Introduction
Colorectal cancer (CRC) is recognized as one of the
most common malignancies, ranking third among all causes of
cancer-related mortality worldwide (1,2). The
available treatment options for CRC in the early stages (I–III) are
surgery and chemoradiotherapy. However, ~1/4 of patients are
diagnosed with advanced CRC (3).
With different stages of the disease being present at the time of
diagnosis and with varying techniques used for CRC treatment, the
median survival time of patients with CRC ranges from <1 to 5
years (4). Efficient treatment
methods and prognostic markers for patients with CRC are currently
lacking; therefore, it is important to investigate the underlying
mechanism of CRC and to develop effective drugs targeting this
disease.
Nintedanib is an orally available tyrosine kinase
inhibitor developed to treat numerous types of cancer (5). Nintedanib inhibits the vascular
endothelial growth factor receptors (VEGFRs) 1–3, platelet-derived
growth factor receptors (PDGFRs) α and β and fibroblast growth
factor receptors (FGFRs) 1–3, which participate in the pathogenic
process of idiopathic pulmonary fibrosis (6,7).
Moreover, nintedanib is clinically used for the treatment of
hepatic failure, liver cancer, ovarian cancer, prostate cancer and
CRC. It has been reported that treatment with nintedanib decreases
the risk of acute exacerbations and that it can stabilize disease
progression (8).
MicroRNAs (miRNAs/miRs) serve important roles in the
development and progression of malignancies. For example, miR-429
has been associated with the regulation of various types of
cellular processes (9). It has been
reported that miR-429 is downregulated in non-small cell lung
cancer cells resistant to nintedanib (10). Moreover, miR-429 may inhibit the
invasion and migration of CRC cells by targeting the PAK6/cofilin
signaling pathway (11). Using the
software ENCORI database (http://starbase.sysu.edu.cn/index.php), preliminary
analysis revealed that miR-429 can interact with dual specificity
protein phosphatase 4 (DUSP4), which is a member of the dual
specificity phosphatase family and specifically dephosphorylates
the MAP kinases ERK1/2, p38 and JNK (12,13).
In addition, miR-429 has been reported to promote the
epithelial-mesenchymal transition of gastric cancer cells to
enhance their resistance to adriamycin (14).
The aim of the present study was to investigate
whether overexpression of miR-429 can increase the sensitivity of
CRC cells to nintedanib by downregulating DUSP4.
Materials and methods
Cell culture and construction of
resistant cells
The CRC cell lines, SW480, LoVo and HCT116 used in
the present study were purchased from the American Type Culture
Collection. Cells were cultured in RPMI-1640 medium (Gibco; Thermo
Fisher Scientific, Inc.) supplemented with 10% FBS (Gibco; Thermo
Fisher Scientific, Inc.) at 37°C with 5% CO2. These cell
lines were amplified and frozen, and one aliquot of each was thawed
for subsequent experimentation. All cells were routinely screened
for the absence of mycoplasma (10).
CRC cells in logarithmic growth were taken and
placed in a cell culture dish at a concentration of
2×106 cells/ml. The cells were treated with 0.01 µM
nintedanib at 37°C for 48 h and medium was replaced by normal
culture solution. After 2 to 3 weeks of continuous culture, the
concentration of nintedanib was increased step by step, and CRC
cells could grow steadily in 0.1 µM nintedanib-containing medium,
thus the CRC-resistant to nintedanib (CRC-R) cell line was
obtained.
Cell treatment
Cell suspensions (5,000 cells/well) were seeded into
96-well plates and treated with increasing concentrations of
nintedanib (0, 0.01, 0.1, 1, 10 and 100 µM; Selleck Chemicals) at
37°C for 72 h to verify the construction of resistant cells.
Cell transfection
SW480-resistant to nintedanib (SW480-R) cells seeded
into a 24-well plate (5×104/well) were transfected with
miR-429 mimic (100 nM) and miR-negative control (miR-NC) (100 nM),
and/or DUSP4 overexpression plasmid (Oe-DUSP4; 50 nM) and the
negative control plasmid (Oe-NC; 50 nM; Guangzhou RiboBio Co.,
Ltd.) at 37°C for 48 h; untreated cells were set as a control
group. DUSP4 sequence was cloned into pcDNA3.1 vector (Invitrogen;
Thermo Fisher Scientific, Inc.).
Transfection was performed using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) according to the manufacturers instructions. The
miR-429 mimic sequence was 5′-TAATACTGTCTGGTAAAACCGT-3′ and the
miR-NC sequence was 5′-UUGAGGCUUCAAUCGACGUTT-3′.
Cell Counting Kit-8 (CCK-8) assay
CRC cells and CRC-R cells in logarithmic growth
phase were placed in a 96-well plate (103 cells/well)
and treated with different concentrations of nintedanib (0.01, 0.1,
1, 10 and 100 µM). SW480-R cells in logarithmic growth phase were
placed in a 96-well plate (103 cells/well) and
transfected with mimic NC or miR-429 mimic. After 72 h of
continuous culture, 10 µl CCK-8 reagent (Dojindo Molecular
Technologies, Inc.) was added to the wells and cells were incubated
for 72 h at 37°C. The absorbance at 450 nm was determined using a
microplate reader (Bio-Rad Laboratories, Inc.). Inhibition rate was
calculated as follows: Inhibition rate (%) = (control group -
experimental group)/(control group - blank group) × 100.
IC50 value was calculated using GraphPad Prism version
6.0 (GraphPad Software, Inc.).
Reverse transcription-quantitative PCR
(RT-qPCR) analysis
Total RNA was extracted from CRC cells and CRC-R
cells using TRIzol® reagent (Thermo Fisher Scientific,
Inc.). Total RNA was then reverse-transcribed into cDNA using a
TransScript® First-Strand cDNA Synthesis SuperMix kit
(TransGen Biotech Co., Ltd.) according to manufacturers protocol.
qPCR was performed using iTaq™ Universal SYBR® Green
Supermix (Bio-Rad Laboratories, Inc.) on an ABI 7500 instrument
(Applied Biosystems; Thermo Fisher Scientific, Inc.). PCR
thermocycling conditions were 90 sec at 95°C, 30 sec at 95°C, 20
sec at 65°C and 30 sec at 72°C, for 40 cycles. Primers were miR-429
forward, 5′-GGGGGTAATACTGTCTGGT-3′ and reverse,
5′-TGCGTGTCGTGGAGTC-3′; U6 forward, 5′-GCTTCGGCAGCACATATACTAA-3′
and reverse, 5′-CGAATTTGCGTGTCATCCTT-3′; DUSP4 forward,
5′-TCACGGCTCTGTTGAATGTC-3′ and reverse, 5′-GATGTCGGCCTTGTGGTTAT-3′;
GAPDH forward, 5′-CGAATTTGCGTGTCATCCTT-3′ and reverse,
5′-CGAATTTGCGTGTCATCCTT-3′. The relative expression levels of miRNA
or mRNA were normalized to U6 or GAPDH, and were calculated based
on the 2−ΔΔCq method (15).
Western blotting
Cellular proteins were extracted from CRC cells and
CRC-R cells lysed using lysis buffer (CWBio). Protein concentration
was quantified by BCA assay. Equal amounts of protein (20 µg) were
separated by SDS-PAGE (Beyotime Institute of Biotechnology) on 10%
gels and were then electrotransferred onto polyvinylidene
difluoride (PVDF) membranes (EMD Millipore). After blocking in
Tris-buffered saline-Tween-20 (0.05%) containing 5% non-fat dry
milk for 1 h at room temperature, the membranes were incubated with
primary antibodies overnight at 4°C. Membranes were then incubated
with horseradish peroxidase-conjugated anti-rabbit IgG secondary
antibody (cat. no. 7074; 1:1,000; Cell Signaling Technology, Inc.)
for the detection of primary antibodies at room temperature for 2
h, and the bands were visualized using an enhanced
chemiluminescence detection kit (EMD Millipore) and analyzed by
ImageJ software (v.1.52; National Institutes of Health). The
following primary antibodies were used: Bcl-2 (cat. no. ab32124;
1:1,000; Abcam), Bax (cat. no. ab182734; 1:1,000; Abcam), cleaved
caspase-3 (cat. no. ab2302; 1:1,000; Abcam), caspase-3 (cat. no.
ab13847; 1:1,000; Abcam), phosphorylated (p)-JNK (cat. no.
ab215208; 1:1,000; Abcam), c-Jun (cat. no. ab40766; 1:1,000;
Abcam), JNK (cat. no. ab199380; 1:1,000; Abcam), multi-drug
resistance protein (MDR1; cat. no. 13342; 1:1,000; Cell Signaling
Technology, Inc.), DUSP4 (cat. no. ab216576; 1:1,000; Abcam) and
GAPDH (cat. no. ab9485; 1:1,000; Abcam).
TUNEL assay
SW480 cells, SW480-R cells and the corresponding
transfected cells were seeded into a 24-well plate
(5×104/well) and cultured at 37°C with 1 µM nintedanib
for 72 h. After the supernatant was discarded, cells were washed
with PBS, fixed with 4% paraformaldehyde for 20 min at room
temperature and washed three more times with PBS. Cells were
treated with 75% ethanol at 4°C overnight, and were washed once
with PBS the following day. According to the manufacturer's
instructions of the TUNEL apoptosis kit (Roche Diagnostics),
apoptosis was observed using a fluorescence microscope
(magnification, ×200; Olympus Corporation).
Dual-luciferase reporter assay
ENCORI database (http://starbase.sysu.edu.cn/index.php) predicted that
DUSP4 could combine with miR-429. DUSP4 3-untranslated region
luciferase reporter gene plasmid was constructed by Shanghai
GenePharma Co., Ltd. DUSP4 mutation was constructed using the
Quick-Change Site-Directed Mutagenesis kit (Agilent Technologies,
Inc.). DUSP4-wild-type (WT) and DUSP4-mutant (MUT) plasmids were
generated via subcloning downstream of the luciferase vector. SW480
cells were seeded into a 24-well plate (5×104/well) and
co-transfected with miR-429 mimic (100 nM) + DUSP4-WT (50 nM) or
miR-429 mimic (100 nM) + DUSP4-MUT (50 nM) using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) at 37°C. After 48 h, cells were assessed using
the Dual-Luciferase Reporter Gene Assay kit (Beyotime Institute of
Biotechnology) according to the manufacturers instructions.
Luminescent signals were quantified with a luminometer (Centro XS3
LB 960; Titertek-Berthold), and firefly luciferase activity was
normalized to Renilla luciferase activity.
Statistical analysis
Data are presented as the mean ± SD obtained from
three independent experiments. Statistical analysis was conducted
using GraphPad Prism version 6.0 (GraphPad Software, Inc.).
Statistical comparisons among multiple groups were performed using
one-way ANOVA followed by Tukeys post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Establishment of nintedanib-resistant
CRC cells
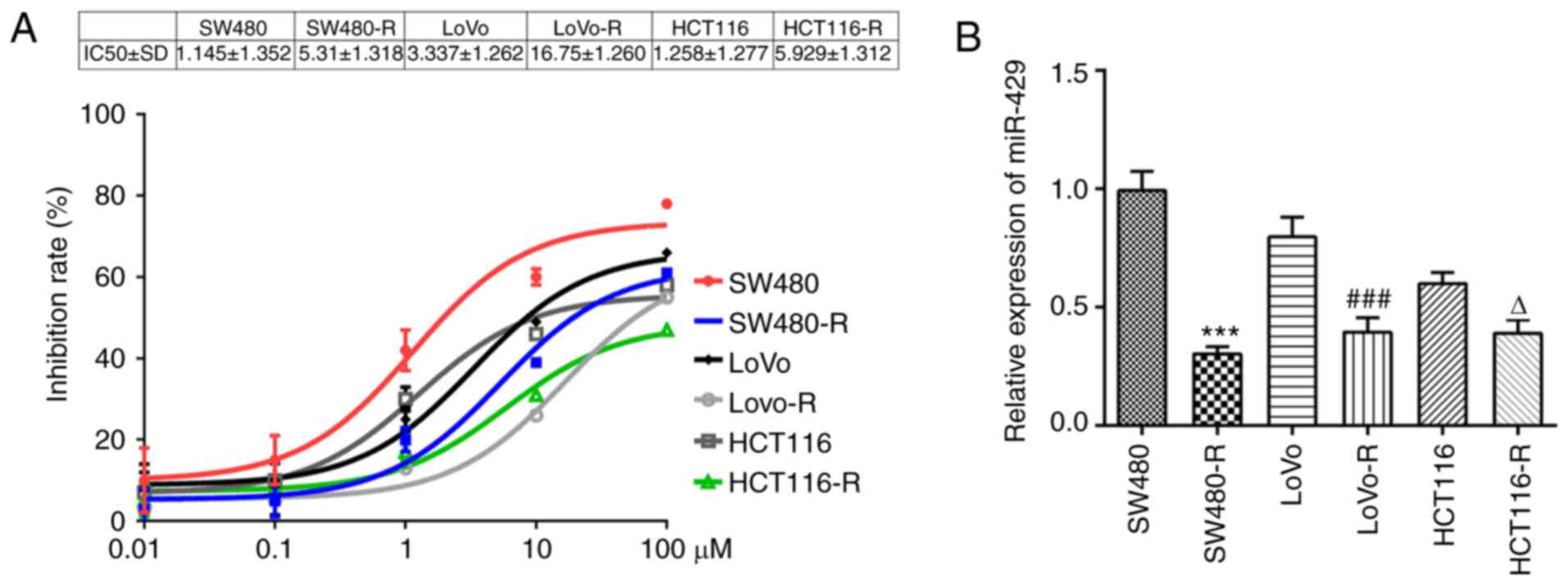
The inhibition rate of cells treated with different
concentrations of nintedanib was measured using a CCK-8 assay. It
was observed that the inhibition rate of SW480 cells pretreated
with nintedanib for 72 h was decreased to a larger extent compared
with the other cells lines and the IC50 of SW480 cells
was 1.145±1.352 (Fig. 1A).
Moreover, miR-429 expression was significantly downregulated in the
SW480-R cells compared with SW480 cells (Fig. 1B). Thus, SW480-R and SW480 cells
were chosen for the subsequent experiments. SW480-R cells were
resistant to nintedanib, whereas SW480 cells were not.
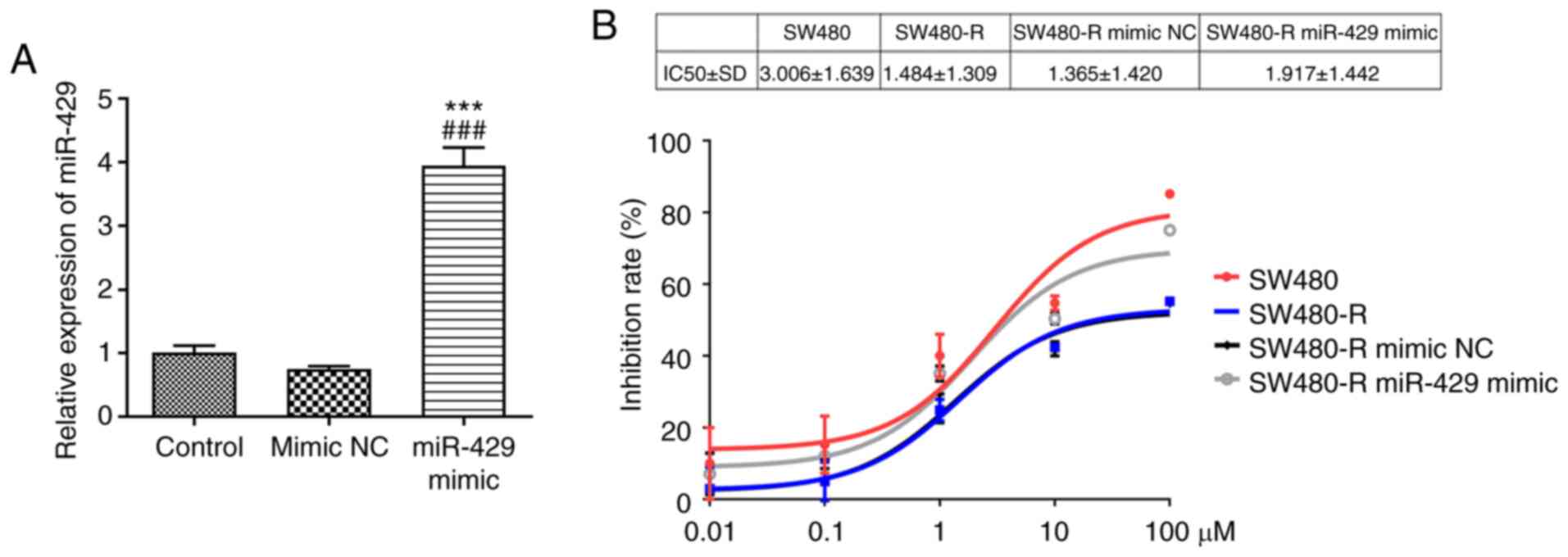
miR-429 mimic increases the
sensitivity of resistant CRC cells to nintedanib
After transfection with the miR-429 mimic, the
successful transfection efficiency of miR-429 mimic was confirmed
via RT-qPCR (Fig. 2A). After
treatment of the cells with different concentrations of nintedanib,
SW480-R cells transfected with the miR-429 mimic demonstrated an
elevated inhibition rate compared with SW480-R cells transfected
with mimic NC (Fig. 2B).
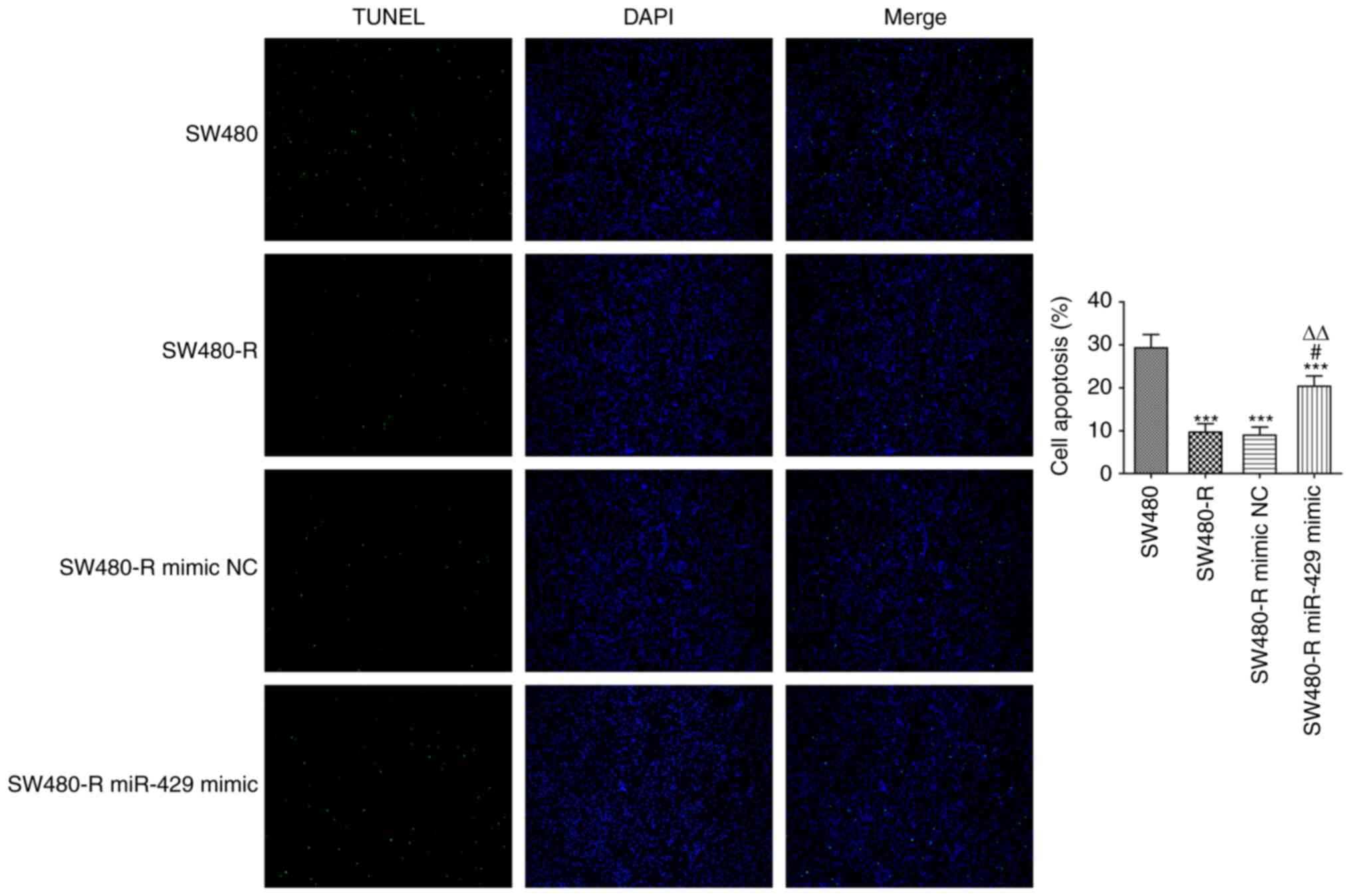
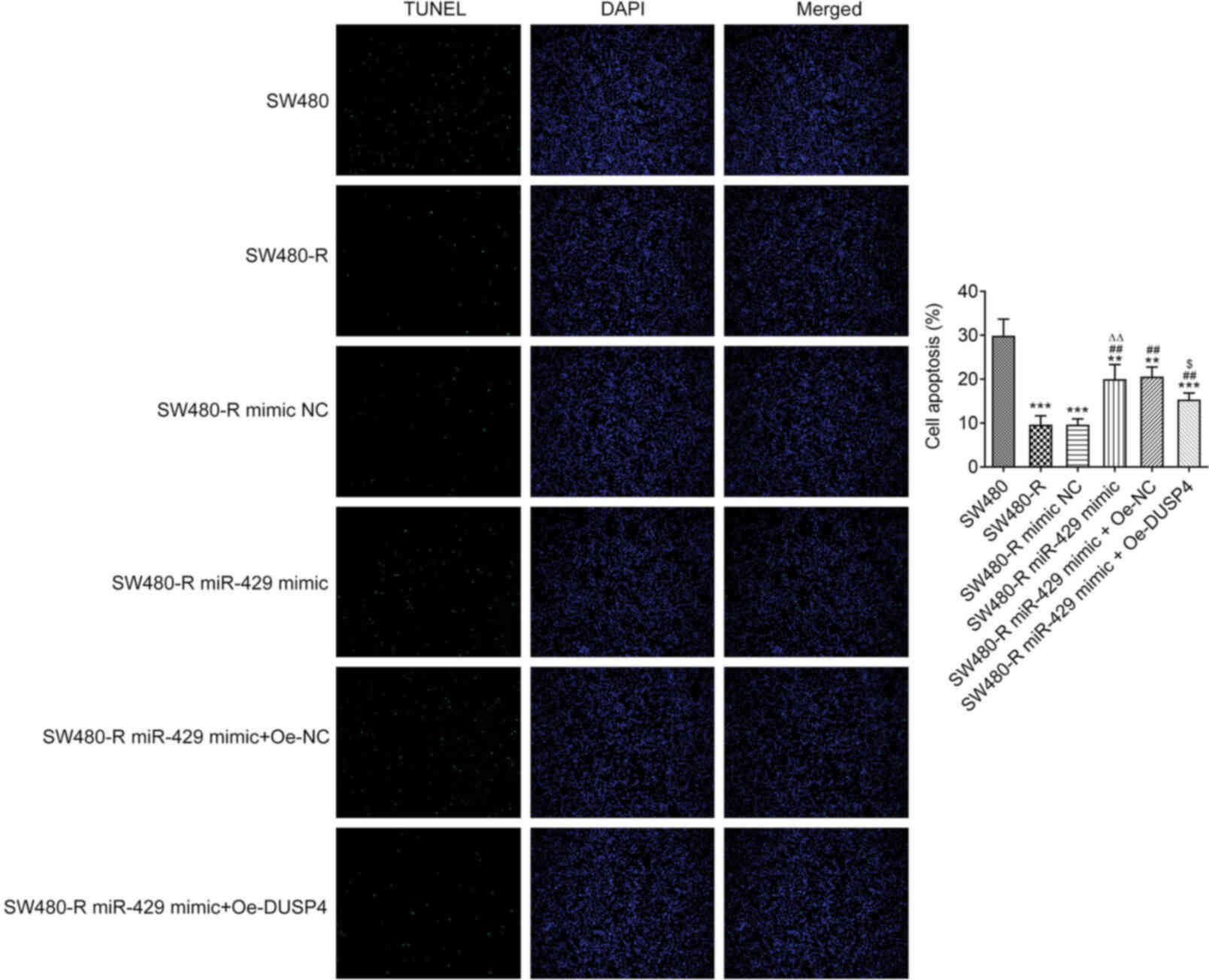
To further confirm the role of miR-429 in SW480-R
cells, cell apoptosis was evaluated. It was revealed that the
apoptosis of SW480-R cells was decreased compared with that of
SW-480 cells, and the miR-429 mimic increased the apoptosis of
SW480-R cells (Fig. 3). Western
blotting of apoptosis-related proteins is shown in Fig. 4A. The protein expression levels of
Bcl-2 were increased, whereas the expression levels of Bax and
cleaved caspase-3 were decreased in SW480-R cells, and the miR-429
mimic reversed the expression of these proteins (Fig. 4B-D). Caspase-3 expression was not
markedly changed in the four groups (Fig. 4E). Therefore, it was suggested that
the miR-429 mimic may promote the sensitivity of CRC cells to
nintedanib.
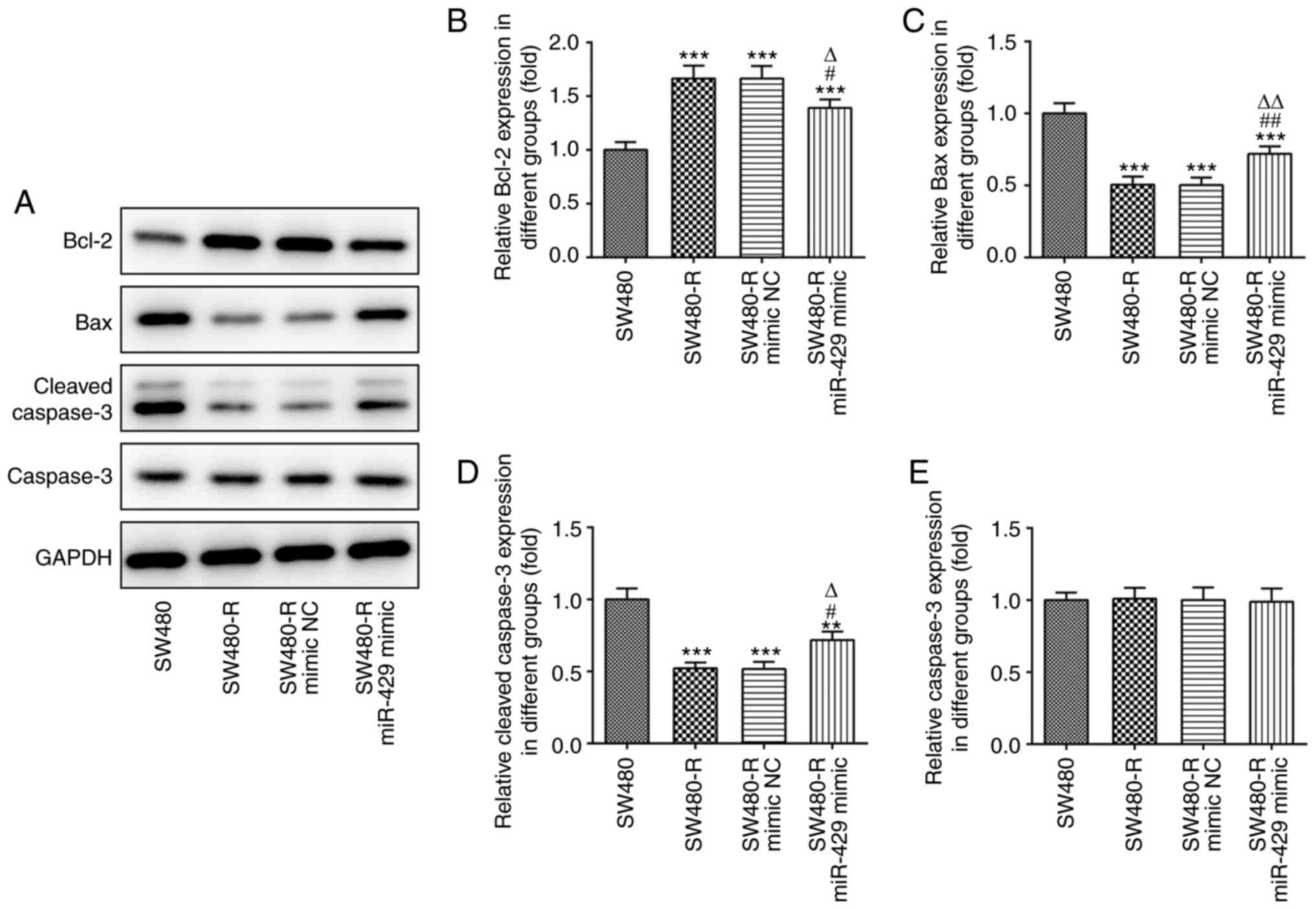 | Figure 4.miR-429 mimic affects the expression
levels of apoptosis-related proteins in resistant CRC cells treated
with nintedanib. (A) Western blotting of apoptosis-related proteins
in resistant CRC cells treated with nintedanib. The expression
levels of apoptosis-related proteins, including (B) Bcl-2, (C) Bax,
(D) cleaved caspase-3 and (E) caspase-3 in CRC cells were detected
by western blotting. **P<0.01, ***P<0.001 vs. SW480;
#P<0.05, ##P<0.01 vs. SW480-R;
∆P<0.05, ∆∆P<0.01 vs. SW480-R mimic NC.
CRC, colorectal cancer; miR-429, microRNA-429; NC, negative
control; -R, resistant to nintedanib. |
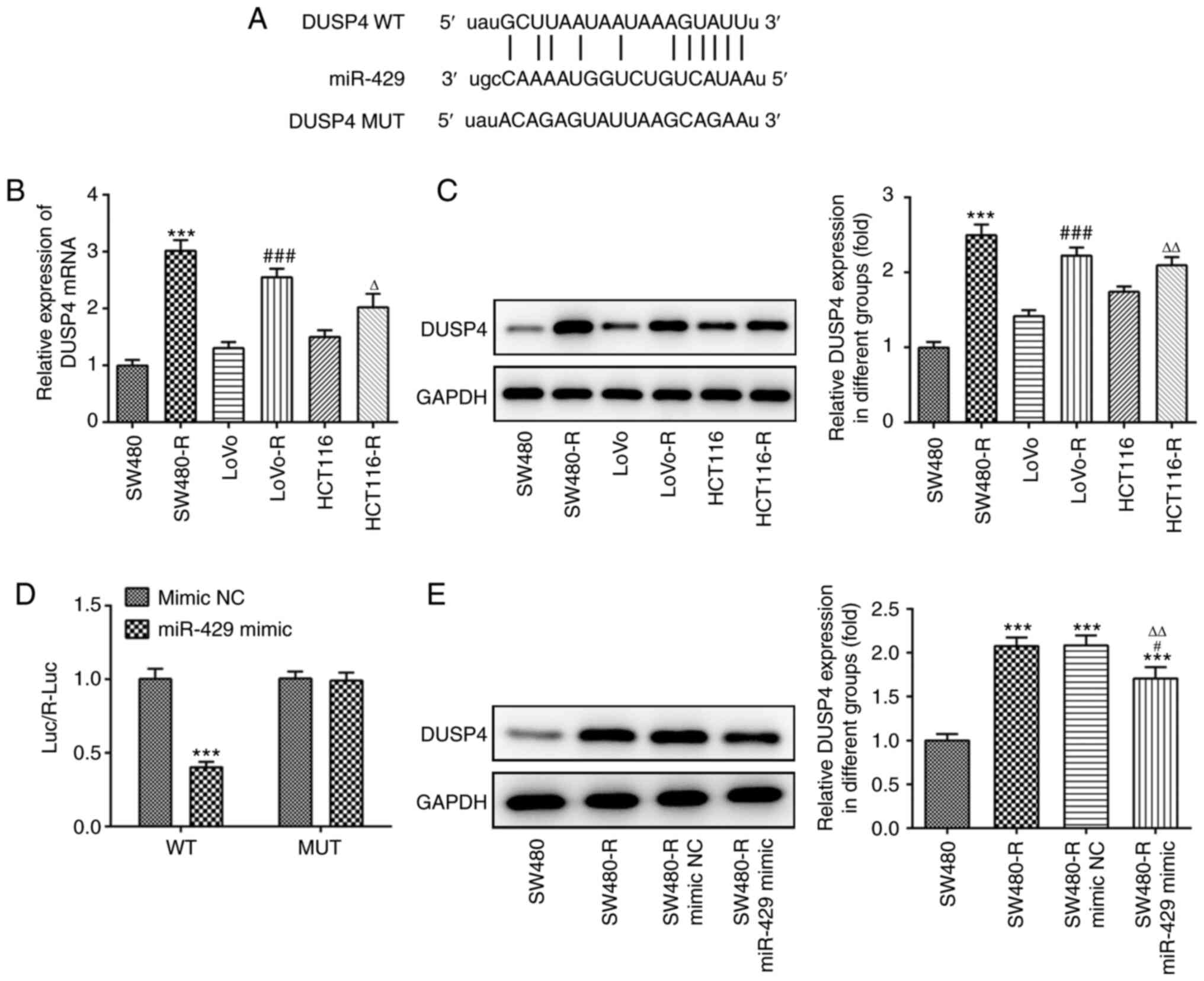
DUSP4 is a target of miR-429
ENCORI software was used to predict miR-429 target
genes; binding sites between miR-429 and DUSP4 were identified
(Fig. 5A). Subsequently, western
blotting and RT-qPCR were used to detect the expression levels of
DUSP4. DUSP4 expression was increased to a greater extent in
SW480-R cells compared with the other cell lines (Fig. 5B and C). Moreover, compared with the
mimic NC + DUSP4-WT group, the luciferase reporter activity of the
miR-429 mimic + DUSP4-WT group was decreased (Fig. 5D). Western blotting was performed to
measure the expression levels of DUSP4. The results demonstrated
that the SW480-R group exhibited increased expression of DUSP4
compared with the SW480 group, and that SW480-R cells transfected
with the miR-429 mimic exhibited decreased expression levels of
DUSP4 compared with the SW480-R mimic NC group (Fig. 5E). These findings indicated that the
expression of DUSP4 was inversely associated with that of
miR-429.
Overexpression of DUSP4 reverses the
effect of miR-429 overexpression on the sensitivity of CRC cells
resistant to nintedanib
To assess whether DUSP4 expression can affect the
sensitivity of CRC cells to nintedanib, Oe-DUSP4 was constructed.
The results of RT-qPCR and western blotting confirmed the
successful transfection of cells with Oe-DUSP4 (Fig. 6A and B). Subsequently, a CCK-8 assay
was used to detect the inhibition rate of SW480-R cells pretreated
with different concentrations of nintedanib for 72 h. The miR-429
mimic increased the inhibition rate of SW480-R cells (Fig. 6C). Compared with the SW480-R miR-429
mimic + Oe-NC group, SW480-R cells transfected with miR-429 mimic
and Oe-DUSP4 exhibited a decreased inhibition rate. The apoptosis
of SW480-R cells and the expression levels of apoptosis-related
proteins were also evaluated. Overexpression of DUSP4 decreased the
apoptosis of miR-429 mimic-transfected SW480-R cells (Fig. 7). Western blotting of
apoptosis-related proteins is shown in Fig. 8A. Overexpression of DUSP4 increased
the expression levels of Bcl-2, and decreased the expression levels
of Bax and cleaved caspase-3 in SW480-R cells transfected with
miR-429 mimic (Fig. 8B-D).
Caspase-3 expression was not obviously changed in the six groups
(Fig. 8E).
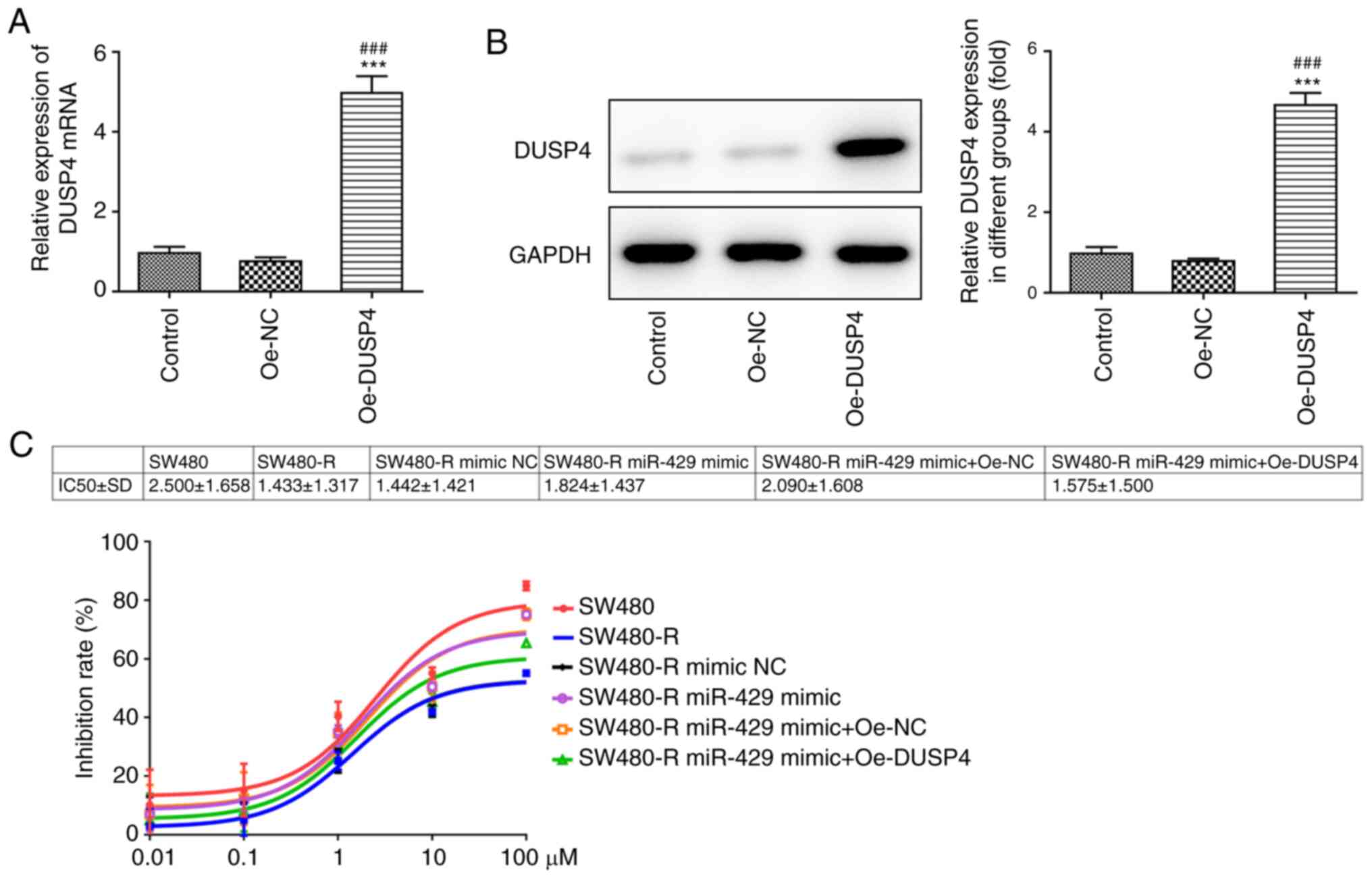 | Figure 6.Overexpression of DUSP4 reverses the
effect of miR-429 overexpression on the inhibition rate of
resistant CRC cells treated with nintedanib. (A) Transfection
efficiency of Oe-DUSP4, as determined by reverse
transcription-quantitative PCR. ***P<0.001 vs. control;
###P<0.001 vs. Oe-NC. (B) Transfection efficiency of
Oe-DUSP4, as determined by western blotting. ***P<0.001 vs.
Control; ###P<0.001 vs. Oe-NC. Control group
consisted of untransfected cells. (C) IC50 value and
inhibition rate of resistant CRC cells treated with different
concentrations of nintedanib post-transfection, as determined by
Cell Counting Kit-8 assay. CRC, colorectal cancer; miR-429,
microRNA-429; DUSP4, dual specificity protein phosphatase 4; NC,
negative control; Oe-DUSP4, DUSP4 overexpression plasmid; Oe-NC, NC
overexpression plasmid; -R, resistant to nintedanib. |
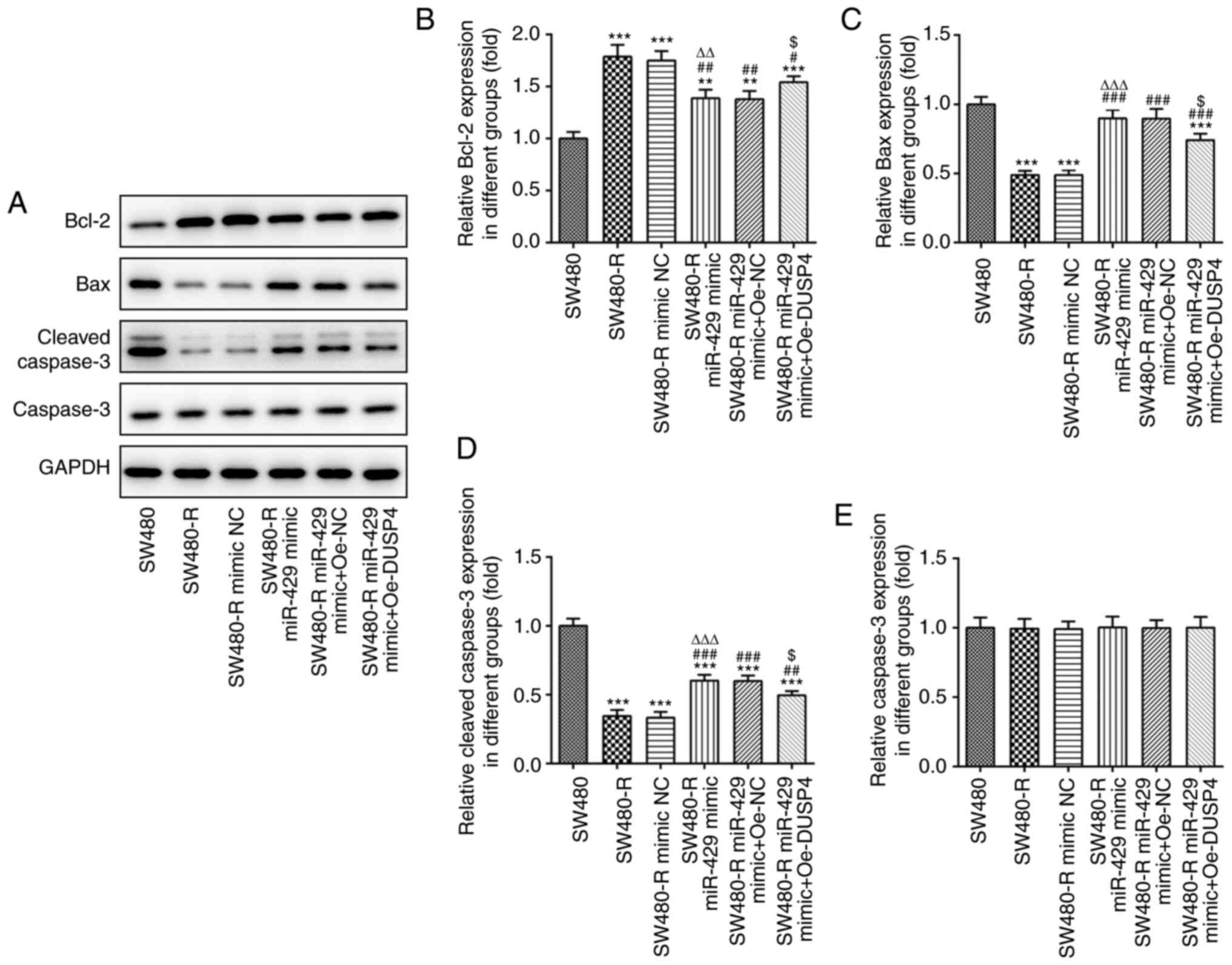 | Figure 8.Overexpression of DUSP4 reverses the
inductive effect of miR-429 overexpression on the expression of
apoptosis-related proteins in resistant CRC cells treated with
nintedanib. (A) Western blotting of apoptosis-related proteins in
resistant CRC cells treated with nintedanib. The expression levels
of apoptosis-related proteins, including (B) Bcl-2, (C) Bax, (D)
cleaved caspase-3 and (E) caspase-3 in CRC cells were detected by
western blotting. **P<0.01, ***P<0.001 vs. SW480;
#P<0.05, ##P<0.01,
###P<0.001 vs. SW480-R; ∆∆P<0.01,
∆∆∆P<0.001 vs. SW480-R mimic NC;
$P<0.05 vs. SW480-R miR-429 mimic + Oe-NC. CRC,
colorectal cancer; miR-429, microRNA-429; DUSP4, dual specificity
protein phosphatase 4; NC, negative control; Oe-DUSP4, DUSP4
overexpression plasmid; Oe-NC, NC overexpression plasmid; -R,
resistant to nintedanib. |
It has been reported that DUSP4 can dephosphorylate
the MAP kinases ERK1/2, p38 and JNK (12). Thus, western blotting was performed
to detect the expression levels of proteins associated with the JNK
signaling pathway (Fig. 9A).
Oe-DUSP4 significantly decreased the protein expression levels of
p-JNK, p/t-JNK and the JNK inhibitor c-Jun compared with those in
SW480-R cells transfected with the miR-429 mimic (Fig. 9B, D and E), but increased the
expression levels of MDR1 (Fig.
9F). There was no significant difference of JNK expression
between the six groups (Fig. 9C).
These findings suggested that overexpression of DUSP4 may reverse
the inducing effect of miR-429 overexpression on the sensitivity of
CRC cells to nintedanib and inhibit the JNK signaling pathway.
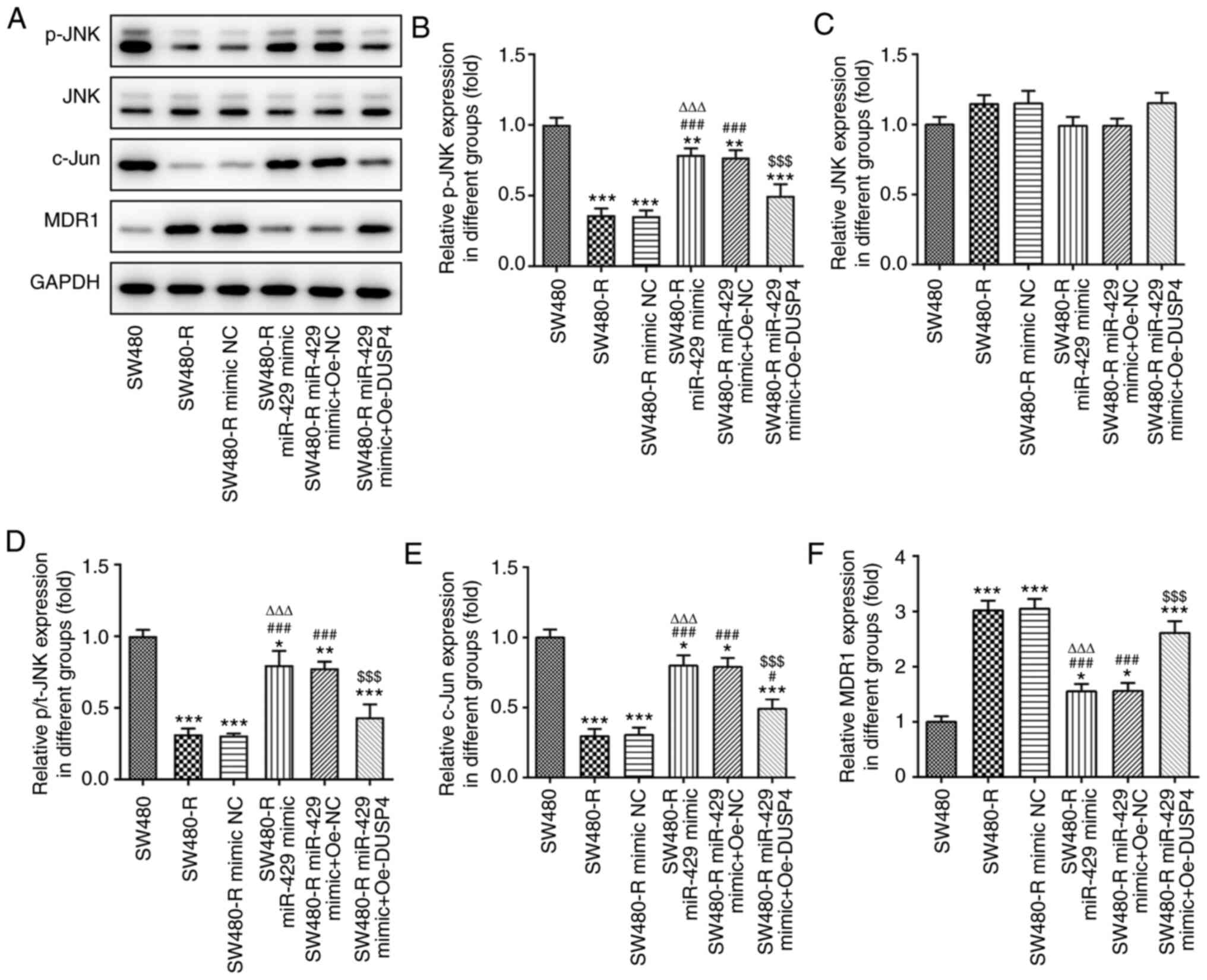 | Figure 9.Overexpression of DUSP4 reverses the
inductive effect of miR-429 overexpression on the expression of
JNK-related proteins in resistant colorectal cancer cells treated
with nintedanib. (A) Western blotting of JNK-related proteins.
Expression levels of JNK-related proteins, including (B) p-JNK, (C)
JNK, (D) p/t-JNK, (E) c-Jun and (F) MDR1 were detected by western
blotting. *P<0.05, **P<0.01, ***P<0.001 vs. SW480;
#P<0.05, ###P<0.001 vs. SW480-R;
∆∆∆P<0.001 vs. SW480-R mimic NC;
$$$P<0.001 vs. SW480-R miR-429 mimic + Oe-NC.
miR-429, microRNA-429; DUSP4, dual specificity protein phosphatase
4; NC, negative control; Oe-DUSP4, DUSP4 overexpression plasmid;
Oe-NC, NC overexpression plasmid; -R, resistant to nintedanib; p-,
phosphorylated; t-, total; MDR1, multidrug resistance protein. |
Discussion
Nintedanib, an intracellular inhibitor of tyrosine
kinases, such as FGFR, PDGFR and VEGFR, has been approved for the
treatment of idiopathic pulmonary fibrosis and various types of
cancer (16,17). However, nintedanib resistance often
leads to unsatisfactory efficacy in the treatment of diseases and
the effect of nintedanib monotherapy on patients with various
diseases is limited (18). It is
widely known that drug resistance is a major limitation to
effective disease treatment, and numerous miRNAs have been reported
to be associated with chemoresistance and poor prognosis (10,19).
Thus, the present study investigated the association between miRNA
expression and drug sensitivity. Notably, a previous study reported
that the low expression levels of miR-200b and miR-141 were
associated with the resistance of non-small cell lung cancer cells
to nintedanib (10).
miRNAs have been discovered in multiple organisms
and are considered participants in the regulation of gene
expression (20). In addition to
their involvement in various biological processes, miRNAs may act
as oncogenes or tumor suppressors to facilitate or delay tumor
progression (21,22). miR-429 is a member of the miR-200
family, which has been reported to serve a key role as a tumor
suppressor in numerous types of carcinoma, such as bladder cancer,
gastric cancer and hepatocellular carcinoma (23–25).
Moreover, miR-429 was shown to be downregulated in metastatic
lesions and act as a tumor suppressor in nasopharyngeal carcinoma
(26). However, two previous
studies reported a contradictory role of miR-429 to that found in
the present study; in these previous studies, miR-429 promoted the
progression of CRC and non-small cell lung cancer (27,28).
In the present study, it was identified that miR-429 increased the
sensitivity of SW480 cells to nintedanib and promoted the apoptosis
of these cells, demonstrating the potential of miR-429 in the
treatment of CRC. Moreover, miR-429 has been shown to be negatively
correlated with metastatic potential in nasopharyngeal carcinoma
cell lines (29).
It was predicted by ENCORI that DUSP4 and miR-429
shared binding sites. Moreover, the present results confirmed that
the expression levels of DUSP4 and miR-429 were inversely
associated. Subsequent experiments further indicated that DUSP4 may
reverse the promoting effect of miR-429 on the sensitivity of SW480
cells to nintedanib, as cells transfected with both the miR-429
mimic and Oe-DUSP4 displayed a lower inhibition rate and apoptotic
rate compared with those transfected with the miR-429 mimic
only.
It has been reported that most DUSPs negatively
regulate the JNK signaling pathway via dephosphorylation (30). Moreover, JNK was previously reported
to enhance the catalytic ability of DUSP4, and the expression of
DUSP4 has been shown to be upregulated in malignant tissues but
downregulated in healthy tissues (31,32).
The JNK signaling pathway is closely associated with a number of
diseases, particularly cancer. The activity of the JNK signaling
pathway is high in most cancer cell lines, and depletion of
individual JNKs can inhibit tumorigenesis (33,34).
Thus, in recent years, JNKs have been increasingly identified as
effective molecular targets for the treatment of various
malignancies, and the use of a number of JNK inhibitors has been
considered as an effective treatment option for tumors (33,35).
However, some previous studies have proposed that JNK proteins are
implicated in tumor suppression (33,36).
Based on this dispute, in the present study, DUSP4 was
overexpressed in SW480 cells to examine whether it acted as an
inhibitory or enhancing molecule of the JNK signaling pathway. The
present results suggested that DUSP4 dephosphorylated JNK, thereby
inhibiting the JNK signaling pathway.
In conclusion, the present study identified the role
of the miR-429/DUSP4 axis in regulating the sensitivity of CRC
cells to nintedanib. Moreover, overexpression of miR-429 may
increase the sensitivity of CRC cells to nintedanib through
inhibiting the JNK signaling pathway by downregulating DUSP4. These
findings may enable the development of prevention strategies for
the resistance of CRC cells to nintedanib.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors contributions
XS contributed to the conception and design of this
study. GC performed the experiments, collected the data and
performed statistical analysis with the help of YL and ZL. GC
drafted the manuscript, which was corrected and revised by XS. All
authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Siegel RL, Miller KD and Jemal A: Cancer
statistics, 2019. CA Cancer J Clin. 69:7–34. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Fanali C, Lucchetti D, Farina M, Corbi M,
Cufino V, Cittadini A and Sgambato A: Cancer stem cells in
colorectal cancer from pathogenesis to therapy: Controversies and
perspectives. World J Gastroenterol. 20:923–942. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Malapelle U: USP11 role in colorectal
cancer growing and metastatisation. EBioMedicine. 48:5–6. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferlay J, Steliarova-Foucher E,
Lortet-Tieulent J, Rosso S, Coebergh JW, Comber H, Forman D and
Bray F: Cancer incidence and mortality patterns in Europe:
Estimates for 40 countries in 2012. Eur J Cancer. 49:1374–1403.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Rodríguez-Portal JA: Efficacy and safety
of Nintedanib for the rreatment of idiopathic pulmonary fibrosis:
An update. Drugs R D. 18:19–25. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Richeldi L, Costabel U, Selman M, Kim DS,
Hansell DM, Nicholson AG, Brown KK, Flaherty KR, Noble PW, Raghu G,
et al: Efficacy of a tyrosine kinase inhibitor in idiopathic
pulmonary fibrosis. N Engl J Med. 365:1079–1087. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Crestani B, Huggins JT, Kaye M, Costabel
U, Glaspole I, Ogura T, Song JW, Stansen W, Quaresma M, Stowasser
S, et al: Long-term safety and tolerability of nintedanib in
patients with idiopathic pulmonary fibrosis: Results from the
open-label extension study, INPULSIS-ON. Lancet Respir Med.
7:60–68. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Collard HR, Richeldi L, Kim DS, Taniguchi
H, Tschoepe I, Luisetti M, Roman J, Tino G, Schlenker-Herceg R,
Hallmann C, et al: Acute exacerbations in the INPULSIS trials of
nintedanib in idiopathic pulmonary fibrosis. Eur Respir J.
49:16013392017. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Xue H and Tian GY: MiR-429 regulates the
metastasis and EMT of HCC cells through targeting RAB23. Arch
Biochem Biophys. 637:48–55. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Nishijima N, Seike M, Soeno C, Chiba M,
Miyanaga A, Noro R, Sugano T, Matsumoto M, Kubota K and Gemma A:
miR-200/ZEB axis regulates sensitivity to nintedanib in non-small
cell lung cancer cells. Int J Oncol. 48:937–944. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Tian X, Wei Z, Wang J, Liu P, Qin Y and
Zhong M: MicroRNA-429 inhibits the migration and invasion of colon
cancer cells by targeting PAK6/cofilin signaling. Oncol Rep.
34:707–714. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Low HB and Zhang Y: Regulatory roles of
MAPK phosphatases in cancer. Immune Netw. 16:85–98. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Gaggianesi M, Turdo A, Chinnici A, Lipari
E, Apuzzo T, Benfante A, Sperduti I, Di Franco S, Meraviglia S, Lo
Presti E, et al: IL4 primes the dynamics of breast cancer
progression via DUSP4 inhibition. Cancer Res. 77:3268–3279. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Kang X, Li M, Zhu H, Lu X, Miao J, Du S,
Xia X and Guan W: DUSP4 promotes doxorubicin resistance in gastric
cancer through epithelial-mesenchymal transition. Oncotarget.
8:94028–94039. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Wollin L, Wex E, Pautsch A, Schnapp G,
Hostettler KE, Stowasser S and Kolb M: Mode of action of nintedanib
in the treatment of idiopathic pulmonary fibrosis. Eur Respir J.
45:1434–1445. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Kolb M, Richeldi L, Behr J, Maher TM, Tang
W, Stowasser S, Hallmann C and du BoisRM: Nintedanib in patients
with idiopathic pulmonary fibrosis and preserved lung volume.
Thorax. 72:340–346. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhao YY, Feng JB, Peng J, Meng LH, Zhang
HX and Wu ML: Signal mining and post-marketing evaluation of
adverse drug reactions of nintedanib. Zhongguo Yiyuan Yaoxue Zazhi.
39:1655–1658. 2019.(In Chinese).
|
|
19
|
Januszyk P, Januszyk K, Wierzbik-Strońsk
M, Boroń D and Grabarek B: Analysis of the differences in the
expression of mRNAs and miRNAs associated with drug resistance in
endometrial cancer cells treated with salinomycin. Curr Pharm
Biotechnol. June 29–2020.https://doi.org/10.2174/1389201021666200629151008
|
|
20
|
Lagos-Quintana M, Rauhut R, Lendeckel W
and Tuschl T: Identification of novel genes coding for small
expressed RNAs. Science. 294:853–858. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Ambros V: MicroRNA pathways in flies and
worms: Growth, death, fat, stress, and timing. Cell. 113:673–676.
2003. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Wu CL, Ho JY, Hung SH and Yu DS: miR-429
expression in bladder cancer and its correlation with tumor
behavior and clinical outcome. Kaohsiung J Med Sci. 34:335–340.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Zhang M, Dong BB, Lu M, Zheng MJ, Chen H,
Ding JZ, Xu AM and Xu YH: miR-429 functions as a tumor suppressor
by targeting FSCN1 in gastric cancer cells. Onco Targets Ther.
9:1123–1133. 2016.PubMed/NCBI
|
|
25
|
Guo C, Zhao D, Zhang Q, Liu S and Sun MZ:
miR-429 suppresses tumor migration and invasion by targeting CRKL
in hepatocellular carcinoma via inhibiting Raf/MEK/ERK pathway and
epithelial-mesenchymal transition. Sci Rep. 8:23752018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Wang F, Jiang C, Sun Q, Yan F, Wang L, Fu
Z, Liu T and Hu F: Downregulation of miR-429 and inhibition of cell
migration and invasion in nasopharyngeal carcinoma. Mol Med Rep.
13:3236–3242. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Han Y, Zhao Q, Zhou J and Shi R: miR-429
mediates tumor growth and metastasis in colorectal cancer. Am J
Cancer Res. 7:218–233. 2017.PubMed/NCBI
|
|
28
|
Xiao P, Liu W and Zhou H: miR-429 promotes
the proliferation of non-small cell lung cancer cells via targeting
DLC-1. Oncol Lett. 12:2163–2168. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Wang Z, Zhu Z, Lin Z, Luo Y, Liang Z,
Zhang C, Chen J and Peng P: miR-429 suppresses cell proliferation,
migration and invasion in nasopharyngeal carcinoma by
downregulation of TLN1. Cancer Cell Int. 19:1152019. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Ha J, Kang E, Seo J and Cho S:
Phosphorylation dynamics of JNK signaling: Effects of
dual-specificity phosphatases (DUSPs) on the JNK pathway. Int J Mol
Sci. 20:61572019.doi: 10.3390/ijms20246157. View Article : Google Scholar
|
|
31
|
Chen P, Hutter D, Yang X, Gorospe M, Davis
RJ and Liu Y: Discordance between the binding affinity of
mitogen-activated protein kinase subfamily members for MAP kinase
phosphatase-2 and their ability to activate the phosphatase
catalytically. J Biol Chem. 276:29440–29449. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Muhammad T, Zhang J, Ma Y, Li Y, Zhang F,
Zhang Y and Liang Y: Overexpression of a mitogen-activated protein
kinase SlMAPK3 positively regulates tomato tolerance to cadmium and
drought stress. Molecules. 24:5562019.https://doi.org/10.3390/molecules24030556 View Article : Google Scholar
|
|
33
|
Wagner EF and Nebreda AR: Signal
integration by JNK and p38 MAPK pathways in cancer development. Nat
Rev Cancer. 9:537–549. 2009. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Raitano AB, Halpern JR, Hambuch TM and
Sawyers CL: The Bcr-Abl leukemia oncogene activates Jun kinase and
requires Jun for transformation. Proc Natl Acad Sci USA.
92:11746–11750. 1995. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Siddiqui MA and Reddy PA: Small molecule
JNK (c-Jun N-terminal kinase) inhibitors. J Med Chem. 53:3005–3012.
2010. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Davis RJ: Signal transduction by the JNK
group of MAP kinases. Cell. 103:239–252. 2000. View Article : Google Scholar : PubMed/NCBI
|