Introduction
Liver cancer is the sixth most common types of
cancer and the fourth leading cause of cancer-associated morality
worldwide (1,2). China had the highest number of primary
liver cancer cases globally, with an incidence rate of 17.8
cases/100,000 inhabitants in 2014 (3). Despite significant progress in liver
cancer therapeutics, the recognition of cancer cell invasion into
the surrounding environment and metastatic spread remains a major
research challenge and clinical problem. Moreover, the underlying
molecular mechanisms that initiate cancer cell invasion and
metastases remain poorly understood. It has been reported that cell
polarity defects, which are associated with cell viability,
motility and adhesion ability, can serve as initiators of cancer
cell invasion and metastatic spread (4–8).
Furthermore, epithelial-mesenchymal transition (EMT), which can
mutually interact with the actin cytoskeleton and cell polarity, is
critical during the early steps of metastasis and invasion
(9–12). These processes appear to be
associated with altered expression of adhesion molecules and
dysregulation of growth factor signaling.
Nerve growth factor (NGF) is a prototypical
neurotrophic factor that is essential for neuronal cell growth and
survival (13,14). NGF can interact with its receptor
tropomyosin receptor kinase A (TrkA) with a high affinity, whereas
it interacts with p75 neurotrophin receptor (p75NTR) with a low
affinity (15). Binding of NGF to
TrkA results in intracellular signaling and leads to cell
differentiation and survival. Conversely, the interaction of NGF
with p75NTR activates Jun-N-terminal kinase and ceramide to promote
apoptosis (16,17). Although NGF is undetectable in adult
and developing livers, its expression is markedly elevated in liver
cancer (18,19). In recent years, several studies have
reported that NGF, together with TrkA and p75NTR, are involved in
aspects of tumor biology, including growth, invasion and metastasis
(20–23). NGF has also been implicated as a
marker of tumor progression and is a potential target for novel
therapeutic approaches in cancer (24,25).
HepG2 cells are non-tumorigenic cells with high
proliferation rates and have been used to evaluate cell polarity
and motility as an in vitro model in several studies
(26,27). The present study generated a
NGF-overexpressing HepG2 cell line to investigate the functional
potential of NGF in liver cancer, and subsequently examined the
regulatory mechanism of NGF on cell motility, polarity and EMT, as
well as the underlying effects on cytoskeleton rearrangements and
apoptosis. The present results could elucidate the possible role of
NGF in hepatic carcinogenesis and provide novel insights into the
treatment of liver cancer.
Materials and methods
Cell culture
The HepG2 cell line used in this study was purchased
from the China Center for Type Culture Collection and was
authenticated by short tandem repeat profiling. The cells were
cultured in DMEM containing 10% FBS (Thermo Fisher Scientific,
Inc.), 100 U/ml penicillin G and 100 µg/ml streptomycin at 37°C
with 5% CO2 and 95% O2.
Plasmid transfections
Lipofectamine® 2000 (Invitrogen; Thermo
Fisher Scientific, Inc.) was used to transfect cells according to
the manufacturer's protocol. In brief, 4 µg pcDNA3 vector
(pcDNA3-control) or pcDNA3-NGF plasmid (gift from Professor Philip
Barker, McGill University, Montreal, Canada) was mixed with 10 µl
Lipofectamine for 20 min at room temperatures and the mixture was
transfected into 90% confluent HepG2 cells for 1 h. The transfected
cells were cultured at 37°C in DMEM for 6 h, and then in DMEM with
10% FBS for 48 h at 37°C with 5% CO2 and 95%
O2. To select stable transfectants, cells were cultured
at 37°C in DMEM with 600 µg/ml G418 for 4 weeks to generate a
stable NGF-overexpressing HepG2 cell line. After a single colony of
stable cells was selected for further culture, the concentration of
G418 was subsequently reduced by half and maintained in
cultivation. One pcNA3-control and two different pcDNA3-NGF stable
cell lines were selected for subsequent studies.
Wound healing assays
In brief, cultured cells with DMEM containing 10%
FBS were grown to 100% confluence on plastic dishes or coverslips
(for microscopic studies) and scratched using a 10 µl pipette tip.
Debris was removed from the wound and washed out with PBS. The
cells were then cultured with DMEM containing 10% FBS and the
images were acquired at 0, 24 and 48 h using an inverted light
microscope (IX83; Olympus Corporation; magnification, ×10) after
cells were wounded. Wound closure was quantitatively analyzed using
ImageJ Fiji software (version 1.53g 4; National Institutes of
Health). Each test was performed in triplicate. A total of 10 mg/l
CEP701 (Sigma-Aldrich Merck KGaA) was added after cells were
scratched and maintained in the culture medium at 37°C until images
at different time points were acquired.
Golgi reorientation polarity
assays
As previously described (28,29),
the wounded cells were fixed with cold 4% paraformaldehyde at 4°C
for 10 min and stained with the cis-Golgi matrix protein of 130 kDa
(GM130) to visualize Golgi positioning after 16 h. A total of 7
µg/ml anti-GM130 antibody (cat. no. ab169276; Abcam) were incubated
with cells at 4°C overnight. The appropriate secondary antibody
conjugated with rhodamine were incubated for 1 h at room
temperature. Then, 4′6-diamidino-2-phenyl-indole (DAPI) staining
was performed as previously described (29). Cell images were acquired using a
Nikon TE2000S fluorescence microscope (magnification, ×20) and were
analyzed using ImageJ Fiji software (version 1.53g 4; National
Institutes of Health). Cell orientation was determined only for
cells at the wound edge. The cell was divided into three 120°
regions, with one region facing the wound edge. The cell was
recognized to possess an aligned Golgi only when its Golgi
realigned to the 120° region facing the wound edge. The cell
positioning angle was calculated between a line along the long axis
of the nucleus and a line tracing the wound front. For example,
cells aligned perpendicular to the leading edge demonstrated a
nearly 90° orientation, whereas cells aligned parallel to the wound
front had a 0° orientation. For each experiment, ≥20 cells were
examined.
Western blot analysis
Whole-cell protein was extracted with cell lysis
buffer (Cell Signaling Technology, Inc.). Protein concentrations
were determined via bicinchoninic acid protein assay kit (Pierce;
Thermo Fisher Scientific, Inc.). A total of 50 µg protein from
whole-cell lysates were solubilized in SDS sample buffer and
separated on SDS 12.5% polyacrylamide gels. The proteins were
transferred to polyvinylidene difluoride membranes and incubated
with blocking solution (Tris buffer containing 0.1% Tween-20 and 5%
non-fat dry milk) at room temperature for 1 h. The membrane was
then incubated with the primary antibody at 4°C overnight and the
secondary antibody at room temperature for 1 h. The primary
antibodies against NGF (1:1,000; cat. no. sc32300; Santa Cruz
Biotechnology Inc.), E-cadherin (1:1,000; cat. no. 14472; Cell
Signaling Technology, Inc.), N-cadherin (1:1,000; cat. no. 4061;
Cell Signaling Technology, Inc.), vimentin (1:1,000; cat. no. 3932;
Cell Signaling Technology, Inc.), F-actin (1:1,000; cat. no.
ab130935; Abcam) and β-actin (1:1,000; cat. no. sc69879; Santa Cruz
Biotechnology Inc.) were used for different proteins with
horseradish peroxidase (HRP)-conjugated secondary antibodies (all
1:2,000; cat. nos. ab205718, ab205719 and ab205720; all Abcam).
β-actin protein was detected as a loading control for whole-cell
protein. An enhanced chemiluminescent substrate for detection of
HRP (cat. no. 32209; Thermo Fisher Scientific, Inc.) was used to
visualize the bands with ChemiDoc imaging system (Bio-Rad
Laboratories, Inc.). The bands were analyzed with ImageJ Fiji
software (version 1.53g 4; National Institutes of Health).
Immunofluorescence and confocal
imaging
Briefly, HepG2 and HepG2-NGF cells were plated onto
sterile coverslips and incubated in a humidified chamber at 37°C. A
total of 10 mg/l CEP701 (Sigma-Aldrich; Merck KGaA) was added at
37°C 24 h before fixation. After 24 h, the cells were washed, fixed
with cold 4% paraformaldehyde at 4°C for 10 min and permeabilized
with 0.2% Triton X-100. After each experiment, cells were washed
three times for 5 min in PBS, then blocked with 5% BSA (Abcam) in
PBS for 30 min at room temperature and incubated with anti-NGF
(1:250; cat. no. ab52918; Abcam) or 5 µg/ml anti-F-actin antibody
(cat. no. ab130935; Abcam) at 4°C overnight. Cells were then
treated with 10 µg/ml corresponding secondary antibody conjugated
with FITC (cat. no. F-2765) or rhodamine (cat. no. R-6393; both
Invitrogen; Thermo Fisher Scientific, Inc.) for 1 h at room
temperature. Nuclear staining was performed by incubating cells
with 0.4 µmol/l DAPI for 2 min at room temperature. Subsequently,
cells were examined under a confocal microscope with 10× or 60× oil
objectives (Olympus Corporation).
Anoikis assay
The anoikis assay was performed as described by
Frisch and Francis (30) by plating
cells into ultra-low attachment plates. Cells were plated at a
density of 100×106 cells, onto 60-mm polyHEMA (10
mg/ml)-coated Petri dishes. After culturing for 24 h, images were
obtained using an inverted light microscope (cat. no. IX83; Olympus
Corporation; magnification, ×10) and cells were collected for flow
cytometry (Attune NxT; Thermo Fisher Scientific, Inc.). BD FACS
Diva software 6.0 (BD Biosciences) was used to analysis the
apoptosis ratio. In order to investigate the effect of CEP701, the
cells were treated at 37°C with 10 mg/l CEP701 for 24 h and images
were acquired.
Statistical analysis
Data are presented as the mean ± SEM (n≥3) P-values
were calculated using an ordinary one-way ANOVA, which was followed
by a Tukey's test. P<0.05 was considered to indicate a
statistically significant difference.
Results
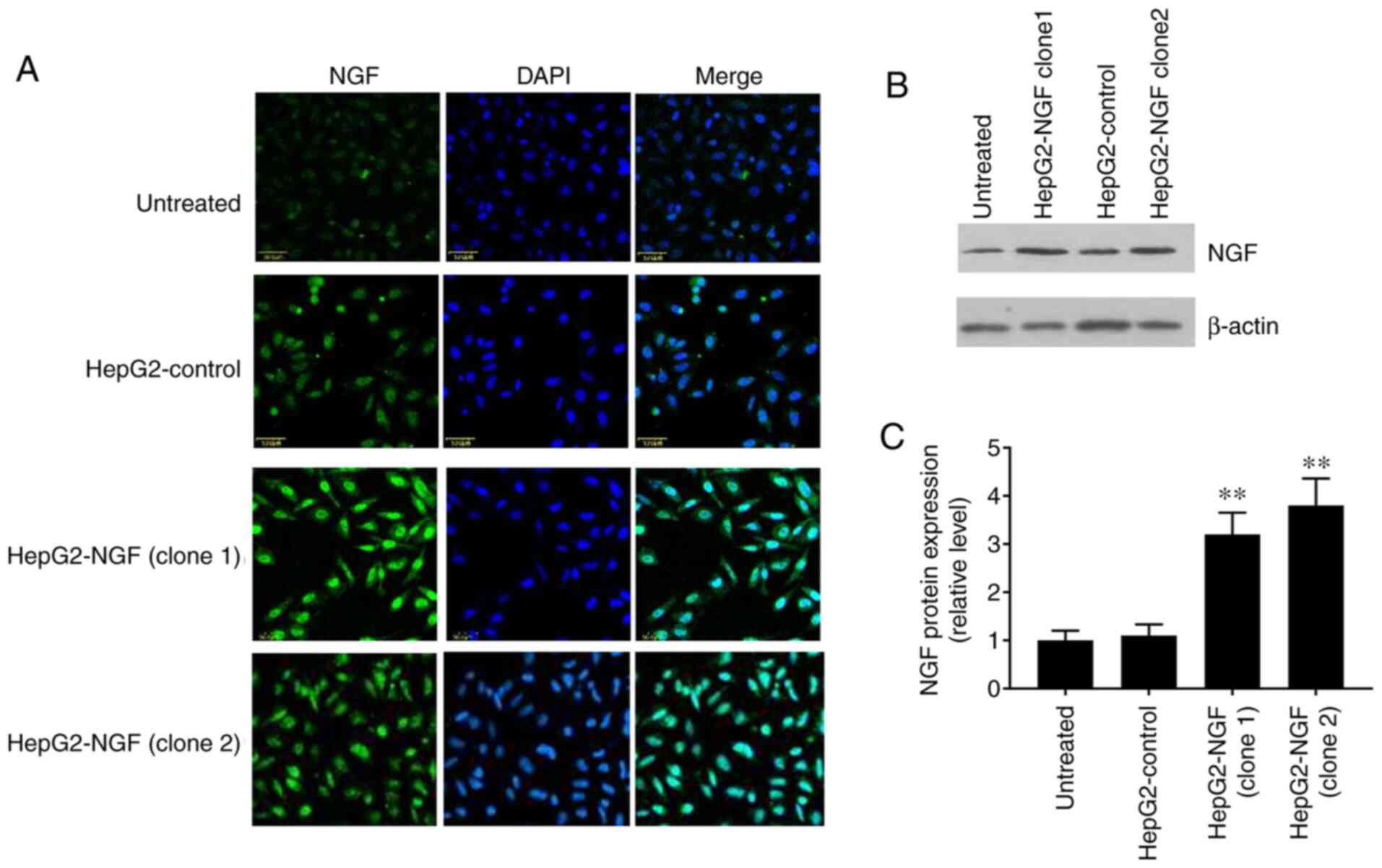
NGF expression in HepG2-pcDNA3-NGF
cells
The pcDNA3-control and pcDNA3-NGF were stably
transfected into HepG2 cells and NGF expression was detected via
western blotting and immunofluorescence. The fluorescence intensity
level of NGF was notably higher in the two NGF-overexpressing HepG2
clones (HepG2-NGF clone 1 and clone 2) compared with that in the
uninfected cells or pcDNA3-control cells (HepG2-control) (Fig. 1A). Furthermore, western blotting
demonstrated that the NGF protein expression level in HepG2-NGF
cells was increased by >3 fold when compared with that observed
in the control group (HepG2-control) (Fig. 1B and C). These results indicated
that the NGF was successfully transfected into HepG2 cells.
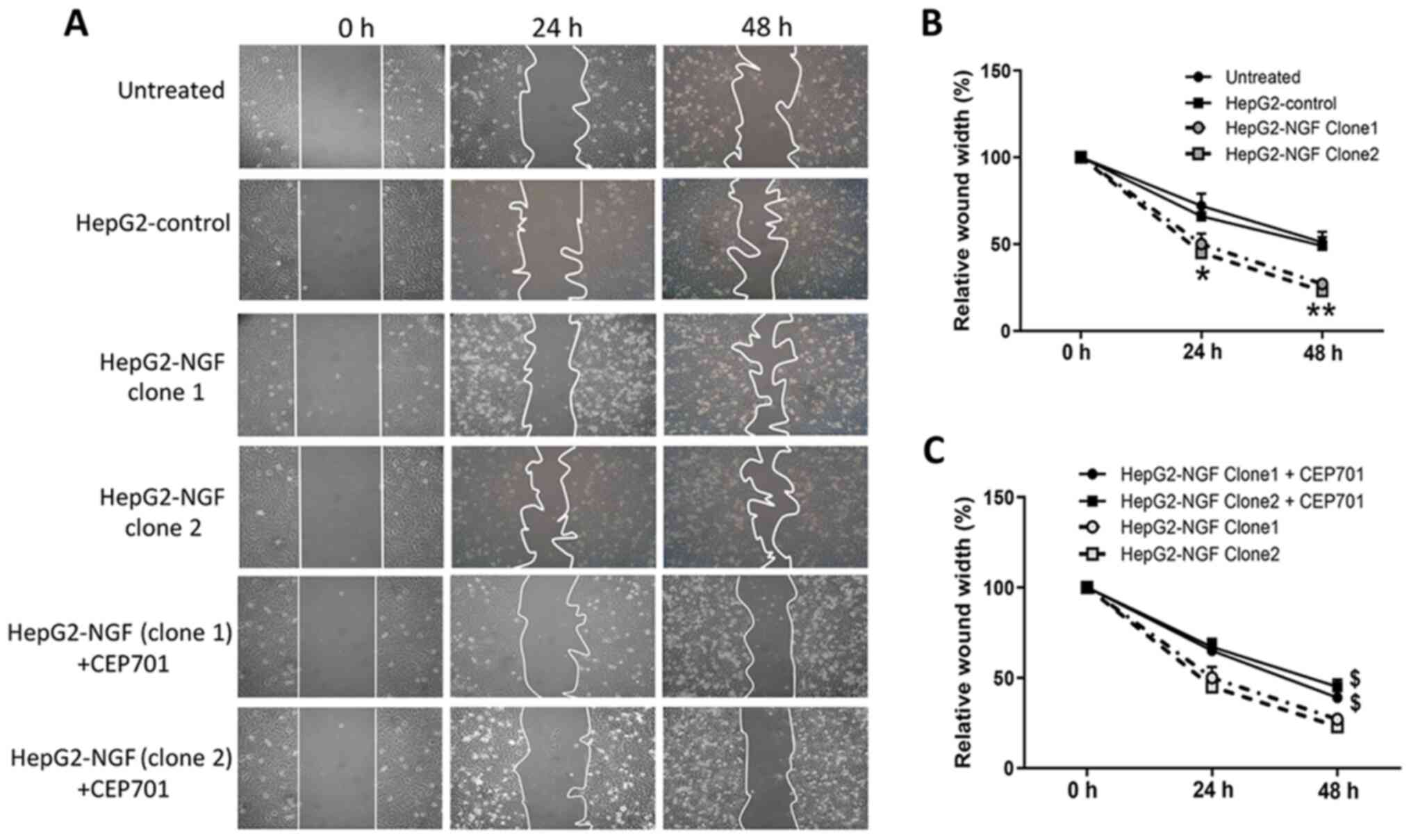
Effect of NGF regulation on cell
motility and polarity
The effects of NGF on cell motility and polarity
were subsequently examined after establishing the HepG2-NGF stable
cell line. As presented in Fig. 2,
48 h after the cells were scratched, the relative wound width of
control cells was ~50% of the original scratch width, compared with
27 and 23% in the two different HepG2-NGF clones, indicating that
NGF overexpression in both HepG2-NGF cell clones can significantly
promote HepG2 cell motility.
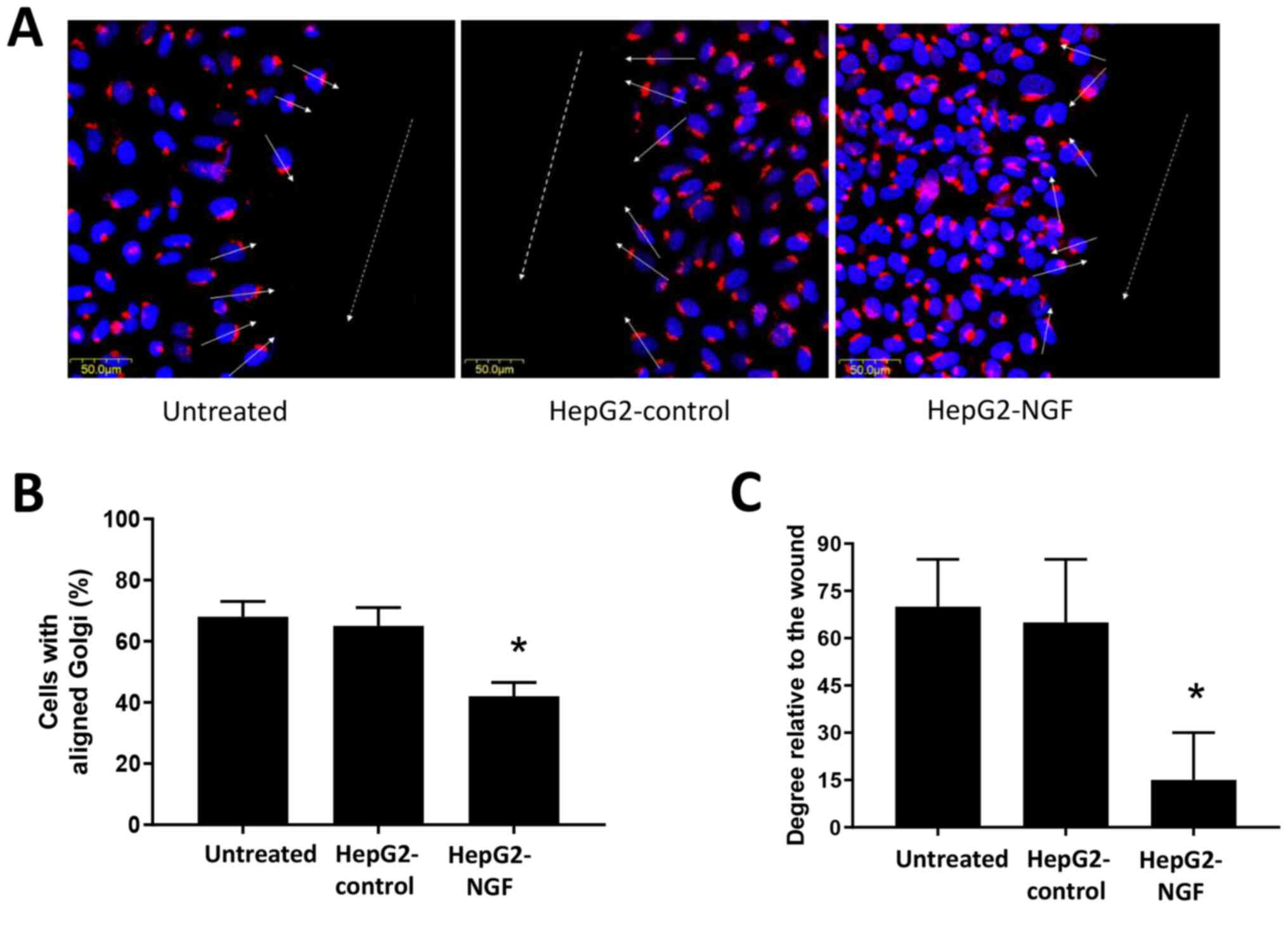
The Golgi serves an important role in protein
trafficking to the leading cell edge and can function as a cell
polarization marker (29,31,32).
Therefore, Golgi reorientation was examined in HepG2-NGF cells when
cell polarity was stimulated. A total of 16 h after cells were
scratched, they were fixed and stained for the protein GM130. The
majority of untreated HepG2 cells and pcDNA3-control cells were
polarized in a direction perpendicular to the wound (the average
orientation was nearly 70° to the wound). Moreover, ~70% of
untreated HepG2 cells and pcDNA3-control cells demonstrated proper
orientation (reoriented in front of the nucleus) and were realigned
to the 120° region facing the direction of movement after
scratching was performed (Fig. 3).
By contrast, HepG2-NGF cells presented only 42% of cells with
proper Golgi positioning and only a 30° orientation relative to the
wound, indicating defective cell polarity after NGF overexpression
in these cells (Fig. 3).
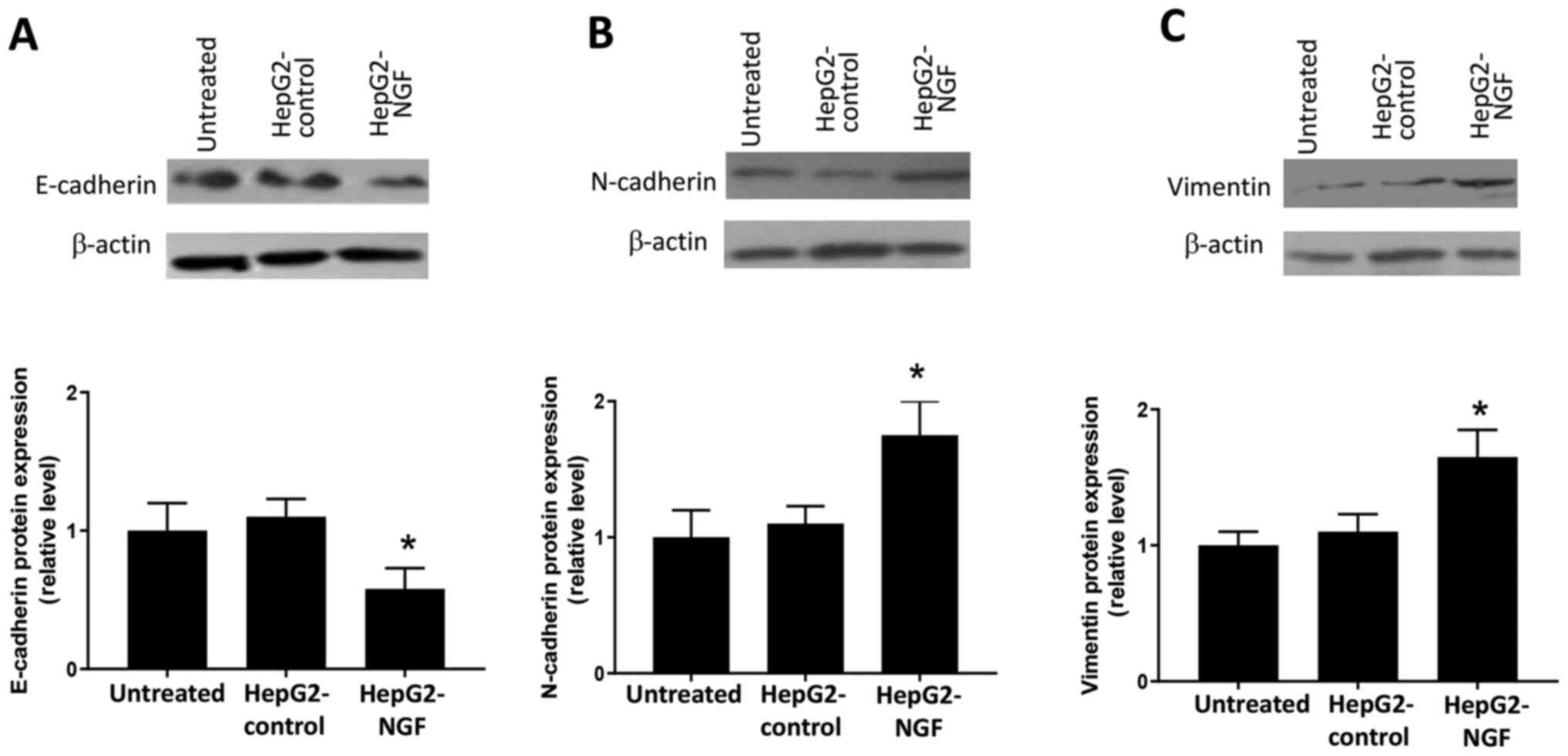
NGF overexpression initiates EMT
In cancer cells, loss of the apical-basal polarity
and acquisition of the migratory phenotype is considered a subtype
of EMT, which is suggested to promote cancer cell migration and
invasion (9–11). Based on the results from Figs. 2 and 3, it was considered that there may be a
potential relationship between NGF and the EMT process. Hence, the
effects of NGF on EMT were subsequently examined. Western blotting
results indicated that NGF overexpression induced the loss of
E-cadherin (Fig. 4A), as well as
the production of N-cadherin (Fig.
4B) and vimentin (Fig. 4C) in
HepG2 cells. These results suggested that NGF could induce a
cadherin switch and initiate EMT in HepG2 cells.
Rearrangement of the actin
cytoskeleton in NGF-overexpressing HepG2 cells
In addition to disrupting cell-cell adhesions and
the overall loss of epithelial homeostasis, the altered functions
of the polarity determinants can result in cytoskeleton
rearrangements and regulate actin dynamics (33,34).
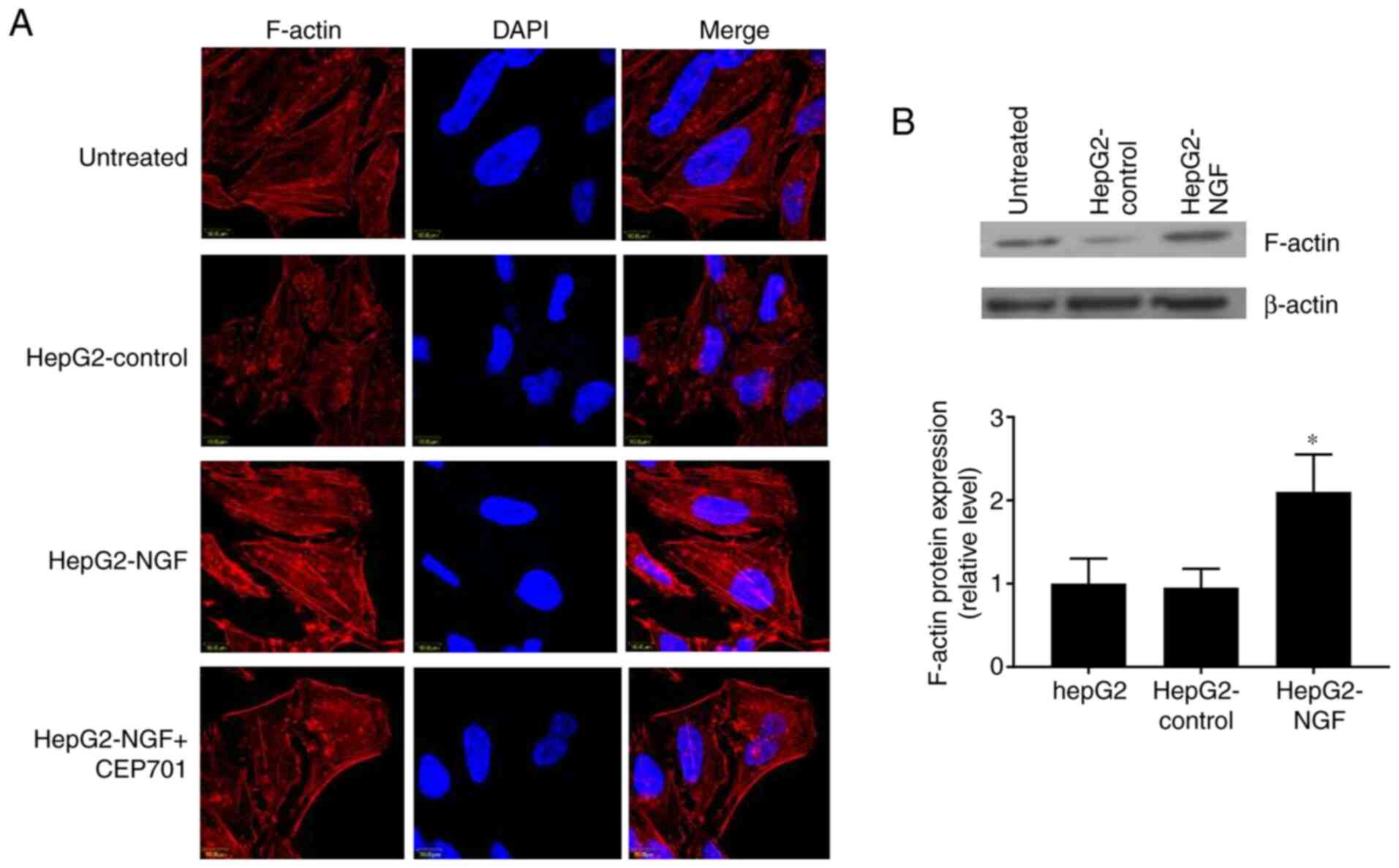
Herein, it was detected whether NGF can affect the F-actin
cytoskeletal arrangement and protein expression. In control cells,
F-actin was organized in a circular pattern and formed
circumferential bundles with visible slim central fibers, as
visualized using immunofluorescence and confocal laser microscopy
(Fig. 5A). However, in HepG2-NGF
cells, F-actin was redistributed into strong central fibers (stress
fibers) and these stress fibers were arranged parallel to the
elongated shape of a cell. Furthermore, the F-actin protein
expression level was increased in NGF-overexpressing HepG2 cells
(Fig. 5B). These results indicated
that NGF overexpression can change the actin cytoskeleton
arrangement in HepG2 cells, even in the absence of stress or
stretch induction.
Effect of NGF overexpression on
anoikis resistance and apoptosis
In a previous study, NGF signaling was reported to
alter cell death and survival in various cancer cells (15,35).
Therefore, the effect of NGF on cell anoikis resistance and
apoptosis was examined in HepG2 cells cultured in a suspension
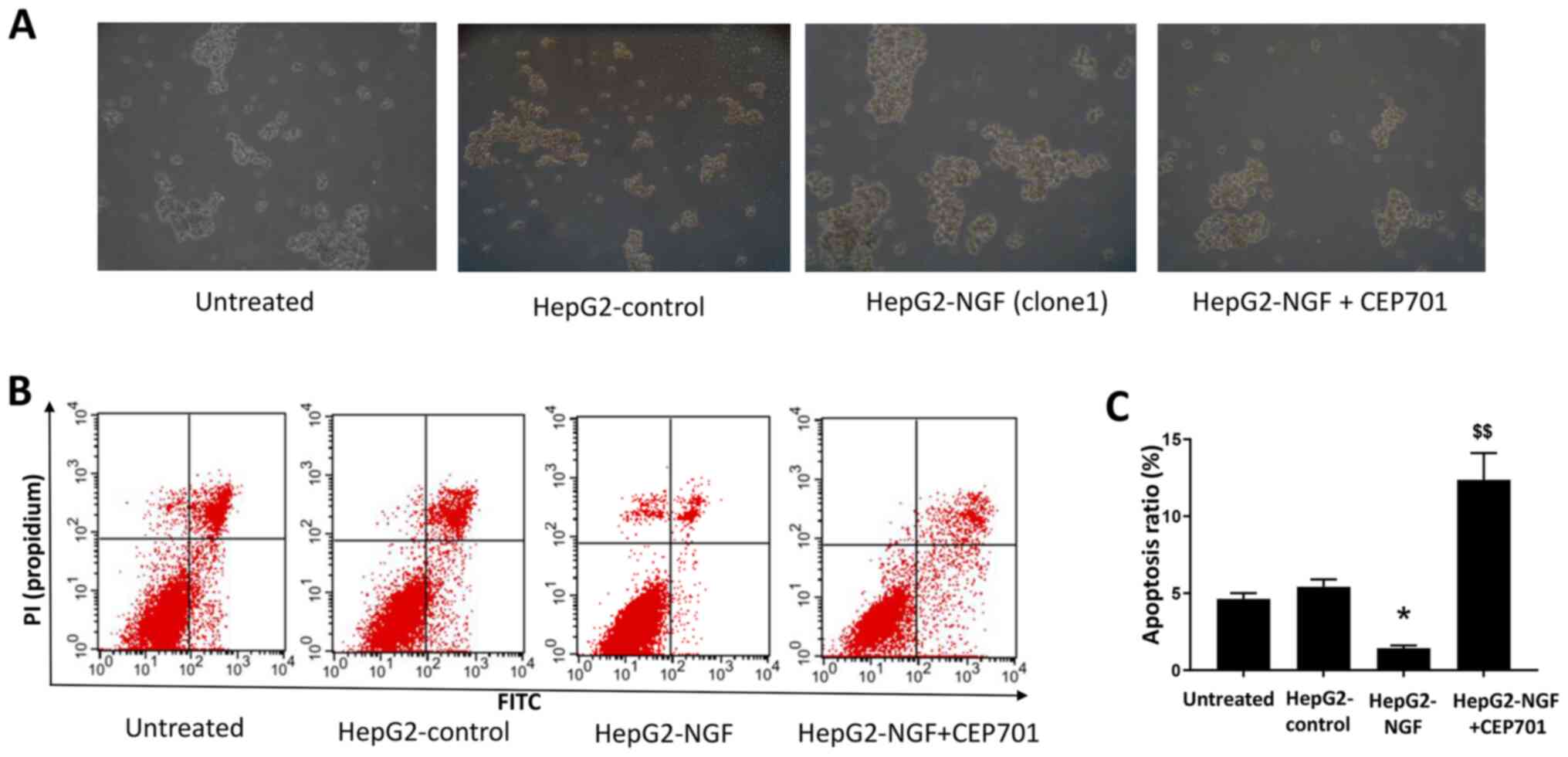
culture model. As presented in Fig.
6, compared with HepG2-control cells, after culturing for 24 h
in ultra-low attachment plates, the diameters of HepG2-NGF cell
colonies were considerably larger and the apoptosis ratios were
lower in HepG2-NGF cells. This indicated that NGF overexpression
could enhance anoikis resistance and prevent apoptosis in HepG2
cells.
Effects of the TrkA receptor inhibitor
CEP701 on cell motility and apoptosis
To determine whether NGF regulates cell motility by
interacting with its receptors, the TrkA receptor inhibitor CEP701
(10 mg/l) was used in wound healing assays in which cells were
scratched, and cell motility was evaluated. A total of 48 h after
cells were wounded, the relative wound width of both HepG2-NGF
clones was considerably higher compared with that of untreated
cells, suggesting that CEP701 can prevent NGF-promoted cell
motility (Fig. 2). Moreover, NGF
overexpression-induced F-actin rearrangement can be prevented by
CEP701 (Fig. 5). The effect of
CEP701 on cell anoikis resistance and apoptosis was further
examined in HepG2-NGF cells cultured in the suspension culture
model. Notably, it was identified that 10 mg/l CEP701 prevented
anoikis resistance and increased the cell apoptosis ratio (Fig. 6).
Discussion
The primary aim of the present study was to
determine the association of NGF, which is reportedly involved in
breast and prostate cancer cell death and survival (15,35),
with liver cancer progression. In the present study, it was
observed that NGF overexpression in HepG2 cells could disrupt cell
polarity and promote cell motility. Additionally, NGF
overexpression could induce EMT and actin cytoskeleton
rearrangement in HepG2 cells. Furthermore, NGF could enhance
anoikis resistance and prevent the apoptosis of HepG2 cells.
Collectively, these data support the hypothesis that NGF signaling
serves a critical role in the invasion and metastasis of liver
cancer.
Cell polarization is required for several cellular
processes, including differentiation, migration, morphogenesis and
motility (33,36). Disruption of cell polarity can
disrupt normal cell behavior, resulting in the cancer initiation
and progression (11,33). Furthermore, disruption of cell
polarity can initiate EMT, which is required for cancer cell
migration and invasion (12,36).
Conversely, EMT can alter the function of polarity complexes and
induce the loss of epithelial polarity (37–39).
Similar to numerous other growth factors and cytokines (40–42),
the current in vitro model data obtained from
scratch-induced migration experiments demonstrated that NGF
overexpression in HepG2 cells could promote cell motility and
induce defective cell polarity. Consistent with these findings, NGF
could induce a cadherin switch and vimentin expression in HepG2
cells, indicating that NGF can initiate EMT. To the best of our
knowledge, the present study was the first to report that NGF was
involved in liver cancer progression, especially in cancer cell
invasion and metastasis, in addition to cell death and
survival.
Previously, it has been reported that cell polarity
proteins can regulate actin dynamics and cytoskeleton organization
(33,34). Conversely, increasing evidence
suggests that the cytoskeleton can regulate cell polarity and
provide the structural design and mechanical strength necessary for
EMT (38,39). Consistent with the polarity defect
and EMT initiation in NGF-overexpressing HepG2 cells, the present
study demonstrated that NGF overexpression could also induce
F-actin redistribution and actin cytoskeleton development from
circumferential bundles (circular actin pattern) to a system of
parallel stress fibers. This rearrangement of the actin
cytoskeleton is a prerequisite for cancer cell migration and
invasion (39).
The mechanism via which NGF overexpression in HepG2
cells induces the loss of the apical-basal polarity and acquires
the migratory phenotype remains unknown. Reportedly, neurotrophins,
their receptors Trk and p75NTR and related signaling pathways serve
an important role in the development of digestive cancer types
(19,24). Moreover, Zhou et al (43) revealed that NGF receptor knockdown
can elevate the expression level of endogenous p53 and result in
hepatoprotective effects in HepG2 cells, while Indo5, which can
inhibit the kinase activities of TrkA and TrkB in HepG2 cells, can
suppress the growth of liver cancer (44). Although the expression of NGF is
undetectable in healthy hepatocytes, NGF and Trk mRNA expression
levels are significantly elevated in the liver tissue of the
majority of patients with liver cancer, as well as in metastatic
liver cancer cell lines, compared with those in healthy tissues or
cell lines (18,25). In agreement with previous studies
(45,46), the present study observed that a
TrkA receptor inhibitor can prevent NGF-induced cell motility,
F-actin rearrangement and anoikis resistance, thus supporting an
autocrine role for NGF signaling via its receptors in hepatocytes
(47). However, the present study
only used one exogenous overexpression system HepG2 cell line to
investigate the role of NGF and its receptor in liver cancer
progression. Due to this limitation, to use a different liver
cancer cell line or primary cultured hepatocytes as another
experimental model, to knockdown NGF or transfect a mutated NGF in
HepG2 cell line or to study the detailed signaling downstream of
NGF/Trk will help to clarify the role of NGF in regulating the cell
motility and polarity.
In conclusion, the present study demonstrated that
NGF overexpression could induce defective liver cancer cell
polarity, EMT initiation and cell cytoskeleton rearrangement, which
are required for tumor progression. The use of NGF as a biomarker
or potential new target could lead to the development of new
factors for diagnosis or for improving therapeutic strategies in
liver cancer.
Acknowledgements
Not applicable.
Funding
This study received funding from the Scientific and
Technological Innovation Foundation of Yantian District of Shenzhen
City (grant no. 20190104).
Availability of data and materials
All data generated or analyzed during this study are
included in this published article.
Authors' contributions
HL was involved in study conceptualization, funding
and design, biochemical experiments, data analysis and
interpretation, and study coordination and manuscript preparation.
HH was involved in experimental design, data interpretation and
analysis, and manuscript preparation. YY, WC, SZ and YZ were
involved in the biochemical experiments, and data interpretation
and analysis. All authors read and approved the final version of
the manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
Glossary
Abbreviations
Abbreviations:
|
NGF
|
nerve growth factor
|
|
EMT
|
epithelial-mesenchymal transition
|
|
Trk
|
tropomyosin receptor kinase
|
|
p75NTR
|
p75 neurotrophin receptor
|
References
|
1
|
Nagtegaal ID, Odze RD, Klimstra D, Paradis
V, Rugge M, Schirmacher P, Washington KM, Carneiro F and Cree IA;
WHO Classification of Tumours Editorial Board, : The 2019 WHO
classification of tumours of the digestive system. Histopathology.
76:182–188. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Bray F, Ferlay J, Soerjomataram I, Siegel
RL, Torre LA and Jemal A: Global cancer statistics 2018: GLOBOCAN
estimates of incidence and mortality worldwide for 36 cancers in
185 countries. CA Cancer J Clin. 68:394–424. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zheng R, Qu C, Zhang S, Zeng H, Sun K, Gu
X, Xia C, Yang Z, Li H, Wei W, et al: Liver cancer incidence and
mortality in China: Temporal trends and projections to 2030. Chin
Jl Cancer Res. 30:571–579. 2018. View Article : Google Scholar
|
|
4
|
Fukata M, Nakagawa M and Kaibuchi K: Roles
of Rho-family GTPases in cell polarisation and directional
migration. Curr Opin Cell Biol. 5:590–597. 2003. View Article : Google Scholar
|
|
5
|
Woodham EF and Machesky LM: Polarised cell
migration: Intrinsic and extrinsic drivers. Chin J Cancer Res.
30:25–32. 2014.
|
|
6
|
Royer C and Lu X: Epithelial cell
polarity: A major gatekeeper against cancer? Cell Death Differ.
18:1470–1477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Jung HY, Fattet L, Tsai JH, Kajimoto T,
Chang Q, Newton AC and Yang J: Apical-basal polarity inhibits
epithelial-mesenchymal transition and tumour metastasis by
PAR-complex-mediated SNAI1 degradation. Nat Cell Biol. 21:359–371.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Lee M and Vasioukhin V: Cell polarity and
cancer-cell and tissue polarity as a non-canonical tumor
suppressor. J Cell Sci. 121:1141–1150. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Larue L and Bellacosa A:
Epithelial-mesenchymal transition in development and cancer: Role
of phosphatidylinositol 3′ kinase/AKT pathways. Oncogene.
24:7443–7454. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Tsai JH and Yang J: Epithelial-mesenchymal
plasticity in carcinoma metastasis. Genes Dev. 27:2192–2206. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Yang J and Weinberg RA:
Epithelial-mesenchymal transition: At the crossroads of development
and tumor metastasis. Dev Cell. 14:818–829. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Moreno-Bueno G, Portillo F and Cano A:
Transcriptional regulation of cell polarity in EMT and cancer.
Oncogene. 27:6958–6969. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Lewin GR and Barde YA: Physiology of the
neurotrophins. Annu Rev Neurosci. 19:289–317. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Hetman M and Xia Z: Signaling pathways
mediating anti-apoptotic action of neurotrophins. Acta Neurobiol
Exp (Wars). 60:531–545. 2000.PubMed/NCBI
|
|
15
|
Bradshaw RA, Pundavela J, Biarc J,
Chalkley RJ, Burlingame AL and Hondermarck H: NGF and ProNGF:
Regulation of neuronal and neoplastic responses through receptor
signaling. Adv Biol Regul. 58:16–27. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Yoon SO, Casaccia-Bonnefil P, Carter B and
Chao MV: Competitive signaling between TrkA and p75 nerve growth
factor receptors determines cell survival. J Neurosci.
18:3273–3281. 1998. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Frade JM, Rodríguez-Tébar A and Barde YA:
Induction of cell death by endogenous nerve growth factor through
its p75 receptor. Nature. 383:166–168. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Tokusashi Y, Asai K, Tamakawa S, Yamamoto
M, Yoshie M, Yaginuma Y, Miyokawa N, Aoki T, Kino S, Kasai S, et
al: Expression of NGF in hepatocellular carcinoma cells with its
receptors in non-tumor cell components. Int J Cancer. 114:39–45.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Kishibe K, Yamada Y and Ogawa K:
Production of nerve growth factor by mouse hepatocellular carcinoma
cells and expression of TrkA in tumor-associated arteries in mice.
Gastroenterology. 122:1978–1986. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Garrido MP, Torres I, Avila A, Chnaiderman
J, Valenzuela-Valderrama M, Aramburo J, Oróstica L, Durán-Jara E,
Lobos-Gonzalez L and Romero C: NGF/TRKA decrease miR-145-5p levels
in epithelial ovarian cancer cells. Int J Mol Sci. 21:76572020.
View Article : Google Scholar
|
|
21
|
Faulkner S, Griffin N, Rowe CW, Jobling P,
Lombard JM, Oliveira SM, Walker MM and Hondermarck H: Nerve growth
factor and its receptor tyrosine kinase TrkA are overexpressed in
cervical squamous cell carcinoma. FASEB Bioadv. 2:398–408. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Blondy S, Christou N, David V, Verdier M,
Jauberteau MO, Mathonnet M and Perraud A: Neurotrophins and their
involvement in digestive cancers. Cell Death Dis. 10:1232019.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Yu X, Liu Z, Hou R, Nie Y and Chen R:
Nerve growth factor and its receptors on onset and diagnosis of
ovarian cancer. Oncol Lett. 14:2864–2868. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Demir IE, Tieftrunk E, Schorn S, Friess H
and Ceyhan GO: Nerve growth factor & TrkA as novel therapeutic
targets in cancer. Biochim Biophys Acta. 1866:37–50.
2016.PubMed/NCBI
|
|
25
|
Berretta M, Cavaliere C, Alessandrini L,
Stanzione B, Facchini G, Balestreri L, Perin T and Canzonieri V:
Serum and tissue markers in hepatocellular carcinoma and
cholangiocarcinoma: Clinical and prognostic implications.
Oncotarget. 8:14192–14220. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Han P, Fu Y, Liu J, Wang Y, He J, Gong J,
Li M, Tan Q, Li D, Luo Y, et al: Netrin-1 promotes cell migration
and invasion by down-regulation of BVES expression in human
hepatocellular carcinoma. Am J Cancer Res. 5:1396–1409.
2015.PubMed/NCBI
|
|
27
|
Yan W, Han P, Zhou Z, Tu W, Liao J, Li P,
Liu M, Tian D and Fu Y: Netrin-1 induces epithelial-mesenchymal
transition and promotes hepatocellular carcinoma invasiveness. Dig
Dis Sci. 59:1213–1221. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Etienne-Manneville S and Hall A: Cdc42
regulates GSK-3beta and adenomatous polyposis coli to control cell
polarity. Nature. 421:753–756. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Zhang S, Schafer-Hales K, Khuri FR, Zhou
W, Vertino PM and Marcus AI: The tumor suppressor LKB1 regulates
lung cancer cell polarity by mediating cdc42 recruitment and
activity. Cancer Res. 68:740–748. 2008. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Frisch SM and Francis H: Disruption of
epithelial cell-matrix interactions induces apoptosis. J Cell Biol.
124:619–626. 1994. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yadav S, Puri S and Linstedt AD: A primary
role for golgi positioning in directed secretion, cell polarity,
and wound healing. Mol Biol Cell. 20:1728–1736. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Ravichandran Y, Goud B and Manneville JB:
The Golgi apparatus and cell polarity: Roles of the cytoskeleton,
the Golgi matrix, and Golgi membranes. Curr Opin Cell Biol.
62:104–113. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Piroli ME, Blanchette JO and Jabbarzadeh
E: Polarity as a physiological modulator of cell function. Front
Biosci (Landmark Ed). 24:451–462. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Elias BC, Das A, Parekh DV, Mernaugh G,
Adams R, Yang Z, Brakebusch C, Pozzi A, Marciano DK, Carroll TJ and
Zent R: Cdc42 regulates epithelial cell polarity and cytoskeletal
function during kidney tubule development. J Cell Sci.
128:4293–4305. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Melck D, De Petrocellis L, Orlando P,
Bisogno T, Laezza C, Bifulco M and Di Marzo V: Suppression of nerve
growth factor Trk receptors and prolactin receptors by
endocannabinoids leads to inhibition of human breast and prostate
cancer cell proliferation. Endocrinology. 141:118–126. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Gandalovičová A, Vomastek T, Rosel D and
Brábek J: Cell polarity signaling in the plasticity of cancer cell
invasiveness. Oncotarget. 7:25022–25049. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Fuertes-Alvarez S, Maeso-Alonso L,
Villoch-Fernandez J, Wildung M, Martin-Lopez M, Marshall C,
Villena-Cortes AJ, Diez-Prieto I, Pietenpol JA, Tissir F, et al:
p73 regulates ependymal planar cell polarity by modulating actin
and microtubule cytoskeleton. Cell Death Dis. 9:11832018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Lomakin AJ, Lee KC, Han SJ, Bui DA,
Davidson M, Mogilner A and Danuser G: Competition for actin between
two distinct F-actin networks defines a bistable switch for cell
polarization. Nat Cell Biol. 17:1435–445. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Olson MF and Sahai E: The actin
cytoskeleton in cancer cell motility. Clin Exp Metastasis.
26:273–287. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Witsch E, Sela M and Yarden Y: Roles for
growth factors in cancer progression. Physiology (Bethesda).
25:85–101. 2010.PubMed/NCBI
|
|
41
|
Okamoto M, Koma YI, Kodama T, Nishio M,
Shigeoka M and Yokozaki H: Growth differentiation factor 15
promotes progression of esophageal squamous cell carcinoma via
TGF-β type II receptor activation. Pathobiology. 87:100–113. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
West NR, McCuaig S, Franchini F and Powrie
F: Emerging cytokine networks in colorectal cancer. Nat Rev
Immunol. 15:615–629. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Zhou X, Hao Q, Liao P, Luo S, Zhang M, Hu
G, Liu H, Zhang Y, Cao B, Baddoo M, et al: Nerve growth factor
receptor negates the tumor suppressor p53 as a feedback regulator.
Elife. 5:e150992016. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Luo T, Zhang SG, Zhu LF, Zhang F, Li W,
Zhao K, Wen XX, Yu M, Zhan YQ, Chen H, et al: A selective c-Met and
Trks inhibitor Indo5 suppresses hepatocellular carcinoma growth. J
Exp Clin Cancer Res. 38:1302019. View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Lagadec C, Meignan S, Adriaenssens E,
Foveau B, Vanhecke E, Romon R, Toillon RA, Oxombre B, Hondermarck H
and Le Bourhis X: TrkA overexpression enhances growth and
metastasis of breast cancer cells. Oncogene. 28:1960–1970. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Festuccia C, Muzi P, Gravina GL,
Millimaggi D, Speca S, Dolo V, Ricevuto E, Vicentini C and Bologna
M: Tyrosine kinase inhibitor CEP-701 blocks the NTRK1/NGF receptor
and limits the invasive capability of prostate cancer cells in
vitro. Int J Oncol. 30:193–200. 2007.PubMed/NCBI
|
|
47
|
Tsai MS, Lee PH, Sun CK, Chiu TC, Lin YC,
Chang IW, Chen PH and Kao YH: Nerve growth factor upregulates
sirtuin 1 expression in cholestasis: A potential therapeutic
target. Exp Mol Med. 50:e4262018. View Article : Google Scholar : PubMed/NCBI
|