Introduction
Intracerebral hemorrhage (ICH), a primary hemorrhage
in the parenchymal region, is a debilitating disease causing damage
to the blood-brain barrier (BBB) and cerebral edema (1–3). ICH
not only damages surrounding brain tissues, but can also cause
secondary complications involving pro-inflammatory reactions, such
as the infiltration of inflammatory cells into the central nervous
system and stimulation of microglial cells (4–6).
Upon stimulation of microglial cells and astrocytes,
the inflammatory response leads to the damage of BBB and the onset
of edema in the brain (6).
Subsequently, leukocytes are recruited to the site of injury,
leading to the activation of pro-inflammatory mediators that can be
potentially toxic to neurons (3–7).
Synergistic effects of these events eventually lead to massive
death of neuronal cells and further neurological damages (7). ICH causes both chronic and acute
damages to brain tissues, through primary mechanical injury caused
by hematomas and secondary complications induced by inflammatory
mediators (8). The management of
neuronal damage after ICH onset relies on the prevention of
secondary brain damage (9).
MicroRNAs (miRNAs) are short (18–25 nucleotides in
length) non-coding RNA transcripts evolutionarily conserved in
eukaryotic cells, and are essential in gene regulation (10). By binding to the 3′-untranslated
regions (3′-UTR) of their target genes, miRNAs can suppress gene
expression through translational inhibition or mRNA degradation
(11). In addition, miRNAs play a
critical role in tumorigenesis. miRNAs(miR)-145 and miR-143,
located on chromosome 5 in humans, are downregulated in colon
cancer (12). A previous study
showed that silencing of miR-143 is implicated in the initiation of
immune responses (13). In
addition, a previous study demonstrated that the expression of
miR-143 is suppressed in colonic cancer (14). Moreover, miR-143 dysregulation is
essential for the induction of inflammation in colon cancer
(12–14).
A single nucleotide polymorphism (SNP), rs41291957,
in the sequence of pre-miR-143 has been shown to alter the
transcription and expression of miR-143, thereby participating in
the process of tumorigenesis (15).
However, further studies are required to understand the effect of
this SNP on miR-143 expression. Since miR-143 is part of a
bicistronic cluster of miRNA genes, the SNPs in the promoter of
miR-143 were analyzed and identified, including rs4705341,
rs353293, rs353292 and rs41291957. These SNPs were revealed to be
significantly associated with an increased risk of colorectal
cancer (15,16). By contrast, the SNPs rs3733846,
rs3733845, rs17796757 and rs4705343 played a protective role
against the pathogenesis of colorectal cancer (17). SNP rs41291957 is located in the
promoter region of miR-143 and can reduce the transcription
efficiency of the miR-143 promoter, thereby decreasing the
expression of miR-143 (18).
Toll-like receptor 2 (TLR2) was found to be a direct target of
miR-143 (19). Furthermore, TLR2
expression is associated with the severity of inflammation and may
affect the prognosis of patients with ICH (20,21).
The present study investigated the association between SNP
rs41291957 and the prognosis of patients with ICH.
Materials and methods
Human subjects and sample
collection
A total of 182 patients with ICH (age, 44–77 years;
109 male patients and 73 female patients) were enrolled in the
present study between September 2015 and August 2017. Serum and CSF
samples were collected from all patients. The patients were divided
into two groups based on the prognosis: i) 150 patients with ICH
were discharged from the hospital after treatment and were
allocated to the group of survived patients; and ii) 32 patients
with ICH, who died in the hospital, were allocated to the group of
deceased patients. In addition, the 182 patients were also divided
into three groups based on rs41291957 SNP genotype: i) CC,
presenting a cytosine in both chromosome (n=28), ii) CT, presenting
both variants (n=67) and iii) TT, presents a thymine in both
chromosomes (n=87). All patients with ICH were diagnosed by
pathological examination. The study was approved by The First
Affiliated Hospital of Chongqing Medical University Ethics
Committee. All patients and their families had given the informed
consent.
SNP sequencing
The rs41291957 SNP genotypes in the 182 patients
were determined by PCR and denaturing high performance liquid
chromatography (DHPLC) using the serum samples collected from each
patient. The primers used for PCR amplification with Pfu DNA
polymerase (Promega Corporation) were designed using Primer Premier
5 Oligo™ 6 software (www.premierbiosoft.com/primerdesign). The primer
sequences were as follows: Allele C forward,
5′-AATTACAACAGCCTCTCGG-3′; allele T forward,
5′-GAATTACAACAGCCTCTTGG-3′ and allele C/T reverse,
5′-GCACTGCACCTCAGGC-3′. The thermocycling conditions included an
initial denaturation at 94°C for 2 min, followed by 35 cycles of
94°C for 1 min, 52°C for 1 min, and 72°C for 1 min. Subsequently,
PCR products (50 µl) were extended for 5 min at 72°C, and the
genotypes of rs41291957 SNP were measured by DHPLC using the WAVE™
DNA Fragment Analysis system (ADS Biotec Ltd.) under partially
denaturing conditions. The column (4.6×250 mm; 5 µm; ADS Biotec
Ltd.) was maintained at 59.3°C and the mobile phase (water and
methanol) flow rate was 0.9 ml/min.
RNA isolation and reverse
transcription-quantitative (RT-qPCR)
The expression levels of miR-143, TLR2 mRNA and
interleukin-16 (IL-16) mRNA in clinical samples and cultured cells
were measured by RT-qPCR. Total RNA of the samples was extracted
using a TRIzol kit, according to the manufacturer's instructions
(Invitrogen, Thermo Fisher Scientific, Inc.). RNA concentration was
calculated using the 260/280 absorbance ratio. Reverse
transcription to cDNA was carried out using an RT kit (Promega
Corporation) according to the manufacturer's protocol. The primers
for the PCR reactions were designed using Primer 5.0 software
(www.premierbiosoft.com/primerdesign) according to the
gene sequences of miR-143, TLR2 mRNA and IL-16 mRNA obtained from
the GenBank database (db.cngb.org).
The following primer pairs were used for qPCR: miR-143 forward,
5′-GCAGTGCTGCATCTCTG-3′ and reverse, 5′-GAACATGTCTGCGTATCTC-3′;
TLR2 forward, 5′-CTTCACTCAGGAGCAGCAAGCA-3′ and reverse,
5′-ACACCAGTGCTGTCCTGTGACA-3′; IL-16 forward,
5′-TTGGACACAGGGTTCTCGCTCA-3′ and reverse,
5′-AGCAGGGAGATAACGGACTGAC-3′; and U6 forward,
5′-TTATGGGTCCTAGCCTGAC-3′ and reverse, 5′-CACTATTGCGGGTCTGC-3′. The
total volume of the RT-qPCR reaction system was 20 µl, which
contained 10 µl SYBR Premix Ex Taq (Takara Bio Inc.), 0.8 µl
forward primer, 0.8 µl reverse primer, 0.4 µl ROX reference dye II
(Invitrogen; Thermo Fisher Scientific, Inc.), 2 µl DNA templates
and 6 µl distilled water. The thermocycling conditions were as
follows: Initial denaturation at 95°C for 30 sec, followed by 40
cycles of 95°C for 5 sec and 60°C for 30 sec. The PCR results were
verified by the melting curve using actin as a reference control.
The relative level of target gene expression was quantified using
the 2−ΔΔCq method (22)
and normalized to the internal reference gene U6.
Cell culture and transfection
THP-1 (human monocytic cells) and human pulmonary
artery smooth muscle cells (HPASMC) obtained from American Type
Culture Collection were cultured in DMEM (Gibco; Thermo Fisher
Scientific, Inc.) supplemented with 10% fetal bovine serum
(Invitrogen; Thermo Fisher Scientific, Inc.). At a confluence rate
of 80–90%, the cells were trypsinized with a 0.25% EDTA-trypsin
solution, precipitated by centrifugation at 167.7 × g for 15 min at
4°C, resuspended and seeded into 24-well plates at a density of
4×105 cells per well. After an overnight incubation at
37°C and 5% CO2, the cells were transfected with 50 µM
miR-143 precursor (5′-UGAGAUGAAGCACUGUAGCUC-3′), TLR2 siRNA
(5′-GGAUCCUCGUGGAUAUCAAUU-3′), IL-16 siRNA
(5′-TCACCTCAACTCCAGTCCGTA-3′) or miRNA scramble negative controls
(5′-UGGGCGUAUAGACGUGUUACAC-3′; all purchased from Santa Cruz
Biotechnology, Inc.) using Lipofectamine® 2000
(Invitrogen; Thermo Fisher Scientific, Inc.), according to the
manufacturer's instructions. After 48 h of cell transfection at
4°C, the cells were collected for subsequent experimentation. Each
transfection experiment was repeated ≥3 times.
Luciferase assay
To investigate the regulatory relationship between
miR-143 and IL-16, and between miR-143 and TLR2, TargetScan
(version 7.2; (targetscan.org) bioinformatics
software was used to predict the binding sites of miR-143 in IL-16
and TLR2. The 3′UTRs of IL-16 and TLR2 containing the binding sites
of miR-143 were amplified by PCR and cloned into pGL3 vectors
(Promega Corporation). The amplification was performed on a PTC-100
thermocycler (Bio-Rad Laboratories, Inc.) using the following
thermocycling conditions: 95°C for 30 sec, 40 cycles of 30 sec at
95°C, 2 min at 58°C and 30 sec at 68°C, 72°C for 5 min. Taq DNA
polymerase (Sigma-Aldrich; Merck KGaA) was used for PCR. In
addition, site-directed mutagenesis was carried out in the putative
binding sites of miR-143 in the 3′UTRs of IL-16 (forward,
5′-TTGGACACAGGGTTCTCGCTCA-3′ and reverse,
5′-AGCAGGGAGATAACGGACTGAC-3′) and TLR2 (forward,
5′-CTTCACTCAGGAGCAGCAAGCA-3′ and reverse,
5′-ACACCAGTGCTGTCCTGTGACA-3′), and the mutated sequences were also
amplified by PCR and cloned into pcDNA3.1 vectors (Promega
Corporation) according to the aforementioned protocol. THP-1 and
HPASMC cells were co-transfected with miR-143 mimic and plasmids
carrying the wild-type or mutant IL-16 or TLR2 using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). After 48 h of transfection, cells were harvested
and the luciferase activity of transfected cells was measured by a
dual luciferase reporter assay system (Promega Corporation) on a
luminometer (TD20/20; Turner Designs) at 4°C. Firefly luciferase
activity was obtained and normalized to the value of Renilla
luciferase activity. Each experiment was repeated ≥3 times.
Western blot analysis
Total protein was extracted using a urea buffer
containing a protease inhibitor cocktail (Bio-Rad Laboratories,
Inc.). Total protein in each sample was measured using a
bicinchoninic acid assay kit (Invitrogen; Thermo Fisher Scientific,
Inc.), according to the manufacturer's protocol. Equal amounts of
protein (40 µg) were resolved using 10% SDS-PAGE gel
electrophoresis and electro-transferred onto a PVDF membrane.
Subsequently, the membrane was blocked for 60 min at room
temperature with TBST containing 5% skim milk. The membrane was
incubated overnight at 4°C with the following primary antibodies:
Anti-TLR2 (cat. no. ab16894; 1:1,000; Abcam), anti-IL-16 (cat. no.
ab207181; 1:1,000; Abcam) and anti-β-actin (cat. no. ab115777;
1:1,000; Abcam). After washing with PBS, the membrane was incubated
with horseradish peroxide-labeled secondary antibodies (cat. no.
7076s; 1:10,000; Cell Signaling Technology Inc.) for 1 h at 37°C
and was subsequently developed using an ECL method with the ECL™
Western Blotting Detection Reagents (Sigma-Aldrich; Merck KGaA).
The membrane was visualized using a Gel Doc EZ Imager (Bio-Rad
Laboratories, Inc.) and the protein bands were analyzed using
ImageJ software (version 1.8.0; National Institutes of Health)
using β-actin as the loading control.
ELISA
The collected clinical and cellular samples were
centrifuged at 4°C for 10 min at 1509.3 × g, and the supernatant
was transferred into sterile centrifuge tubes. ELISA kits including
the human TNF-α ELISA kit (cat. no. RAB1089; Sigma-Aldrich; Merck
KGaA), NF-κB p65 ELISA kit (cat. no. ab176648; Abcam), human IFN-γ
ELISA kit (cat. no. RAB0223; Sigma-Aldrich; Merck KGaA), human IL-6
ELISA kit (cat. no. RAB0306; Sigma-Aldrich; Merck KGaA) and human
IL-10 ELISA kit (cat. no. RAB0244; Sigma-Aldrich; Merck KGaA) were
used according to the manufacture's protocol. The samples were
diluted and added into a microtiter plate precoated with
anti-TNF-α, anti-NF-κB, anti-IFN, anti-IL6 and anti-IL10
antibodies. After being incubated for 30 min at 37°C, 50 µl of
ELISA reactive solution was added to each well of the microplate
and incubated for another 30 min at 37°C. Subsequently, 50 µl of
chromogenic reagent A was added into each well, followed by the
addition of 50 µl of chromogenic reagent B. The microplate was then
gently shaken for 30 sec, and was subsequently incubated in the
dark for 15 min at 37°C. After the microplate was removed from the
incubation, the wells presented a blue color. If the signal
intensity was too low, the incubation time was slightly extended by
30 min. At the end of incubation, 50 µl Alkaline Phosphatase stop
solution (Sigma-Aldrich; Merck KGaA) was added into each well to
stop the reaction, and the color in the wells turned to yellow.
Within 15 min of reaction termination, the optical density (OD)
value of each well was measured at a wavelength of 450 nm. A
standard curve was then drawn for each target substance using OD
and concentration values. Each experiment was repeated ≥3
times.
Statistical analysis
Data are presented as the mean ± SD. The data were
analyzed using SPSS software (version 21.0; IBM Corp.). Student's
t-test was used for comparison between two groups. One-way ANOVA
test was used for comparison between multiple groups, followed by
Tukey's test. The survival rates of patients with ICH in different
groups were compared using the Kaplan-Meier analysis. P<0.05 was
considered to indicate a statistically significant difference.
Results
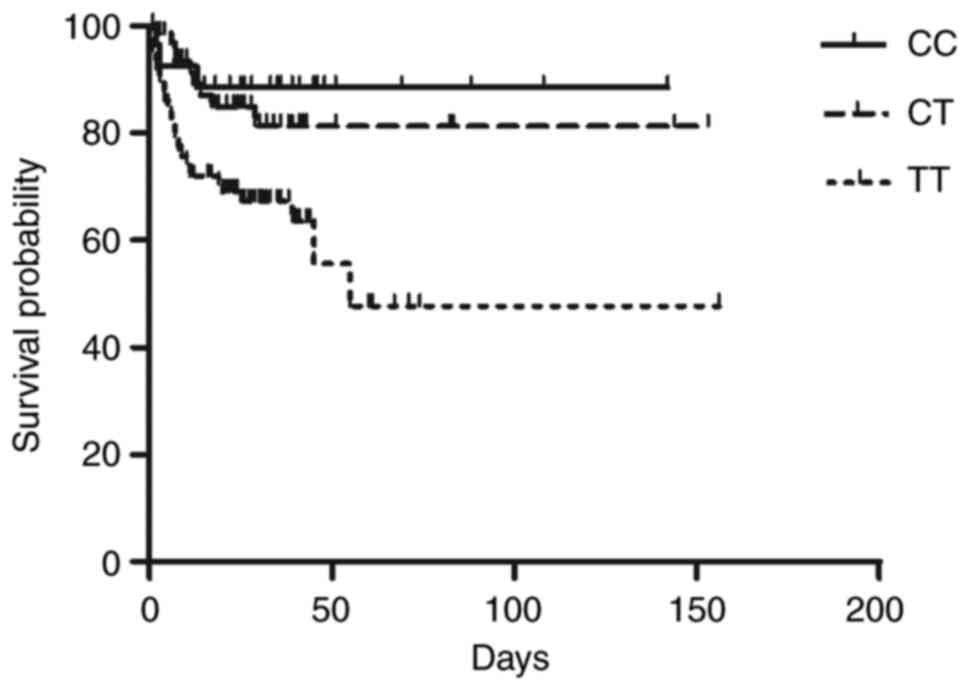
Patients carrying the TT genotype are
associated with the lowest survival rate
The present study recruited 182 subjects with ICH,
genotyped as CC (n=28), CT (n=67) and TT (n=87) according to the
SNP rs41291957. Demographic and clinicopathological characteristics
of all participants were presented in Table I. Kaplan-Meier survival curves were
plotted for and are presented in Fig.
1. Upon the assessment of post-ICH mortality in patients
followed-up for ≥6 months after their discharge, it was found that
patients carrying the TT genotype of SNP rs41291957 were associated
with the lowest survival rate, while the patients carrying the CC
genotype of SNP rs41291957 were associated with the highest
survival rate.
 | Table I.Demographic and clinicopathological
characteristics of patients with ICH. |
Table I.
Demographic and clinicopathological
characteristics of patients with ICH.
| Clinical
variable | CC + CT (n=95) | TT (n=87) | P-value |
|---|
| Age, years | 60.5±15.3 | 61.1±16.2 | 0.8215 |
| Female (%) | 39 (41.3) | 34 (40.6) | 0.6325 |
| Hypertension
(%) | 77 (81.3) | 69 (78.1) | 0.4425 |
| History of stroke
(%) | 11 (12.0) | 13 (15.6) | 0.1254 |
| History of ICH
(%) | 6
(6.7) | 8
(9.3) | 0.2571 |
| Hematoma size,
cc | 23.5±3.6 | 25.8±6.5 | 0.6251 |
| Admission
intraventricular hemorrhage (%) | 20 (22.0) | 27 (31.3) | 0.8248 |
| Admission
hydrocephalus (%) | 55 (58.7) | 46 (53.1) | 0.1364 |
| Intraventricular
hemorrhage score |
7.2±4.1 |
7.8±6.6 | 0.6481 |
| Midline shift,
mm |
2.3±0.8 |
2.6±1.4 | 0.1351 |
| Hematoma evacuation
(%) | 11 (12.0) | 10 (12.5) | 0.5124 |
| Ventricular
drainage (%) | 64 (68.0) | 54 (62.5) | 0.1354 |
| Intrathecal tissue
plasminogen activator (%) | 5
(5.3) | 5
(6.3) | 0.8452 |
| Ventricular shunt
(%) | 6
(6.0) | 8
(9.3) | 0.2458 |
| Pneumonia (%) | 33 (34.7) | 33 (37.5) | 0.4518 |
|
Ventriculitis/meningitis (%) | 3
(3.3) | 3
(3.1) | 0.8479 |
| Sepsis (%) | 3
(3.3) | 3
(3.1) | 0.8479 |
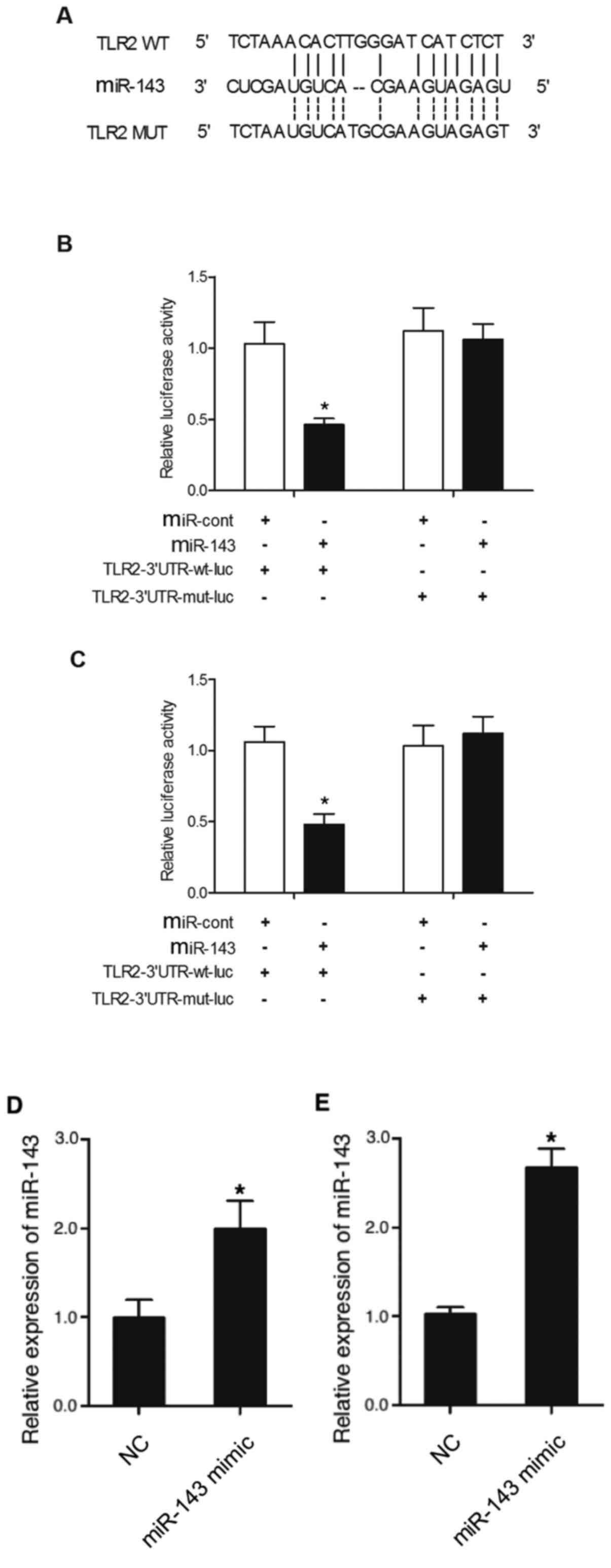
TLR2 is a direct target gene of
miR-143
To predict the regulatory relationship between
miR-143 and TLR2, a computational analysis was conducted, and TLR2
was identified as a candidate target gene of miR-143, and a
possible binding site of miR-143 was found in the 3′UTR of TLR2
(Fig. 2A). The luciferase activity
of THP-1 and HPASMC cells co-transfected with wild-type or mutant
TLR2 mRNA in conjunction with miR-143 or miRNA controls was
investigated. Relative luciferase activity of THP-1 cells
co-transfected with wild-type TLR2 mRNA and miR-143 was
significantly downregulated compared with the THP-1 cells
co-transfected with wild-type TLR2 mRNA and miRNA controls
(Fig. 2B). However, cells
transfected with mutant TLR2 mRNA exhibited a similar level of
luciferase activity in the presence of miR-143 or miRNA controls
(Fig. 2B). Similar results were
obtained in HPASMC cells (Fig. 2C).
The transfection of miR-143 mimics alone also significantly
increased the expression level of miR-143 in THP-1 (Fig. 2D) and HPASMC (Fig. 2E) cells, indicating the successful
transfection of miR-143 mimics. Collectively, the present results
suggested that TLR2 mRNA was a target of miR-143.
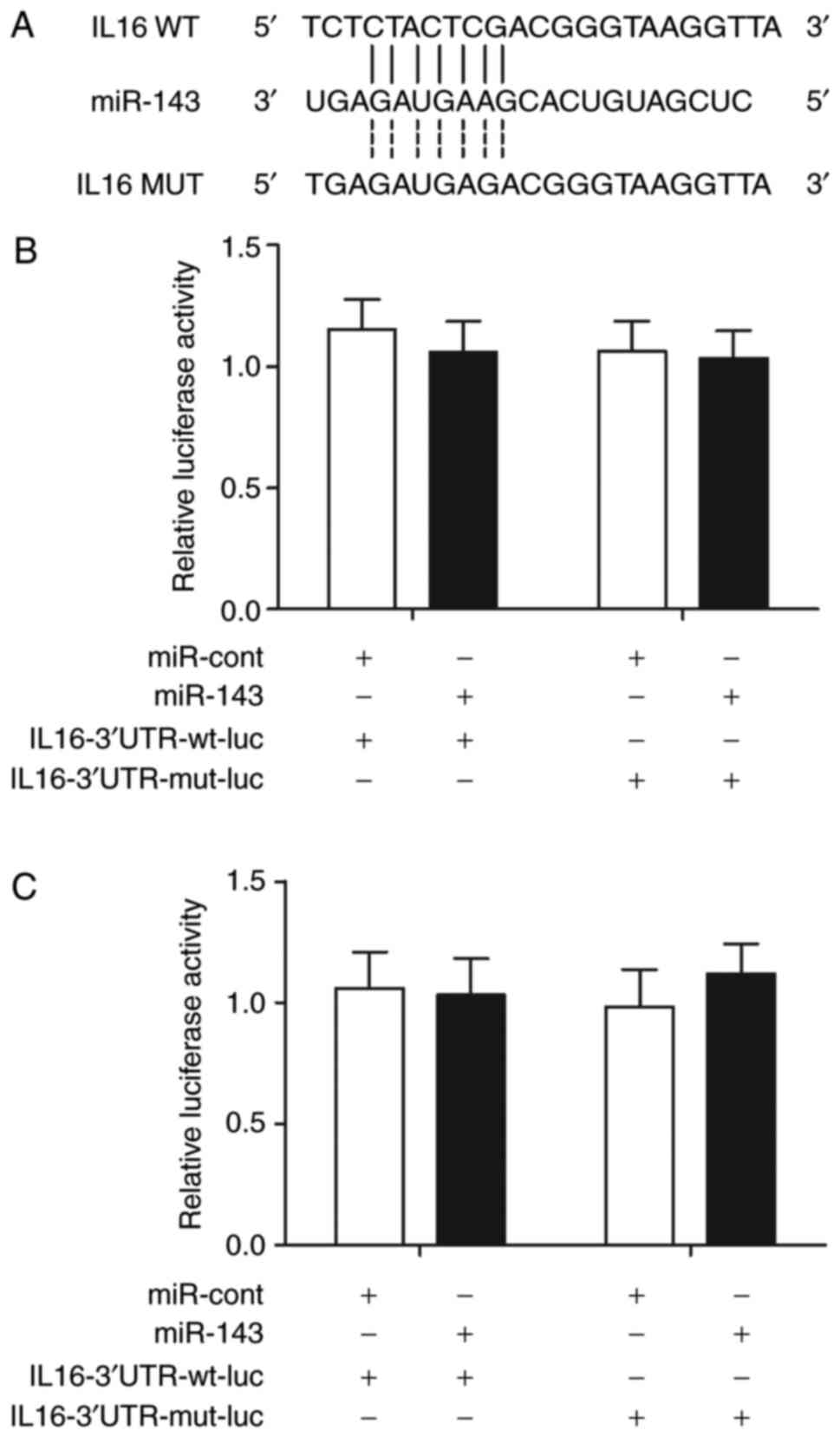
IL-16 mRNA is not a target of miR-143
and are not associated
IL-16 is involved in post-ICH inflammation. The
TargetScan online bioinformatics tool was used to locate the
possible binding site of miR-143 in the 3′UTR of IL-16 mRNA
(Fig. 3A). A luciferase assay was
performed by co-transfecting THP-1 and HPASMC cells with wild-type
or mutant IL-16 mRNA in conjunction with miR-143 or miRNA controls.
No significant difference was observed between THP-1 cells
co-transfected with wild-type or mutant IL-16 mRNA in conjunction
with miR-143 or miRNA controls (Fig.
3B). Similar results were also obtained in HPASMC cells
(Fig. 3C), indicating the absence
of a regulatory relationship between miR-143 and IL-16 mRNA.
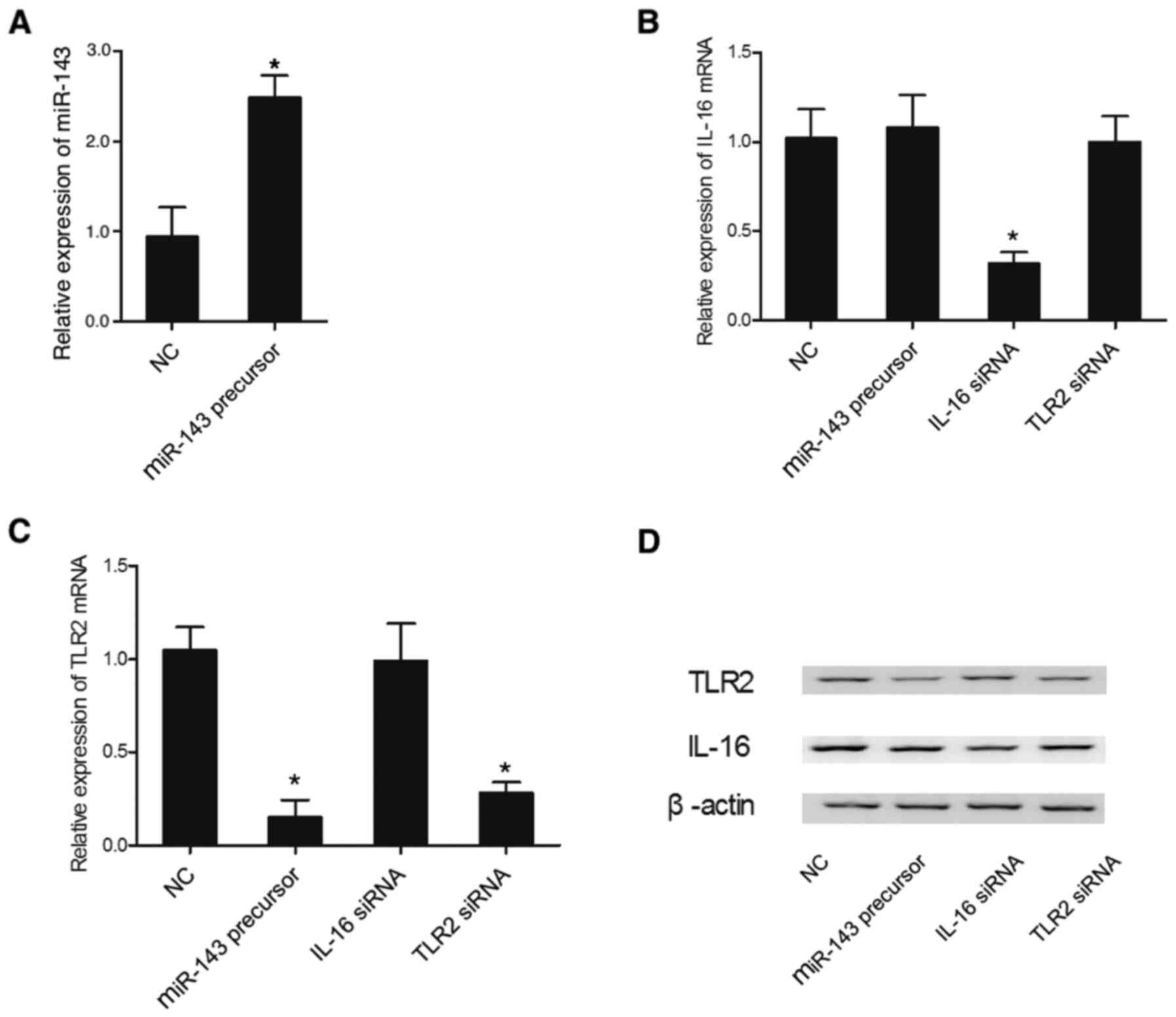
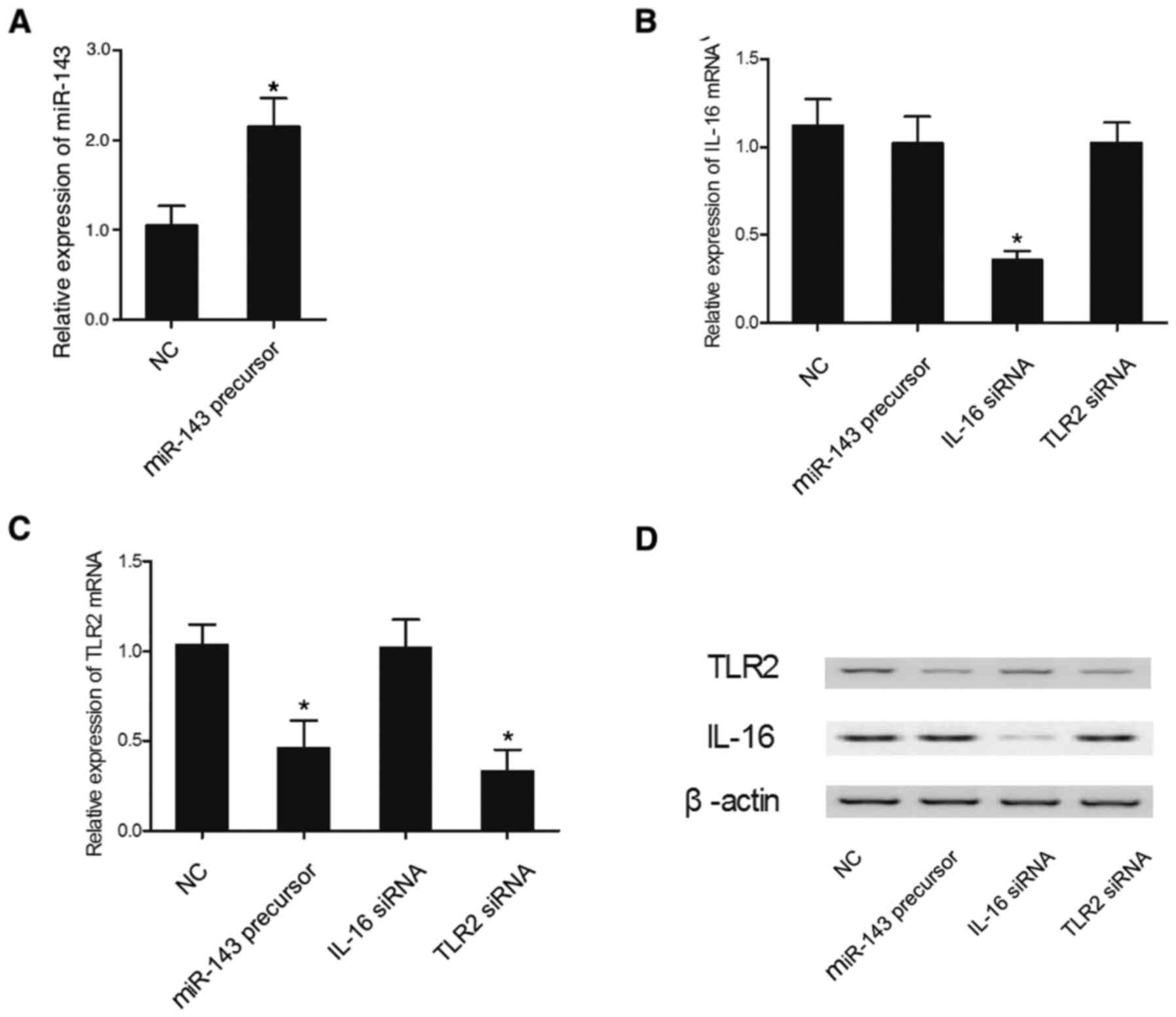
Effect of miR-143 overexpression on
the expression levels of IL-16 and TLR2
The expression levels of IL-16 mRNA and TLR2 mRNA
were measured in THP-1 and HPASMC cells transfected with miR-143
precursor, IL-16 siRNA or TLR2 siRNA. The relative expression of
miR-143 in THP-1 cells was significantly increased in the presence
of miR-143 precursor, demonstrating successful transfection of
miR-143 precursor in THP-1 cells (Fig.
4A). The relative expression of IL-16 mRNA in THP-1 cells
(Fig. 4B) was suppressed by IL-16
siRNA, while other treatments failed to affect the relative
expression of IL-16. However, the relative expression of TLR2 mRNA
was markedly reduced in the THP-1 cells (Fig. 4C) transfected with either miR-143
precursor or TLR2 siRNA. In addition, the expression levels of
IL-16 and TLR2 proteins were altered in the presence of IL-16 siRNA
and miR-143 precursor or TLR2 siRNA, respectively (Fig. 4D). Similar results were also
obtained in HPASMC cells (Fig. 5),
which further indicated the presence of a negative regulatory
relationship between miR-143 and TLR2.
Rs41291957 polymorphism affects the
production of inflammatory factors in patients with ICH
The expression of other inflammatory factors,
including TNFα, IFN, IL-6, IL-10 and NF-κB, were compared in
patients with ICH carrying different genotypes of rs41291957 SNP.
The expression levels of TNFα, IFN, IL-6, IL-10 and NF-κB in the
CSF samples were highest in patients with ICH and the CC genotype,
but were significantly reduced in patients with CT and TT genotypes
(Fig. 6A-E). In contrast, the level
of miR-143 in the CSF samples was lowest in patients with ICH and
the CC genotype, while miR-143 levels were increased in patients
with CT and TT genotypes (Fig. 6F).
Similar results were also obtained in serum samples (Fig. 7), indicating that the SNP rs41291957
located in the promoter region of miR-143 could affect the
transcription efficiency of miR-143. Therefore, the increased
miR-143 expression in the patients carrying the TT genotype of SNP
rs41291957 could increase the expression of pro-inflammatory factor
TLR2, thus resulting in a poorer prognosis of ICH.
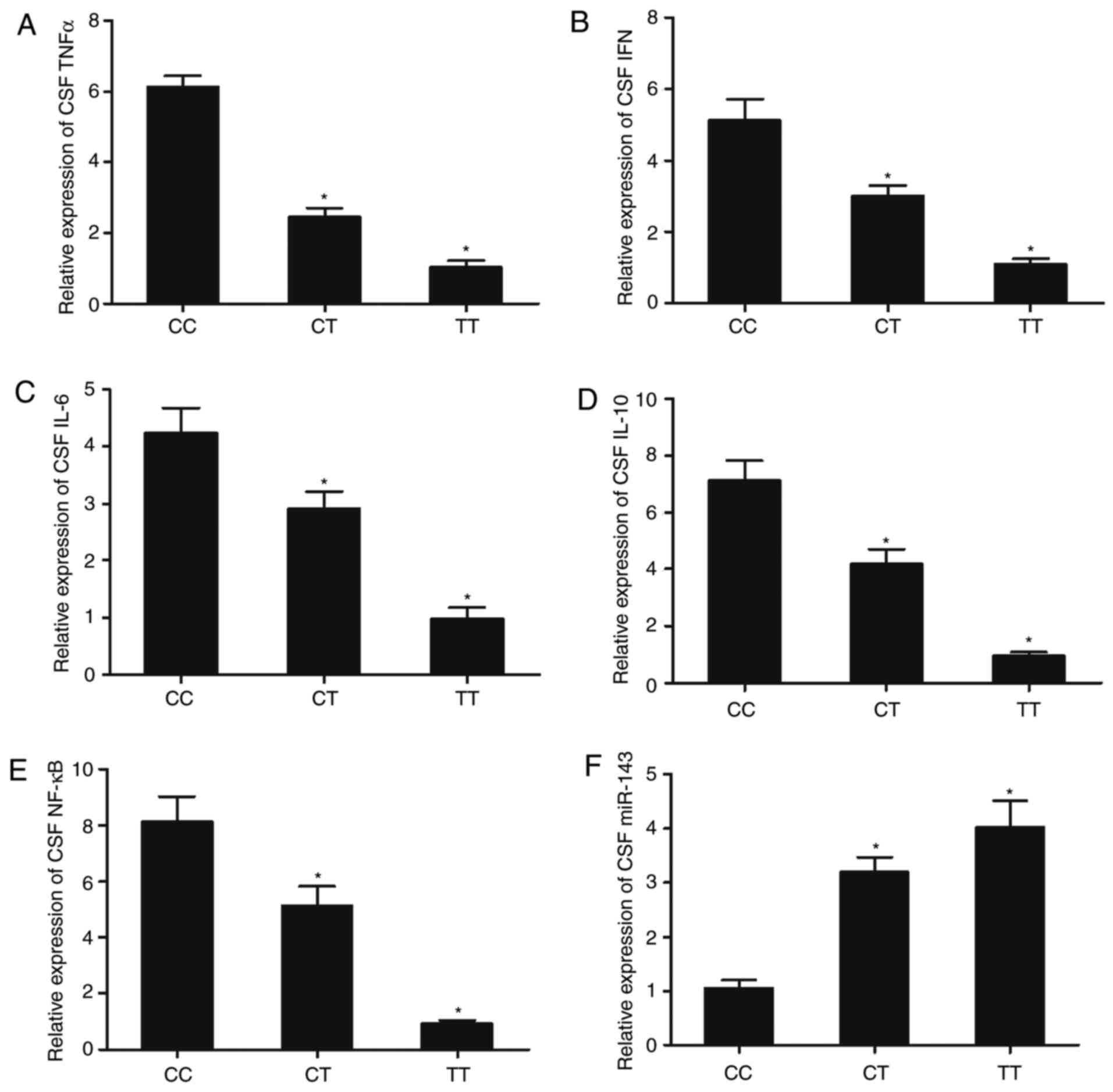 | Figure 6.Expression of miR-143 and
inflammatory factors, including TNFα, IFN, IL-6, IL-10, and NF-κB,
in CSF samples genotyped as CC, CT and TT. Expression of (A) TNFα,
(B) IFN, (C) IL-6, (D) IL-10, (E) NF-κB and (F) miR-143 in CSF
samples from patients genotyped as CC, CT and TT. n=3. *P<0.05
vs. CC group. miR-143, microRNA-143; TNFα, tumor necrosis factor α;
IFN, interferon; IL, interleukin; CSF, cerebrospinal fluid. |
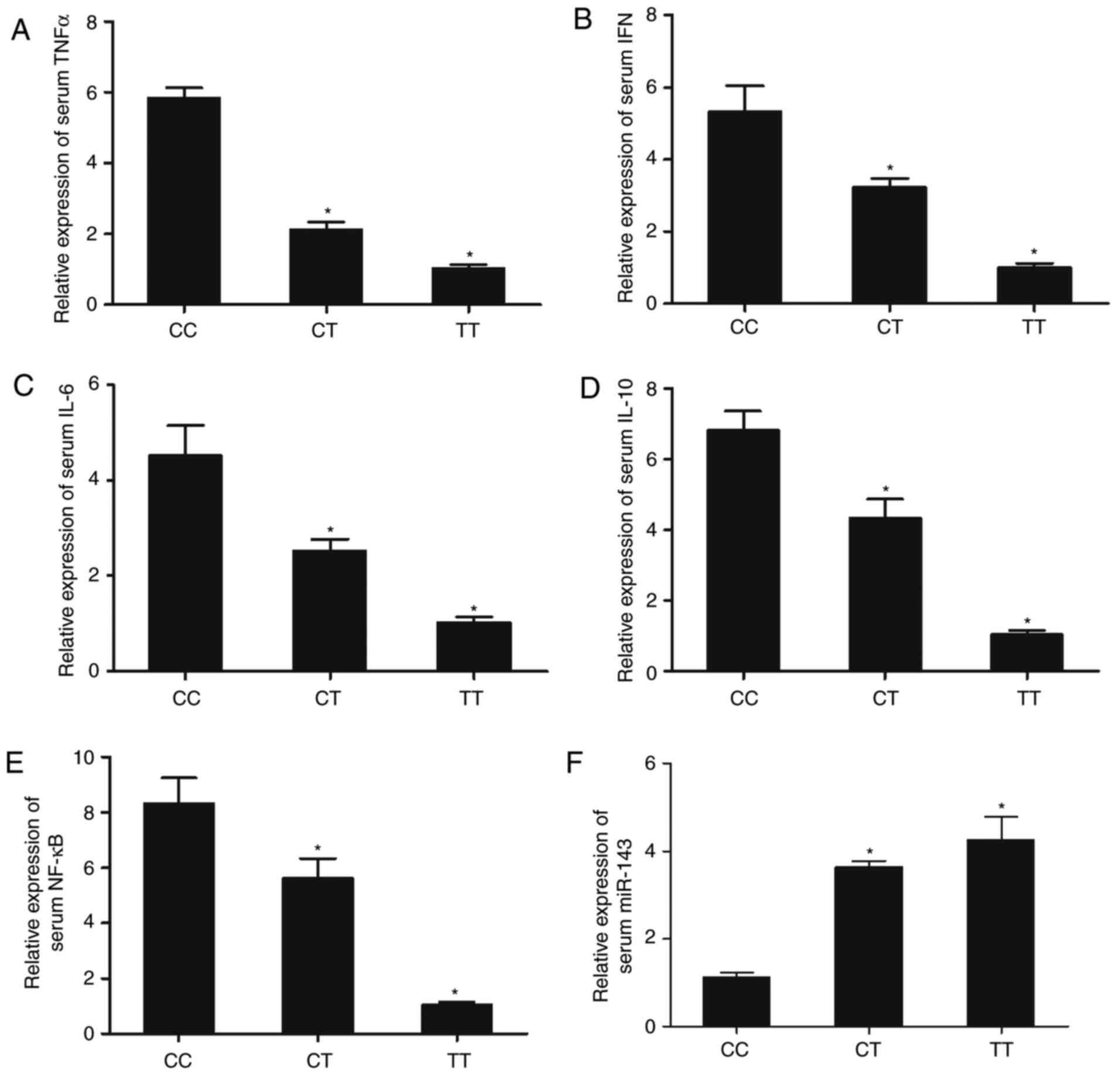 | Figure 7.Expression of miR-143 and
inflammatory factors, including TNFα, IFN, IL-6, IL-10, and NF-κB,
in serum samples genotyped as CC, CT and TT. Expression of (A)
TNFα, (B) IFN, (C) IL-6, (D) IL-10, (E) NF-κB and (F) miR-143 in
serum samples from patients genotyped as CC, CT and TT. n=3.
*P<0.05 vs. CC group. miR-143, microRNA-143; TNFα, tumor
necrosis factor α; IFN, interferon; IL, interleukin; CSF,
cerebrospinal fluid. |
Effect of miR-143 transfection in the
presence or absence of TLR2 overexpression on the production of
inflammatory factors in THP-1 cells
Transfection of miR-143 precursors downregulated the
production of TNFα, IFN, IL-6, IL-10 and NF-κB, while the
overexpression of TLR2 promoted the production of these
inflammatory cytokines (Fig.
8).
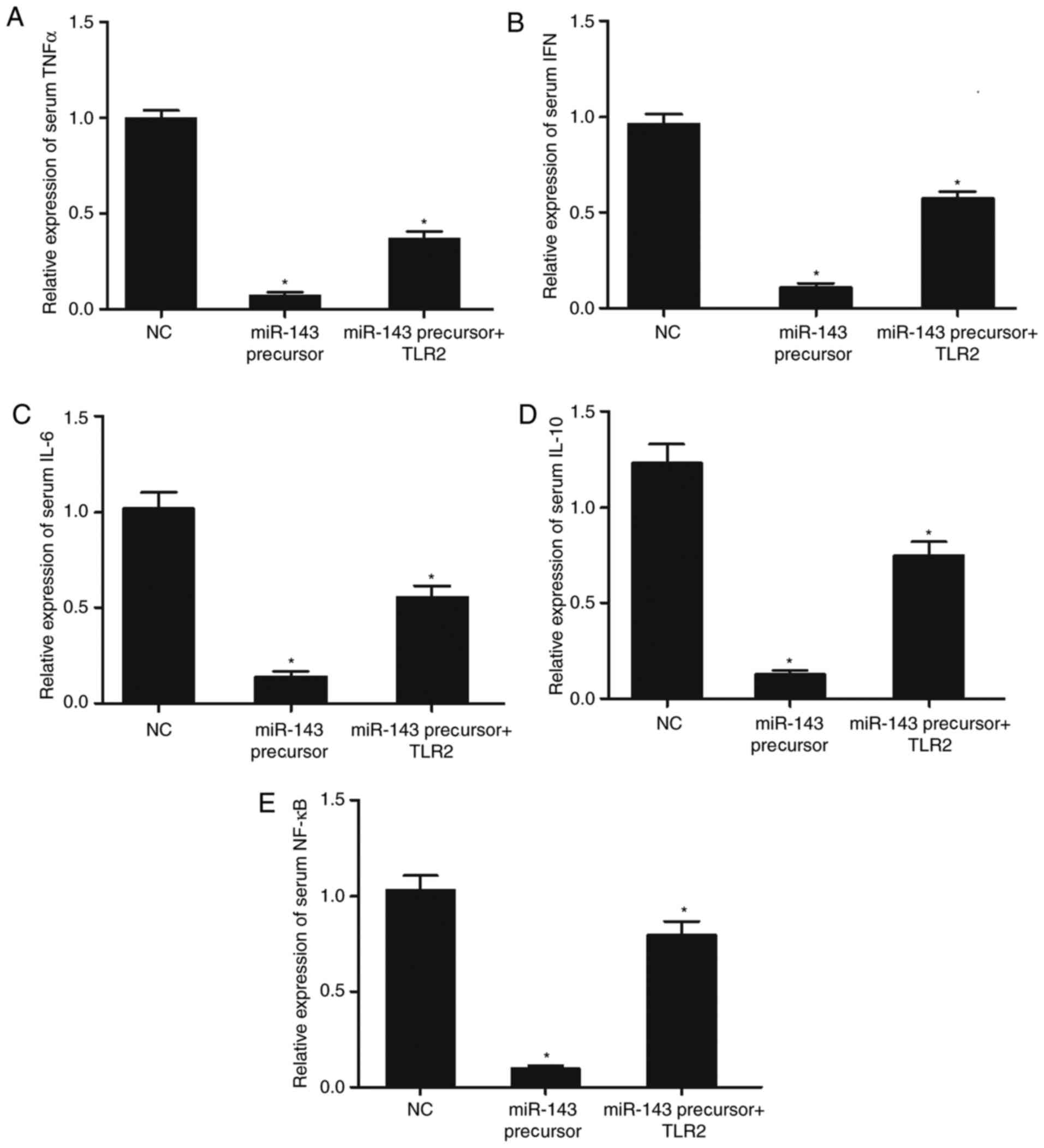 | Figure 8.Effect of miR-143 transfection in the
absence or presence of TLR2 overexpression on the production of
inflammatory factors in THP-1 cells. (A) Transfection of miR-143
reduced the production of TNFα, whereas the presence of TLR2
increased the production of TNFα. (B) Transfection of miR-143
reduced the production of IFN, whereas the presence of TLR2
increased the production of IFN. (C) Transfection of miR-143
reduced the production of IL-6, whereas the overexpression of TLR2
increased the production of IL-6. (D) Transfection of miR-143
reduced the production of IL-10, whereas the presence of TLR2
increased the production of IL-10. (E) Transfection of miR-143
reduced the production of NF-κB, whereas the overexpression of TLR2
increased the production of NF-κB. n=3. *P<0.05 vs. NC group.
miR-143, microRNA-143; TLR2, toll-like receptor 2; TNFα, tumor
necrosis factor α; IFN, interferon; IL, interleukin; CSF,
cerebrospinal fluid; NC, negative control. |
Discussion
ICH is a serious disease associated with high
mortality (23). However, there are
no effective treatments currently available to treat ICH (23). In addition, the chronic and acute
damages caused by ICH, such as primary mechanical injuries in the
brain tissues and secondary complications induced by inflammatory
responses, further reduced the efficacy of ICH treatments (8). Therefore, an anti-inflammatory
approach may be favorable for ICH treatment, by reducing
inflammation-induced secondary brain damages, to improve the
quality of life of patients (9).
Previous studies have suggested that the inflammation of brain
tissues not only affects post-ICH brain recovery, but also plays
essential roles in the recovery from other brain traumas (24,25).
In addition, the pro-inflammatory factors secreted by endothelial
cells, neuronal cells, astrocytes and microglia during brain injury
can activate cellular adhesion molecules, thereby enhancing the
recruitment of leukocytes into brain parenchyma and aggravating
post-ICH brain injury (5,24,25).
A previous study has shown that miR-143 can play a
role in the prevention of ischemia-induced brain injury (26). The present study investigated the
regulatory relationship between miR-143 and TLR2 via computational
analysis and luciferase assay. TLR2 was identified as a target gene
of miR-143, with a miR-143 binding site located in the 3′UTR of
TLR2. In addition, the downregulated luciferase activity in the
cells co-transfected with wild-type TLR2 mRNA and miR-143 suggested
that TLR2 mRNA was a target of miR-143. Furthermore, no regulatory
relationship was found between IL-16 and miR-143.
TLR2 is a transmembrane receptor with the ability to
recognize peptidoglycan and lipoteichoic acid secreted by bacteria
(27). In addition, TLR2 can induce
the production of inflammatory signals (28). Previous studies have shown that TLR2
acts as a receptor for inflammatory factors, including high
mobility group box-1, soluble CD14, hyaluronan and heat shock
proteins, generated by pathogens and endogenously (27–32).
In addition, several studies have hypothesized that the activation
of TLR2 by these factors could be implicated in the generation of
inflammatory responses in various of neurological diseases
(33,34). The present study measured the
expression levels of IL-16 mRNA and TLR2 mRNA in the cells
transfected with miR-143 precursor, IL-16 siRNA and TLR2 siRNA. The
present results showed that the relative expression levels of TLR2
mRNA was inhibited in the cells treated with miR-143 precursor or
TLR2 siRNA. However, the relative expression of IL-16 mRNA was only
reduced in the cells treated with IL-16 siRNA. The present results
were also validated by western blot analysis. In a previous study,
TLR2 inhibition in mice was shown to reduce post-ICH neutrophil
infiltration. Neutrophil infiltration can induce tissue damage via
the release and production of reactive oxygen species and
proteases, thus affecting the induction of myeloperoxidase (MPO)
activity after the onset of ICH (35). Furthermore, the oxidative stress
caused by MPO can trigger neuronal apoptosis (36–38).
Therefore, a reduction in neutrophil infiltration may consecutively
attenuate the severity of ICH-induced brain injury. These previous
studies demonstrated that the occurrence of post-ICH secondary
brain injury is closely related to TLR2 activation as well as
subsequent matrix metallopeptidase 9 stimulation and BBB disruption
(35–38). The present study measured the
expression of inflammatory factors, including TNFα, IFN, IL-6,
IL-10 and NF-κB, as well as the expression of miR-143 in the
CSF/serum samples collected from patients with ICH. The expression
of TNFα, IFN, IL-6, IL-10 and NF-κB was increased in subjects
carrying the CC genotype, and decreased in CT and TT genotypes of
SNP rs41291957. Meanwhile, the level of miR-143 showed an opposite
trend as that of above inflammatory factors.
In previous studies, miR-143 was shown to function
as a tumor suppressor in the pathogenesis of a wide array of cancer
types (17,39–42).
Li et al (16) showed that
the rs41291957 SNP in pri-miR-143 could alter the susceptibility to
colorectal cancer. A previous study showed that, by targeting
adducin3, miR-143 played an essential role in the morphogenesis of
cardiac chamber in zebrafish (43).
Miyasaka et al (44)
demonstrated that cardiogenesis is mediated by miR-143. The present
study examined Kaplan-Meier survival curves for 182 patients with
ICH carrying CC, CT and TT genotypes of SNP rs41291957. The
patients carrying the TT genotype were associated with the lowest
survival rate, while the patients carrying the CC genotype were
associated with the highest survival rate. rs41291957 SNP was found
to be associated with the expression level of miR-143.
Additionally, miR-143 was identified to target TLR2, whose
expression could be associated with the severity of inflammatory
conditions and the prognosis of ICH. Therefore, rs41291957 SNP may
be used as a novel biomarker to predict the prognosis of ICH.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XY and XS designed the study. ZG and FC evaluated
the literature. XY, ZG, FC, ZT and ZH collected and analyzed the
data. XY and XS wrote the manuscript. All authors approved the
final manuscript.
Ethics approval and consent to
participate
The Human Research Ethics Committees of The First
Affiliated Hospital of Chongqing Medical University provided
approval for the present study. All methods were performed in
accordance with the Declaration of Helsinki. Written informed
consent was obtained from all patients or their first-degree
relatives prior to surgery.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Wang MD, Wang Y, Xia YP, Dai JW, Gao L,
Wang SQ, Wang HJ, Mao L, Li M, Yu SM, et al: High serum miR-130a
levels are associated with severe perihematomal edema and predict
adverse outcome in acute ICH. Mol Neurobiol. 53:1310–1321. 2016.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Rincon F, Friedman DP, Bell R, Mayer SA
and Bray PF: Targeted temperature management after intracerebral
hemorrhage (TTM-ICH): Methodology of a prospective randomized
clinical trial. Int J Stroke. 9:646–651. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Zhang Y, Chen Y, Wu J, Manaenko A, Yang P,
Tang J, Fu W and Zhang JH: Activation of dopamine D2 receptor
suppresses neuroinflammation through αB-crystalline by inhibition
of NF-κB nuclear translocation in experimental ICH mice model.
Stroke. 46:2637–2646. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Xu C, Wang T, Cheng S and Liu Y: Increased
expression of T cell immunoglobulin and mucin domain 3 aggravates
brain inflammation via regulation of the function of
microglia/macrophages after intracerebral hemorrhage in mice. J
Neuroinflammation. 10:1412013. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Chen M, Li X, Zhang X, He X, Lai L, Liu Y,
Zhu G, Li W, Li H, Fang Q, et al: The inhibitory effect of
mesenchymal stem cell on blood-brain barrier disruption following
intracerebral hemorrhage in rats: Contribution of TSG-6. J
Neuroinflammation. 12:612015. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lei B, Dawson HN, Roulhac-Wilson B, Wang
H, Laskowitz DT and James ML: Tumor necrosis factor α antagonism
improves neurological recovery in murine intracerebral hemorrhage.
J Neuroinflammation. 10:1032013. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Harder LM, Bunkenborg J and Andersen JS:
Inducing autophagy: A comparative phosphoproteomic study of the
cellular response to ammonia and rapamycin. Autophagy. 10:339–355.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Wakisaka Y, Chu Y, Miller JD, Rosenberg GA
and Heistad DD: Spontaneous intracerebral hemorrhage during acute
and chronic hypertension in mice. J Cereb Blood Flow Metab.
30:56–69. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Rincon F and Mayer SA: Novel therapies for
intracerebral hemorrhage. Curr Opin Crit Care. 10:94–100. 2004.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Bartel DP: MicroRNAs: Target recognition
and regulatory functions. Cell. 136:215–233. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Nadal E, Truini A, Nakata A, Lin J, Reddy
RM, Chang AC, Ramnath N, Gotoh N, Beer DG and Chen G: A novel serum
4-microRNA signature for lung cancer detection. Sci Rep.
5:124642015. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Akao Y, Nakagawa Y and Naoe T: MicroRNAs
143 and 145 are possible common onco-microRNAs in human cancers.
Oncol Rep. 16:845–850. 2006.PubMed/NCBI
|
|
13
|
Starczynowski DT, Kuchenbauer F,
Argiropoulos B, Sung S, Morin R, Muranyi A, Hirst M, Hogge D, Marra
M, Wells RA, et al: Identification of miR-145 and miR-146a as
mediators of the 5q-syndrome phenotype. Nat Med. 16:49–58. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Xie H, Lim B and Lodish HF: MicroRNAs
induced during adipogenesis that accelerate fat cell development
are downregulated in obesity. Diabetes. 58:1050–1057. 2009.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Cordes KR, Sheehy NT, White MP, Berry EC,
Morton SU, Muth AN, Lee TH, Miano JM, Ivey KN and Srivastava D:
miR-145 and miR-143 regulate smooth muscle cell fate and
plasticity. Nature. 460:705–710. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Li L, Pan X, Li Z, Bai P, Jin H, Wang T,
Song C, Zhang L and Gao L: Association between polymorphisms in the
promoter region of miR-143/145 and risk of colorectal cancer. Hum
Immunol. 74:993–997. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Wu D, Huang P, Wang L, Zhou Y, Pan H and
Qu P: MicroRNA-143 inhibits cell migration and invasion by
targeting matrix metalloproteinase 13 in prostate cancer. Mol Med
Rep. 8:626–630. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Yin X, Sun S, Zhao J, Yang J, Lei X, Xu C
and Li K: Rs4705342 polymorphism is involved in the tumorigenesis
of HBV positive HCC by altering the binding affinity of HBV induced
NF-κB with the promoter region of microRNA-143. J Cell Biochem.
119:5233–5242. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Liu X, Gong J and Xu B: miR-143
down-regulates TLR2 expression in hepatoma cells and inhibits
hepatoma cell proliferation and invasion. Int J Clin Exp Pathol.
8:12738–12747. 2015.PubMed/NCBI
|
|
20
|
Min H, Hong J, Cho IH, Jang YH, Lee H, Kim
D, Yu SW, Lee S and Lee SJ: TLR2-induced astrocyte MMP9 activation
compromises the blood brain barrier and exacerbates intracerebral
hemorrhage in animal models. Mol Brain. 8:232015. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Wang YC, Zhou Y, Fang H, Lin S, Wang PF,
Xiong RP, Chen J, Xiong XY, Lv FL, Liang QL and Yang QW: Toll-like
receptor 2/4 heterodimer mediates inflammatory injury in
intracerebral hemorrhage. Ann Neurol. 75:876–889. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Burns JD, Fisher JL and
Cervantes-Arslanian AM: Recent advances in the acute management of
intracerebral hemorrhage. Neurosurg Clin N Am. 29:263–272. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Bernales S, Schuck S and Walter P:
ER-phagy: Selective autophagy of the endoplasmic reticulum.
Autophagy. 3:285–287. 2007. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Bampton ET, Goemans CG, Niranjan D,
Mizushima N and Tolkovsky AM: The dynamics of autophagy visualized
in live cells: From autophagosome formation to fusion with
endo/lysosomes. Autophagy. 1:23–36. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Bai Y, Zhang Y, Han B, Yang L, Chen X,
Huang R, Wu F, Chao J, Liu P, Hu G, et al: Circular RNA DLGAP4
ameliorates ischemic stroke outcomes by targeting miR-143 to
regulate endothelial-mesenchymal transition associated with
blood-brain barrier integrity. J Neurosci. 38:32–50. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Park JS, Svetkauskaite D, He Q, Kim JY,
Strassheim D, Ishizaka A and Abraham E: Involvement of toll-like
receptors 2 and 4 in cellular activation by high mobility group box
1 protein. J Biol Chem. 279:7370–7377. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Huang QQ, Sobkoviak R, Jockheck-Clark AR,
Shi B, Mandelin AM II, Tak PP, Haines GK III, Nicchitta CV and Pope
RM: Heat shock protein 96 is elevated in rheumatoid arthritis and
activates macrophages primarily via TLR2 signaling. J Immunol.
182:4965–4973. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Ohashi K, Burkart V, Flohé S and Kolb H:
Cutting edge: Heat shock protein 60 is a putative endogenous ligand
of the toll-like receptor-4 complex. J Immunol. 164:558–561. 2000.
View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Vabulas RM, Ahmad-Nejad P, Ghose S,
Kirschning CJ, Issels RD and Wagner H: HSP70 as endogenous stimulus
of the Toll/interleukin-1 receptor signal pathway. J Biol Chem.
277:15107–15112. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Termeer C, Benedix F, Sleeman J, Fieber C,
Voith U, Ahrens T, Miyake K, Freudenberg M, Galanos C and Simon JC:
Oligosaccharides of hyaluronan activate dendritic cells via
toll-like receptor 4. J Exp Med. 195:99–111. 2002. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Bsibsi M, Bajramovic JJ, Van
Duijvenvoorden E, Persoon C, Ravid R, Van Noort JM and Vogt MH:
Identification of soluble CD14 as an endogenous agonist for
Toll-like receptor 2 on human astrocytes by genome-scale functional
screening of glial cell derived proteins. Glia. 55:473–482. 2007.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Hanke ML and Kielian T: Toll-like
receptors in health and disease in the brain: Mechanisms and
therapeutic potential. Clin Sci (Lond). 121:367–387. 2011.
View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Lehnardt S: Innate immunity and
neuroinflammation in the CNS: The role of microglia in Toll-like
receptor-mediated neuronal injury. Glia. 58:253–263.
2010.PubMed/NCBI
|
|
35
|
Weiss SJ: Tissue destruction by
neutrophils. N Engl J Med. 320:365–376. 1989. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Albers DS and Beal MF: Mitochondrial
dysfunction and oxidative stress in aging and neurodegenerative
disease. J Neural Transm Suppl. 59:133–154. 2000.PubMed/NCBI
|
|
37
|
Franklin JL: Redox regulation of the
intrinsic pathway in neuronal apoptosis. Antioxid Redox Signal.
14:1437–1448. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Valencia A and Morán J: Reactive oxygen
species induce different cell death mechanisms in cultured neurons.
Free Radic Biol Med. 36:1112–1125. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Guo H, Chen Y, Hu X, Qian G, Ge S and
Zhang J: The regulation of Toll-like receptor 2 by miR-143
suppresses the invasion and migration of a subset of human
colorectal carcinoma cells. Mol Cancer. 12:772013. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Ma Q, Jiang Q, Pu Q, Zhang X, Yang W, Wang
Y, Ye S, Wu S, Zhong G, Ren J, et al: MicroRNA-143 inhibits
migration and invasion of human non-small-cell lung cancer and its
relative mechanism. Int J Biol Sci. 9:680–692. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Ni Y, Meng L, Wang L, Dong W, Shen H, Wang
G, Liu Q and Du J: MicroRNA-143 functions as a tumor suppressor in
human esophageal squamous cell carcinoma. Gene. 517:197–204. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
42
|
Zhang Y, Wang Z, Chen M, Peng L, Wang X,
Ma Q, Ma F and Jiang B: MicroRNA-143 targets MACC1 to inhibit cell
invasion and migration in colorectal cancer. Mol Cancer. 11:232012.
View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Deacon DC, Nevis KR, Cashman TJ, Zhou Y,
Zhao L, Washko D, Guner-Ataman B, Burns CG and Burns CE: The
miR-143-adducin3 pathway is essential for cardiac chamber
morphogenesis. Development. 137:1887–1896. 2010. View Article : Google Scholar : PubMed/NCBI
|
|
44
|
Miyasaka KY, Kida YS, Banjo T, Ueki Y,
Nagayama K, Matsumoto T, Sato M and Ogura T: Heartbeat regulates
cardiogenesis by suppressing retinoic acid signaling via expression
of miR-143. Mech Dev. 128:18–28. 2011. View Article : Google Scholar : PubMed/NCBI
|