zhang X and Dai P: Clinical and molecular
characterization of POU3F4 mutations in multiple DFNX2 Chinese
families. BMC Med Genet. 19:1572018. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Du W, Han MK, Wang DY, Han B, Zong L, Lan
L, Yang J, Shen Q, Xie LY, Yu L, et al: A POU3F4 mutation causes
nonsyndromic hearing loss in a Chinese X-linked recessive family.
Chin Med J (Engl). 130:88–92. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Wu D, Huang W, Xu Z, Li S, Zhang J, Chen
X, Tang Y, Qiu J, Wang Z, Duan X and Zhang L: Clinical and genetic
study of 12 Chinese Han families with nonsyndromic deafness. Mol
Genet Genomic Med. 8:e11772020. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Aristidou C, Theodosiou A, Bak M, Mehrjouy
MM, Constantinou E, Alexandrou A, Papaevripidou I,
Christophidou-Anastasiadou V, Skordis N, Kitsiou-Tzeli S, et al:
Position effect, cryptic complexity, and direct gene disruption as
disease mechanisms in de novo apparently balanced translocation
cases. PLoS One. 13:e02052982018. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Anderson EA, Ozutemiz C, Miller BS, Moss
TJ and Nascene DR: Hypothalamic hamartomas and inner ear
diverticula with X-linked stapes gusher syndrome-new associations?
Pediatr Radiol. 50:142–145. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Siddiqui A, D'Amico A, Colafati GS, Cicala
D, Talenti G, Rajput K, Pinelli L and D'Arco F: Hypothalamic
malformations in patients with X-linked deafness and incomplete
partition type 3. Neuroradiology. 61:949–952. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Henry BM, Skinningsrud B, Saganiak K,
Pękala PA, Walocha JA and Tomaszewski KA: Development of the human
pancreas and its vasculature-An integrated review covering
anatomical, embryological, histological, and molecular aspects. Ann
Anat. 221:115–124. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Zhou Q and Melton DA: Pancreas
regeneration. Nature. 557:351–358. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Gromada J, Franklin I and Wollheim CB:
Alpha-cells of the endocrine pancreas: 35 years of research but the
enigma remains. Endocr Rev. 28:84–116. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Takeda Y, Fujita Y, Sakai K, Abe T,
Nakamura T, Yanagimachi T, Sakagami H, Honjo J, Abiko A, Makino Y
and Haneda M: Expression of transcription factors in
MEN1-associated pancreatic neuroendocrine tumors. Endocrinol
Diabetes Metab Case Rep. 2017:17–0088. 2017.PubMed/NCBI
|
|
52
|
Li F, Su Y, Cheng Y, Jiang X, Peng Y, Li
Y, Lu J, Gu Y, Zhang C, Cao Y, et al: Conditional deletion of Men1
in the pancreatic β-cell leads to glucagon-expressing tumor
development. Endocrinology. 156:48–57. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Bramswig NC and Kaestner KH:
Transcriptional regulation of α-cell differentiation. Diabetes Obes
Metab. 13 (Suppl 1):S13–S20. 2011. View Article : Google Scholar
|
|
54
|
Tan SY, Mei Wong JL, Sim YJ, Wong SS,
Mohamed Elhassan SA, Tan SH, Ling Lim GP, Rong Tay NW, Annan NC,
Bhattamisra SK and Candasamy M: Type 1 and 2 diabetes mellitus: A
review on current treatment approach and gene therapy as potential
intervention. Diabetes Metab Syndr. 13:364–372. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Zamfirov K and Philippe J: Musculoskeletal
complications in diabetes mellitus. Rev Med Suisse. 13:917–921.
2017.(in French). PubMed/NCBI
|
|
56
|
Phadnis SM, Joglekar MV, Dalvi MP,
Muthyala S, Nair PD, Ghaskadbi SM, Bhonde RR and Hardikar AA: Human
bone marrow-derived mesenchymal cells differentiate and mature into
endocrine pancreatic lineage in vivo. Cytotherapy. 13:279–293.
2011. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
Sueda R, Imayoshi I, Harima Y and Kageyama
R: High Hes1 expression and resultant Ascl1 suppression regulate
quiescent vs. active neural stem cells in the adult mouse brain.
Genes Dev. 33:511–523. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
58
|
Shimazaki T, Arsenijevic Y, Ryan AK,
Rosenfeld MG and Weiss S: A role for the POU-III transcription
factor Brn-4 in the regulation of striatal neuron precursor
differentiation. EMBO J. 18:444–456. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Tan X, Zhang L, Qin J, Tian M, Zhu H, Dong
C, Zhao H and Jin G: Transplantation of neural stem cells
co-transfected with Nurr1 and Brn4 for treatment of Parkinsonian
rats. Int J Dev Neurosci. 31:82–87. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Tan X, Zhang L, Zhu H, Qin J, Tian M, Dong
C, Li H and Jin G: Brn4 and TH synergistically promote the
differentiation of neural stem cells into dopaminergic neurons.
Neurosci Lett. 571:23–28. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Zhang X, Jin G, Wang L, Hu W, Tian M, Qin
J and Huang H: Brn-4 is upregulated in the deafferented hippocampus
and promotes neuronal differentiation of neural progenitors in
vitro. Hippocampus. 19:176–186. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Shi J, Jin G, Zhu H, Tian M, Zhang X, Qin
J and Tan X: The role of Brn-4 in the regulation of neural stem
cell differentiation into neurons. Neurosci Res. 67:8–17. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Zhang X, Zhang L, Cheng X, Guo Y, Sun X,
Chen G, Li H, Li P, Lu X, Tian M, et al: IGF-1 promotes Brn-4
expression and neuronal differentiation of neural stem cells via
the PI3K/Akt pathway. PLoS One. 9:e1138012014. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Ninkovic J, Steiner-Mezzadri A, Jawerka M,
Akinci U, Masserdotti G, Petricca S, Fischer J, von Holst A,
Beckers J, Lie CD, et al: The BAF complex interacts with Pax6 in
adult neural progenitors to establish a neurogenic cross-regulatory
transcriptional network. Cell Stem Cell. 13:403–418. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Guo J, Cheng X, Zhang L, Wang L, Mao Y,
Tian G, Xu W, Wu Y, Ma Z, Qin J, et al: Exploration of the
Brn4-regulated genes enhancing adult hippocampal neurogenesis by
RNA sequencing. J Neurosci Res. 95:2071–2079. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Zhang L, Zhang X, Zhang Y, Xu N, Wang J,
Zhu Y and Xia C: Brn4 promotes the differentiation of radial glial
cells into neurons by inhibiting CtBP2. Life Sci. 254:1168662020.
View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Dhivya V and Balachandar V: Cell
replacement therapy is the remedial solution for treating
Parkinson's disease. Stem Cell Investig. 4:592017. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Tan XF, Qin JB, Jin GH, Tian ML, Li HM,
Zhu HX, Zhang XH, Shi JH and Huang Z: Effects of Brn-4 on the
neuronal differentiation of neural stem cells derived from rat
midbrain. Cell Biol Int. 34:877–882. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Thomson JA, Itskovitz-Eldor J, Shapiro SS,
Waknitz MA, Swiergiel JJ, Marshall VS and Jones JM: Embryonic stem
cell lines derived from human blastocysts. Science. 282:1145–1147.
1998. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Takahashi K and Yamanaka S: Induction of
pluripotent stem cells from mouse embryonic and adult fibroblast
cultures by defined factors. Cell. 126:663–676. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Takahashi K, Tanabe K, Ohnuki M, Narita M,
Ichisaka T, Tomoda K and Yamanaka S: Induction of pluripotent stem
cells from adult human fibroblasts by defined factors. Cell.
131:861–872. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Jerabek S, Ng CK, Wu G, Arauzo-Bravo MJ,
Kim KP, Esch D, Malik V, Chen Y, Velychko S, MacCarthy CM, et al:
Changing POU dimerization preferences converts Oct6 into a
pluripotency inducer. EMBO Rep. 18:319–333. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Ma K, Deng X, Xia X, Fan Z, Qi X, Wang Y,
Li Y, Ma Y, Chen Q, Peng H, et al: Direct conversion of mouse
astrocytes into neural progenitor cells and specific lineages of
neurons. Transl Neurodegener. 7:292018. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Ma Y, Wang K, Pan J, Fan Z, Tian C, Deng
X, Ma K, Xia X, Huang Y and Zheng JC: Induced neural progenitor
cells abundantly secrete extracellular vesicles and promote the
proliferation of neural progenitors via extracellular
signal-regulated kinase pathways. Neurobiol Dis. 124:322–334. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
75
|
He M, Zhang H, Li Y, Tian C, Tang B, Huang
Y and Zheng J: Direct and selective lineage conversion of human
fibroblasts to dopaminergic precursors. Neurosci Lett. 699:16–23.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Black JB, Adler AF, Wang HG, D'Ippolito
AM, Hutchinson HA, Reddy TE, Pitt GS, Leong KW and Gersbach CA:
Targeted epigenetic remodeling of endogenous Loci by
CRISPR/Cas9-based transcriptional activators directly converts
fibroblasts to neuronal cells. Cell Stem Cell. 19:406–414. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
77
|
Chuang W, Sharma A, Shukla P, Li G, Mall
M, Rajarajan K, Abilez OJ, Hamaguchi R, Wu JC, Wernig M and Wu SM:
Partial reprogramming of pluripotent stem cell-derived
cardiomyocytes into neurons. Sci Rep. 7:448402017. View Article : Google Scholar : PubMed/NCBI
|
|
78
|
Luo C, Lee QY, Wapinski O, Castanon R,
Nery JR, Mall M, Kareta MS, Cullen SM, Goodell MA, Chang HY, et al:
Global DNA methylation remodeling during direct reprogramming of
fibroblasts to neurons. Elife. 8:e401972019. View Article : Google Scholar : PubMed/NCBI
|
|
79
|
Treutlein B, Lee QY, Camp JG, Mall M, Koh
W, Shariati SA, Sim S, Neff NF, Skotheim JM, Wernig M and Quake SR:
Dissecting direct reprogramming from fibroblast to neuron using
single-cell RNA-seq. Nature. 534:391–395. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
80
|
Zhu X, Zhou W, Jin H and Li T: Brn2 alone
is sufficient to convert astrocytes into neural progenitors and
neurons. Stem Cells Dev. 27:736–744. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
81
|
Kim SM, Kim JW, Kwak TH, Park SW, Kim KP,
Park H, Lim KT, Kang K, Kim J, Yang JH, et al: Generation of
integration-free induced neural stem cells from mouse fibroblasts.
J Biol Chem. 291:14199–14212. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
82
|
Kwak TH, Hali S, Kim S, Kim J, La H, Kim
KP, Hong KH, Shin CY, Kim NH and Han DW: Robust and reproducible
generation of induced neural stem cells from human somatic cells by
defined factors. Int J Stem Cells. 13:80–92. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
83
|
Han DW, Tapia N, Hermann A, Hemmer K,
Höing S, Araúzo-Bravo MJ, Zaehres H, Wu G, Frank S, Moritz S, et
al: Direct reprogramming of fibroblasts into neural stem cells by
defined factors. Cell Stem Cell. 10:465–472. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
84
|
Chang YK, Srivastava Y, Hu C, Joyce A,
Yang X, Zuo Z, Havranek JJ, Stormo GD and Jauch R: Quantitative
profiling of selective Sox/POU pairing on hundreds of sequences in
parallel by Coop-seq. Nucleic Acids Res. 45:832–845. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
85
|
Bar-Nur O, Verheul C, Sommer AG, Brumbaugh
J, Schwarz BA, Lipchina I, Huebner AJ, Mostoslavsky G and
Hochedlinger K: Lineage conversion induced by pluripotency factors
involves transient passage through an iPSC stage. Nat Biotechnol.
33:761–768. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
86
|
Velychko S, Kang K, Kim SM, Kwak TH, Kim
KP, Park C, Hong K, Chung C, Hyun JK, MacCarthy CM, et al: Fusion
of reprogramming factors alters the trajectory of somatic lineage
conversion. Cell Rep. 27:30–39.e4. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
87
|
Zou Q, Yan Q, Zhong J, Wang K, Sun H, Yi X
and Lai L: Direct conversion of human fibroblasts into neuronal
restricted progenitors. J Biol Chem. 289:5250–5260. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
88
|
Potts MB, Siu JJ, Price JD, Salinas RD,
Cho MJ, Ramos AD, Hahn J, Margeta M, Oldham MC and Lim DA: Analysis
of Mll1 deficiency identifies neurogenic transcriptional modules
and Brn4 as a factor for direct astrocyte-to-neuron reprogramming.
Neurosurgery. 75:472–482. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
89
|
Yu Q, Chen J, Deng W, Cao X, Wang Y, Zhou
J, Xu W, Du P, Wang Q, Yu J and Xu X: Direct reprogramming of mouse
fibroblasts into neural cells via Porphyra yezoensis polysaccharide
based high efficient gene co-delivery. J Nanobiotechnology.
15:822017. View Article : Google Scholar : PubMed/NCBI
|
|
90
|
Huffman JL and Harmer B: End of Life Care.
StatPearls [Internet]. StatPearls Publishing; Treasure Island, FL:
2020
|
|
91
|
Faguet GB: Quality end-of-life cancer
care: An overdue imperative. Crit Rev Oncol Hematol. 108:69–72.
2016. View Article : Google Scholar : PubMed/NCBI
|
|
92
|
Bhagirath D, Yang TL, Tabatabai ZL, Majid
S, Dahiya R, Tanaka Y and Saini S: BRN4 is a novel driver of
neuroendocrine differentiation in castration-resistant prostate
cancer and is selectively released in extracellular vesicles with
BRN2. Clin Cancer Res. 25:6532–6545. 2019. View Article : Google Scholar : PubMed/NCBI
|