Introduction
Diabetic cardiomyopathy (DCM) is a cardiovascular
complication of diabetes that is one of the primary causes of
disability or death in patients with diabetes worldwide (1). Myocardial cell and fibroblast death
are fundamental alterations in DCM, which can trigger heart
remodeling and lead to left ventricular dysfunction, resulting in
the occurrence of DCM and heart failure (2). The abnormal glucose metabolism caused
by high glucose (HG) is a key initiating factor of DCM (3). As the disease progresses, oxidative
stress damage of the myocardial tissue is increased, and excessive
active oxygen increases the expression levels of various
inflammatory factors and induces the inflammatory response
(4). The long-term inflammatory
environment induces tissue and cell damage (4). However, the exact mechanism underlying
DCM is not completely understood. Recent progress in the field of
diabetes research has suggested that pyroptosis may be closely
associated with the disease (5).
Pyroptosis is essential in the development of
organs, cell renewal, differentiation and other physiological
processes (6). Pyroptosis requires
nucleotide oligomerization domain-like receptor protein (NLRP)3 to
recognize pathogen-associated molecules and damage-associated
molecules to induce caspase-1 and caspase-4/5/11 expression
(7,8). Caspase expression results in cleavage
of the downstream gasdermin D (GSDMD) protein, leading to cell
membrane rupture, and the release of cell contents and inflammatory
factors, including IL-1 and IL-18 (7,8). The
production of inflammasomes, which contain large multiprotein
complexes consisting of caspase-1, apoptosis-associated speck-like
protein (ASC) and NLRP, in cardiac fibroblasts is indispensable
(9).
The field of epigenetics has gradually become a
research hotspot for the investigation of various diseases, and its
subcategories primarily involve the fields of microRNAs
(miRNAs/miRs), long non-coding RNAs (lncRNAs) and DNA methylation
(10,11). Recently, Zhang et al
(12) reported that
metastasis-associated lung adenocarcinoma transcript-1 (MALAT1)
expression was increased significantly in heart tissues derived
from diabetic model rats, whereas MALAT1 knockdown enhanced left
ventricular function by reducing cardiomyocyte apoptosis. MALAT1
was originally reported to be overexpressed in tumor tissue
samples, and to participate in the regulation of tumor cell
proliferation, invasion, migration and metabolism (13). Further research on lncRNAs and
vascular diseases by Michalik et al (14) demonstrated that MALAT1 was also
involved in regulating the biological functions of vascular
endothelial cells, including phenotypic switching, basal sprouting
and migration. However, whether MALATl also serves a critical role
in high glucose (HG)-induced H9C2 cardiomyocyte pyroptosis has not
been previously reported. In recent years, numerous studies have
demonstrated that miR-141-3p is closely associated with DCM
development (15,16). In an in vitro
hypoxia/reoxygenation model, miR-141-3p expression levels were
significantly reduced and miR-141-3p overexpression significantly
decreased HR-induced cardiomyocyte apoptosis (16). Furthermore, the database prediction
indicated that miR-141-3p is one of the downstream targets of
MALAT1 (17). However, the roles
and mechanisms underlying the lncRNA-MALAT1/miR-141-3p axis in
HG-induced cardiomyocyte pyroptosis are not completely
understood.
Based on the aforementioned studies, the present
study established MALAT1 and miR-141-3p knockdown and
overexpression in HG-treated H9C2 cardiomyocytes to investigate
whether lncRNA-MALAT1 could target miR-141-3p and regulate
HG-induced H9C2 cardiomyocyte pyroptosis. The results of the
present study may provide a novel molecular target and research
direction for the potential treatment of DCM.
Materials and methods
Cell culture and groups
Rat cardiomyocytes (H9C2; The Cell Bank of Type
Culture Collection of The Chinese Academy of Sciences) were
cultured in DMEM (HyClone; GE Healthcare Life Sciences)
supplemented with 10% fetal bovine serum (FBS; cat. no. S9030;
Beijing Solarbio Science & Technology Co., Ltd.) and 1%
penicillin-streptomycin in a constant temperature incubator at 37°C
with 5% CO2. Cells were subcultured every 2–3 days and
used for subsequent experiments. According to a previous study
(18), cells were divided into the
following two groups: i) Normal glucose; cells were incubated with
normal glucose (NG; 5.5 mM); and ii) high glucose; cells were
incubated with high glucose (HG; 30 mM) at 37°C until they reached
~70–80% confluence.
Reverse transcription-quantitative PCR
(RT-qPCR)
RT-qPCR was performed to measure MALAT1 and
miR-141-3p expression cells. Cells were centrifuged at 4°C for 15
min at 850 × g. Total RNA was extracted using TRIzol®
(cat. no. N065; Nanjing Jiancheng Bioengineering Institute). The
RNA purity was assessed using a spectrophotometer (UV-1600PC;
Shanghai Mapada Instruments Co., Ltd.). Subsequently, total RNA was
reverse transcribed into cDNA using a PrimeScript™ RT reagent kit
(cat. no. RR047A; Takara Bio, Inc.). Subsequently, qPCR was
performed using SYBR Green PCR Master Mix (MedChem Express) on a
Mastercycler® nexus X2 (Roche Diagnostics) according to
the manufacturer's protocol. The following thermocycling conditions
were used for qPCR: 95°C for 15 sec; followed by 35 cycles at 60°C
for 60 sec and 72°C for 40 sec, then 72°C for 10 min. The following
primers (Shanghai Shenggong Biology Engineering Technology Service,
Ltd.) were used for qPCR: MALAT1 forward,
5′-CTTCCCTAGGGGATTTCAGG-3-′ and reverse,
5′-GATGCAAATGCCTCTGAGTG-3′; GAPDH forward,
5′-TGACTTCAACAGCGACACCCA-3′ and reverse,
5′-CACCCTGTTGCTGTAGGCCAAA-3′; miR-141-3p forward,
5′-CTCAAGGCAACCTACCGAAAAG-3′ and reverse,
5′-TATCGGACCCATCACGGAGTGG-3′; U6 forward, 5′-CTCGCTTCGGCAGCACA-3′
and reverse, 5′-AACGCTTCACGAATTTGCGT-3′. mRNA expression levels
were quantified using the 2−ΔΔCq method (19) and normalized to the internal
reference genes U6 and GAPDH, respectively.
Cell transfection and grouping
The pcDNA3.1-MALAT1, MALAT1 siRNA (si-MALAT1) and
their corresponding controls were purchased from Shanghai
GenePharma Co., Ltd. H9C2 cells (2×105 cells/ml) were
transfected at 37°C for 24 h with Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.) as previously
described (20). Cells were divided
into the following groups: i) NG, cells were cultured in normal
glucose (5.5 mM) for 24 h; ii) HG, cells were cultured in high
glucose (30 mM) for 24 h; iii) HG + MALAT1 overexpression negative
control (NC1), cells were transfected with 50 nM control
non-targeting adenovirus vector for 24 h, then cultured in HG
medium for 24 h; iv) HG + MALAT1 overexpression (MALAT1), cells
were transfected with 50 nM pcDNA3.1-MALAT1 for 24 h, then cultured
in HG medium for 24 h; v) HG + MALAT1 siRNA scramble control (NC2)
cells were transfected with 50 nM MALAT1 siRNA scramble control
(5′-UUCUCCGAACGUGUCACGUTT-3′) for 24 h then cultured in HG medium
for 24 h; and vi) HG + MALAT1 siRNA (si-MALAT1), cells were
transfected with 50 nM MALAT1 siRNA (5′-GATCCATAATCGGTTTCAA-3′) for
24 h then cultured in HG medium for 24 h. Transfection efficiency
was assessed via RT-qPCR.
Dual luciferase reporter assay
The target gene prediction between lnc MALAT1 and
miR-141-3p was performed using TargetScan software 3.0 (www.targetscan.org). Wild-type (WT) and mutant (MUT)
3′-untranslated regions (UTRs) of MALAT1 were cloned into the
pGL3/luciferase vector (Promega Corporation). The sequences were
inserted downstream of the luciferase gene and cloned as previously
described (17). Cells
(2×105 cells/ml) were co-transfected with 50 ng MALAT1
WT 3′UTR or MALAT1 MUT 3′UTR, then co-transfected with 20 nM
miR-141-3p mimic (5′-UAACACUGUCUGGUAAAGAUGG-3′) or miR-NC
(5′-ACGUGACACGUUCGGAGAATT-3′) into H9C2 cells using Lipofectamine
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). At 48 h
post-transfection, luciferase activities were measured using the
Dual Luciferase Reporter System (Promega Corporation). Firefly
luciferase activity was normalized to Renilla luciferase activity
for each transfected cell sample.
Detection of pyroptosis by TUNEL
staining
Cells were fixed with 4% paraformaldehyde at room
temperature for 30 min and washed with phosphate buffer saline
(PBS) three times, 5 min each time. Subsequently, cells were
incubated with PBS containing 0.3% Triton X-100 on ice for 5 min at
room temperature. Following washing twice with PBS, cells were
incubated with 50 µl TUNEL detection solution (Beyotime Institute
of Biotechnology) for 1 h at 37°C in the dark and then washed three
times with PBS. Cell nuclei were stained with DAPI (1 mg/ml) at
room temperature for 10 min in the dark. After washing with PBS,
the plate was sealed with anti-fluorescence quenching liquid and
observed using a BX51 fluorescence microscope (Olympus
Corporation). A total of five fields were viewed under ×400
magnification. The TUNEL positive cell rate (%) was calculated
according to the following formula: (Number of positive cells/total
number of cells) ×100.
Immunofluorescence staining
GSDMD-N expression was detected by performing
immunofluorescence staining. Cells (2×104 cells/ml) were
seeded on the slide fixed with 4% paraformaldehyde at room
temperature for 30 min and washed with PBS three times, 5 min each
time. Following drying of the slide using absorbent paper, the
slides were blocked with 5% goat serum (MP20008; Yuanye, Shanghai,
China) for 30 min at room temperature. The slides were incubated
with a rabbit anti-rat GSDMD-N polyclonal primary antibody (1:200;
cat. no. AF4013; Affinity Biosciences) at 4°C for overnight in
dark. Subsequently, the slides were incubated at room temperature
for 1 h, washed four times with PBS and incubated with 50 µl
FITC-labeled goat anti-rabbit IgG secondary antibody (1:500, cat.
no. bs-0295G; BIOSS) for 1 h at room temperature in dark. Following
washing, cells were counterstained with DAPI (1 mg/ml) at room
temperature for 10 min in dark. Anti-fluorescence quenching liquid
was used for sealing. Stained cells were observed using a
fluorescent microscope (BX51; Olympus Corporation). A total of five
fields were viewed under 400 magnification.
Western blotting
The protein expression levels of ASC, GSDMD-N,
caspase-1, NLRP3 and GSDMD were measured via western blotting.
Cells were lysed using ice-cold RIPA lysis buffer (Beyotime
Institute of Biotechnology) and centrifuged at 850 × g for 20 min
at 4°C to obtain the supernatant. Protein concentrations were
determined using a BCA kit (cat. no. A045-4-2; Nanjing Jiancheng
Bioengineering Institute). Proteins (40 µg) were separated via 10%
SDS-PAGE and transferred to PVDF membranes (cat. no. 3010040001;
Roche Diagnostics) for 30 min. Following blocking in tris-buffered
saline containing 2% Tween-20 (TBST) solution supplemented with 5%
BSA (Beijing Solarbio Science & Technology Co., Ltd.) for 1 h
at 4°C, the membranes were incubated overnight at 4°C with primary
antibodies (diluted in TBST supplemented with 3% BSA) targeted
against the following: ASC (1:1,000; cat. no. DF7540; Affinity
Biosciences), GSDMD-N (1:500; cat. no. AF4013; Affinity
Biosciences), caspase-1 (1:1,000; cat. no. ET1608-69; HUABIO),
NLRP3 (1:500; cat. no. ab91413; Abcam), GSDMD (1:500; cat. no.
ab245565; Abcam) and β-actin (1:1,000; cat. no. ET1702-67; HUABIO).
Subsequently, the membranes were incubated with a HRP-conjugated
goat anti-rabbit IgG secondary antibody (1:2,000; cat. no.
bs-0295G; BIOSS) for 1 h at room temperature. Following washing
with PBS, protein bands were visualized by incubating the membranes
with ECL developer (Thermo Fisher Scientific, Inc.) in the dark,
followed by detection using the Gel Doc XR System gel imaging
analysis system (Bio-Rad Laboratories, Inc.). Protein expression
levels were semi-quantified using ImageJ software 5.0 (National
Institutes of Health) with β-actin as the loading control.
Verification experiment
The miR-141-3p siRNA, miR-141-3p mimic, and their
corresponding controls were purchased from Shanghai GenePharma Co.,
Ltd. Cells (2×105 cells/ml) were transfected with
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.) and divided into the following groups: i) NG; ii)
HG; iii) HG + MALAT1 overexpression; iv) HG + miR-141-3p siRNA
scramble control (NC3), cells were transfected with 10 nM
miR-141-3p siRNA scramble control (5′-ACGUGACACGUUCGGAGAATT-3′) at
37°C for 24 h, according to previous study (21), then cultured in HG medium for 24 h;
v) HG + miR-141-3p siRNA (si-miR), cells were transfected with 10
nM miR-141-3p siRNA (5′-CCAUCUUUACCAGACAGUGUUA-3′) at 37°C for 24
h, then cultured in HG medium for 24 h; vi) HG + MALAT1
overexpression + miR-141-3p overexpression scramble control (MALAT1
+ NC4), cells were transfected with 10 nM miR-141-3p mimic scramble
control (5′-ACGUGACACGUUCGGAGAATT-3′) and 50 nM pcDNA3.1-MALAT1 at
37°C for 24 h, then cultured in HG medium for 24 h; and vii) HG +
MALAT1 overexpression + miR-141-3p overexpression (MALAT1 + miR),
cells were transfected with 10 nM miR-141-3p mimic
(5′-UAACACUGUCUGGUAAAGAUGG-3′) and 50 nM pcDNA3.1-MALAT1 at 37°C
for 24 h, then cultured in HG medium for 24 h. Transfection
efficiency was determined via RT-qPCR.
Statistical analysis
Statistical analyses were performed using SPSS
software (version 19.0; IBM Corp.). A total of three experimental
repeats were performed and data are presented as the mean ± SD.
Comparisons among multiple groups were analyzed using one-way ANOVA
followed by Tukey's post hoc test. P<0.05 was considered to
indicate a statistically significant difference.
Results
MALAT1 and miR-141-3p expression
levels in H9C2 cells
MALAT1 mRNA expression levels were significantly
increased, whereas miR-141-3p expression levels were markedly
decreased in the HG group compared with the NG group (Fig. 1A and B). In the MALAT1 group, MALAT1
mRNA expression levels were notably increased, whereas miR-141-3p
expression levels were significantly decreased compared with the HG
group (Fig. 1C and D). The opposite
effects on MALAT1 and miR-141-3p expression levels were observed in
the si-MALAT1 group compared with the HG group.
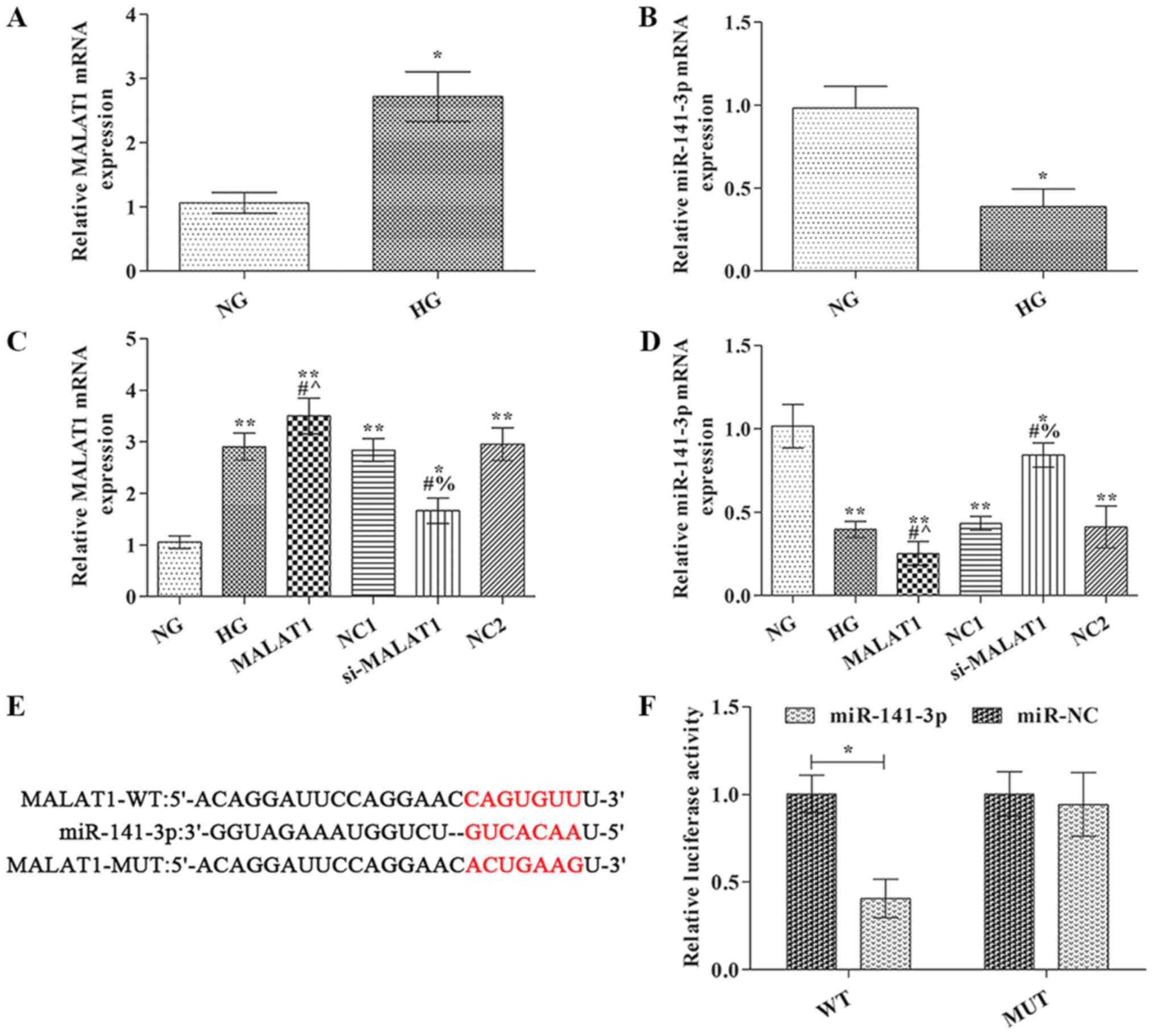 | Figure 1.MALAT1 and miR-141-3p expression
levels were detected via reverse transcription-quantitative PCR.
(A) MALAT1 and (B) miR-141-3p expression levels. (C) Transfection
efficiency of MALAT1 overexpression and knockdown. (D) Effect of
MALAT1 knockdown and overexpression on miR-141-3p expression. (E)
TargetScan was used to predict the binding site between miR-141-3p
and MALAT1. (F) Dual luciferase reporter assays were conducted to
verify the interaction between miR-141-3p and MALAT1. *P<0.05
and **P<0.01 vs. NG; #P<0.05 vs. HG;
^P<0.05 vs. NC1; %P<0.05 vs. NC2.
MALAT1, metastasis-associated lung adenocarcinoma transcript-1;
miR, microRNA; NG, normal glucose; HG, high glucose; NC, negative
control; NC1, MALAT1 overexpression NC group; NC2, MALAT1 knockdown
NC group; WT, wild-type; MUT, mutant; si, small interfering
RNA. |
MALAT1 was identified as a target of miR-141-3p
using TargetScan (www.targetscan.org) (Fig.
1E). To further verify whether miR-141-3p targeted MALAT1, a
dual luciferase reporter system was used (Fig. 1F). The results indicated that
compared with miR-NC, miR-141-3p mimic significantly reduced the
luciferase activity of MALAT1 WT 3′UTR, but did not significantly
alter the luciferase activity of MALAT1 MUT 3′UTR.
Effects of MALAT1 on cell pyroptosis
and GSDMD-N expression
Blue staining indicated a standard cell nucleus,
whereas green staining indicated a TUNEL label-positive nucleus
(Fig. 2A). Pyroptosis was not
observed in the NG group. The rate of TUNEL positive cells in the
HG group was markedly higher compared with the NG group
(P<0.05). Compared with the HG group, the rate of TUNEL positive
cells was significantly increased in the MALAT1 group, but
significantly reduced in the si-MALAT1 group (P<0.05). GSDMD-N
expression in the NG group was notably lower compared with the HG
group, with protein expression localized on the cell membrane
(P<0.01; Fig. 2B). Compared with
the HG group, GSDMD-N expression was significantly increased in the
MALAT1 group, but significantly decreased in the si-MALAT1 group
(P<0.01).
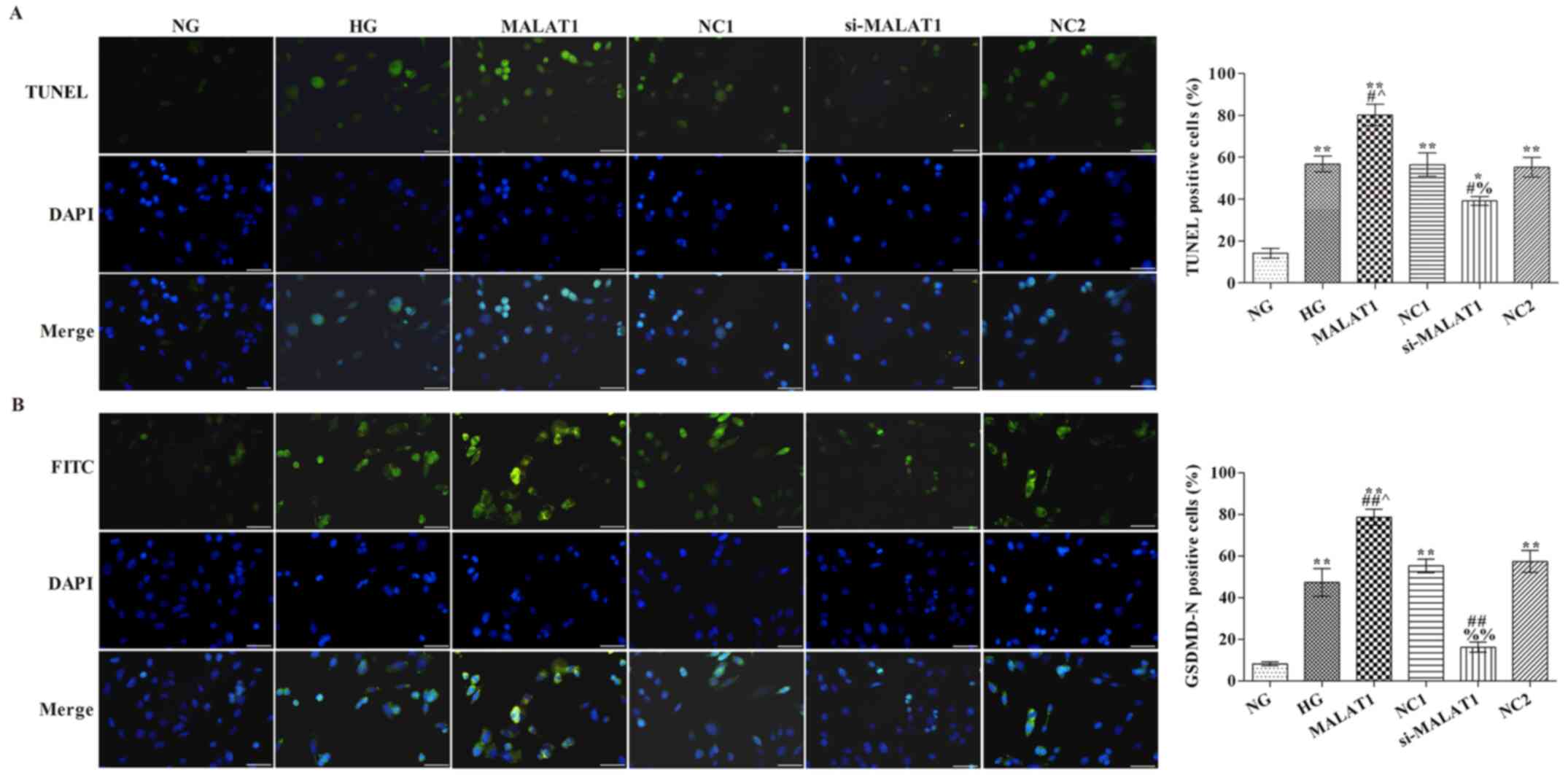 | Figure 2.Effects of MALAT1 on cell pyroptosis
and GSDMD-N expression. (A) TUNEL staining was performed to detect
the levels of pyroptosis (scale bar, 50 µm). (B) GSDMD-N expression
levels were detected via immunofluorescence staining (scale bar, 50
µm). *P<0.05 and **P<0.01 vs. NG; #P<0.05 and
##P<0.01 vs. HG; ^P<0.05 vs. NC1;
%P<0.05, %%P<0.01 vs. NC2. MALAT1,
metastasis-associated lung adenocarcinoma transcript-1; GSDMD,
gasdermin D; NG, normal glucose; HG, high glucose; NC, negative
control; NC1, MALAT1 overexpression NC group; NC2, MALAT1 knockdown
NC group; si, small interfering RNA. |
Effect of MALAT1 on
pyroptosis-associated protein expression levels
Compared with the NG group, ASC, GSDMD-N, caspase-1
and NLRP3 protein expression levels were significantly increased,
but GSDMD protein expression levels were significantly decreased in
the HG group (all P<0.05; Fig.
3). Compared with the HG group, ASC, GSDMD-N, caspase-1 and
NLRP3 protein expression levels were significantly increased,
whereas GSDMD protein expression levels were significantly
decreased in the MALAT1 group (all P<0.05). However, the
opposite effects on protein expression were observed in the
si-MALAT1 group compared with the HG group (all P<0.05).
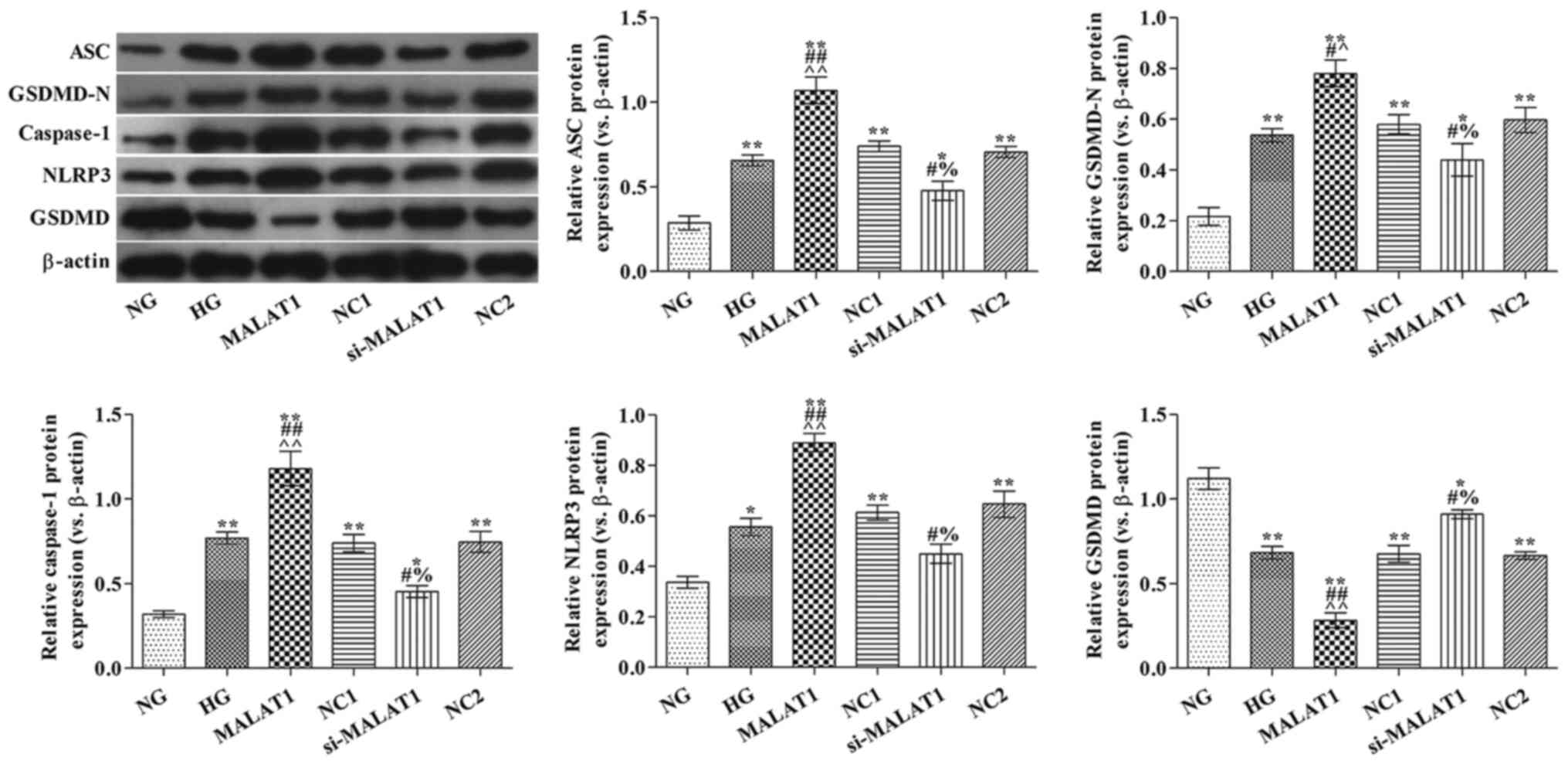 | Figure 3.Effects of MALAT1 on
pyroptosis-associated protein expression levels. ASC, GSDMD-N,
caspase-1, NLRP3 and GSDMD protein expression levels were measured
via western blotting. *P<0.05 and **P<0.01 vs. NG;
#P<0.05 and ##P<0.01 vs. HG;
^P<0.05 and ^^P<0.01 vs. NC1;
%P<0.05 vs. NC2. MALAT1, metastasis-associated lung
adenocarcinoma transcript-1; ASC, apoptosis-associated speck-like
protein; GSDMD, gasdermin D; NLRP3, nucleotide oligomerization
domain-like receptor protein 3; NG, normal glucose; HG, high
glucose; NC, negative control; NC1, MALAT1 overexpression NC group;
NC2, MALAT1 knockdown NC group; si, small interfering RNA. |
Effects of MALAT1 on miR-141-3p
expression, cell pyroptosis and GSDMD-N expression
To assess the transfection efficiency of miR-141-3p
mimic and inhibitor, the expression levels of miR-141-3p were
analyzed via RT-qPCR (Fig. 4A).
Compared with the NG group, miR-141-3p expression was significantly
increased by miR-141-3p mimic and decreased by miR-141-3p siRNA
(P<0.01). Furthermore, cells were transfected with MALAT1 mimic
(Fig. 4B). miR-141-3p expression
levels in all other groups were significantly reduced compared with
the NG group (Fig. 4B). Compared
with the HG group, miR-141-3p knockdown or MALAT1 overexpression
significantly decreased miR-142-3p expression (both P<0.05).
Compared with the MALAT1 group, miR-142-3p expression was
significantly increased in the MALAT1 + miR group (P<0.05).
GSDMD-N expression levels were significantly increased in all other
groups compared with the NG group (all P<0.01; Fig. 4D). The rate of TUNEL positive cells
(all P<0.05; Fig. 4C) and
GSDMD-N expression levels (all P<0.01; Fig. 4D) were significantly increased in
the MALAT1, si-miR and MALAT1 + NC4 groups compared with the HG
group. In addition, the number of TUNEL positive cells and GSDMD-N
expression levels were also significantly reduced in the MALAT1 +
miR group compared with the MALAT1 group (both P<0.01).
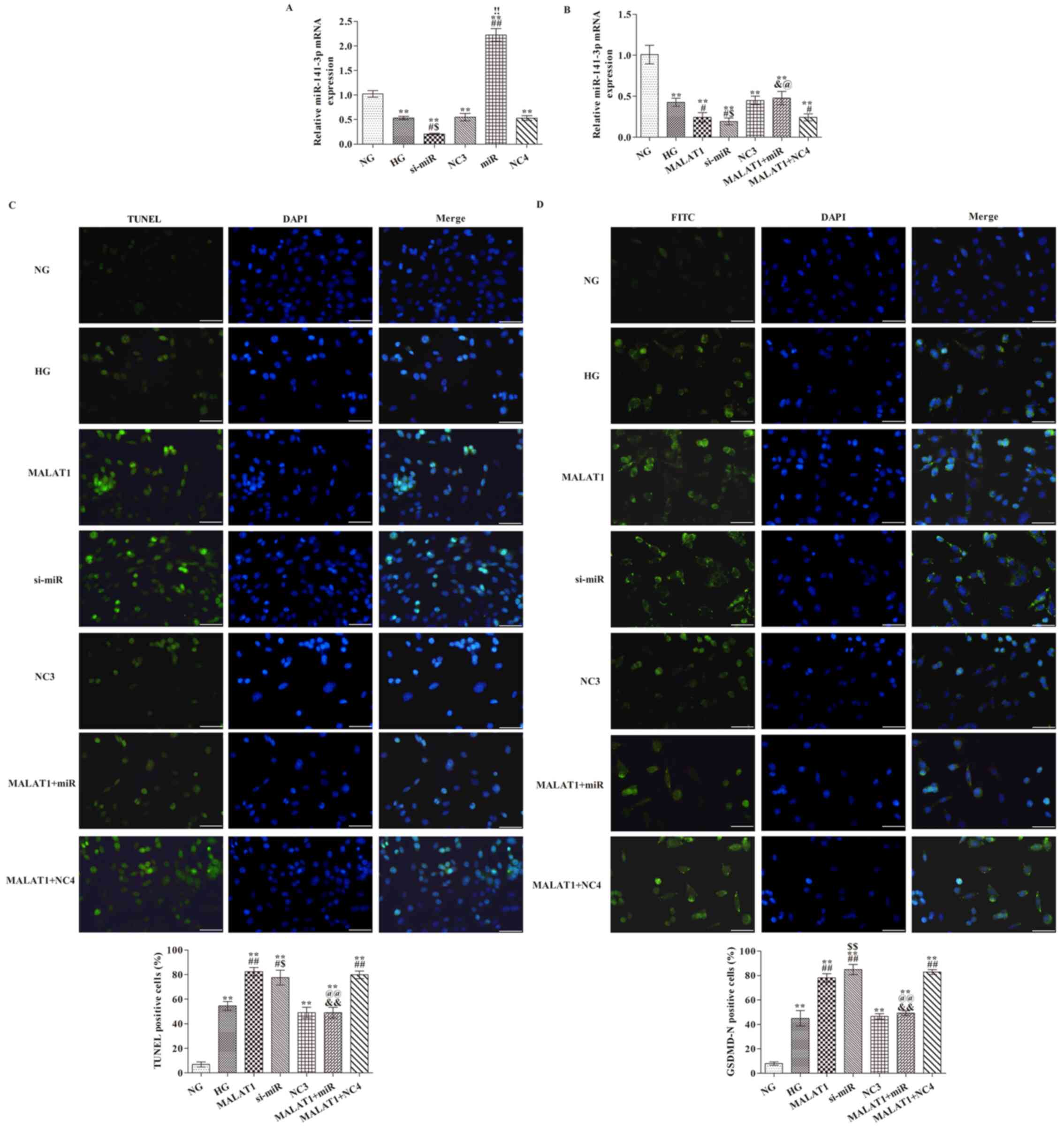 | Figure 4.Effects of MALAT1 on cell pyroptosis
and GSDMD-N expression are mediated via targeting miR-141-3p. (A)
Transfection efficiency of miR-141-3p overexpression and knockdown.
(B) Effect of miR-141-3p overexpression and knockdown on miR-141-3p
expression levels following HG treatment. (C) TUNEL staining was
performed to detect the levels of pyroptosis (scale bar, 50 µm).
(D) GSDMD-N expression levels were detected via immunofluorescence
staining (scale bar, 50 µm). **P<0.05 vs. NG;
#P<0.05 and ##P<0.05 vs. HG;
&P<0.05 and &&P<0.01 vs.
MALAT1; $P<0.05 and $$P<0.01 vs. NC3;
!!P<0.01 vs. NC4; @P<0.05 and
@@P<0.01 vs. MALAT1 + NC4. MALAT1,
metastasis-associated lung adenocarcinoma transcript-1; GSDMD,
gasdermin D; miR, microRNA; HG, high glucose; NG, normal glucose;
NC, negative control; NC3, miR-141-3p knockdown NC group; NC4,
miR-141-3p overexpression NC group; si, small interfering RNA. The
cell nuclei were stained with DAPI; The positive nucleus were
stained with FITC. |
MALAT1 alters pyroptosis-related
protein expression levels via targeting miR-141-3p
ASC, GSDMD-N, caspase-1 and NLRP3 protein expression
levels were significantly increased in all other groups, whereas
GSDMD protein expression levels were significantly decreased in all
other groups compared with the NG group (all P<0.05; Fig. 5). ASC, GSDMD-N, caspase-1 and NLRP3
protein expression levels in the MALAT1, si-miR and MALAT1 + NC4
groups were significantly increased, whereas GSDMD protein
expression levels were significantly decreased compared with the HG
group (all P<0.05). ASC, GSDMD-N, caspase-1 and NLRP3 protein
expression levels in the MALAT1 + miR group were significantly
reduced, whereas GSDMD protein expression levels were significantly
increased compared with the MALAT1 group (all P<0.05).
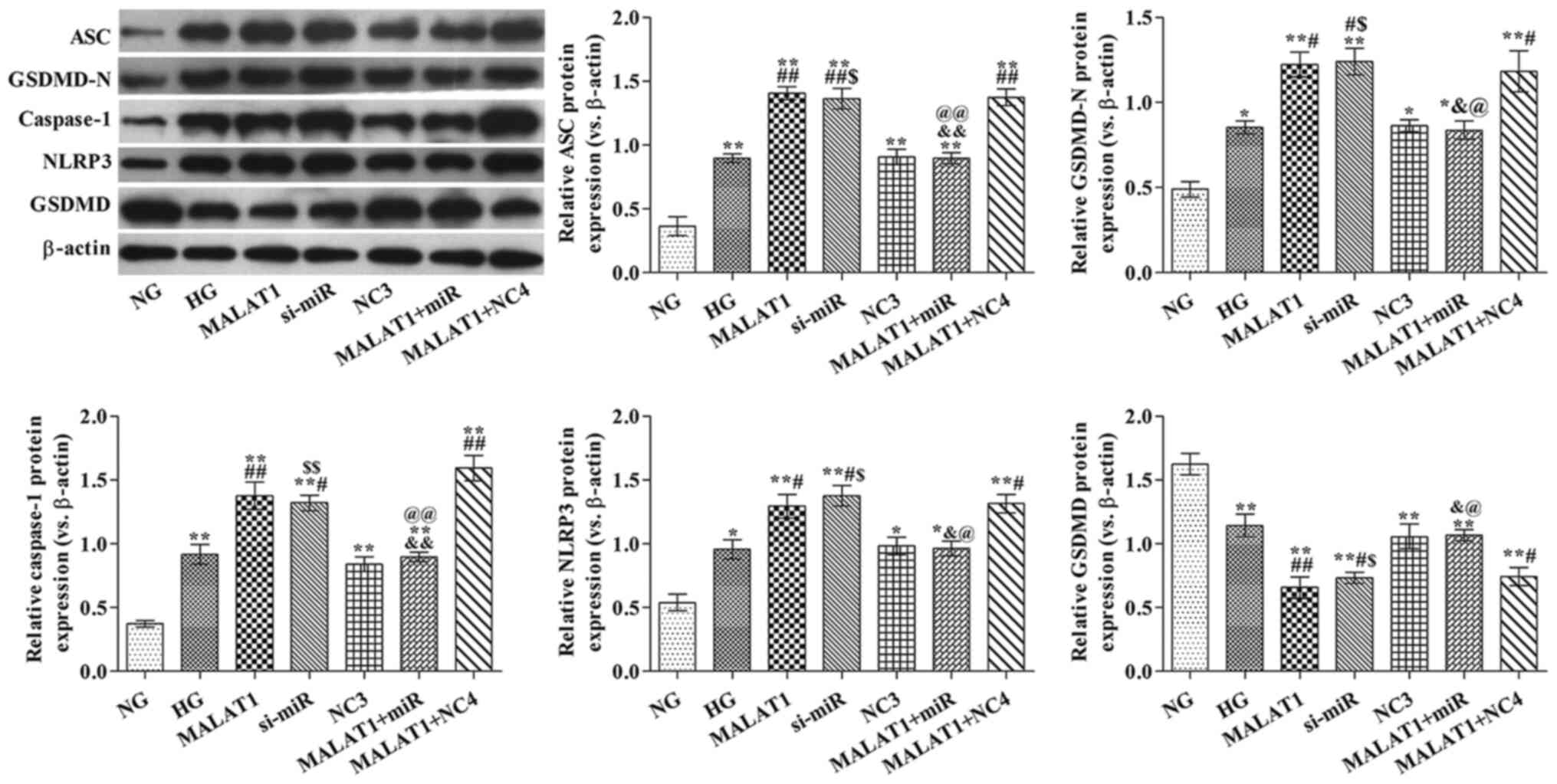 | Figure 5.Effects of MALAT1 on
pyroptosis-associated protein expression levels are mediated via
targeting miR-141-3p. ASC, GSDMD-N, caspase-1, NLRP3 and GSDMD
protein expression levels were measured via western blotting.
*P<0.05 and **P<0.05 vs. NG; #P<0.05 and
##P<0.05 vs. HG; &P<0.05 and
&&P<0.01 vs. MALAT1; $P<0.05
and $$P<0.05 vs. NC3; @P<0.05 and
@@P<0.01 vs. MALAT1 + NC4. MALAT1,
metastasis-associated lung adenocarcinoma transcript-1; miR,
microRNA; ASC, apoptosis-associated speck-like protein; GSDMD,
gasdermin D; NLRP3, nucleotide oligomerization domain-like receptor
protein 3; NG, normal glucose; HG, high glucose; NC, negative
control; NC3, miR-141-3p knockdown NC group; NC4, miR-141-3p
overexpression NC group; si, small interfering RNA. |
Discussion
The development of diabetic myocardial damage is a
complex process; although, hyperglycemia is typically considered as
the primary risk factor leading to diabetic myocardial damage
(22), the specific molecular
mechanism that induces cell damage is not completely understood. A
previous study indicated that pyroptosis was closely associated
with the occurrence and development of DCM (23). In addition, caspase-1 mRNA and
protein expression levels were significantly increased in the
cardiomyocytes of diabetic model rats (24). An additional study demonstrated that
NLRP3 activated caspase-1-mediated pyroptosis in DCM and served a
significant role in the development of the disease (25). Therefore, the molecular mechanism
underlying HG-induced cardiomyocyte pyroptosis could be used to
identify novel molecular therapeutic targets for DCM. In the
present study, pyroptosis-associated protein expression levels,
including ASC, GSDMD-N, caspase-1, NLRP3 and GSDMD, were assessed
in NG- and HG-treated cells. The results demonstrated that
pyroptosis-associated protein expression levels were significantly
increased in the HG group compared with the NG group, which
suggested that HG induced H9C2 cardiomyocyte pyroptosis.
In addition, lncRNAs interact with miRNAs, which
influences the development of several diseases, including DCM
(12,26). In a previous study, the involvement
of MALAT1 in DCM was examined, and the results indicated that
MALAT1 participated in the regulation of cardiomyocyte apoptosis
(12). Gong et al (17) demonstrated that lncRNA MALAT1 bound
to miR-141-3p, which decreased miR-141-3p expression levels during
the development of atherosclerosis. Therefore, the present study
explored the effect of lncRNA-MALAT1 on HG-induced H9C2
cardiomyocyte pyroptosis. The results suggested that the effects of
MALAT1 were mediated via targeting miR-141-3p. MALAT1 expression
levels were significantly increased, whereas miR-141-3p expression
levels were significantly decreased in HG-treated H9C2 cells
compared with the NG group. Furthermore, compared with the HG
group, MALAT1 overexpression significantly reduced miR-141-3p
expression levels, increased the rate of TUNEL positive cells and
upregulated pyroptosis-associated protein expression levels,
whereas MALAT1 knockdown displayed the opposite effects. Moreover,
the rate of TUNEL positive cells, and the expression levels of
GSDMD-N and pyroptosis-associated proteins were all significantly
reduced by miR-141-3p overexpression in MALAT1-overexpression H9C2
cells.
At present, >100,000 lncRNAs have been identified
in the human body (27). However,
despite the vast number of lncRNAs and their complex regulatory
networks, the functions of the majority of lncRNAs are not
completely understood, which includes lncRNAs involved in the
regulation of glucose and the pathogenesis of diabetes. In the
present study, there were several limitations. MALAT1 and
miR-141-3p in rats with diabetic cardiomyopathy needs further
exploration, as do mechanisms of MALAT1 and miR-141-3p.
Collectively, the present study provided novel insight for the
diagnosis and targeted therapy of DCM. The results suggested that
it might be possible to prevent the damage caused by HG-induced
pyroptosis in the early stages, thereby preventing the occurrence
and development of DCM.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
AW and WS designed the study, performed the
experiments, collected data and drafted the manuscript. WS and FM
analyzed and interpreted the experimental data. AW and FM assessed
the raw data and were responsible for confirming the legitimacy of
the data. FM participated in the coordination of the study and
revised the manuscript. All authors read and approved the final
manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lee WS and Kim J: Diabetic cardiomyopathy:
Where we are and where we are going. Korean J Intern Med.
32:404–421. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Shen L, Li L, Li M, Wang W, Yin W, Liu W
and Hu Y: Silencing of NOD2 protects against diabetic
cardiomyopathy in a murine diabetes model. Int J Mol Med.
42:3017–3026. 2018.PubMed/NCBI
|
|
3
|
Wakisaka M, Kamouchi M and Kitazono T:
Lessons from the trials for the desirable effects of sodium glucose
Co-transporter 2 inhibitors on diabetic cardiovascular events and
renal dysfunction. Int J Mol Sci. 20:56682019. View Article : Google Scholar
|
|
4
|
Hu X, Bai T, Xu Z, Liu Q, Zheng Y and Cai
L: Pathophysiological fundamentals of diabetic cardiomyopathy.
Compr Physiol. 7:693–711. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
American, Diabetes, Association: 2.
Classification and diagnosis of diabetes: Standards of medical care
in diabetes-2019. Diabetes Care. 42 (Suppl 1):S13–S28. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Pasparakis M and Vandenabeele P:
Necroptosis and its role in inflammation. Nature. 517:311–320.
2015. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Xu B, Jiang M, Chu Y, Wang W, Chen D, Li
X, Zhang Z, Zhang D, Fan D, Nie Y, et al: Gasdermin D plays a key
role as a pyroptosis executor of non-alcoholic steatohepatitis in
humans and mice. J Hepatol. 68:773–782. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Shi J, Gao W and Shao F: Pyroptosis:
Gasdermin-mediated programmed necrotic cell death. Trends Biochem
Sci. 42:245–254. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Yue RC, Lu SZ, Luo Y, Wang T, Liang H,
Zeng J, Liu J and Hu HX: Calpain silencing alleviates myocardial
ischemia-reperfusion injury through the NLRP3/ASC/Caspase-1 axis in
mice. Life Sci. 233:1166312019. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Biswas S, Thomas AA and Chakrabarti S:
LncRNAs: Proverbial genomic ‘Junk’ or key epigenetic regulators
during cardiac fibrosis in diabetes? Front Cardiovasc Med.
5:282018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Asrih M and Steffens S: Emerging role of
epigenetics and miRNA in diabetic cardiomyopathy. Cardiovasc
Pathol. 22:117–125. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Zhang M, Gu H, Xu W and Zhou X:
Down-regulation of lncRNA MALAT1 reduces cardiomyocyte apoptosis
and improves left ventricular function in diabetic rats. Int J
Cardiol. 203:214–216. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ji P, Diederichs S, Wang W, Böing S,
Metzger R, Schneider PM, Tidow N, Brandt B, Buerger H, Bulk E, et
al: MALAT-1, a novel noncoding RNA, and thymosin beta4 predict
metastasis and survival in early-stage non-small cell lung cancer.
Oncogene. 22:8031–8041. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Michalik KM, You X, Manavski Y,
Doddaballapur A, Zörnig M, Braun T, John D, Ponomareva Y, Chen W,
Uchida S, et al: Long noncoding RNA MALAT1 regulates endothelial
cell function and vessel growth. Circ Res. 114:1389–1397. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Qin Q, Cui L, Zhou Z, Zhang Z, Wang Y and
Zhou C: Inhibition of microRNA-141-3p reduces hypoxia-induced
apoptosis in H9c2 rat cardiomyocytes by activating the
RP105-Dependent PI3K/AKT signaling pathway. Med Sci Monit.
25:7016–7025. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yao B, Wan X, Zheng X, Zhong T, Hu J, Zhou
Y, Qin A, Ma Y and Yin D: Critical roles of microRNA-141-3p and
CHD8 in hypoxia/reoxygenation-induced cardiomyocyte apoptosis. Cell
Biosci. 10:202020. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Gong D, Zhao ZW, Zhang Q, Yu XH, Wang G,
Zou J, Zheng XL, Zhang DW, Yin WD and Tang CK: The long noncoding
RNA metastasis-associated lung adenocarcinoma Transcript-1
Regulates CCDC80 expression by targeting miR-141-3p/miR-200a-3p in
vascular smooth muscle cells. J Cardiovasc Pharmacol. 75:336–343.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Zhang J, Jiang T, Liang X, Shu S, Xiang X,
Zhang W, Guo T, Xie W, Deng W and Tang X: lncRNA MALAT1 mediated
high glucose-induced HK-2 cell epithelial-to-mesenchymal transition
and injury. J Physiol Biochem. 75:443–452. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Ye Y, Zhang F, Chen Q, Huang Z and Li M:
LncRNA MALAT1 modified progression of clear cell kidney carcinoma
(KIRC) by regulation of miR-194-5p/ACVR2B signaling. Mol Carcinog.
58:279–292. 2019. View
Article : Google Scholar : PubMed/NCBI
|
|
21
|
Xing Y, Jing H, Zhang Y, Suo J and Qian M:
MicroRNA-141-3p affected proliferation, chemosensitivity, migration
and invasion of colorectal cancer cells by targeting EGFR. Int J
Biochem Cell Biol. 118:1056432020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Adameova A and Dhalla NS: Role of
microangiopathy in diabetic cardiomyopathy. Heart Fail Rev.
19:25–33. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Luo B, Li B, Wang W, Liu X, Liu X, Xia Y,
Zhang C, Zhang Y, Zhang M and An F: Rosuvastatin alleviates
diabetic cardiomyopathy by inhibiting NLRP3 inflammasome and MAPK
pathways in a type 2 diabetes rat model. Cardiovasc Drugs Ther.
28:33–43. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Shi J, Zhao Y, Wang K, Shi X, Wang Y,
Huang H, Zhuang Y, Cai T, Wang F and Shao F: Cleavage of GSDMD by
inflammatory caspases determines pyroptotic cell death. Nature.
526:660–665. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Fann DY, Lee SY, Manzanero S, Tang SC,
Gelderblom M, Chunduri P, Bernreuther C, Glatzel M, Cheng YL,
Thundyil J, et al: Intravenous immunoglobulin suppresses NLRP1 and
NLRP3 inflammasome-mediated neuronal death in ischemic stroke. Cell
Death Dis. 4:e7902013. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Feng Y, Xu W, Zhang W, Wang W, Liu T and
Zhou X: LncRNA DCRF regulates cardiomyocyte autophagy by targeting
miR-551b-5p in diabetic cardiomyopathy. Theranostics. 9:4558–4566.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Taylor DH, Chu ET, Spektor R and Soloway
PD: Long non-coding RNA regulation of reproduction and development.
Mol Reprod Dev. 82:932–956. 2015. View Article : Google Scholar : PubMed/NCBI
|



















