Introduction
Intervertebral disc degeneration (IDD), the leading
cause of low back pain and spinal disabilities (1–3), poses
a significant social and medical burden on adults worldwide.
Various diseases secondary to IDD, namely, degenerative disc
diseases (DDD), occur in approximately 71% of males and 77% of
females older than age 50 and in more than 90% of the older
population (4,5).
Traditional surgical interventions that remove
degenerative nucleus pulposus (NP) tissue and are dependent on
internal fixation are not able to fully restore spinal function and
may result in various complications, including deep infection and
neurological impairment (1,2,6–9). New
stem cell-based therapies are promising alternatives to traditional
interventions targeting early to middle-stage IDD before its
irreversible progression requires open intervention. Specifically,
stem cell-based therapies for IDD introduce exogenous or autologous
mesenchymal stem cells (MSCs) into the target intervertebral disc
via injection (8,10–12).
The rationale for this treatment is two-fold. First, MSCs can
differentiate into NP-like cells, express NP marker genes (e.g.,
SOX9, type II collagen, and aggrecan), and stimulate the formation
of the extracellular matrix (ECM), which partially enforces the
biomechanical property of the disc and further protects the MSCs
from the harsh intra-disc microenvironment. Second, the functioning
of degenerative NPCs needs to be restored under the influence of
cytokines and growth factors produced by the MSCs.
The potential of MSCs to treat disc degeneration has
been proven in many in vitro (6,8,13) and
in vivo (9–11) studies. Adipose-derived mesenchymal
stem cells (ASCs) can be induced to differentiate into NP-like
cells with significantly higher expression of specific markers,
including aggrecan, SOX9, and type II collagen (14). Animal studies have confirmed such
in vitro evidence and shown that ASCs injected into injured
lumbar discs increase secretion of the ECM and decrease the
ossification of damaged cartilage in the NP compared to untreated
controls (15). Accumulating
encouraging data on ASCs as well as their relatively ample
availability and clinical safety make them one of the most
promising candidates for stem cell therapy for IDD (16). However, the underlying cellular
mechanisms of cross-talking between ASCs and degenerative target
cells at the gene level have not yet been comprehensively
investigated.
Long non-coding RNAs (lncRNAs) and microRNAs
(miRNAs) have been increasingly investigated because of their
regulatory functions in gene expression (17,18).
lncRNAs are involved in various biological processes with high
tissue specificity and low sequence conservation (19,20),
and differentially expressed lncRNAs have previously been
highlighted in the disc degeneration process (21,22).
Previous findings showed that lncRNAs were differentially expressed
by degenerative nucleus pulposus cells (NPCs) when they were
co-cultured with ASCs (22).
Furthermore, miRNAs regulate gene expression, influence various
biological activities [e.g., cell proliferation, differentiation,
and metabolism (23)], and play
noteworthy roles in the disc degeneration process (24–27).
For instance, miR-27b (24) and
miR-93 (25) specifically promote
matrix degradation and hasten disc degeneration. However, to the
best of our knowledge, no study has underlined the specific roles
of lncRNAs and miRNAs within a co-culture of ASCs and degenerative
NPCs that mimics the microenvironments of clinical ASCs in the
treatment of IDD.
Advanced gene expression microarray and
bioinformatics modalities were used to identify lncRNA and mRNA
mappings during the differentiation of ASCs into NP-like cells and
to further show specifically involved signaling pathways and gene
regulation networks.
Materials and methods
Ethics approval and patient
consent
All human tissue was obtained and used with
patients' informed consent and under the approval of the
institutional review broad of Shanghai General Hospital, Shanghai
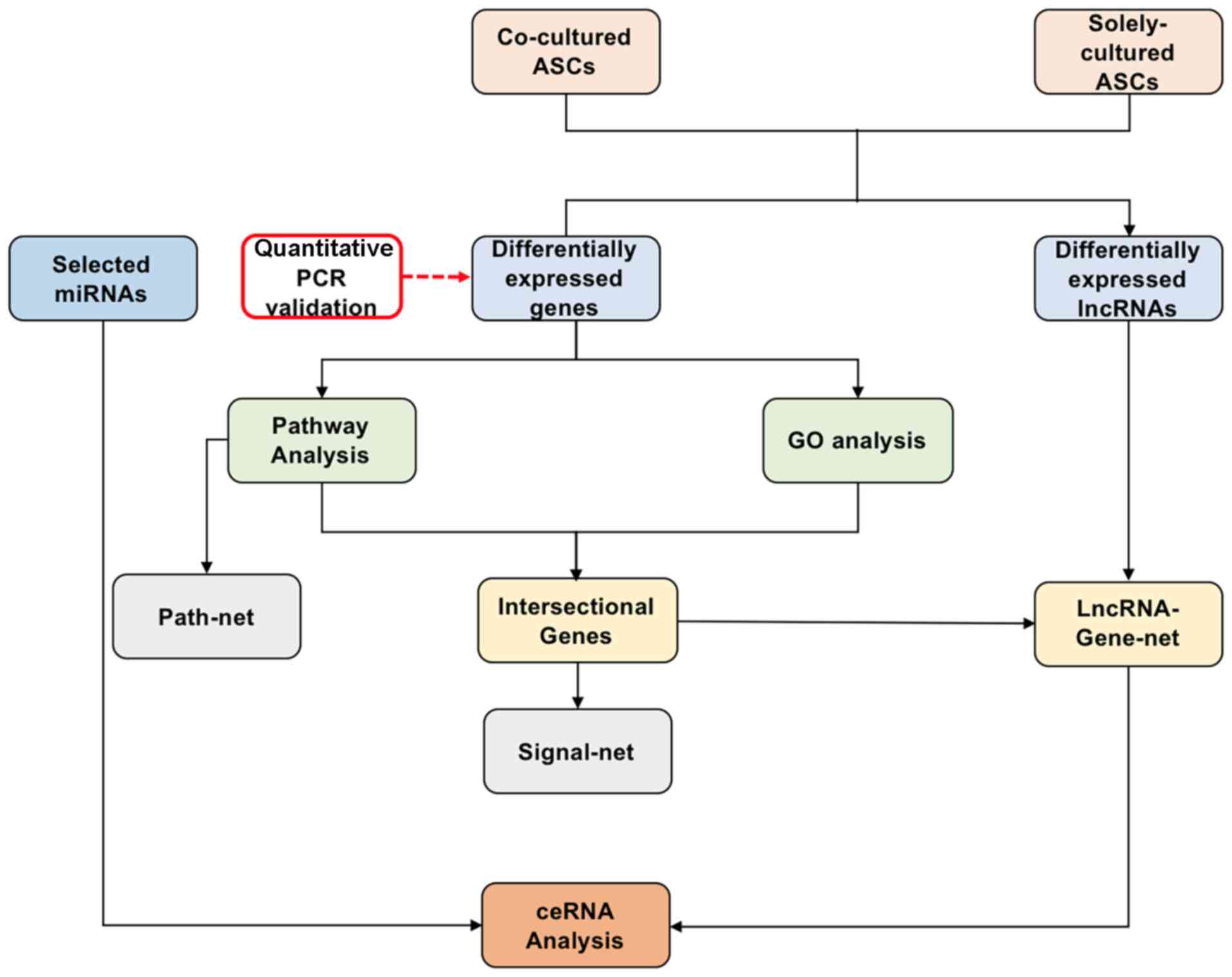
Jiao Tong University, Shanghai, China. A schematic of the study is
shown in Fig. 1.
Isolation and culture of NPCs
The human NP tissue was obtained surgically from
degenerative discs [grades III–IV according to the Pfirrmann
grading system (12)] of three
patients diagnosed with lumbar spondylosis. These patients (one
female and two males, 46- to 58-years of age, with a mean age of
52) were enrolled and the tissue was collected at Shanghai General
Hospital, Shanghai, China. NPCs were isolated according to a method
previously reported (6,8,12).
Briefly, the NP tissue was stored in cold phosphate-buffered saline
(PBS) and then transferred to the lab. After being washed in PBS to
remove all blood and annulus fibrous tissue, the NP tissue was cut
into 1 mm sections amid serum-free medium, i.e., Dulbecco's
modified Eagle's medium/F12 medium (DMEM/F12; HyClone),
supplemented with 1% penicillin/streptomycin. Then the tissue was
digested with 0.4 mg/ml collagenase II (Sigma-Aldrich; Merck KGaA)
at 37°C for 5 h before normal medium was used to end the process.
Thereafter, the cell suspension was passed consecutively through
filters (100, 70, 40 µm) to remove debris, then centrifuged at 400
× g for 5 min in room temperature to form the cell pellets. The
supernatant was discarded, and cells were resuspended and cultured
in DMEM supplemented with 10% fetal bovine serum (FBS) in an
incubator at 37°C with 5% CO2 then passaged at 80%
confluency. The medium was changed every 2–3 days.
Isolation, culture, and identification
of ASCs
ASCs were identified and isolated from adipose
tissue based on reported methods (12,28,29).
Briefly, adipose tissue was harvested from identical patients as
the NP tissue, and then reserved in DMEM. After being carefully
washed with PBS to remove blood, fat tissue was minced into 1
mm2 sections, and then treated with 0.1% type I
collagenase (Sigma-Aldrich; Merck KGaA) at 37°C for 60 min on a
shaking bench. Subsequently, the cell tissue mixture was filtered
as mentioned previously, followed by centrifugation (400 × g for 5
min at room temperature) and removal of the supernatant. The cell
pellets were resuspended with and cultured in DMEM/F12 (HyClone)
containing 10% FBS and 1% penicillin/streptomycin in an incubator
at 37°C with 5% CO2. The medium was changed every 2–3
days, and the cells were passaged when confluency reached 80%.
Multilineage differentiation and flow cytometry were
performed to identify the stemness of the isolated cells. Briefly,
adipogenic medium was used to induce differentiation of candidate
cells at passage 3. Adipogenic induction was performed by seeding
cells in adipogenic medium supplemented with 0.5 mM isobutyl
methylxanthine, 0.5 mM dexamethasone, and 60 mM indomethacin
(Sigma-Aldrich; Merck KGaA) and culturing them for 7 days. ASCs
were cultured with osteogenic medium (STEMCELL) for 2 weeks, then
fixed with 4% paraformaldehyde for subsequent staining; the
chondrogenic potential of isolated cells was tested by seeding and
culturing them with a specific chondrogenic medium for 14 days
(STEMCELL) and then fixing them for evaluation. All culture media
were changed every 2–3 days.
In vitro differentiation of ASCs was
evaluated by histological staining for adipogenesis (Oil Red O
staining), osteogenesis (Alizarin Red staining), and chondrogenesis
(Alcian blue and type II collagen staining). Furthermore,
expression of surface markers, including CD105, CD73, CD29, CD34,
CD45, and human leukocyte antigen (HLA)-DR, was investigated as
described previously (12).
Co-culture of ASCs and NPCs
To imitate stem cell therapy in vitro, we
established a cell-cell co-culture system using a 6-well plate with
a 0.4 µm pore size membrane. A total of 6.0×104
degenerative NPCs were seeded onto the base of the well, and the
ASCs were seeded onto the upper surface of the membrane. Both NPCs
and ASCs at passage 3 were used. The cell ratio was 1:1 and it was
chosen based on the previous studies (6,8). The
cells were co-cultured for 7 days in DMEM/F12 at 37°C and 5%
CO2 in a humidified atmosphere, and the medium was
changed every 2 days. The co-cultured ASCs (n=3) formed the
experimental group, whereas ASCs cultured alone in the same
conditions (n=3) acted as controls.
RNA isolation and quality control
Total RNA from co-cultured ASCs and ASCs was
cultured alone using TRIzol (Invitrogen), then purified it with the
RNase Kit (Bio-Rad Laboratories, Inc.). The quantity of RNA was
measured using a spectrophotometer (NanoDrop-1000, Thermo
Scientific Fisher) and tested its quality with 1% agarose gel
electrophoresis as previously described (30).
Microarray analysis
A multistep strategy was used for the gene
microarray and subsequent analyses (Fig. 1). ASCs co-cultured with degenerative
NPCs (the experimental group) and ASCs cultured alone (the control
group) were used to identify differentially expressed RNAs.
Microarray analysis was performed by GMINIX Informatics Ltd. Co.
(Shanghai, China). The data have been uploaded to the NCBI Gene
Expression Omnibus and can be accessed via the GEO Series accession
GSE118927 (ncbi.nlm.nih.gov/geo/query/acc.cgi?acc=GSE118927).
Differential expression of mRNAs and lncRNAs between
the co-cultured ASCs and ASCs cultured alone was determined with a
random variance model (RVM) t-test to increase the degrees of
freedom in the small data sets. P<0.05 as the threshold for
significance, these differentially expressed RNAs were further
selected and evaluated with false discovery rate analysis.
Unsupervised hierarchical clustering was performed to generate a
cluster map.
Quantitative (q)PCR validation
To verify the microarray data, six RNAs were
randomly selected for qPCR validation. Briefly, cDNA was generated
with the Taqman Reverse Transcription kit (Invitrogen; Thermo
Fisher Scientific, Inc.) following the manufacturer's protocol.
Gene expression was investigated by qPCR with the SYBR-Green
Mastermix on a CFX96 Touch Real-time PCR Detection System (both
Bio-Rad Laboratories, Inc.). The thermocycling conditions (totally
35 cycles) consisted of the initial denaturation of 30 sec at 95°C,
followed by 60 sec annealing at 65°C, and the extension step lasted
for 1 min/kb at 68°C. Human GAPDH was used as the housekeeping gene
to determine the relative expression of selected genes, and
relative changes in gene expression were compared to those of
untreated cells by the 2−ΔΔCq method (12). All reactions were performed in
triplicate, and the primer sequences are provided in Table I.
 | Table I.Primers used in qPCR. |
Table I.
Primers used in qPCR.
| Genes | Forward sequence
5′-3′ | Reverse sequence
5′-3′ |
|---|
| IGF1 |
GCTCTTCAGTTCGTGTGTGGA |
GCCTCCTTAGATCACAGCTCC |
| XIST |
TCTAGTCCCCCAACACCCTT |
GGAGGACGTGTCAAGAAGACA |
| TTTY15 |
CTCATCACCTGGAGTCCGTG |
CAACGGCAAATACTTTAGGTTTTCT |
| FABP3 |
TGGAGTTCGATGAGACAACAGC |
CTCTTGCCCGTCCCATTTCTG |
| TGFB2 |
CAGCACACTCGATATGGACCA |
CCTCGGGCTCAGGATAGTCT |
| GPC3 |
ATTGGCAAGTTATGTGCCCAT |
TTCGGCTGGATAAGGTTTCTTC |
| GAPDH |
GGAGCGAGATCCCTCCAAAAT |
GGCTGTTGTCATACTTCTCATGG |
Gene ontology (GO) analysis
Using the GO Database (GO Consortium, geneontology.org), we performed GO analysis to explore
the functions of the differentially expressed mRNAs using the
two-sided Fisher's exact test and Chi-square test. Each
differentially expressed gene was evaluated independently and
marked as up- or downregulated. P-values of all differentially
expressed genes were computed in all GO categories, and P<0.01
was considered to be significant.
Pathway analysis
The significance levels of pathways associated with
differentially expressed genes were analyzed based on the Kyoto
Encyclopedia of Genes and Genomes database (KEGG; genome.jp/kegg/).
Fisher's exact test and the Chi-square test were used to select the
key pathways according to a significance threshold of P<0.05.
Additionally, a Path-net, the interaction network consisting of the
significant pathways, was generated based on information for those
pathways from the KEGG database.
Signal-net analysis
Using the KEGG database, we constructed a Signal-net
to reveal the gene-gene interplay among the significant
differentially expressed genes in both the GO analysis and pathway
analysis. The Signal-net shows molecular networks consisting of
nodes and lines representing genes and regulatory patterns of
activation or phosphorylation. We calculated the importance of each
gene by counting the numbers of upstream and downstream genes in
the form of in-degrees and out-degrees. The betweenness centrality
of each gene was recorded according to its in-degree and
out-degree; a higher betweenness centrality suggesting more
importance in the regulation of the gene network.
lncRNA-mRNA-net analysis
The co-expression network of lncRNAs and mRNAs was
investigated based on differentially expressed lncRNAs and
intersectional mRNAs with both GO analysis and pathway
analysis.
Competing endogenous RNA (ceRNA)
analysis
miRNAs are single-strand non-coding RNAs shorter
than 22 nucleotides responsible for regulating gene expression by
binding to miRNA response elements (MREs) on mRNAs (31,32).
lncRNAs are also non-coding RNAs but are longer than 200
nucleotides; they both harbor MREs and compete with RNAs for miRNA
binding. lncRNAs regulate the number of miRNA by sequestering and
binding to them (33); thus, they
can participate in post-transcriptional regulation as ceRNAs.
We selected 25 miRNAs associated with disc
degeneration that were differentially expressed by ASCs co-cultured
with degenerative NPCs and investigated their roles in the
regulation of mRNAs and lncRNAs (Table SI). The target mRNAs of
miRNAs were predicted based on TargetScan (www.targetscan.org/) and miRanda (www.microrna.org/microrna/home.do) for
the ceRNAs analysis. The ceRNA network was further constructed by
those negatively regulated intersectional lncRNAs and mRNAs.
Statistical analysis
Data are presented as the mean ± SD. Differences
between groups were evaluated with unpaired Student's t-test in
SPSS 20.0 (IBM). Statistical significance was set at P<0.05.
Results
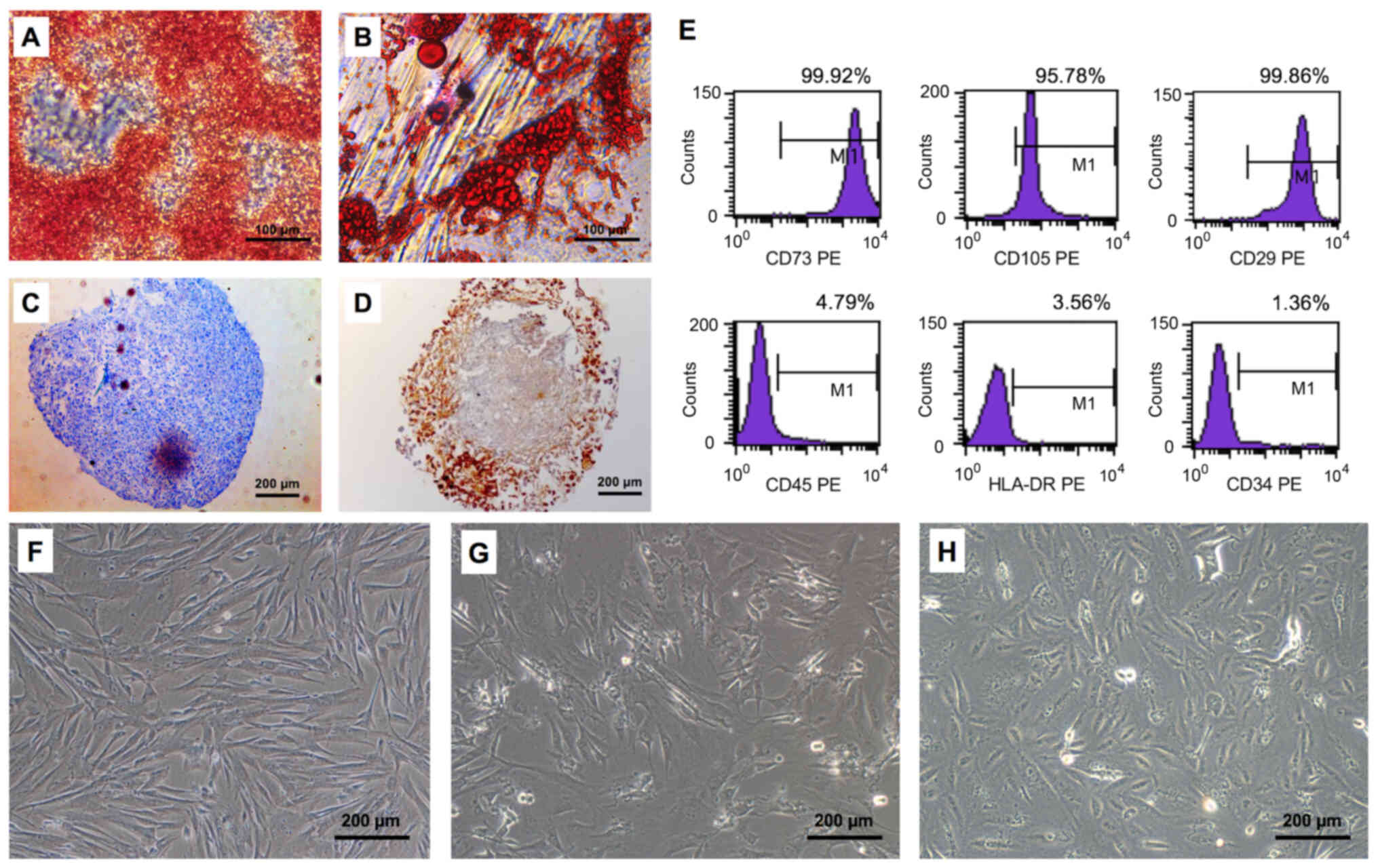
Multilineage differentiation and
identification of ASCs
The cells isolated from the human adipose tissue
were demonstrated to be MSCs with the stemness of multilineage
differentiation (Fig. 2A-D) and
mesenchymal markers (Fig. 2E). In
particular, the ASCs differentiated into osteocytes and adipose
cells as shown by positive Alizarin Red (Fig. 2A) and Oil Red O (Fig. 2B) staining, respectively. In
addition, the ASCs differentiated into chondrocytes (Fig. 2C) and produced type II collagen in
the chondrogenic environment (Fig.
2D). Furthermore, MSC surface markers expressed by ASCs
included CD73, CD105, and CD29; no hematopoietic stem cell markers
(e.g. CD45, CD34 or HLA-DR) were detected (Fig. 2E).
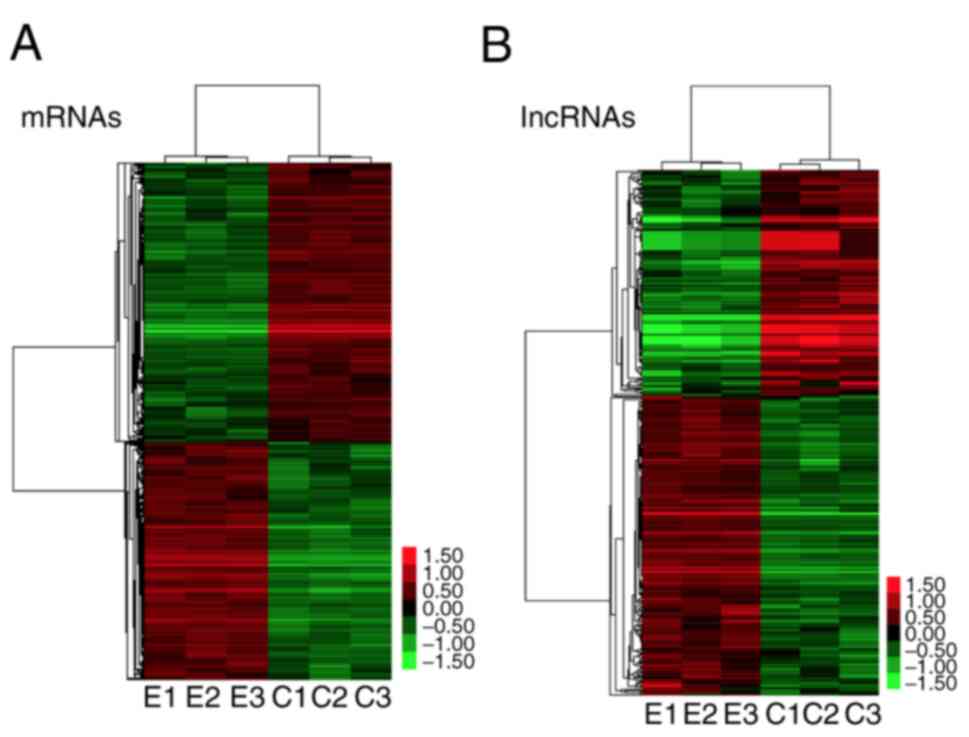
Differentially expressed mRNAs and
lncRNAs in co-cultured ASCs
A total of 1757 genes were found to be
differentially expressed by ASCs, of which carboxypeptidase X (M14
family) member 1 (CPXM1) had the largest increase in expression
(28-fold change; P<0.001) and mRNA coding cartilage oligomeric
matrix protein (COMP) had the largest decrease in expression
(-61-fold change; P<0.001; Fig.
3A).
A total of 360 lncRNAs were identified as
differentially expressed by ASCs, of which XIST (X-inactive
specific transcript) was maximally upregulated (23-fold change) and
SNAR-E was greatly downregulated (12-fold change; Fig. 3B).
qPCR validation
The microarray data were further validated through
six differentially expressed RNAs (IGF1, XIST, TTTY15, FABP3,
TGFB2, GPC3) that were randomly selected for qPCR analysis. Of
these RNAs, upregulation of IGF1, XIST, TGFB2, and GPC3 in the
microarray was validated (all P<0.05; Fig. 4). Similarly, in good agreement with
gene-chip data, TTTY15 and FABP3 were demonstrated to be
downregulated (both P<0.05; Fig.
4A). In addition, the expression levels of SOX9, type II
collagen, and aggrecan were significantly higher in the
Experimental group than these in the Control group (all P<0.05,
Fig. 4B).
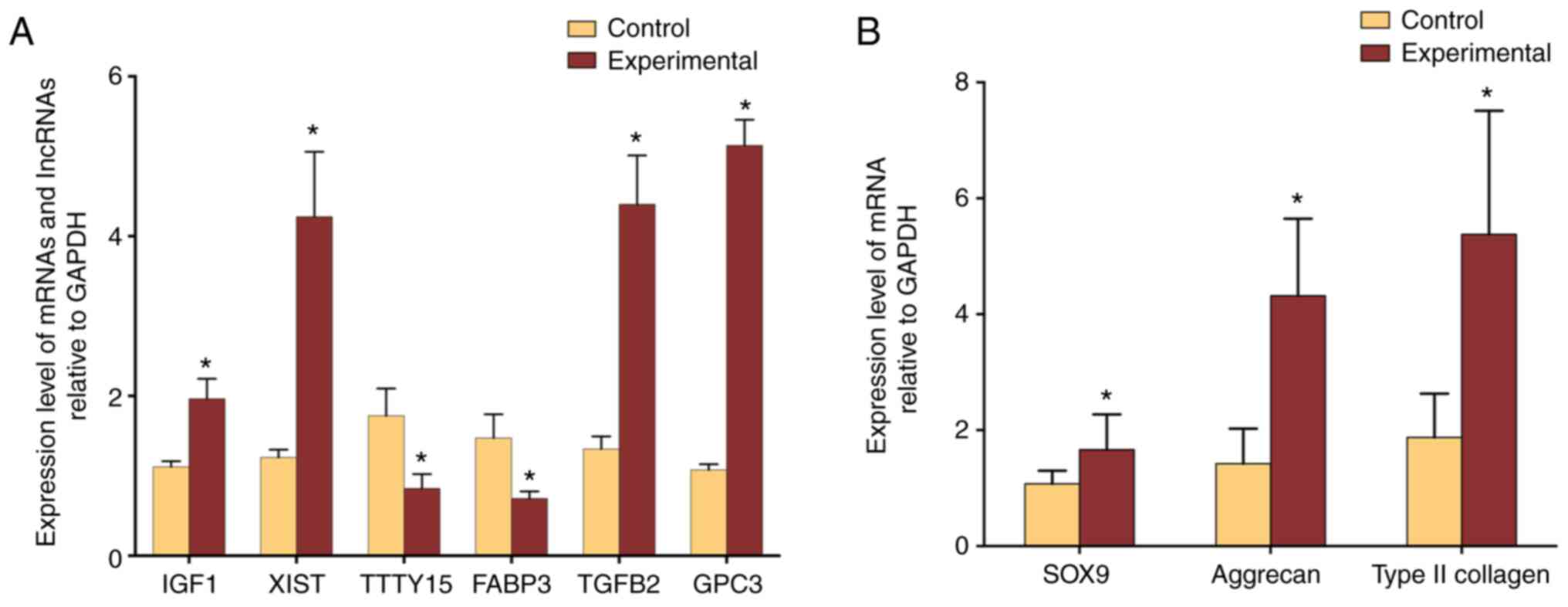 | Figure 4.Validation of microarray data with
real-time PCR. (A) Expression of IGF1, XIST, TGFB2, and GPC3 was
significantly upregulated in the experimental group of ASCs
co-cultured with degenerated NPCs compared to control ASCs cultured
alone, whereas the expression of TTTY15 and FABP3 was significantly
downregulated in the experimental group. (B) Expression levels of
NPC specific maker genes. The expression of SOX9, type II collagen,
and aggrecan were significantly higher in the Experimental group
than these in the Control group (P=0.0153, P=0.0003, P<0.0001,
respectively). *P<0.05. IGF1, insulin-like growth factor 1;
XIST, X inactive-specific transcript; TTTY15, testis specific
transcript, Y-linked 15; FABP3, fatty acid binding protein 3;
TGFB2, transforming growth factor, beta 2; GPC3, glypican-3. |
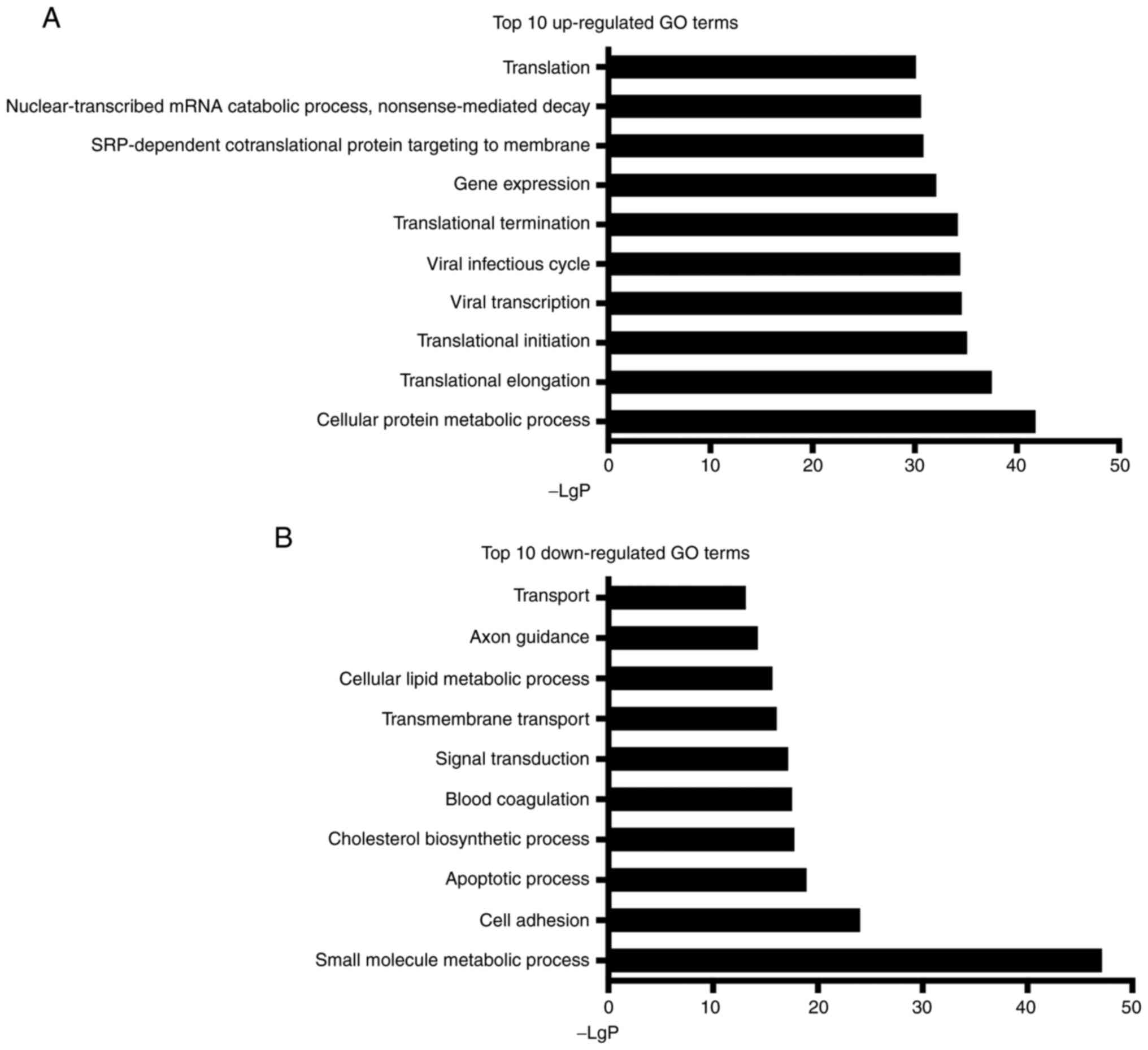
GO analysis
The GO analysis identified 589 upregulated and 661
downregulated GO terms among all differentially expressed mRNAs. In
particular, cellular protein metabolic process (GO:0044267) was
most significantly upregulated (-LgP=41.8; Fig. 5), and the small molecule metabolic
process (GO:0044281) was most considerably downregulated
(-LgP=47.1). Note that various GO terms related to gene translation
could be observed among the 10 most upregulated terms, including
translational elongation (GO:0006414), translational initiation
(GO:0006413), and translational termination (GO:0006415).
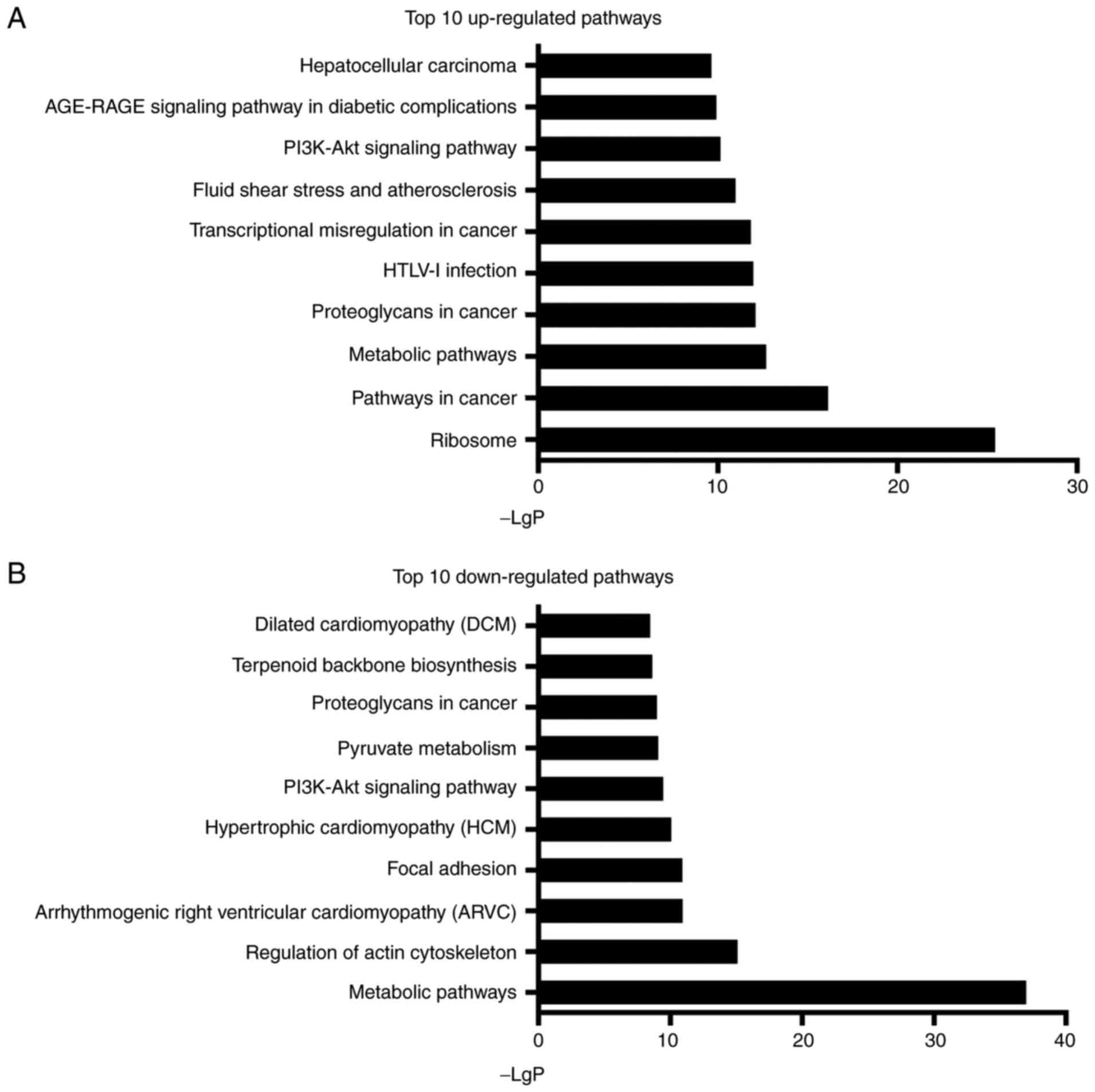
Pathway analysis
A total of 299 pathways were identified as
significantly enriched during the co-culture of ASCs and
degenerative NPCs, of which 144 pathways were significantly
upregulated and 155 were significantly downregulated (all
P<0.05). Specifically, ribosome (Path ID: 03010) was the most
upregulated, with a -LgP of 25.4, whereas the metabolic pathways
(Path ID: 01100) were the most significantly downregulated, with a
-LgP of 37.0 (Fig. 6).
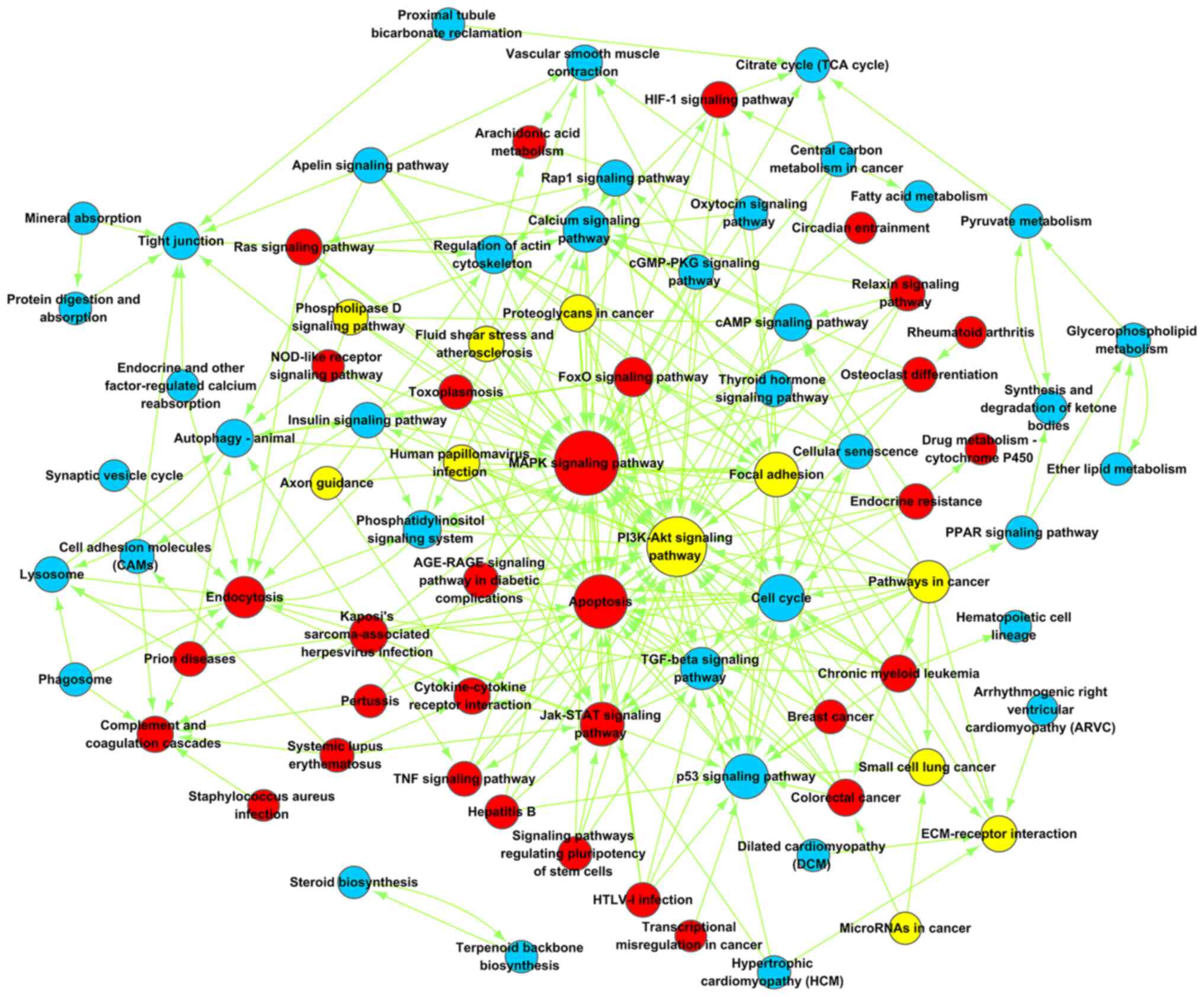
The Path-net was constructed to elucidate the
complex interactions among all significantly enriched pathways and
to represent all directly and systematically involved pathways
during the co-culture of ASCs with NPCs (Fig. 7; Table
II). The mitogen-activated protein kinase (MAPK) signaling
pathway was found to play an important role in the differentiation
of ASCs into NP-like cells, with the highest betweenness centrality
at 44. The PI3K-AKT signaling pathway was both up- and
downregulated during differentiation with the second highest
betweenness centrality (degree=38). The cell cycle pathway was the
most significantly downregulated signaling pathway.
 | Table II.Top 10 pathways in path-net
analysis. |
Table II.
Top 10 pathways in path-net
analysis.
| Path name | Style | Degree | Indegree | Outhegree |
|---|
| MAPK signaling
pathway | up | 44 | 40 | 4 |
| PI3K-Akt signaling
pathway | up, down | 38 | 30 | 8 |
| Apoptosis | up | 30 | 25 | 5 |
| Cell cycle | Down | 21 | 19 | 2 |
| Calcium signaling
pathway | Down | 19 | 16 | 3 |
| Focal adhesion | Down, up | 18 | 11 | 7 |
| p53 signaling
pathway | Down | 18 | 15 | 3 |
| Jak-STAT signaling
pathway | Up | 17 | 12 | 5 |
| Pathways in
cancer | Up, down | 15 | 0 | 15 |
| TGF-beta signaling
pathway | Down | 15 | 12 | 3 |
Signal-net
The Signal-net was constructed to explore the
interactions of all genes with significant alterations in both the
GO and pathway analyses (Fig. 8,
Table III). The KRAS gene
was found to play the most important role (downregulated;
degree=20) within the differentiation of ASCs into NP-like cells,
followed by phospholipase C β-1 (PLCB1) and PLCB4 (both
downregulated; degree=17). By contrast, FOS showed the most
significant upregulation (degree=15). In good agreement with the
results of the pathway analysis, MAPK family genes were also
identified as playing critical roles in the Signal-net, including
MAPK8 (downregulated; degree=16) and MAPK13 (upregulated;
degree=15).
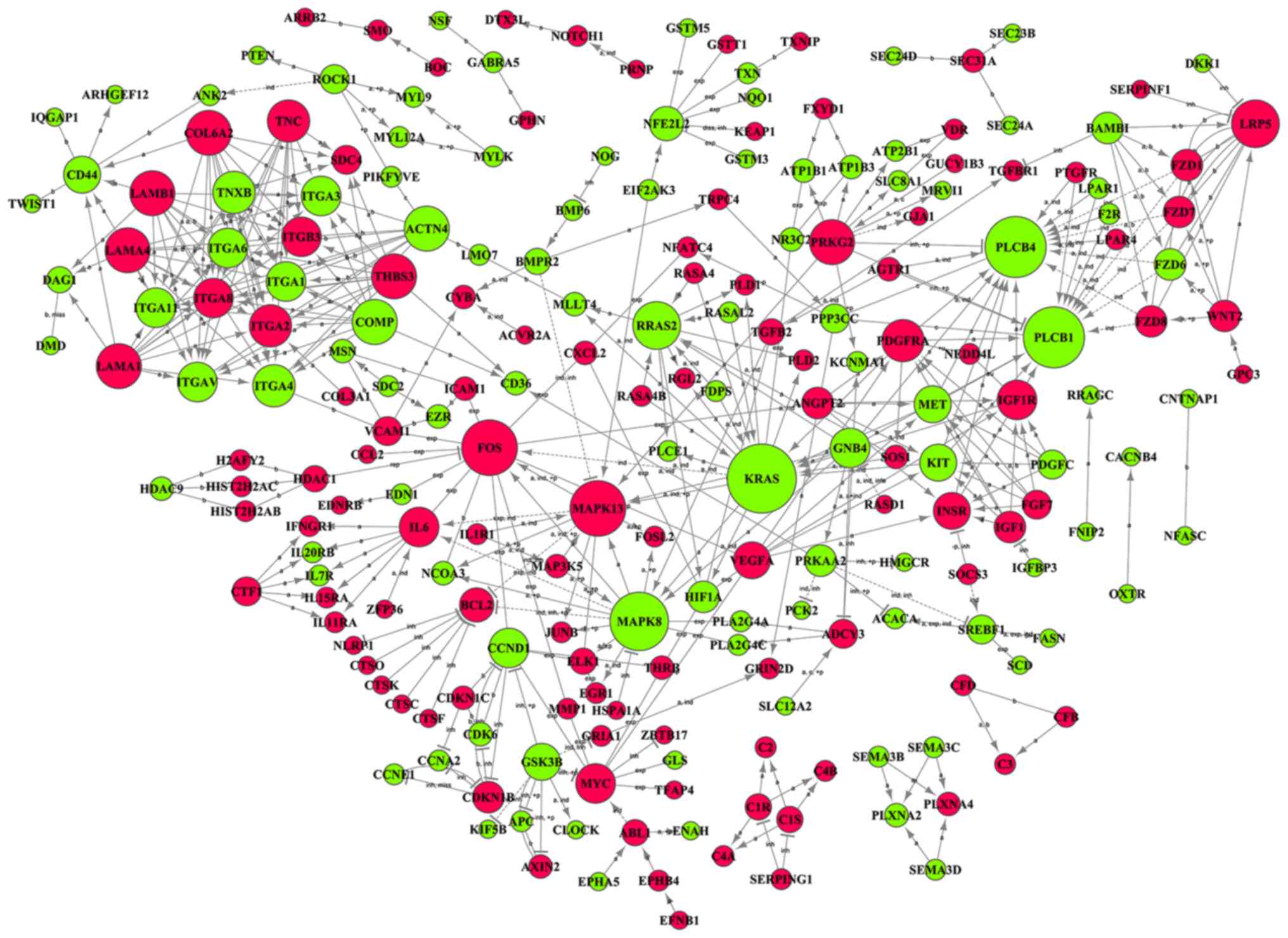 | Figure 8.Signal-net showing the interaction
among 218 differentially expressed genes. The KRAS gene, which was
downregulated, was the most critical player in the Signal-net
(degree=20), and FOS was the most important upregulated gene in the
network (degree=15). Up- and downregulated genes are indicated by
red and green circles, respectively. The circle size represents the
number of other genes interacting with the specific gene.
Inter-gene interactions are reported as activation (a),
binding/association (b), compound (c), phosphorylation (+p),
repression (rep), ubiquitination (+u), inhibition (inb), expression
(exp), dephosphorylation (-p), indirect effect (ind), missing
interaction (miss), and dissociation (diss). |
 | Table III.Top 10 genes in Signal-Net
analysis. |
Table III.
Top 10 genes in Signal-Net
analysis.
| Gene symbol | Style | Degree | Indegree | Outhegree |
|---|
| KRAS | Down | 20 | 11 | 9 |
| PLCB1 | Down | 17 | 16 | 1 |
| PLCB4 | Down | 17 | 16 | 1 |
| MAPK8 | Down | 16 | 5 | 11 |
| FOS | Up | 15 | 5 | 10 |
| MAPK13 | Up | 15 | 8 | 7 |
| LRP5 | Up | 12 | 8 | 4 |
| RRAS2 | Down | 12 | 6 | 6 |
| ACTN4 | Down | 11 | 1 | 10 |
| COL6A2 | Up | 11 | 0 | 11 |
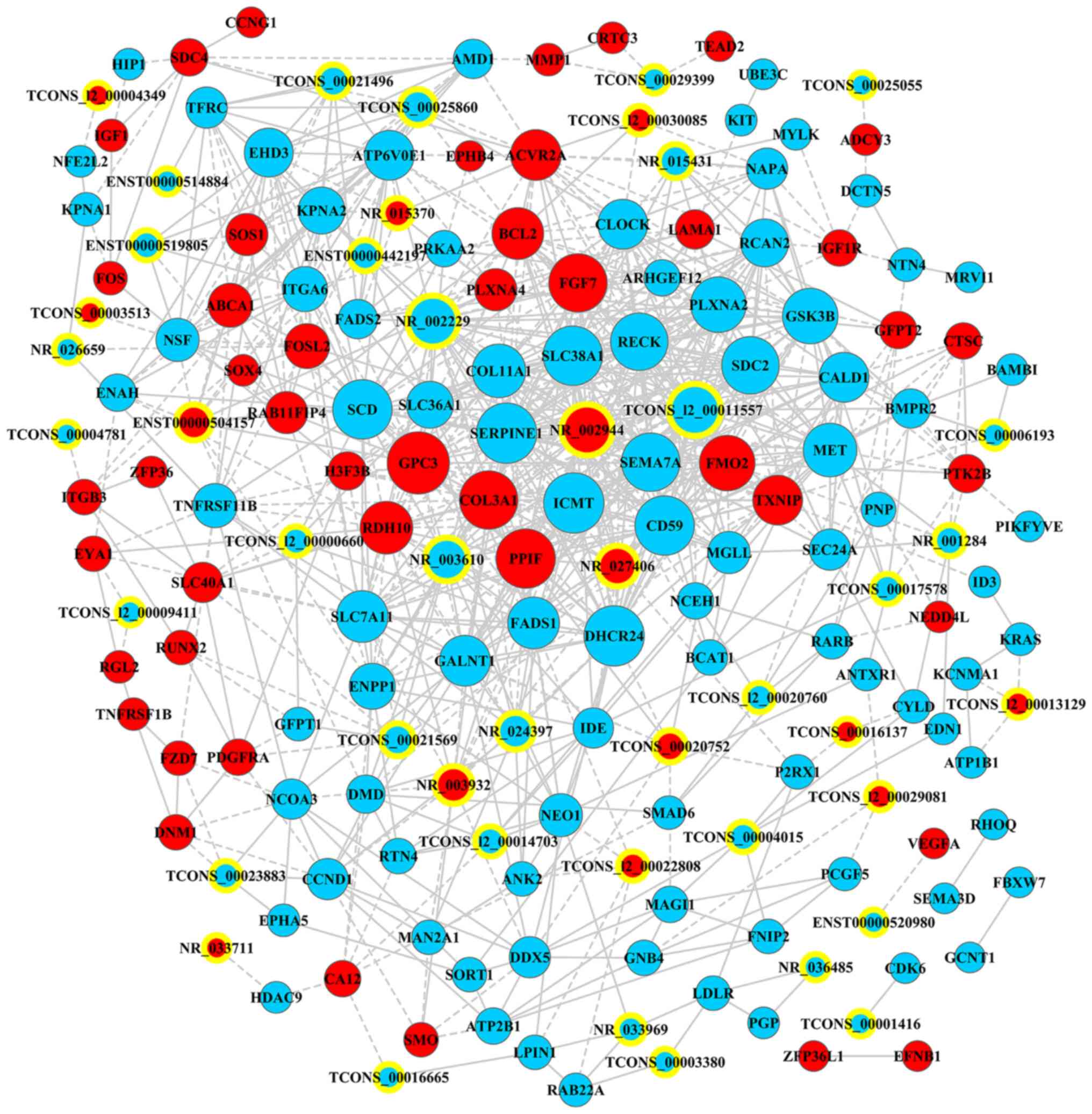
lncRNA-mRNA co-expression network
A lncRNA-mRNA co-expression network was established
on the basis of 178 RNAs (44 lncRNAs and 134 mRNAs) converting into
875 pairs of co-expressed lncRNA-mRNA (Fig. 9; Table
IV). In the co-expression network, GPC3 (glypican-3; coding;
downregulated; degree=34) had the highest number of interactions,
followed by SERPINE1 (serpin peptidase inhibitor, clade E, member
1; coding; downregulated; degree=32) and ICMT (isoprenylcysteine
carboxyl methyltransferase; coding; downregulated; degree=32),
whereas TCONS_l2_00011557 had the highest interaction (degree=29)
among all non-coding RNAs.
 | Table IV.Top 10 mRNAs and lncRNAs in
lncRNA-mRNA co-expression network. |
Table IV.
Top 10 mRNAs and lncRNAs in
lncRNA-mRNA co-expression network.
| Gene symbol | Biotype | Style | Degree |
|---|
| GPC3 | Coding | up | 34 |
| SERPINE1 | Coding | Down | 32 |
| ICMT | Coding | Down | 32 |
| DHCR24 | Coding | Down | 32 |
| SLC38A1 | Coding | Down | 31 |
| PPIF | Coding | up | 31 |
| CD59 | Coding | Down | 31 |
| SCD | Coding | Down | 31 |
| FGF7 | Coding | up | 30 |
| SEMA7A | Coding | Down | 30 |
|
TCONS_l2_00011557 | Noncoding | Down | 29 |
| NR_002229 | Noncoding | Down | 28 |
| NR_002944 | Noncoding | up | 28 |
| NR_003610 | Noncoding | Down | 20 |
| NR_027406 | Noncoding | up | 17 |
| NR_003932 | Noncoding | up | 14 |
| NR_024397 | Noncoding | Down | 13 |
|
ENST00000504157 | Noncoding | up | 12 |
| TCONS_00025860 | Noncoding | Down | 12 |
| NR_015431 | Noncoding | Down | 10 |
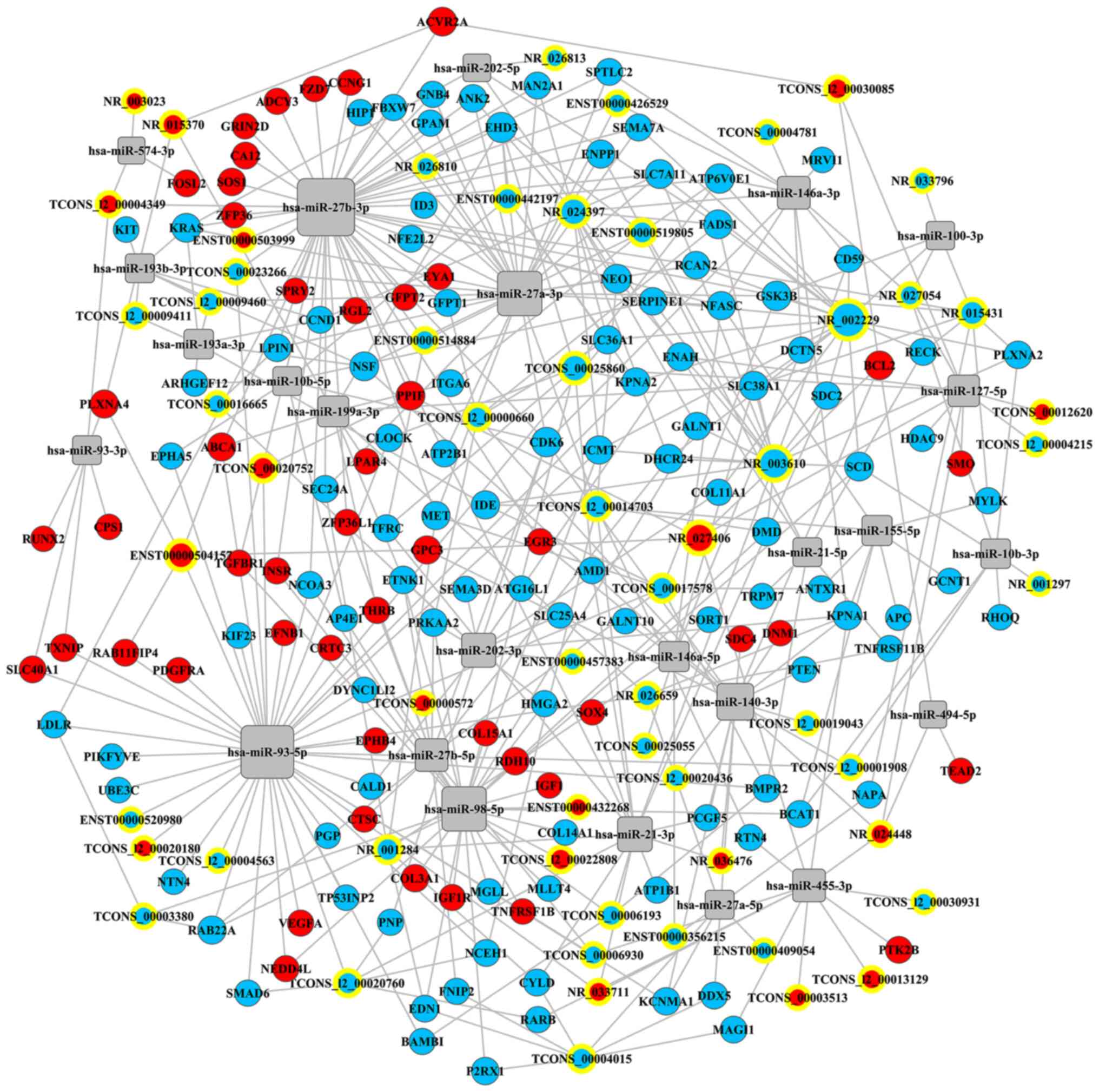
ceRNA network
A ceRNA network was constructed from 25 IDD-related
miRNAs to investigate their regulatory interplay with
differentially expressed mRNAs and lncRNAs during the
differentiation of ASCs into NP-like cells (Fig. 10; Table
V). It is noteworthy that hsa-miR-27b-3p (degree of
interaction=50) was the most important miRNA, followed by
hsa-miR-93-5p (degree of interaction=43) and hsa-miR-27a-3p (degree
of interaction=30). NR_002229, NR_003610, and NR_024397 were the
three most important lncRNAs in the regulatory network, with
degrees of interaction of 19, 16, and 11, respectively.
 | Table V.Top10 mRNAs, microRNAs and lncRNAs in
ceRNA network. |
Table V.
Top10 mRNAs, microRNAs and lncRNAs in
ceRNA network.
| RNA symbol | Type | Style | Degree |
|---|
| CDK6 | mRNA | Down | 7 |
| EHD3 | mRNA | Down | 6 |
| GSK3B | mRNA | Down | 6 |
| KPNA1 | mRNA | Down | 6 |
| ATP6V0E1 | mRNA | Down | 5 |
| FADS1 | mRNA | Down | 5 |
| ICMT | mRNA | Down | 5 |
| KRAS | mRNA | Down | 5 |
| NEO1 | mRNA | Down | 5 |
| NFASC | mRNA | Down | 5 |
| hsa-miR-27b-3p | miRNA | NO | 50 |
| hsa-miR-93-5p | miRNA | NO | 43 |
| hsa-miR-27a-3p | miRNA | NO | 30 |
| hsa-miR-98-5p | miRNA | NO | 30 |
| hsa-miR-140-3p | miRNA | NO | 18 |
| hsa-miR-21-3p | miRNA | NO | 15 |
| hsa-miR-202-3p | miRNA | NO | 14 |
|
hsa-miR-146a-3p | miRNA | NO | 12 |
| hsa-miR-27b-5p | miRNA | NO | 12 |
| hsa-miR-455-3p | miRNA | NO | 12 |
| NR_002229 | lncRNA | Down | 19 |
| NR_003610 | lncRNA | Down | 16 |
| NR_024397 | lncRNA | Down | 11 |
| NR_015431 | lncRNA | Down | 8 |
| TCONS_00025860 | lncRNA | Down | 8 |
|
TCONS_l2_00014703 | lncRNA | Down | 8 |
| TCONS_00004015 | lncRNA | Down | 7 |
| TCONS_00017578 | lncRNA | Down | 7 |
| NR_001284 | lncRNA | Down | 6 |
|
TCONS_l2_00020760 | lncRNA | Down | 6 |
Discussion
We identified 1,757 differentially expressed genes
and 360 differentially expressed lncRNAs in ASCs co-cultured with
degenerative NPCs. Among all the differentially expressed genes,
559 significantly upregulated and 661 significantly downregulated
GO terms were identified involving 298 significantly enriched
pathways. These results highlight the sophisticated interplay of
these mRNAs and lncRNAs with miRNAs.
The MAPK and PI3K-Akt signaling pathways were
identified as playing crucial roles within NPC-directed
differentiation of ASCs. Three distinct MAPK subsets have been
identified in mammalian cells: Classical MAPK (ERK), p38 kinase,
and C-Jun N-terminal kinase/stress-activated protein kinase
(JNK/SAPK) (34). In general, the
MAPK signaling pathway is involved in many complex cellular
processes, including proliferation, differentiation, development,
and apoptosis. It can convey, amplify, and integrate external
bio-signals and elicit cellular responses (34). Regarding IDD, the MAPK signaling
pathway participates in many pathophysiological mechanisms and
contributes to disc degeneration (35,36).
Tian et al reported the increasing degradation of aggrecan
with modulated expression of ADAMTS-4 (A dis-integrin and
metalloproteinase with thrombospondin motifs 4) in NPCs through
activation of MAPK and NF-κB signaling (37).
However, our data indicate that the MAPK signaling
pathway was upregulated when the ASCs differentiated into NP-like
cells in vitro, which suggests that this pathway is also
important in the regeneration of a degenerated disc. Such a
discrepancy between previous studies and the present one is
possibly attributable to the diverse roles this signaling pathway
plays in various cellular behaviors, including both metabolic
(e.g., proliferation and differentiation) and catabolic (e.g.,
apoptosis) activity. Similar positive roles of the MAPK signaling
pathway have been previously reported. For instance, the p38 MAPK
signaling pathway is activated after estrogen-induced enhancement
of matrix synthesis in NPCs, whereas inhibiting p38 MAPK signaling
significantly abolishes such effects of estrogen on matrix
synthesis in NPCs (38).
The most important finding of the Signal-net
analysis is that the COMP gene was closely associated with
integrins (ITG) family members, including ITGB3, ITGA2, and ITGA3.
COMP can interact with ECM proteins, bridging those molecules in a
mesh pattern (39). Specifically,
it binds to type II collagen in the presence of Zn2+ at
four defined sites on the collagen molecule (40), and it also connects to aggrecan in a
concentration-dependent manner (41). The present study emphasizes a tight
correlation between COMP and the ITG family, which is partially
consistent with findings from Chen et al, where COMP
mediates chondrocyte attachment through its interactions with
integrins (42). In addition, COMP
interacts with growth factors and acts as a ‘lattice’ to present
them for utilization by the cells (39). In the present study, expression
levels of COMP decreased in co-cultured ASCs may be biological
because the ASCs stopped proliferation and began differentiation
when they were co-cultured with NPCs, while the cell number and
activity may have been suppressed by differentiation compared with
solely-cultured ASCs. However, the pathological decrease of COMP
may hinder the ECM assembly and further weaken the cell attachment.
Therefore, the role COMP plays in ASCs differentiation towards
NP-like cell remains to be further elucidated.
LncRNAs have recently gained widespread attention as
novel and crucial players in biological regulation. Wan et
al investigated the differentially expressed lncRNAs in human
intervertebral disc degeneration with lncRNA-mRNA microarray
analysis (22). They found 1,052
lncRNAs and 1,314 mRNAs were differentially expressed between
healthy human NP and degenerative NP tissues, of which the
ENST00000461676 was the most significantly upregulated lncRNA with
the fold-change at 136 and the NR_003716 was the top downregulated
one with the fold-change at 149, despite the specific function of
these lncRNAs in the IVDD not having been studied. Furthermore, we
previously addressed the aberrantly expressed mRNAs and lncRNAs in
degenerative NPCs co-cultured with ASCs, to determine the mRNAs and
lncRNAs which may play a critical role during stem-cell-based
therapy of IDD (12). In
particular, we found that the secreted phosphoprotein 1 (SPP1),
metallothionein 1F (MT1F) and ectonucleotide pyrophosphatase 1
(ENPP1) presented the top three differentially expressed mRNAs by
the degenerative NPCs, after they were co-cultured with ASCs.
Our data identified XIST (fold change=23) as the
most significantly altered lncRNA in the co-culturing process.
XIST, a 17–20 kb non-coding RNA, is essential for whole-chromosome
silencing (43) via its
inactivation of the X chromosome (44,45).
Additionally, XIST provides one of few tangible readouts of the
stem cell quality and influences the pluripotent stem cell
population (46). However, further
investigations are required for a detailed functional
characterization of XIST and other lncRNAs involved in NPC-directed
differentiation of ASCs. Furthermore, the upregulation of IGF-1 and
TGFβ was proven to protect the degeneration of IVD (47,48);
the IGF-1 and TGFβ-2 was upregulated in the co-cultured ASCs in the
present study (Fig. 4), indicating
the protective function of ASCs to the NPCs. The expression of GPC3
was also upregulated in ASCs after co-culturing. GPC3 is known as a
potential marker due to its higher gene expression and protein
expression in NPCs compared with that in chondrocytes (49,50).
Therefore, the increment of GPC3 expression suggests ASCs
differentiated towards NP-like cells.
Additionally, the TTTY15 was significantly
downregulated in the current study (Fig. 4A), yet its role in intervertebral
disc degeneration has not been investigated. TTTY15 had been
demonstrated to regulate hypoxia-induced injury of vascular
endothelial cells (51). Similarly,
Huang et al found suppression of TTTY15 could mitigate
hypoxia-induced injury in cardiomyocytes. Therefore, we speculate
that the decrease of TTTY15 expression may indicate the tolerance
of co-cultured ASCs to hypoxia, the condition that NPCs regularly
undergo within intervertebral discs (52). The FABP3 expression was decreased
after co-culturing (Fig. 4A). FABP3
binds to fatty acid for the intracellular lipid droplet
accumulation, and it increases MSC survival (53,54).
It decreased possibly because the ASCs differentiated into NP-like
cells after co-culturing. However, the contribution of these RNAs
in the NPC-oriented differentiation of ASCs is currently unknown,
and further investigation is required.
Regarding the ceRNA network, miR-27b-3p plays the
most critical role in the network (degree of interaction=50),
followed by hsa-miR-93-5p (degree of interaction=43). Specifically,
miR-27b targets directly matrix metalloproteinase-13 (24), which is excessively expressed in
degenerative disc tissue (55), and
its downregulation leads to type II collagen loss and thus the
development of IDD. Our data underline the critical role of miR-93
in the differentiation of ASCs into NP-like cells, which is in
agreement with previous evidence. miR-93 was previously found to be
expressed less in human degenerative NP tissue in a disc
degeneration-dependent manner (25). These two miRNAs, miR-27b and miR-93,
which are significantly altered during disc degeneration may also
be markedly differentially expressed in NPC-guided differentiation
of ASCs; however, further investigations are needed to validate any
possible associations.
This study has several limitations. First, we
investigated numerous RNAs via gene microarray assay but only
validated results selectively by qPCR because of practicality and
cost. In addition, we selectively interpreted the results based on
previous studies and clinical implications. To the best of our
knowledge, the present study was the second and the independent
part of one project, which also included our previous study
(12), which reported the
differentially expressed mRNAs and lncRNAs between the NPCs
cocultured with ASCs and the solely-cultured NPCs, thus the NPCs
were not included as the control. Second, no further specific
evaluations of particular roles were performed for NPC-directed
differentiation of ASCs using overexpression or RNA interference.
Similarly, the Path-net and the Signal-net were only outlined on
the basis of a bioinformatics database, and comprehensive
understanding of the specific key pathways or factors requires
further sophisticated in vitro or in vivo
studies.
In conclusion, findings of the present study
revealed differentially expressed mRNAs and lncRNAs in ASCs
co-cultured with degenerative NPCs and highlights sophisticated
cross-talking among mRNAs, lncRNAs, and miRNAs. Our data provide
valuable information for the further development and clinical
translation of stem cell-based approaches to treating IDD.
Acknowledgements
Not applicable.
Funding
This work was supported by the National Natural
Science Foundation of China (no. 81301580).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
ZH, QW and JWu conceptualized and designed the
study. ZH, QW, RG, XW and JWa performed the experiments and
collected the data. ZH, LG, RG and JWu analyzed and interpreted the
data. ZH, LG, XW and JWa drafted the manuscript. JWu supervised the
study and revised the manuscript. All authors read and approved the
final manuscript.
Ethics approval and consent to
participate
All human tissues were obtained and used with the
patients' informed consent and under the approval of Institutional
Review Broad of the Shanghai General Hospital, Shanghai Jiao Tong
University, Shanghai, China (approval no. 2019SQ098).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Han Z, Gao L, Shi Q, Chen L and Chen C:
Quantitative magnetic resonance imaging for diagnosis of
intervertebral disc degeneration of the cervico-thoracic junction:
A pilot study. Am J Transl Res. 10:925–935. 2018.PubMed/NCBI
|
|
2
|
Chen P, Wu C, Huang M, Jin G, Shi Q, Han Z
and Chen C: Apparent diffusion coefficient of diffusion-weighted
imaging in evaluation of cervical intervertebral disc degeneration:
An observational study with 3.0 T magnetic resonance imaging.
Biomed Res Int. 2018:68430532018.PubMed/NCBI
|
|
3
|
Chen C, Jia Z, Han Z, Gu T, Li W, Li H,
Tang Y, Wu J, Wang D, He Q and Ruan D: Quantitative T2 relaxation
time and magnetic transfer ratio predict endplate biochemical
content of intervertebral disc degeneration in a canine model. BMC
Musculoskelet Disord. 16:1572015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Urban JP and Roberts S: Degeneration of
the intervertebral disc. Arthritis Res Ther. 5:120–130. 2003.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Andersson GB: Epidemiological features of
chronic low-back pain. Lancet. 354:581–585. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Ruan D, Zhang Y, Wang D, Zhang C, Wu J,
Wang C, Shi Z, Xin H, Xu C, Li H and He Q: Differentiation of human
Wharton's jelly cells toward nucleus pulposus-like cells after
coculture with nucleus pulposus cells in vitro. Tissue Eng Part A.
18:167–175. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Zeng Y, Chen C, Liu W, Fu Q, Han Z, Li Y,
Feng S, Li X, Qi C, Wu J, et al: Injectable microcryogels
reinforced alginate encapsulation of mesenchymal stromal cells for
leak-proof delivery and alleviation of canine disc degeneration.
Biomaterials. 59:53–65. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Han Z, Zhang Y, Gao L, Jiang S and Ruan D:
Human wharton's jelly cells activate degenerative nucleus pulposus
cells in vitro. Tissue Eng Part A. 24:1035–1043. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Ying J, Han Z, Zeng Y, Du Y, Pei S, Su L,
Ruan D and Chen C: Evaluation of intervertebral disc regeneration
with injection of mesenchymal stem cells encapsulated in
PEGDA-microcryogel delivery system using quantitative T2 mapping: A
study in canines. Am J Transl Res. 11:2028–2041. 2019.PubMed/NCBI
|
|
10
|
Ying J, Han Z, Pei S, Su L and Ruan D:
Effects of stromal cell-derived factor-1α secreted in degenerative
intervertebral disc on activation and recruitment of nucleus
pulposus-derived stem cells. Stem Cells Int. 2019:91478352019.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Zhang Y, Tao H, Gu T, Zhou M, Jia Z, Jiang
G, Chen C, Han Z, Xu C, Wang D, et al: The effects of human
Wharton's jelly cell transplantation on the intervertebral disc in
a canine disc degeneration model. Stem Cell Res Ther. 6:1542015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Han Z, Wang J, Gao L, Wang Q and Wu J:
Aberrantly expressed messenger RNAs and long noncoding RNAs in
degenerative nucleus pulposus cells co-cultured with
adipose-derived mesenchymal stem cells. Arthritis Res Ther.
20:1822018. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun Z, Liu ZH, Zhao XH, Sun L, Chen YF,
Zhang WL, Gao Y, Zhang YZ, Wan ZY, Samartzis D, et al: Impact of
direct cell co-cultures on human adipose-derived stromal cells and
nucleus pulposus cells. J Orthop Res. 31:1804–1813. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Sakai D, Mochida J, Iwashina T, Watanabe
T, Nakai T, Ando K and Hotta T: Differentiation of mesenchymal stem
cells transplanted to a rabbit degenerative disc model: Potential
and limitations for stem cell therapy in disc regeneration. Spine
(Phila Pa 1976). 30:2379–2387. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Chun HJ, Kim YS, Kim BK, Kim EH, Kim JH,
Do BR, Hwang SJ, Hwang JY and Lee YK: Transplantation of human
adipose-derived stem cells in a rabbit model of traumatic
degeneration of lumbar discs. World Neurosurg. 78:364–371. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Sivakamasundari V and Lufkin T: Stemming
the degeneration: IVD stem cells and stem cell regenerative therapy
for degenerative disc disease. Adv Stem Cells.
2013:7245472013.PubMed/NCBI
|
|
17
|
Hung T and Chang HY: Long noncoding RNA in
genome regulation: Prospects and mechanisms. RNA Biol. 7:582–585.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Mercer TR, Dinger ME and Mattick JS: Long
non-coding RNAs: Insights into functions. Nat Rev Genet.
10:155–159. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
19
|
Yang L, Froberg JE and Lee JT: Long
noncoding RNAs: Fresh perspectives into the RNA world. Trends
Biochem Sci. 39:35–43. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Batista PJ and Chang HY: Long noncoding
RNAs: Cellular address codes in development and disease. Cell.
152:1298–1307. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Liu X, Che L, Xie YK, Hu QJ, Ma CJ, Pei
YJ, Wu ZG, Liu ZH, Fan LY and Wang HQ: Noncoding RNAs in human
intervertebral disc degeneration: An integrated microarray study.
Genom Data. 5:80–81. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wan ZY, Song F, Sun Z, Chen YF, Zhang WL,
Samartzis D, Ma CJ, Che L, Liu X, Ali MA, et al: Aberrantly
expressed long noncoding RNAs in human intervertebral disc
degeneration: A microarray related study. Arthritis Res Ther.
16:4652014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Ambros V: The functions of animal
microRNAs. Nature. 431:350–355. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Li HR, Cui Q, Dong ZY, Zhang JH, Li HQ and
Zhao L: Downregulation of MIR-27b is involved in loss of Type II
collagen by directly targeting matrix metalloproteinase 13 (MMP13)
in human intervertebral disc degeneration. Spine (Phila Pa 1976).
41:E116–E123. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Jing W and Jiang W: MicroRNA-93 regulates
collagen loss by targeting MMP3 in human nucleus pulposus cells.
Cell Prolif. 48:284–292. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Li Z, Yu X, Shen J, Chan MT and Wu WK:
MicroRNA in intervertebral disc degeneration. Cell Prolif.
48:278–283. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Yan N, Yu S, Zhang H and Hou T: Lumbar
disc degeneration is facilitated by MiR-100-mediated FGFR3
suppression. Cell Physiol Biochem. 36:2229–2236. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Russo V, Yu C, Belliveau P, Hamilton A and
Flynn LE: Comparison of human adipose-derived stem cells isolated
from subcutaneous, omental, and intrathoracic adipose tissue depots
for regenerative applications. Stem Cells Transl Med. 3:206–217.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Schaffler A and Buchler C: Concise review:
Adipose tissue-derived stromal cells-basic and clinical
implications for novel cell-based therapies. Stem Cells.
25:818–827. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Eischen-Loges M, Oliveira KMC, Bhavsar MB,
Barker JH and Leppik L: Pretreating mesenchymal stem cells with
electrical stimulation causes sustained long-lasting pro-osteogenic
effects. PeerJ. 6:e49592018. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Chen J, Xu J, Li Y, Zhang J, Chen H, Lu J,
Wang Z, Zhao X, Xu K, Li Y, et al: Competing endogenous RNA network
analysis identifies critical genes among the different breast
cancer subtypes. Oncotarget. 8:10171–10184. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Jonas S and Izaurralde E: Towards a
molecular understanding of microRNA-mediated gene silencing. Nat
Rev Genet. 16:421–433. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Li CY, Liang GY, Yao WZ, Sui J, Shen X,
Zhang YQ, Peng H, Hong WW, Ye YC, Zhang ZY, et al: Integrated
analysis of long non-coding RNA competing interactions reveals the
potential role in progression of human gastric cancer. Int J Oncol.
48:1965–1976. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhang W and Liu HT: MAPK signal pathways
in the regulation of cell proliferation in mammalian cells. Cell
Res. 12:9–18. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Zhang K, Ding W, Sun W, Sun XJ, Xie YZ,
Zhao CQ and Zhao J: Beta1 integrin inhibits apoptosis induced by
cyclic stretch in annulus fibrosus cells via ERK1/2 MAPK pathway.
Apoptosis. 21:13–24. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Niu CC, Lin SS, Yuan LJ, Chen LH, Wang IC,
Tsai TT, Lai PL and Chen WJ: Hyperbaric oxygen treatment suppresses
MAPK signaling and mitochondrial apoptotic pathway in degenerated
human intervertebral disc cells. J Orthop Res. 31:204–209. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Tian Y, Yuan W, Fujita N, Wang J, Wang H,
Shapiro IM and Risbud MV: Inflammatory cytokines associated with
degenerative disc disease control aggrecanase-1 (ADAMTS-4)
expression in nucleus pulposus cells through MAPK and NF-κB. Am J
Pathol. 182:2310–2321. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Li P, Xu Y, Gan Y, Wang L, Ouyang B, Zhang
C, Luo L, Zhao C and Zhou Q: Estrogen enhances matrix synthesis in
nucleus pulposus cell through the estrogen receptor β-p38 MAPK
pathway. Cell Physiol Biochem. 39:2216–2226. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Acharya C, Yik JH, Kishore A, Van Dinh V,
Di Cesare PE and Haudenschild DR: Cartilage oligomeric matrix
protein and its binding partners in the cartilage extracellular
matrix: Interaction, regulation and role in chondrogenesis. Matrix
Biol. 37:102–111. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Rosenberg K, Olsson H, Morgelin M and
Heinegard D: Cartilage oligomeric matrix protein shows high
affinity zinc-dependent interaction with triple helical collagen. J
Biol Chem. 273:20397–20403. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Chen FH, Herndon ME, Patel N, Hecht JT,
Tuan RS and Lawler J: Interaction of cartilage oligomeric matrix
protein/thrombospondin 5 with aggrecan. J Biol Chem.
282:24591–24598. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Chen FH, Thomas AO, Hecht JT, Goldring MB
and Lawler J: Cartilage oligomeric matrix protein/thrombospondin 5
supports chondrocyte attachment through interaction with integrins.
J Biol Chem. 280:32655–32661. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Penny GD, Kay GF, Sheardown SA, Rastan S
and Brockdorff N: Requirement for Xist in X chromosome
inactivation. Nature. 379:131–137. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Brockdorff N, Ashworth A, Kay GF, McCabe
VM, Norris DP, Cooper PJ, Swift S and Rastan S: The product of the
mouse Xist gene is a 15 kb inactive X-specific transcript
containing no conserved ORF and located in the nucleus. Cell.
71:515–526. 1992. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Clemson CM, McNeil JA, Willard HF and
Lawrence JB: XIST RNA paints the inactive X chromosome at
interphase: Evidence for a novel RNA involved in nuclear/chromosome
structure. J Cell Biol. 132:259–275. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Minkovsky A, Patel S and Plath K: Concise
review: Pluripotency and the transcriptional inactivation of the
female mammalian X chromosome. Stem Cells. 30:48–54. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Kennon JC, Awad ME, Chutkan N, DeVine J
and Fulzele S: Current insights on use of growth factors as therapy
for intervertebral disc degeneration. Biomol Concepts. 9:43–52.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Gruber HE, Hoelscher GL, Ingram JA, Bethea
S and Hanley EN: IGF-1 rescues human intervertebral annulus cells
from in vitro stress-induced premature senescence. Growth Factors.
26:220–225. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Yuan M, Yeung CW, Li YY, Diao H, Cheung
KMC, Chan D, Cheah K and Chan PB: Effects of nucleus pulposus
cell-derived acellular matrix on the differentiation of mesenchymal
stem cells. Biomaterials. 34:3948–3961. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Minogue BM, Richardson SM, Zeef LA,
Freemont AJ and Hoyland JA: Characterization of the human nucleus
pulposus cell phenotype and evaluation of novel marker gene
expression to define adult stem cell differentiation. Arthritis
Rheum. 62:3695–3705. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Zheng J, Zhuo YY, Zhang C, Tang GY, Gu XY
and Wang F: LncRNA TTTY15 regulates hypoxia-induced vascular
endothelial cell injury via targeting miR-186-5p in cardiovascular
disease. Eur Rev Med Pharmacol Sci. 24:3293–3301. 2020.PubMed/NCBI
|
|
52
|
Huang S, Tao W, Guo Z, Cao J and Huang X:
Suppression of long noncoding RNA TTTY15 attenuates hypoxia-induced
cardiomyocytes injury by targeting miR-455-5p. Gene. 701:1–8. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Wang S, Zhou Y, Andreyev O, Hoyt RF Jr,
Singh A, Hunt T and Horvath KA: Overexpression of FABP3 inhibits
human bone marrow derived mesenchymal stem cell proliferation but
enhances their survival in hypoxia. Exp Cell Res. 323:56–65. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Bensaad K, Favaro E, Lewis CA, Peck B,
Lord S, Collins JM, Pinnick KE, Wigfield S, Buffa FM, Li JL, et al:
Fatty acid uptake and lipid storage induced by HIF-1α contribute to
cell growth and survival after hypoxia-reoxygenation. Cell Rep.
9:349–365. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Weiler C, Nerlich AG, Zipperer J,
Bachmeier BE and Boos N: 2002 SSE Award Competition in Basic
Science: Expression of major matrix metalloproteinases is
associated with intervertebral disc degradation and resorption. Eur
Spine J. 11:308–320. 2002. View Article : Google Scholar : PubMed/NCBI
|