Introduction
Gastric cancer (GC) is considered one of the leading
causes of cancer-associated mortality and the global incidence of
GC has risen in recent years (1).
The onset and progression of GC is complex, and the underlying
mechanisms remain largely unknown (2). Furthermore, the overall survival rate
of patients with GC is poor (1,2).
Although therapeutic approaches for GC have improved due to the
broad usage of endoscopic/surgical resection, the prognosis of GC
remains unfavorable (3,4). The therapeutic outcome of patients
with GC is closely associated with disease stage at diagnosis, and
the mortality rate of patients with advanced GC is high (3–5).
Therefore, it is important to discover novel diagnostic/prognostic
biomarkers for GC; therefore, the treatment for this disease can be
improved.
The nuclear factor I B (NFIB) gene functions as a
transcription factor (6). NFIB
serves essential roles in embryonic development and tissue
differentiation in the fetus (6).
NFIB is widely expressed in embryonic/adult tissues, and is
abundantly detected in the lungs, skeletal muscle and heart
(6). Upregulation of NFIB has been
observed in numerous types of cancer, such as GC, breast, lung and
prostate cancer, and it is considered a common oncogene (6–9).
Furthermore, Wu et al (9)
revealed that NFIB was able to promote GC cell growth and
metastasis by targeting Akt/Stat3 signaling, which could be a
promising molecular mechanism for the treatment of patients with
GC. Circular RNAs (circRNAs) are a novel class of non-coding RNAs,
which form a continuous loop and have a more stable structure than
their linear counterparts (10).
The roles of circRNAs have been widely investigated. It has been
revealed that certain circRNAs function as potential gene
regulators; however, the detailed mechanisms of most circRNAs
remain largely unknown (10).
circRNAs are involved in the pathogenesis of various diseases,
including nervous system disorders and cancer (11–14).
Furthermore, circSFMBT2 and circ100269 have been reported to be
involved in the proliferation of GC cells, consequently affecting
tumor progression in GC (13,14).
Therefore, circRNAs may serve essential roles during the
development of GC; however, the detailed regulatory roles of
circRNAs in GC remain elusive and require further investigation.
Human epidermal growth factor receptor 2 (HER2) is a key regulator
during the development of breast cancer (15). Previous studies have indicated that
HER2 overexpression may be a frequent abnormality in numerous types
of cancer, including GC (15–17).
HER2 molecular targeted therapy has been introduced for patients
with advanced GC, and HER2 status could be essential to select
patients who may benefit from targeted therapy (17).
In the present study, the expression levels of NFIB
were evaluated in GC specimens and cells, and its regulatory roles
were further elucidated. Our data suggested that NFIB could be
considered a promising target for the precise treatment of patients
with GC.
Materials and methods
Clinical specimens
A total of 50 GC (29 men and 21 women; age range,
36–67 years; mean age, 53±14.2 years) tissues and matched
para-carcinoma samples (≥5 cm from tumor margin) were obtained at
the Affiliated Hospital of North Sichuan Medical College (Nanchong,
China) between August 2010 and September 2015. The exclusion
criteria were as follows: Patients that received
radiotherapy/chemotherapy prior to surgery were excluded.
Immediately upon collection, tissues were sectioned into slices,
snap-frozen in liquid nitrogen and stored at −80°C until further
use. Patient biopsies were examined by two independent pathologists
and all samples were anonymized. The experimental protocol was
approved by the Medical Ethics Committee of the Affiliated Hospital
of North Sichuan Medical College. Written informed consent was
obtained from all patients. Upregulation of NFIB was also revealed
by Gene Expression Profiling Interactive Analysis (GEPIA;
http://gepia.cancer-pku.cn/).
Cell culture
Human GC cell lines AGS and MKN-45, and the normal
human gastric epithelial cell line GES-1 were purchased from the
American Type Culture Collection. The cells were cultured in DMEM
containing 10% FBS, 100 µg/ml streptomycin and 100 U/ml penicillin
(all from Cytiva). The cells were maintained at 37°C in a
humidified incubator containing 5% CO2.
Infection and transfection
To generate the knockdown cell models of NFIB and
circMAPK7D1, short hairpin RNAs (shRNAs) targeting NFIB (sh-NFIB)
and circMAPK7D1 (sh-circMAPK7D1), and a negative control (sh-NC)
were purchased from Shanghai Genepharma Co., Ltd. After annealing,
shRNA fragments were integrated into lentiviral pU6-Luc-Puro vector
(Shanghai Genepharma Co., Ltd.). Cells (4×105) were
infected with shRNA lentiviruses (MOI, 50) and incubated at 37°C
for 24 h. To generate a cell model overexpressing circMAPK7D1,
wild-type (oe-circMAPK7D1) or non-targeting (oe-NC) fragments were
amplified using PCR, then inserted into PLCDH-cir vector
(Invitrogen; Thermo Fisher Scientific, Inc.). Cells
(4×105) were transfected with vectors (1 µg) at 37°C for
48 h. Up- or downregulation of the corresponding genes was
confirmed using reverse transcription-quantitative PCR (RT-qPCR).
All transfections were conducted using Lipofectamine®
2000 (Invitrogen; Thermo Fisher Scientific, Inc.). A total of 12 h
post-transfection, the culture supernatant was replaced with fresh
DMEM containing 10% FBS.
RT-qPCR
Total RNA was isolated from clinical specimens or
cells using TRIzol® reagent (Invitrogen; Thermo Fisher
Scientific, Inc.). Extracted RNA was reverse transcribed into cDNA
using PrimeScript™ RT kit (Takara Biotechnology Co., Ltd.). For RT,
the sample was incubated at room temperature for 30 min;
subsequently, the sample was incubated at 42°C for 45 min, 99°C for
5 min and 5°C for 5 min in a PCR cycler. qPCR was carried out using
SYBR Green PCR Master Mix (Takara Biotechnology Co., Ltd.), and the
reaction was performed on an ABI 7500 Real-Time PCR system (Applied
Biosystems; Thermo Fisher Scientific, Inc.) Endogenous GAPDH was
used as a reference gene. The sequences used for qPCR were as
follows: NFIB, forward, 5′-CCTCACTGGTACTGGGGTAT-3′ and reverse,
5′-TGGACATTGGCCGGTAAGAT-3′; circMAPK7D1, forward,
5′-GCGTCGGTGACTAAGCAATC-3′ and reverse,
5′-TCAAGAACAGATGGAAGAATGG-3′; HER2, forward,
5′-GGTCCTGGAAGCCACAAGG-3′ and reverse, 5′-GGTTTTCCCACCACATCCTCT-3′;
GAPDH, forward, 5′-GTCTCCTCTGACTTCAACAGCG-3 and reverse,
5′-ACCACCCTGTTGCTGTAGCCAA-3′. The PCR program used was as follows:
95°C for 5 min, followed by 45 cycles at 95°C for 15 sec, 60°C for
20 sec and 72°C for 10 sec, and a final extension step at 72°C for
10 min. Relative gene expression levels were analyzed using the
2−∆∆Cq method (18).
Western blot analysis
Total protein was isolated from tissues or cells
using RIPA buffer (Beyotime Institute of Biotechnology). The
concentration of the extracted protein was then evaluated using the
BCA assay (Beyotime Institute of Biotechnology). Equal amounts of
protein (40 µg) were separated by SDS-PAGE on 10% gels and were
then transferred onto nitrocellulose membranes (EMD Millipore).
Membranes were blocked with Tris-buffered saline (TBS) containing
5% skimmed milk at room temperature for 1 h, and were incubated
with the following primary antibodies: NFIB (1:1,000; cat. no.
ab186738; Abcam), HER2 (1:2,000; cat. no. 2242; Cell Signaling
Technology, Inc.) or GAPDH (1:2,000; cat. no. sc-32233; Santa Cruz
Biotechnology, Inc.) at 4°C overnight. Subsequently, the membranes
were incubated with HRP-conjugated anti-mouse (1:10,000; cat. no.
sc-2371; Santa Cruz Biotechnology, Inc.) or anti-rabbit IgG
(1:10,000; cat. no. sc-2357; Santa Cruz Biotechnology, Inc.) at
room temperature for 1 h. Protein bands were visualized using ECL
detection kit (Pierce; Thermo Fisher Scientific, Inc.) and the
blots were semi-quantified using ImageJ software (version 1.48;
National Institutes of Health).
Cell Counting Kit-8 (CCK-8) assay
Transfected and infected cells were seeded into a
96-well plate at a density of 3×104 cells/well.
Subsequently, the proliferative activity of cells was determined on
days 1, 2, 3 and 4. Briefly, 10 µl CCK-8 solution (Beyotime
Institute of Biotechnology) was added to each well. After
incubation for 2 h at 37°C, the absorbance was detected at 450 nm
using a microplate reader (Bio-Rad Laboratories, Inc.).
Cell cycle distribution and apoptosis
assay
Transfected and infected cells were seeded into a
6-well plate at a density of 6×105 cells/well. Cells
were then collected through low-speed centrifugation (2,600 × g) at
4°C for 5 min. Cell pellets were washed and resuspended using PBS,
then fixed using pre-chilled ethanol (70%) at room temperature for
15 min and stored at 4°C overnight. Cells were centrifuged (10,000
× g) and then resuspended in propidium iodide (PI; Sigma-Aldrich;
Merck KGaA) staining buffer with 50 µl/ml PI and 250 µl/ml RNase A
at 4°C for 1 h. Cell cycle distribution was evaluated using a flow
cytometer (FACSCalibur; BD Biosciences) and was analyzed using
FlowJo software (version 7.6; FlowJo LLC). In order to examine cell
apoptosis, the cell suspension was incubated with 5 µl Annexin
V-FITC and 10 µl PI solution at 4°C in the dark for 30 min
(Shenzhen Jingmei Biotech Engineering Co. Ltd.), and cell apoptotic
rate was analyzed by flow cytometry (FACSCalibur; BD Biosciences)
and interpreted using FlowJo software. Both early
(FITC+PI−) and late
(FITC+PI+) apoptosis were evaluated.
circRNA microarray
Total RNA was extracted using TRIzol®
reagent (Invitrogen; Thermo Fisher Scientific, Inc.) and a RNeasy
Mini kit (Qiagen GmbH). Fluorescence-labeled targets were produced
for circRNA array. Human circRNA array v2 (CapitalBio Technology
Co., Ltd.) was designed and genes were mounted onto the chip;
target sequences of corresponding circRNAs were obtained from
Circbase (http://www.circbase.org/). Labeled
targets were then hybridized with extracted RNA samples, which were
scanned using the Agilent Microarray Scanner (Agilent Technologies,
Inc.). All data were normalized according to Quantile algorithm
(19). Arrays were conducted using
the protocol supplied by Agilent Technologies, Inc.
RNA pulldown assay
Biotinylated wild-type (WT) or mutant (MUT) NFIB
(NFIB-WT-Bio/NFIB-MUT-Bio) and circMAP7D1
(circMAP7D1-WT-Bio/circMAP7D1-MUT-Bio) probes, and a negative
control (NC-Bio) probe were generated by Shanghai GenePharma Co.,
Ltd. Briefly, for RNA pulldown assay, RNA was extracted from cells,
as aforementioned. Extracted RNA was labeled with biotinylated
probes. Lysates of AGS and MKN-45 cells were prepared using lysis
buffer containing KCl (10 mM), MgCl2 (1.5 mM), Tris-Cl pH 7.5 (10
mM), DTT (5 mM) and SUPERase. In™ (60 U/ml; Thermo Fisher
Scientific, Inc.). Lysates were mixed with pro-labeled RNA in the
aforementioned lysis buffer. The binding reaction was incubated at
room temperature for 1 h and labeled with Dynabeads M-280
Streptavidin, and the mixture was rotated overnight at 4°C (Thermo
Fisher Scientific, Inc.). The beads with immobilized NFIB or
circMAP7D1 were treated with 10 mM ethylenediaminetetraacetic acid.
TRIzol® was used to extract the bound RNAs, which were
further subjected to RT-qPCR, as aforementioned. After pulldown,
the crosslinked molecules were identified using mass spectrometry
as previously described (20).
Assessment of mRNA stability
Cells were seeded into a 6-well plate
(6×105 cells/well) and incubated at 37°C overnight. The
cells were then treated with actinomycin D (ActD; 5 µg/ml;
MedChemExpress) for different durations (15, 30 and 60 min) to
inhibit further RNA synthesis at 37°C. Total RNA was isolated and
analyzed using RT-qPCR, as aforementioned. The remaining mRNA
levels of HER2 at each time point were normalized to the levels
detected at the beginning of the experiment.
Luciferase reporter assay
Fragments containing complementary sequences of NFIB
were cloned into a luciferase plasmid [WT-NFIB; OBiO Technology
(Shanghai) Corp., Ltd.]. A vector carrying mutant fragments of NFIB
was used as a control [MUT-NFIB; OBiO Technology (Shanghai) Corp.,
Ltd.]. AGS cells (4×105/well) were seeded onto a 6-well
plate co-transfected with 1 µg luciferase vectors and
oe-circMAP7D1/oe-NC (synthesized by Shanghai Genepharma Co., Ltd.)
using Lipofectamine 2000. After 48 h, luciferase activity was
determined using dual luciferase reporter system kit (Promega
Corporation). Luciferase activity was normalized to Renilla
luciferase activity.
Immunohistochemistry (IHC)
Immunostaining of HER2 was carried out on
paraffin-embedded tissue sections (thickness, 10 µm); tissues were
fixed with 4% paraformaldehyde at 4°C overnight before embedding.
The sections were dewaxed using xylene, and rehydrated through a
graded ethanol series and water. Antigen retrieval was performed in
a microwave using 10 mM sodium citrate for 15 min. Tissues were
then incubated with FBS at room temperature for 30 min and with a
primary antibody against HER2 (1:100; cat. no. 2242; Cell Signaling
Technology, Inc.) in a humidified incubator at 4°C overnight.
Subsequently, the sections were rinsed in TBS three times and
incubated with biotinylated secondary antibody (1:100; cat. no.
E0432; Dako; Agilent Technologies, Inc.). Protein expression was
then detected using a streptavidin-biotin-peroxidase system (ABC
kit; Dako; Agilent Technologies, Inc.). Stained slides were
observed using a light microscope (magnification, ×200; CX23 model;
Olympus Corporation). Staining intensity was semi-quantified using
ImageJ software (version 1.46).
Statistical analysis
All data are presented as the mean ± standard
deviation and were analyzed using SPSS software (version 17.0;
SPSS, Inc.). All assays were performed in triplicate. The
significance of differences was analyzed using one-way analysis of
variance (ANOVA) or paired/unpaired Student's t-test. Newman-Keuls
test was conducted following ANOVA. The relationship between RNA
expression was evaluated by Pearson's correlation analysis.
Receiver operating characteristic (ROC) curve analysis was
performed to assess the power of NFIB expression in distinguishing
between GC and para-carcinoma tissues, patients with and without
metastasis, and grade III–IV and I–II GC. P<0.05 was considered
to indicate a statistically significant difference.
Results
Expression levels of NFIB are
increased in GC specimens and cells
The expression levels of NFIB were examined in 50
pairs of GC and para-carcinoma tissues using RT-qPCR. The
expression levels of NFIB were significantly elevated in GC
specimens compared with those in the normal control specimens
(Fig. 1A). Furthermore, the
association between NFIB expression and the progression of GC was
investigated. The expression levels of NFIB were significantly
upregulated in patients with GC and metastasis compared with those
in patients with GC without metastasis (Fig. 1B). In addition, the expression of
NFIB was significantly increased in patients with grade III–IV GC
compared with that in patients with grade I–II GC (Fig. 1C). Moreover, ROC curve analyses
suggested that NFIB expression signature exhibited a high area
under the curve value in distinguishing between GC and
para-carcinoma tissues, between GC with and without metastasis, and
between grade III–IV GC and I–II GC (Fig. 1D-F). Consistent with the present
findings, upregulation of NFIB was also revealed by Gene Expression
Profiling Interactive Analysis (GEPIA; http://gepia.cancer-pku.cn/) (Fig. 1G). Furthermore, increased protein
expression levels of NFIB were detected in GC specimens compared
with those in normal control specimens, as determined using western
blotting (Fig. 1H). In addition,
the mRNA and protein expression levels of NFIB were upregulated in
GC cell lines compared with those in normal human gastric
epithelial cells (Fig. 1I). These
findings indicated that the expression levels of NFIB were elevated
in GC and may contribute to tumor development.
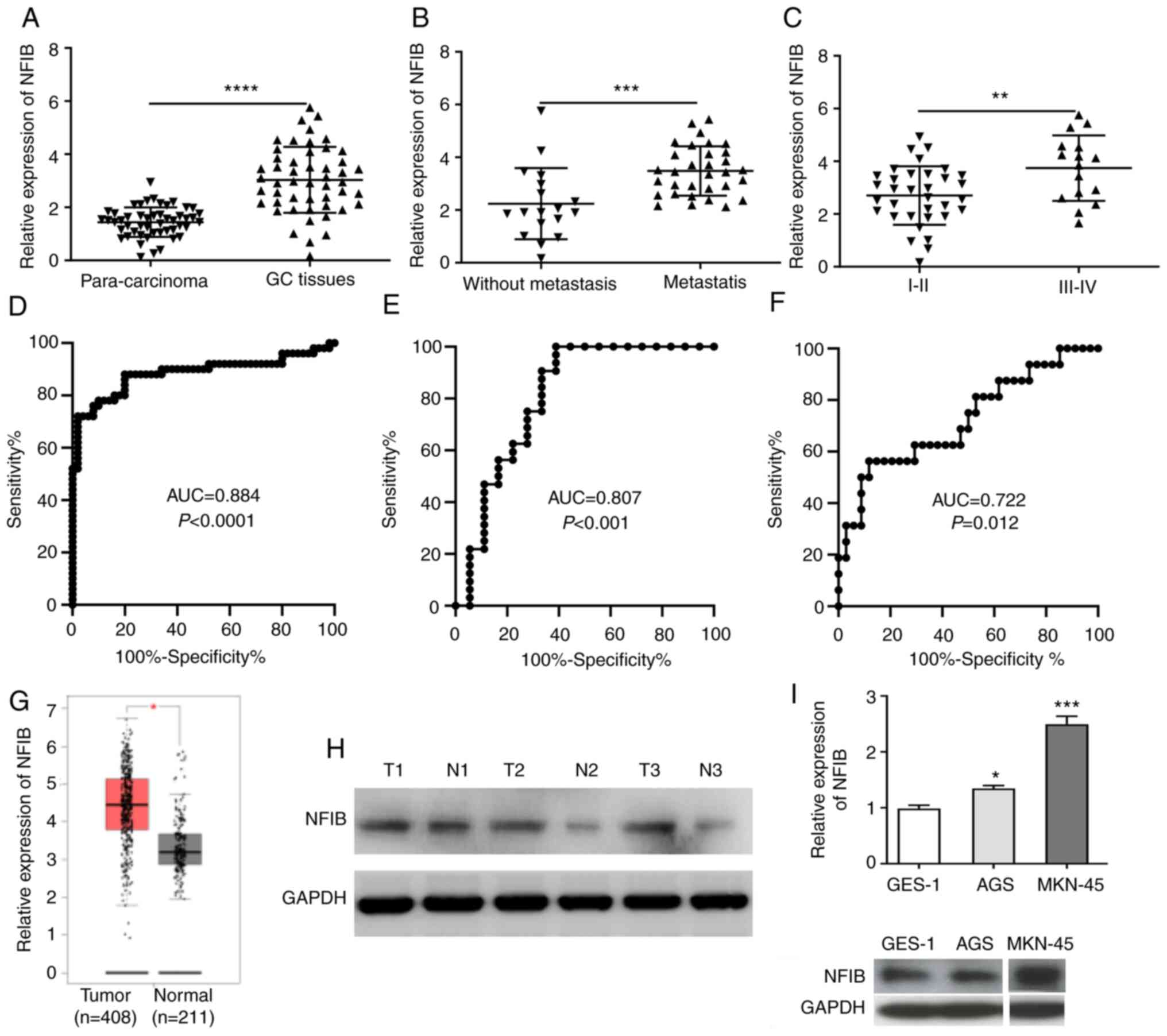 | Figure 1.NFIB expression is upregulated in GC
tissues and cells. (A) Expression levels of NFIB were examined in
50 GC specimens and matched para-carcinoma tissues by RT-qPCR. (B)
NFIB expression was determined in patients with GC with or without
metastasis. (C) Expression levels of NFIB were detected in patients
with GC at different grades. Receiver operating characteristic
curve analyses revealed that the expression profile of NFIB
exhibited high AUC values in distinguishing (D) between GC and
para-carcinoma samples, (E) between patients with and without
metastasis, and (F) between grade III–IV and I–II GC. (G)
Upregulation of NFIB was also revealed by Gene Expression Profiling
Interactive Analysis. (H) Elevated NFIB expression was observed in
GC samples using western blot analysis. (I) RT-qPCR and western
blotting indicated that NFIB mRNA and protein expression levels
were upregulated in GC cell lines compared with in the normal human
gastric epithelial cell line GES-1. Two non-relevant bands were
removed from the blots. *P<0.05, **P<0.01, ***P<0.001,
****P<0.0001 vs. GES-1 or as indicated. AUC, area under curve;
GC, gastric cancer; NFIB, nuclear factor I B; RT-qPCR, reverse
transcription-quantitative polymerase chain reaction; T, tumor; N,
normal. |
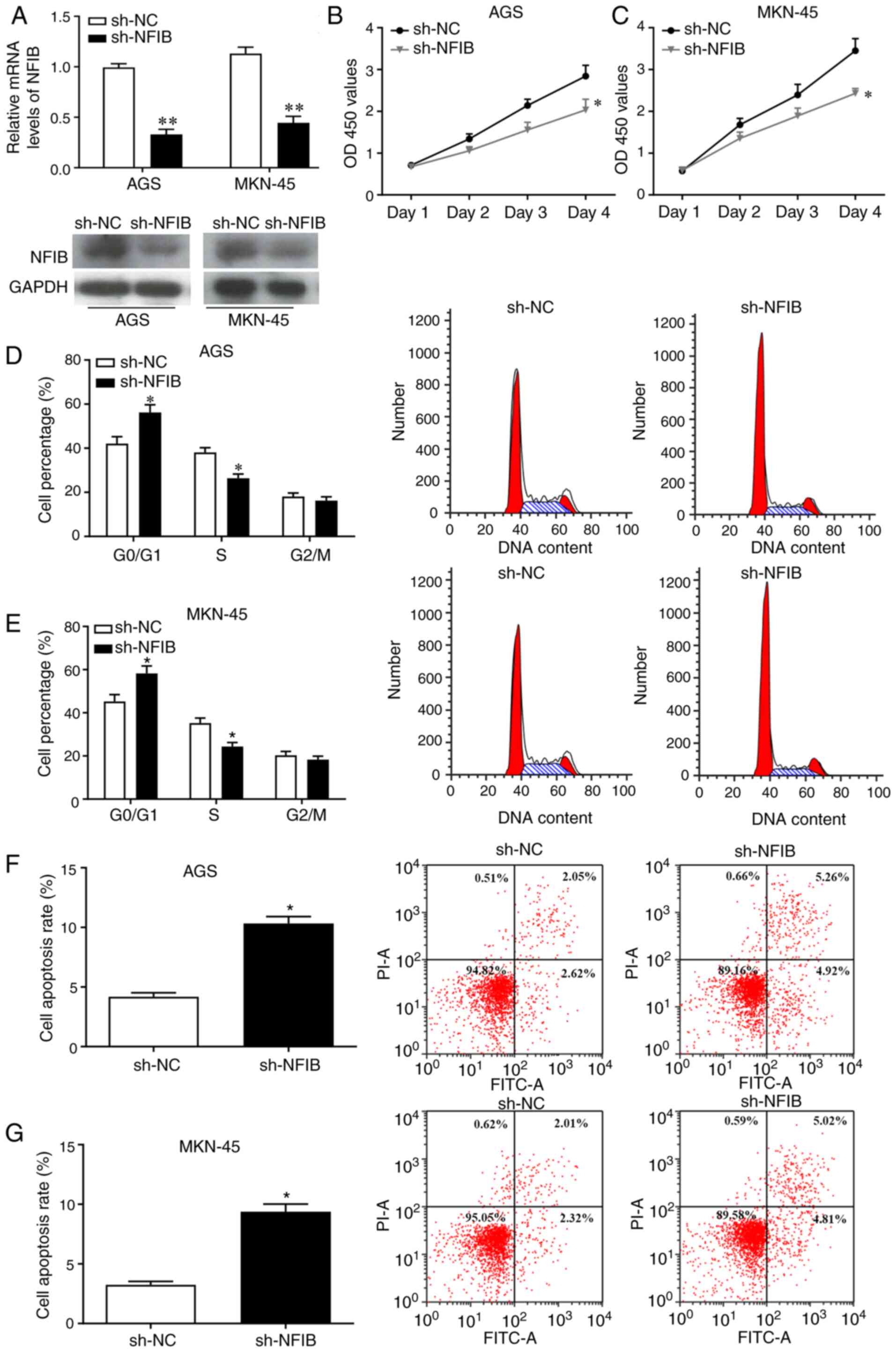
Knockdown of NFIB inhibits the
proliferation of GC cells and promotes apoptosis
To identify the roles of NFIB in tumor progression
of GC, NFIB was knocked down in AGS and MKN-45 cells. Infection
efficiency was confirmed by RT-qPCR and western blotting (Fig. 2A). In addition, CCK-8 assay
suggested that the proliferation of GC cells infected with sh-NFIB
was significantly reduced (Fig. 2B and
C). To further investigate the effects of NFIB on GC cell
proliferation, cell cycle distribution and apoptosis were examined
in GC cells infected with sh-NFIB compared with the control cells.
The results revealed that the percentage of cells in
G0/G1 phase was significantly elevated,
whereas the percentage of cells in S phase was notably reduced
following NFIB knockdown, suggesting that the GC cell cycle was
shifted from S and G2/M phase to
G0/G1 phase (Fig.
2D and E). Moreover, flow cytometry indicated that knockdown of
NFIB enhanced the apoptosis of GC cells (Fig. 2F and G). These results indicated
that knockdown of NFIB may promote cell cycle arrest in
G0/G1 phase and enhance the apoptosis of GC
cells.
circMAP7D1 is a novel target of NFIB
in GC
In order to investigate the mechanisms of
NFIB-mediated signaling, the downstream molecules of NFIB were
predicted using circRNA microarray. The expression profiles of
circRNAs were evaluated in GC cells infected with sh-NC or sh-NFIB.
The expression levels of circMAPK7D1 were markedly downregulated
following NFIB knockdown (Fig. 3A).
Furthermore, RT-qPCR revealed the upregulation of circMAP7D1 in GC
specimens and cells compared with in the normal controls (Fig. 3B and C). RNA pulldown assay
indicated that NFIB was able to interact with circMAP7D1 in GC
cells (Fig. 3D). Furthermore, the
interaction between NFIB and circMAP7D1 was confirmed using a
luciferase assay; overexpressed circMAP7D1 significantly suppressed
luciferase activity of the plasmids containing WT-NFIB, but not the
MUT control (Fig. 3E), indicating
that WT-NFIB could interact with circMAP7D1 in GC cells. In
addition, the expression levels of circMAP7D1 were markedly
decreased in GC cells infected with sh-NFIB (Fig. 3E), and a positive correlation was
observed between NFIB and circMAP7D1 expression in GC tissues
(Fig. 3F). These findings suggested
that circMAP7D1 may be a promising target of NFIB in GC cells.
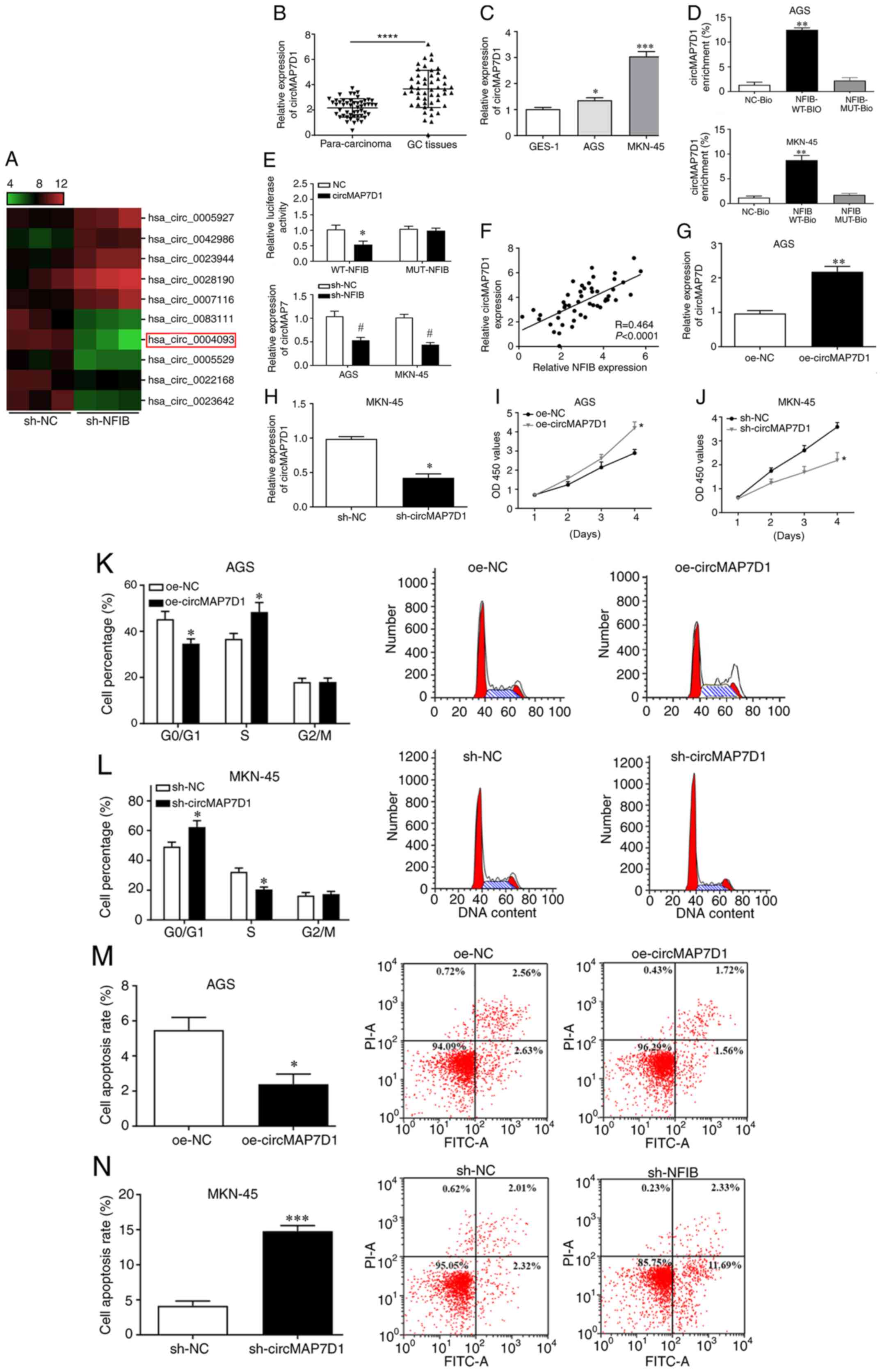 | Figure 3.circMAP7D1 is a promising downstream
molecule of NFIB in GC and is associated with GC cell
proliferation. (A) Expression profiles of circRNAs in GC cells
infected with sh-NC and sh-NFIB were evaluated using a circRNA
microarray. (B) Expression levels of circMAP7D1 were examined in GC
tissues using RT-qPCR. ****P<0.0001 vs. para-carcinoma tissues.
(C) Expression levels of circMAP7D1 were detected in cells using
RT-qPCR. *P<0.05, ***P<0.001, vs. GES-1. (D) RNA pulldown
assay was conducted to evaluate the interaction between NFIB and
circMAP7D1. **P<0.01 vs. NC-Bio. (E) Luciferase activity of
WT-NFIB-treated cells was decreased by circMAP7D1, and the
expression of circMAP7D1 was reduced in GC cells following the
infection with sh-NFIB. *P<0.05 vs. NC, #P<0.05
vs. sh-NC. (F) NFIB and circMAP7D1 expression was positively
correlated in GC specimens. (G) GC cells were transfected with
oe-circMARP7D1A **P<0.01 vs. oe-NC. (H) GC cells were infected
with sh-circMAP7D1. *P<0.05 vs. sh-NC. (I) Cell proliferation
was examined post-transfection with oe-circMARP7D1A. *P<0.05 vs.
oe-NC. (J) Cell proliferation was determined post-infection with
sh-circMAP7D1. *P<0.05 vs. sh-NC. (K) Cell cycle progression and
(M) apoptotic rate of GC cells were determined post-transfection
with oe-circMARP7D1A. *P<0.05 vs. oe-NC. (L) Cell cycle
progression and (N) apoptotic rate of GC cells were evaluated
post-infection with sh-circMAP7D1 using flow cytometry. *P<0.05,
***P<0.001 vs. sh-NC. circRNA, circular RNA; GC, gastric cancer;
MUT, mutant; NC, negative control; PI, propidium iodide; RT-qPCR,
reverse transcription-quantitative polymerase chain reaction; sh,
short hairpin RNA; WT, wild-type. |
circMAP7D1 is involved in the
regulation of GC cell proliferation and apoptosis
To further identify the regulatory roles of
circMAP7D1 in the development of GC, AGS and MKN-45 cells were
transfected with oe-circMARP7D1A and infected with sh-circMAP7D1,
and transfection and infection efficiency was confirmed by RT-qPCR
(Fig. 3G and H). Notably, cell
proliferation was significantly enhanced by oe-circMAP7D1 and
suppressed by sh-circMAP7D1 (Fig. 3I
and J). Moreover, cell cycle arrest and apoptosis were
inhibited post-transfection with oe-circMAP7D1 and were promoted
post-infection with sh-circMAP7D1 (Fig.
3K-N). These results revealed that circMAP7D1 may be involved
in the regulation of GC cell proliferation and apoptosis.
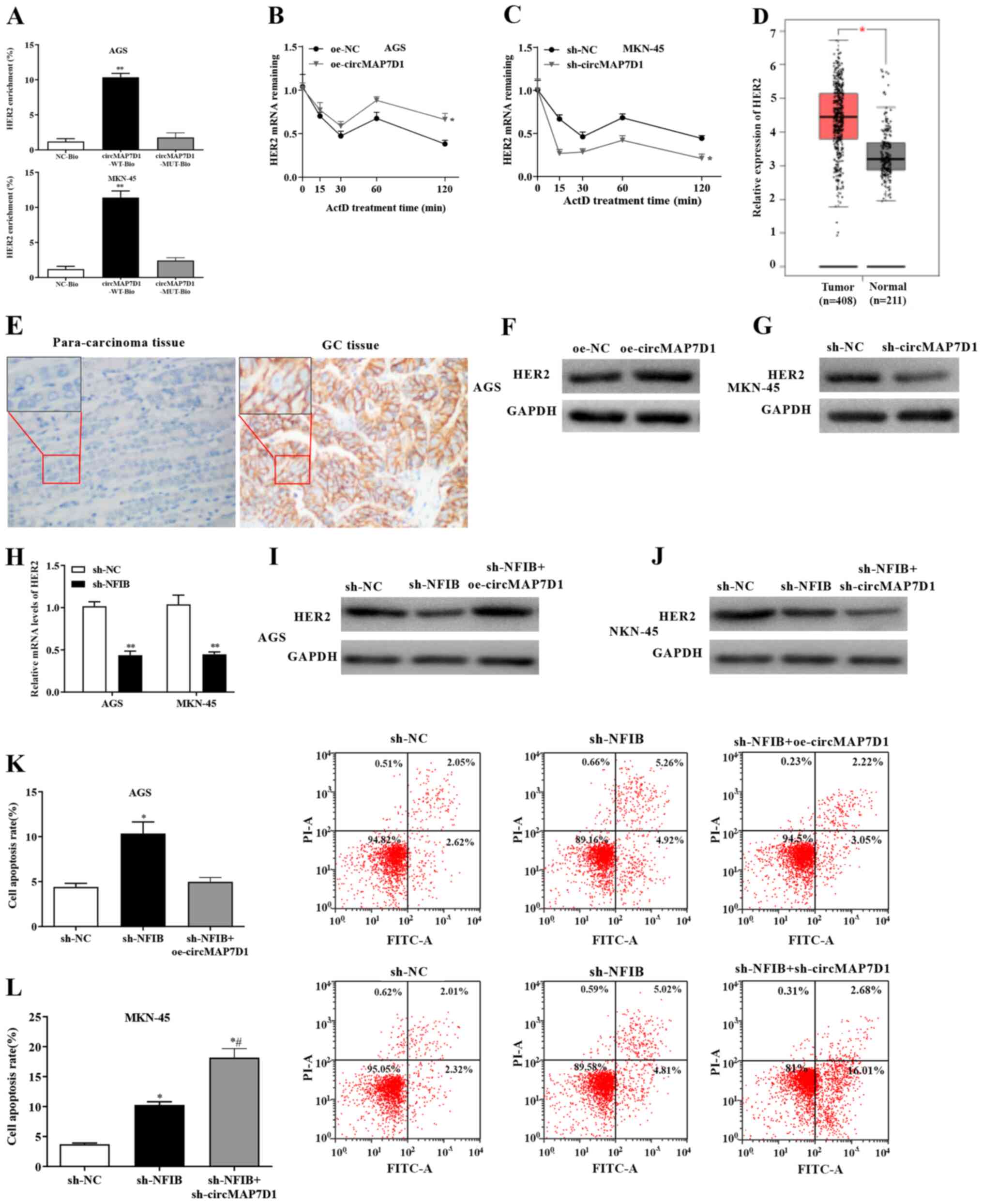
Stability of HER2 mRNA is enhanced by
circMAP7D1
To identify the putative target of circMAP7D1 in GC
cells, a further functional study was performed. The results of RNA
pulldown assay indicated that circMAP7D1 could interact with HER2
in GC cells (Fig. 4A). Briefly,
following pulldown, the crosslinked molecules were identified using
mass spectrometry (data not shown), and HER2 was selected for
further experiments. Notably, overexpression of circMAP7D1
significantly increased the stability of HER2 mRNA, whereas HER2
stability was markedly decreased by the knockdown of circMAP7D1
(Fig. 4B and C). In consistence
with the present results, upregulated levels of HER2 were detected
in GC samples by GEPIA (Fig. 4D).
In addition, IHC indicated that the protein expression levels of
HER2 were markedly increased in GC tissues compared with in
para-carcinoma control tissues (Fig.
4E). Furthermore, the expression levels of HER2 were elevated
in GC cells transfected with oe-circMAP7D1 and reduced
post-infection of cells with sh-circMAP7D1 (Fig. 4F and G). Additionally, the mRNA
expression levels of HER2 were markedly decreased in GC cells
infected with sh-NFIB (Fig. 4H).
These findings indicated that circMAP7D1 may be able to stabilize
HER2 mRNA in GC.
circMAP7D1 is involved in
NFIB-modulated GC cell proliferation and apoptosis
To further identify the roles of circMAP7D1 in the
regulation of GC cell proliferation, a functional analysis was
carried out. Western blotting revealed that the protein expression
levels of HER2 were reduced in GC cells infected with sh-NFIB,
whereas these effects were reversed by overexpression of circMAP7D1
and were strengthened by circMAP7D1 knockdown (Fig. 4I and J). Furthermore, the apoptotic
rates of GC cells were significantly increased post-infection with
sh-NFIB, whereas this effect was abrogated by oe-circMAP7D1 and
enhanced by sh-circMAP7D1 (Fig. 4K and
L). These findings suggested that NFIB was able to promote the
proliferation of GC cells, and that NFIB exerted its regulatory
function by upregulating circMAP7D1, which subsequently stabilized
HER2 mRNA.
Discussion
NFIB is considered to be an oncogenic factor in
tumor progression, and increased levels of NFIB have been detected
in malignancies, such as GC (6–9). In
the present study, upregulation of NFIB was revealed in GC
specimens and cells, and NFIB knockdown markedly suppressed the
proliferation of GC cells. Emerging evidence has indicated that
circRNAs are associated with tumorigenesis in various types of
cancer and dysregulation of circRNAs has also been detected
(13,14,21–25).
Moreover, previous studies have elucidated the important roles of
circRNAs in the pathogenesis of GC (26–28);
however, at present, the detailed function of most circRNAs in GC
are not completely understood.
The present findings revealed that the expression
levels of circMAP7D1 were elevated in GC tissues, and NFIB and
circMAP7D1 expression was positively correlated. Moreover, an RNA
pulldown assay revealed the interaction between NFIB and circMAP7D1
in GC cells. In addition, the inhibitory effects of sh-NFIB on cell
proliferation could be reversed or strengthened by oe-circMAP7D1 or
sh-circMAP7D1, respectively. These findings suggested that
circMAP7D1 may be a promising target of NFIB in GC and could
participate in NFIB-modulated GC cell proliferation.
Recent studies have suggested that upregulation of
HER2 could be associated with the development of various types of
cancer, including GC (15–17). Numerous oncogenes and growth factors
are regulated post-transcriptionally. The key mechanism of
post-transcriptional regulation of oncogenes is through controlling
the rate of mRNA turnover; until now, only a few regulatory factors
have been discovered that could regulate a large pool of target
mRNAs, indicating that a minor perturbation during the controlling
process could lead to marked effects that may result in the
progression of a complex disorder such as cancer (29–31).
The present data revealed the interaction between circMAP7D1 and
HER2 in GC cells, and HER2 mRNA stability was notably increased by
circMAP7D1 overexpression and decreased by circMAP7D1 knockdown.
These findings suggested that HER2 could be a novel downstream
molecule of circMAP7D1 in GC; moreover, NFIB may stabilize HER2
mRNA through upregulating circMAP7D1. It may be hypothesized that
circMAP7D1 could regulate the expression of HER2 through microRNA
sponges, or it may affect the expression of HER2 at the
transcriptional or translational level; this hypothesis requires
further investigation. However, there were limitations in the
present study. For example, cell-counting experiments should be
performed to confirm the effects on proliferation. In future,
further functional studies should be performed to assess the
involvement of circMAP7D1 and HER2 in NFIB-modulated cell cycle
progression and apoptosis.
In conclusion, the present study provided
substantial evidence on the role of NFIB in tumorigenesis and
revealed that NFIB was a novel oncogenic factor during the
pathogenesis of GC. NFIB could promote the proliferation of GC
cells by upregulating circMAP7D1 to stabilize HER2 mRNA,
consequently inducing the tumor progression of GC. Therefore, NFIB
could be considered a putative candidate for targeted therapy of
patients with GC.
Acknowledgements
Not applicable.
Funding
The present study was funded by the Science and
Technology Research Project of Chongqing Education Commission
(grant no. KJQN201900104).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
YJ designed the present study. HY, ZW, XL, MC and XZ
performed the experiments and data analysis. YJ and HY confirm the
authenticity of all the raw data. All authors drafted the
manuscript, and read and approved the final manuscript.
Ethics approval and consent to
participate
The experimental protocol was approved by the
Medical Ethics Committee of the Affiliated Hospital of North
Sichuan Medical College. Written informed consent was obtained from
all patients.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Lyons K, Le LC, Pham YTH, Borron C, Park
JY, Tran CTD, Tran TV, Tran HTT, Vu KT, Do CD, et al: Gastric
cancer: Epidemiology, biology, and prevention: A mini review. Eur J
Cancer Prev. 28:397–412. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Allemani C, Weir HK, Carreira H, Harewood
R, Spika D, Wang XS, Bannon F, Ahn JV, Johnson CJ, Bonaventure A,
et al: Global surveillance of cancer survival 1995–2009: Analysis
of individual data for 25,676,887 patients from 279
population-based registries in 67 countries (CONCORD-2). Lancet.
385:977–1010. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Park YM, Cho E, Kang HY and Kim JM: The
effectiveness and safety of endoscopic submucosal dissection
compared with endoscopic mucosal resection for early gastric
cancer: A systematic review and metaanalysis. Surg Endosc.
25:2666–2677. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Choi MK, Kim GH, Park DY, Song GA, Kim DU,
Ryu DY, Lee BE, Cheong JH and Cho M: Long-term outcomes of
endoscopic submucosal dissection for early gastric cancer: A
single-center experience. Surg Endosc. 27:4250–4258. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Takahashi T, Saikawa Y and Kitagawa Y:
Gastric cancer: Current status of diagnosis and treatment. Cancers
(Basel). 5:48–63. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Becker-Santos DD, Lonergan KM,
Gronostajski RM and Lam WL: Nuclear factor I/B: A master regulator
of cell differentiation with paradoxical roles in cancer.
EBioMedicine. 22:2–9. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Dooley AL, Winslow MM, Chiang DY, Banerji
S, Stransky N, Dayton TL, Snyder EL, Senna S, Whittaker CA, Bronson
RT, et al: Nuclear factor I/B is an oncogene in small cell lung
cancer. Genes Dev. 25:1470–1475. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Nanda JS, Awadallah WN, Kohrt SE, Popovics
P, Cates JM, Mirosevich J, Clark PE, Giannico GA and Grabowska MM:
Nuclear factor I/B increases in prostate cancer to support androgen
receptor activation. BioRxiv. Jun 27–2019.(Epub ahead of print).
doi: 10.1002/pros.24019.
|
|
9
|
Wu C, Zhu X, Liu X, Ruan T, Wan W and Tao
K: NFIB promotes cell growth, aggressiveness, metastasis and EMT of
gastric cancer through the Akt/Stat3 signaling pathway. Oncol Rep.
40:1565–1573. 2018.PubMed/NCBI
|
|
10
|
Memczak S, Jens M, Elefsinioti A, Torti F,
Krueger J, Rybak A, Maier L, Mackowiak SD, Gregersen LH, Munschauer
M, et al: Circular RNAs are a large class of animal RNAs with
regulatory potency. Nature. 495:333–338. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
You X, Vlatkovic I, Babic A, Will T,
Epstein I, Tushev G, Akbalik G, Wang M, Glock C, Quedenau C, et al:
Neural circular RNAs are derived from synaptic genes and regulated
by development and plasticity. Nat Neurosci. 18:603–610. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Su M, Xiao Y, Ma J, Tang Y, Tian B, Zhang
Y, Li X, Wu Z, Yang D, Zhou Y, et al: Circular RNAs in cancer:
Emerging functions in hallmarks, stemness, resistance and roles as
potential biomarkers. Mol Cancer. 18:902019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Sun H, Xi P, Sun Z, Wang Q, Zhu B, Zhou J,
Jin H, Zheng W, Tang W, Cao H and Cao X: Circ-SFMBT2 promotes the
proliferation of gastric cancer cells through sponging miR-182-5p
to enhance CREB1 expression. Cancer Manag Res. 16:5725–5734. 2018.
View Article : Google Scholar
|
|
14
|
Zhang Y, Liu H, Li W, Yu J, Li J, Shen Z,
Ye G, Qi X and Li G: CircRNA_100269 is downregulated in gastric
cancer and suppresses tumor cell growth by targeting miR-630. Aging
(Albany NY). 9:1585–1593. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Iqbal N and Iqbal N: Human epidermal
growth factor receptor 2 (HER2) in cancers: Overexpression and
therapeutic implications. Mol Biol Int. 2014:8527482014. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yan M, Schwaederle M, Arguello D, Millis
SZ, Gatalica Z and Kurzrock R: HER2 expression status in diverse
cancers: Review of results from 37,992 patients. Cancer Metastasis
Rev. 34:157–164. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Abrahao-Machado LF and Scapulatempo-Neto
C: HER2 testing in gastric cancer: An update. World J
Gastroenterol. 22:4619–4625. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Hu J and He X: Enhanced quantile
normalization of microarray data to reduce loss of information in
gene expression profiles. Biometrics. 63:50–59. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yang Y, Wen L and Zhu H: Unveiling the
hidden function of long non-coding RNA by identifying its major
partner-protein. Cell Biosci. 5:592015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lei M, Zheng G, Ning Q, Zheng J and Dong
D: Translation and functional roles of circular RNAs in human
caner. Mol Cancer. 19:302020. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Qiu L, Wang T, Ge Q, Xu H, Wu Y, Tang Q
and Chen K: Circular RNA signature in hepatocellular carcinoma. J
Cancer. 10:3361–3372. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lei B, Tian Z, Fan W and Ni B: Circular
RNA: A novel biomarker and therapeutic target for human cancers.
Int J Med Sci. 16:292–301. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Vo J, Cieslik M, Zhang Y, Shukla S, Xiao
L, Zhang Y, Wu YM, Dhanasekaran SM, Engelke CG, Cao X, et al: The
landscape of circular RNA in cancer. Cell. 176:869–881. 2019.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Kristensen LS, Hansen TB, Venø MT and
Kjems J: Circular RNAs in cancer: Opportunities and challenges in
the field. Oncogene. 37:555–565. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Fang X, Wen J, Sun M, Yuan Y and Xu Q:
CircRNAs and its relationship with gastric cancer. J Cancer.
10:6105–6113. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Shi P, Wan J, Song H and Ding X: The
emerging role of circular RNAs in gastric cancer. Am J Cancer Res.
8:1919–1932. 2018.PubMed/NCBI
|
|
28
|
Zhu Z, Rong Z, Luo Z, Yu Z, Zhang J, Qiu Z
and Huang C: Circular RNA circNHSL1 promotes gastric cancer
progression through the miR-1306-3p/SIX1/vimentin axis. Mol Cancer.
18:1262019. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Perron G, Jandaghi P, Solanki S,
Safisamghabadi M, Storoz C, Karimzadeh M, Papadakis AI, Arseneault
M, Scelo G, Banks RE, et al: A general framework for interrogation
of mRNA stability programs identifies RNA-binding proteins that
govern cancer transcriptomes. Cell Rep. 23:1629–1650. 2018.
View Article : Google Scholar
|
|
30
|
Benjamin D and Moroni C: mRNA stability
and cancer: An emerging link? Expert Opin Biol Ther. 7:1515–1529.
2007. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Sun X, Hu Y, Wu J, Shi L, Zhu L, Xi PW,
Wei JF and Ding Q: RBMS2 inhibits the proliferation by stabilizing
P21 mRNA in breast cancer. J Exp Clin Cancer Res. 37:2982018.
View Article : Google Scholar : PubMed/NCBI
|