Introduction
Atherosclerosis (AS) is a chronic inflammatory
disease of the vascular wall with multiple causes. It is
characterized by the deposition of lipids in the arterial wall,
infiltration of immune cells, and formation of fibrous caps
(1). AS is the leading pathological
cause of cardiovascular disease and stroke. Carotid plaque rupture
and thrombus formation are the main causes of ischemic stroke.
Patients with unstable/vulnerable plaque AS (UA) are prone to
rupture and thrombus formation, leading to cardiovascular or
cerebrovascular events, such as acute coronary syndrome, ischemic
stroke, and peripheral vascular disease (2). Therefore, identifying the properties
of carotid plaques may help improve the prediction and prevention
of cardiovascular and cerebrovascular events. Previous reports
noted that several factors sampled from plasma or serum may
contribute to carotid plaque vulnerability, symptom status, degree
of stenosis and stroke risk. These factors include the serum
protein C-X-C motif chemokine ligand 9, microRNA (miR)-17-92, and
circular RNA (circRNA)-089763 (3–5).
Exosomes are extracellular vesicles secreted by
living cells. They can circulate in biological fluids, permeate
biological barriers (for example, the brain-blood barrier or the
placental barrier), target recipient cells, and thus share
intercellular signaling molecules between cells (6,7).
Exosomes may represent a potential and effective tool for the
diagnosis and treatment of diseases, due to their extensive
presence in the body and relative ease of sampling. Exosomes can
also be a source of disease-related proteins, miRNAs, and circRNAs
(8–10). Transfer of exosome-enclosed
proteins, miRNAs and circRNAs has been identified as an
intercellular communication method in cardiovascular diseases, such
as atherosclerosis (11,12). The signaling molecules carried by
exosomes circulate in the body and can affect the biological
behavior of target cells, thereby acting as key factors in disease
development.
circRNAs are endogenous non-coding RNA molecules
that differ from linear RNA by their structure, a closed covalent
loop lacking 3′ or 5′ polarity (13). circRNAs can sponge miRNAs through
stable complementary binding and regulate gene expression. Numerous
studies have demonstrated that circRNAs are stable and abundant
within exosomes; moreover, they may be key to the mechanisms
through which exosomes promote certain disease (1,14,15).
Emerging evidence suggests that endothelial cell
dysfunction induces an increase in cell proliferation and migration
that leads to re-endothelialization and angiogenesis of carotid
plaques, which in turn promotes plaque destabilization, plaque
rupture, and thrombus formation (16–18).
However, whether exosomal circRNAs in the serum of patients with
carotid plaques are involved in this process remains unknown.
The aim of the present study was to determine
whether exosomal circRNAs were associated with UA formation.
Specifically, the regulatory role of serum exosomal circRNAs on
endothelial cell behavior was examined in patients with UA, in
order to identify novel mechanisms underlying plaque stability.
Materials and methods
Patient recruitment
The present study was approved by The Ethics
Committee of The Eighth Affiliated Hospital of Sun Yat-Sen
University. Written informed consent was obtained from all
participants or their families. A total of 42 patients diagnosed
with carotid plaque under B-ultrasound examination at The Eighth
Affiliated Hospital of Sun Yat-Sen University from March 2017 and
December 2017 were enrolled in this study. The exclusion criteria
were: i) total occlusion lesion; ii) acute coronary syndrome; iii)
cardiac shock; and iv) presence of cardiomyopathy, severe anemia,
severe renal impairment, or malignant tumor.
Carotid plaque images were obtained using a 1.5-T
magnetic resonance system (1.5-T system; Philips Healthcare). A
volume isotropic turbo spin echo acquisition (VISTA) sequence was
used. The iMap parameters were as follows: i) T1-weighted VISTA,
Time of Repetition (TR)/Time of Echo (TE)=400/16 ms, refocusing
angle=60°, thickness=1 mm, field of view=18 cm, matrix=384×384,
SENSE factor=2, number of signal averaging=2); ii) T2-weighted
VISTA, TR/TE=3500/119 ms, refocusing angle=60°, thickness=1 mm,
field of view=18 cm, matrix=384×384, SENSE factor=2, number of
signal averaging=2, and iii) time of flight Magnetic Resonance
Angiography, TR/TE=16/6.9 ms, flip angle=60°, thickness=1.5 mm,
field of view=22 cm, matrix=512×512, SENSE factor=1.8, number of
signal averaging=2. Patients with lipid-rich and necrotic core
plaques were diagnosed with UA (n=20) according to previous studies
(signal intensity ratio ≥1.25 using T1-weighted VISTA) (19,20).
All other patients were diagnosed with SA (n=22). Clinical and
demographic characteristics, as well as information on ongoing
treatments are presented in Table
I.
 | Table I.Baseline clinical and biochemical
characteristics of the enrolled patients. |
Table I.
Baseline clinical and biochemical
characteristics of the enrolled patients.
| Variables | SA (n=22) | UA (n=20) | P-value |
|---|
| Age, years, mean
(SD) | 47.8 (11.08) | 53.1 (5.84) | 0.072 |
| Sex, male, n
(%) | 9 (81.1) | 7 (70) | 0.654 |
| Smoking, n (%) | 9 (81.8) | 6 (60) | 0.507 |
| Asymptomatic, n
(%) | 2 (18.2) | 4 (40) | 0.426 |
| Amaurosis fugax, n
(%) | 5 (45.5) | 4 (40) | 0.863 |
| Transient ischemic
attack, n (%) | 4 (36.4) | 2 (20) | 0.557 |
| SBP, mmHg, mean
(SD) | 120.7 (13.81) | 130.80 (22.29) | 0.314 |
| DBP, mmHg, mean
(SD) | 76.43 (10.22) | 84.60 (18.40) | 0.085 |
| TC, mmol/l, mean
(SD) | 5.32 (1.46) | 4.74 (1.22) | 0.314 |
| TG, mmol/l, median
(IQR) | 1.47 (0.9–1.6) | 1.62 (1.1–1.9) | 0.251 |
| HDL-C, mmol/l,
median (IQR) | 1.03 (1.0–1.1) | 1.01 (0.9–1.1) | 0.918 |
| LDL-C, mmol/l, mean
(SD) | 2.84 (0.15) | 3.33 (0.18) | 0.035 |
| GLU, mmol/l, median
(IQR) | 5.49 (5.1–6.4) | 5.61 (5.6–6.0) | 0.468 |
| GHb,%, median
(IQR) | 5.70 (5.5–5.8) | 5.75 (5.5–6.1) | 0.842 |
| Uric acid, µmol/l,
median (IQR) | 377
(324.0–408.0) | 447
(363.0–536.0) | 0.261 |
| ALT, U/l, median
(IQR) | 38.00
(26.8–42.8) | 31.50
(18.8–55.5) | 0.705 |
| ALB, U/l, median
(IQR) | 42.60
(40.6–43.2) | 39.75
(37.1–42.0) | 0.085 |
| AST, U/l, median
(IQR) | 30.00
(24.6–40.4) | 25.95
(18.2–53.7) | 0.387 |
| BUN, mmol/l, median
(IQR) | 4.60
(3.7–4.8) | 6.08 (5.0–7.5) | 0.085 |
| GGT, U/l, median
(IQR) | 30.60
(22.2–50.4) | 38.80
(24.9–52.4) | 0.654 |
| sCr, µmol/l, median
(IQR) | 71.36
(50.0–143.0) | 77.49
(55–153.0) | 0.332 |
| CRP, mg/l, mean
(IQR) | 10.55
(1.42–33.77) | 17.95
(3.56–40.68) | 0.029 |
Exosome isolation
A volume of 100 µl serum was collected from every
patient. All samples were combined into a UA serum pool and an SA
serum pool. Both serum pools were centrifuged for 20 min at 2,000 ×
g, 30 min at 10,000 × g, then 20 min at 14,000 × g. All
centrifugation steps were performed at 4°C. Exosomes were then
isolated using a GET-Exosome isolation kit (GenExosome
Technologies, Inc.) according to the manufacturer's instructions.
The concentration of exosomes was evaluated using a BCA assay. All
serum-Exos were stored at −80°C or immediately used for further
experiments.
circRNA isolation
Exosomal circRNAs were extracted from serum-Exos
using Trizol® (Thermo Fisher Scientific, Inc.). For
digestion of linear RNAs, 1 mg of total RNA was incubated with 2
U/µg RNase R for 1 h at 37°C (Thermo Fisher Scientific, Inc.). The
purity and concentration of the RNA samples were assessed with a
NanoDrop™ spectrophotometer (NanoDrop™ Technologies; Thermo Fisher
Scientific, Inc.).
circRNA microarray analysis
Following extraction from SA or UA serum-Exos pools,
circRNAs were divided into three parallel samples for each group
(SA-Exos-1, 2 and 3 vs. UA-Exos-1, 2 and 3) and used for circRNA
microarray analysis. The Human CircRNA Array v2 (CapitalBio
Technology Co., Ltd.) was used; with each slide containing four
identical arrays (4×180K format), representing ~170,340 human
circRNAs. A total of 4,974 Agilent control probes were included in
the array, and 7,775 circRNAs were examined. GeneSpring software
(version 13.0; Agilent Technologies, Inc.) was used to analyze,
standardize and control the quality of the circRNA microarray data.
To identify the differentially expressed genes, a fold change
threshold of ≥1.2 or ≤-1.2 was used. Statistical analysis was
carried out using Student's t-test, with P<0.05
indicating a statistically significant difference. Cluster 3.0
(Stanford University) was used for log2 transformation
and median centering of the data, which were further analyzed
through hierarchical clustering using the average linkage
criterion. The miRanda (http://www.miranda.org/) and CircInteractome
(https://circinteractome.nia.nih.gov/)
tools were used to select the target miRNAs of circRNA-0006896 and
construct a circRNA-miRNA network. A competing endogenous RNA
(ceRNA) network map was constructed using Cytoscape.
Cell culture
Human umbilical vein endothelial cells (HUVECs) were
purchased from the American Type Culture Collection and cultured in
RPMI-1640 medium supplemented with 10% FBS (both from Thermo Fisher
Scientific, Inc.) at 37°C in a humidified atmosphere containing 5%
CO2. At 80–90% confluence, cells were sub-cultured at a
dilution ratio of 1:3. All experiments were performed with cells in
the logarithmic growth phase. For experiments, the cells were
cultured to approximately 80% confluence prior to incubation with
serum exosomes from AS patients with UA (UA-Exos) or stable plaque
(SA-Exos).
Exosome tracking
Isolated exosomes were labelled using a GET-Exosome
labeling kit (GenExosome Technologies, Inc.) according to the
manufacturer's protocol. Briefly, 500 µg/ml serum-Exos (or PBS as a
control) were mixed with an equal volume of GenExosome
ExoPKH26 tracking dye for 10 min at 37°C. The free dye
was then removed using GenExosome centrifugal columns (10-kDa
cutoff). Next, 1×105 cell/ml HUVECs were seeded in a
35-mm confocal dish and incubated with 50 µl 500 µg/ml labelled
UA-Exos or SA-Exos (or PBS as a control) in RPMI-1640 medium
supplemented with 1% exosome-free FBS at 37°C for 24 h.
Internalized, labelled exosome signals were visualized under a
confocal fluorescence microscope (magnification, ×400; Leica
Microsystems, Inc.).
Dual-luciferase reporter assay
Dual-luciferase reporter assays were performed using
the psiCHECK2 vector (Promega Corporation). First, the wild-type
(WT) or mutant (MUT) circRNA-0006896 sequences of the putative
binding-site for miR1264 were subcloned into the psiCHECK2 vector.
Next, 293 cells (ATCC) were seeded in 24-well plates at a density
of 5 ×104 cells/well and co-transfected with of
psiCHECK2-circRNA-0006896-WT or -MUT, together with the miR-1264
mimic or mimic-miR-control (0.2 mg luciferase plasmid and 50 nM
miRNA mimic) using Lipofectamine® 3000 (Invitrogen,
Thermo Fisher Scientific, Inc.). The cells were harvested 48 h
after transfection, and luciferase activity was assessed with a
Dual-Luciferase® Reporter Assay System (Promega
Corporation). Results are presented as relative Renilla
luciferase activities, which were normalized to firefly luciferase
activities.
RNA fluorescence in situ hybridization
(FISH)
RNA FISH was performed to determine the location of
circRNA-0006896 and miR1264 in HUVECs. Biotin-labelled circRNA-
0006896
(5′-biotin-TGTATGGGGAGATGTCTCTCTTTGAGTTAGGTCTAAAGATGATGG-3′) and
digoxigenin-labeled miR-1264 probes
(5′-digoxigenin---AACAGGTGCTCAAATAAGACTTG 3′) were designed and
synthesized by Guangzhou RiboBio Co., Ltd. HUVECs were seeded in
6-well plates at a density of 1×105 cells/ml and
incubated with 200 µl UA-Exos (500 µg/ml) or SA-Exos (500 µg/ml) or
PBS (mock group) in 2 ml conditioned medium at 37°C for 24 h. After
immobilization with 4% paraformaldehyde, cells were prehybridized
in prehybridization buffer (1X PBS with 0.5% Triton X-100), then
hybridized in hybridization buffer (40% formamide; 10% Dextran
sulfate; 1X Denhardt's solution; 4X saline sodium citrate buffer;
10 mM DDT, 1 mg/ml yeast transfer RNA; 1 mg/ml sheared salmon sperm
DNA) with 20 nmol biotin-labelled probes specific for
circRNA-0006896 and 20 nmol digoxigenin-labelled locked nucleic
acid miR-1264 probes at 60°C overnight. The signals of
biotin-labelled probes were detected using Cy3-Streptavidin (Thermo
Fisher Scientific, Inc.). The signals of digoxigenin-labelled
locked nucleic acid miR-1264 probes were detected using the
Molecular Probes™ TSA™ kit with Alexa Fluor™ 488 tyramide reagent
(Thermo Fisher Scientific, Inc.). Finally, the cells were incubated
with 4′,6-diamidino-2-phenylindole (DAPI; 1:100, Thermo Fisher
Scientific, Inc.) for 15 min at room temperature. The images were
acquired using a confocal microscope (magnification, ×600; Leica
Microsystems, Inc.).
RNA extraction and
reverse-transcription quantitative (RT-q) PCR
Total RNAs were extracted from pooled serum-Exos
samples, serum-Exos samples from individual patients or HUVECs by
using Trizol® reagent (Thermo Fisher Scientific, Inc.).
The individual serum-Exos samples was isolated from a 500-µl volume
of individual patient serum using a GET-Exosome isolation kit
(GenExosome Technologies, Inc.). circRNAs or DNMT1mRNA were reverse
transcribed to cDNA with a RevertAid™H Minus First-Strand cDNA
Synthesis Kit (Fermentas, Inc.). The reverse-transcription reaction
was carried out for 60 min at 42°C, followed by a second step of 10
min at 70°C and a final hold at 4°C. miR-1264 was reverse
transcribed to cDNA with a Mir-X™ miRNA First-Strand Synthesis Kit
(Clontech Laboratories, Inc.). The RT reaction was carried out for
60 min at 37°C, followed by a second step of 5 min at 85°C and a
final hold at 4°C. All cDNA was synthesized from 1 µg of total RNA.
The qPCR was carried out using a SYBR™ Green PCR Master Mix Kit
(Thermo Fisher Scientific, Inc.).
The sequences of the primers (all from GeneCopoeia)
used were as follows: i) circRNA-0006896 forward,
5′-TTGGGAAGCCTGGAATATGA-3′ and reverse,
5′-TGGGGAGATGTCTCTCTTTGA-3′; ii) circRNA-0012592 forward,
5′-TCGCATCTACTGGTGTGACC-3′ and reverse, 5′-TCAGTGCAGATGTGTGAGCA-3′;
iii) circRNA-0087352 forward, 5′-TGATGCAGGAGATGATGAGG-3′ and
reverse, 5′-ATTATATCCCCCTGGGATGC-3′; iv) circRNA-0073009 forward,
5′-TTGCTATGACTACATTTTGAGGTTTT-3′ and reverse,
5′-TGGCCTCTCCGAAGTAGAAA-3′; v) circRNA-0008517 forward,
5′-GAATCAAACCTTGGGGACCT-3′ and reverse, 5′-AAGGATGGGTTCAGGTAGGG-3′;
vi) circRNA-0009054 forward, 5′-CTTTCCTCCAACAGCCACAC-3′ and
reverse, 5′-CTGCACGCTCTGTAGTCGAG-3′; vii) DNMT1 forward,
5′-CCTAGCCCCAGGATTACAAGG-3′ and reverse,
5′-ACTCATCCGATTTGGCTCTTTC-3′; viii) GAPDH forward,
5′-CAATGACCCCTTCATTGACC-3′ and reverse, 5′-TTGATTTTGGAGGGATCTCG-3′;
ix) miR1264 forward, 5′-CAAGTCTTATTTGAGCACCTGTT-3′ and Universal 5′
primer 5′-GCGAGCACAGAATTAATACGAC-3′; x) U6 forward,
5′-CTCGCTTCGGCAGCACA-3′ and reverse, 5′-AACGCTTCACGAATTTGCGT-3′.
The thermocycling conditions consisted of an initial denaturation
for 2 min at 95°C, followed by 40 cycles of 20 sec at 95°C, 30 sec
at 60°C, and 40 sec at 72°C. All reactions were performed in an ABI
7500 real-time fluorescence qPCR system (Applied Biosystems; Thermo
Fisher Scientific, Inc.). GAPDH and U6 snRNA were used as the
reference gene. Relative gene expression levels were calculated
using the 2−ΔΔCq method (21).
Western blot analysis
Serum-Exos and HUVECs were collected and incubated
on ice for 30 min using RIPA lysis buffer (Beyotime Institute of
Biotechnology) containing protease inhibitor cocktail
(Sigma-Aldrich; Merck KGaA). Protein quantification was performed
by Pierce™ BCA Protein Assay Kit (Thermo Fisher Scientific, Inc.).
The equivalent of 5 µg protein were separated by 8–12% SDS-PAGE
(Beijing Solarbio Science & Technology Co., Ltd.), then
transferred to PVDF (EMD Millipore). After blocking using 5%
non-fat milk for 1 h at room temperature, the membrane was
incubated with 1:1,000 dilution of primary antibodies against CD63
(cat. no. ab134045), CD9 (cat. no. ab92726), TSG101 (cat. no.
ab125011), phosphorylated (p)-STAT3 (cat. no. ab76315), STAT3 (cat.
no. ab68153), SOCS3 (cat. no. ab16030), DNMT1 (cat. no. ab188453)
and GAPDH (cat. no. ab8245; reference protein) overnight at 4°C.
Next, the membrane was rinsed three times with TBS + 0.1% Tween-20
and incubated for 1 h with the horseradish peroxidase-conjugated
goat anti-rabbit (ab205718; 1:5,000) or goat anti-mouse (ab205719;
1:5,000) secondary antibodies at ambient temperature. Finally, The
signal of protein bands were visualized by Pierce™ ECL Western
Blotting Substrate (Thermo Fisher Scientific, Inc.) using the Tanon
4200 system (Tanon). The integrated density values were calculated
using Quantity One v4.6.6 software (Bio-Rad Raboratories). All
antibodies were purchased from Abcam.
MTT assay
HUVECs were seeded in 96-well plates at a density of
5×104 cells/ml in a100-µl volume of conditioned medium.
Experimental samples were treated with 50 µg/ml UA-Exos or SA-Exos
for 24 or 48 h, while mock samples were treated with PBS. Blank
wells were prepared by adding 100 µl of culture medium to one well
per plate. For each group, six replicates were set up in adjacent
wells. After incubation, 20 µl MTT (5 g/l) was added to each well,
and the plate was incubated for an additional 4 h at 37°C. Then,
the medium in each well was removed, and 150 µl of DMSO was added
for 10 min. The absorbance was measured in each well at 490 nm
using a Multiskan Ascent instrument (Thermo Fisher Scientific,
Inc.). Each experiment was repeated three times.
Wound healing assay
HUVECs were seeded in 6-well plates at a density of
1×106 cells/ ml in a 2-ml volume of FBS-free RPMI-1640
medium. Experimental samples were treated with 50 µg/ml UA-Exos or
SA-Exos, while mock samples were treated with PBS. When cell
confluence reached 80–90%, then a wound was created across the
diameter of the well surface with a pipette tip. After 24 h, the
wound area was measured under a light microscope (magnification,
×100). The migration distance was measured at five locations per
sample. The level of migration area was evaluated as follows:
migration area (%)=(A0-A1)/A0 × 100, where A0 is the initial wound
area and A1 is the wound area at 24 h.
Transwell migration assay
Cell migration assays were performed using Transwell
cell culture chambers (Thermo Fisher Scientific, Inc.). HUVECs were
seeded into the upper chamber at a density of 2×104
cells/ml in a 100-µl volume of conditioned medium, to which 50
µg/ml UA-Exos or SA-Exos was added. The HUVECs treated with 50 µl
PBS were used as mock group. The lower chamber was loaded with 0.8
ml medium supplemented with 10% exosome-free FBS. The Transwell
plates were incubated at 37°C and 5% CO2 for 24 h. After
incubation, the filters were stained with 0.1% crystal violet, then
decolorized with 33% acetic acid (22). Finally, the absorbance was measured
at 570 nm using a Multiskan Ascent instrument (Thermo Fisher
Scientific, Inc.) to evaluate the migratory ability of HUVECs.
Statistical analysis
SPSS 16.0 (SPPS, Inc.) was used for statistical
analysis. The distribution of the data was determined using the
Kolmogorov-Smirnov test. Normally distributed data are presented as
the mean ± SD, and were analyzed using an unpaired Student's
t-test, or one-way ANOVA followed by Bonferroni correction. Data
with a skewed distribution are presented as the medians and
interquartile ranges (IQR), and were analyzed using Mann-Whitney's
U test. Categorical variables are presented as counts or
percentages, and were analyzed using the χ2 test.
Correlations between serum-Exo-circRNA-0006986 and clinical
biochemical parameters were evaluated using Spearman's rank
correlation analysis. Scientific graphs were generated using
GraphPad Prism (version 5.0; GraphPad Software, Inc.). P<0.05
was considered to indicate a statistically significant
difference.
Results
Demographic and baseline
characteristics of the study population
A total of 22 patients with SA (age range, 37–57)
and 20 patients with (age range, 41–65) were enrolled in this
study. Their baseline clinical and biochemical characteristics are
summarized in Table I. The
concentration the concentration of low-density lipoprotein
cholesterol (LDL-C) and C-reactive protein (CRP) were significantly
increased in the UA group, compared with the SA group. No
significant differences in age, sex, smoking status, or any other
biochemical parameter were observed between the two groups.
Exosome isolation and
identification
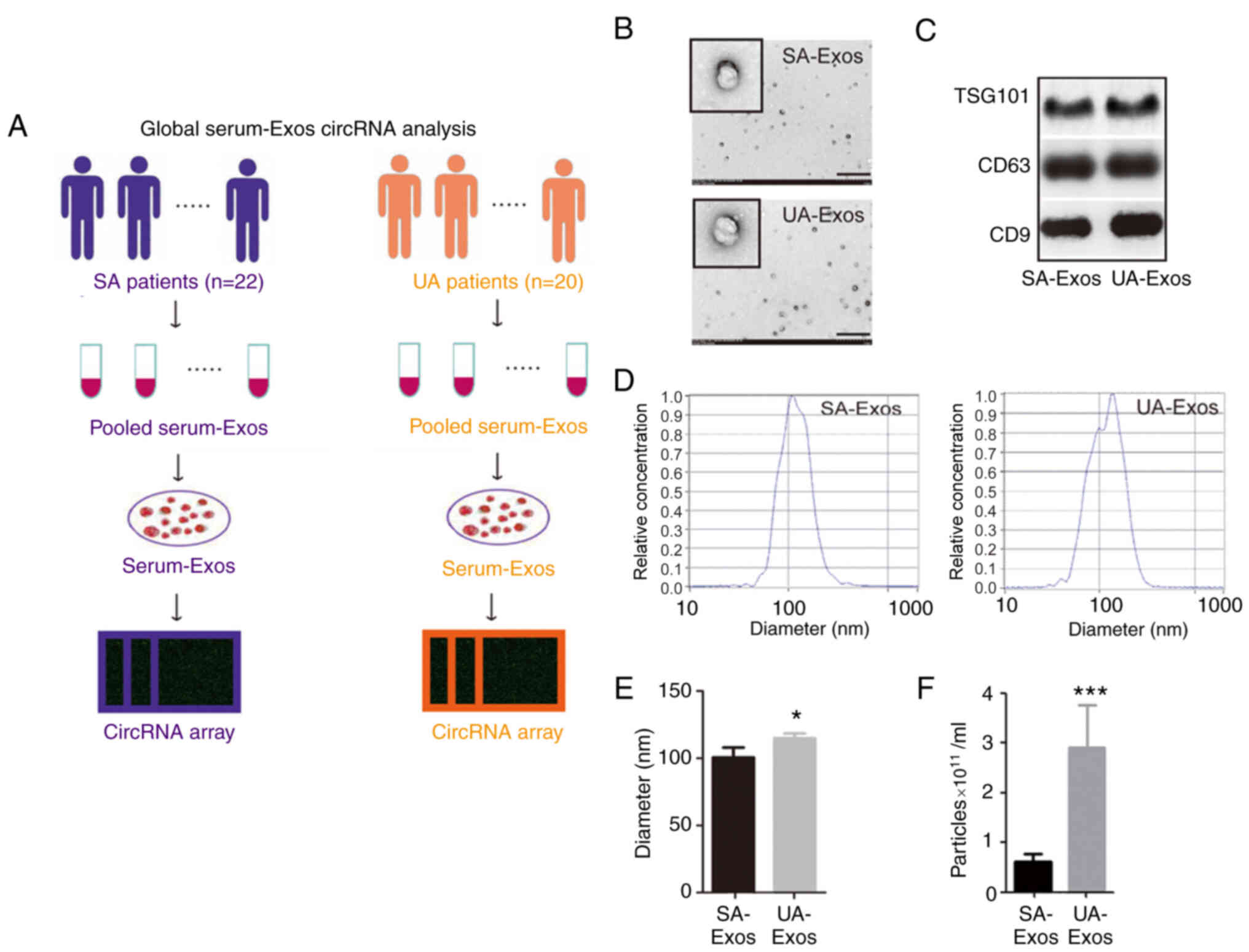
Serum samples from patients with SA or UA were
pooled for exosome extraction (Fig.
1A). Under the transmission electron microscope, the exosomes
exhibited a double-membrane structure in both groups, and the
diameters of most exosomes were <200 nm (Fig. 1B). Moreover, the exosomes expressed
the exosomal markers CD9, CD63, and TSG101 (Fig. 1C). The concentration of exosomes in
the SA group was significantly increased, compared with the UA
group (P<0.001). However, their diameters were only marginally
different in the two groups (P<0.05; Fig. 1D-F).
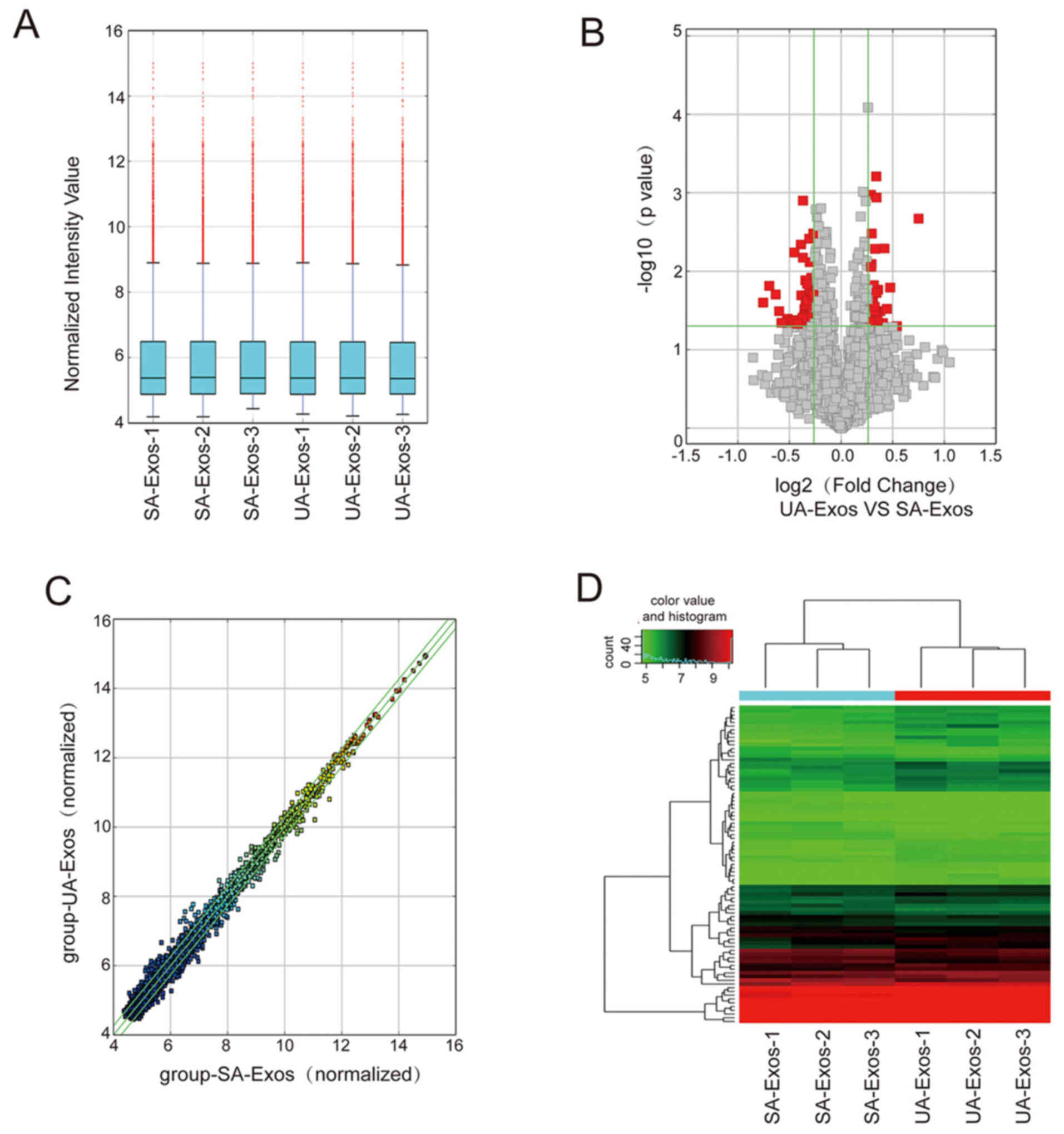
circRNA profile overview
The expression profile of Exos-circRNAs was
determined using microarray analysis. The expression levels of
circRNA molecules were normalized to the same order of magnitude
prior to the statistical analysis. The median expression levels of
different groups nearly the same following normalization,
indicating a great degree of standardization (Fig. 2A). A total of 75 significant
differentially expressed circRNAs in the UA-Exos and SA-Exos groups
are displayed as a volcano plot in Fig.
2B. A scatter plot demonstrated the variability of 5,343
circRNAs expression in UA-Exos and SA-Exos (Fig. 2C). The expression levels of all
5,343 circRNAs in UA-Exos and SA-Exos were quantified and
visualized on a hierarchical clustering heat map. Only 37 circRNAs
were upregulated and 48 were downregulated in the UA-Exos group,
compared with SA-Exos, indicating a strikingly similar expression
pattern of circRNAs between these two groups (Fig. 2D).
RT-qPCR validation, prediction of
mRNA-miRNA-circRNA relationships and enrichment analysis of the
biological functions of circRNA-0006896-co-expressed genes
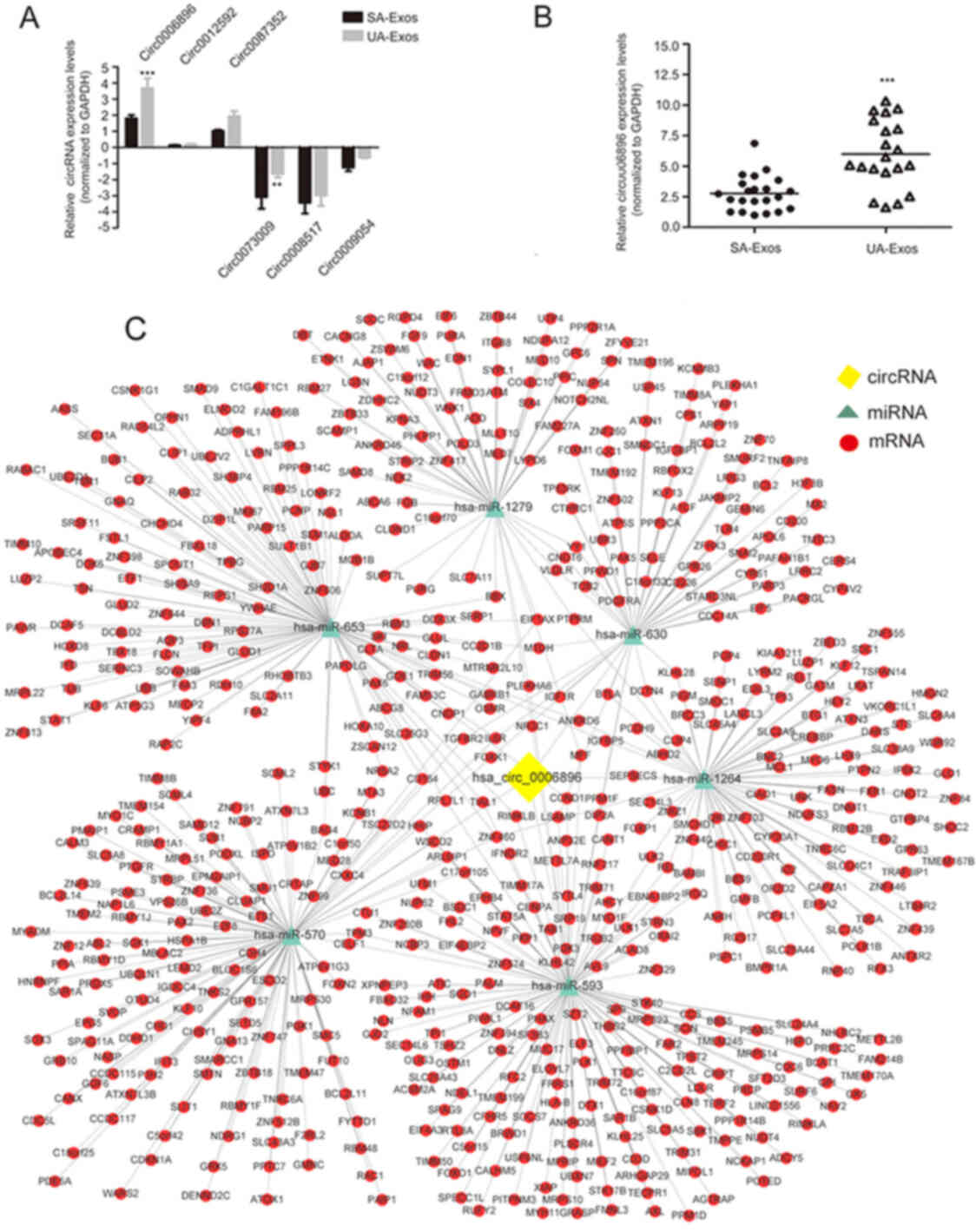
To verify the microarray profiling expression data,
the expression of the three most upregulated (circRNA-0006896,
circRNA-0012592 and circRNA-0087352) or downregulated circRNAs
(circRNA-0073009, circRNA-0008517 and circRNA-0009054) was
validated using RT-qPCR. Among these circRNAs, the circRNA-0006896
was the only one that expressed over ~2-fold UA-Exos samples
relative to SA-Exos (Fig. 3A).
Therefore, circRNA-0006896 was selected as a candidate for
subsequent experiments. Moreover, the expression trend of
circRNA-0006896 was further verified in individual serum-Exos
samples from patients with SA (n=22) and UA (n=20). As shown in
Fig. 3B, the expression of
circRNA-0006896 was significantly increased in serum-Exos from UA
patients, compared with SA patients. These results suggested that
circRNA-0006896 may be involved in plaque instability.
Furthermore, circRNAs can serve as a competitive
endogenous RNA (ceRNA) to sponge miRNAs to regulate the target
mRNAs (14,15). To describe the role of
circRNA-0006896 during the progression of carotid plaque
destabilization, bioinformatics analysis was used to predict the
potential target miRNAs of circRNA-0006896 and construct a
circRNA-miRNA network map associated with AS. This analysis
predicted seven target miRNAs (miR-1264, miR-1279, miR-570,
miR-593, miR-630 and miR-653) for circRNA-0006896. A ceRNA network
map was then constructed, which contained one circRNA, six miRNAs
and 768 mRNAs (Fig. 3C). This
network map revealed that the upregulation of circRNA-0006896 in UA
-Exos might upregulate the expression levels of 122 mRNA molecules
by reducing the expression of miR1264, upregulate 118 mRNA targets
by reducing the expression of miR1279, upregulate 70 mRNA targets
by reducing the expression of miR570, upregulate 285 mRNA targets
by reducing the expression of miR593, upregulate 96 mRNA targets by
reducing the expression of miR630 and upregulate 194 mRNA targets
by reducing the expression of miR653 (Fig. 3C).
Correlations between circRNA-006896
serum-Exos levels with biochemical parameters in AS patients
To further examine the role of circRNA-0006896 in
the progression of carotid plaque instability, Spearman's rank
correlation analysis was carried out to determine whether exosomal
circRNA-0006896 levels were associated with biochemical parameters
in patients with SA or UA. circRNA-006896 levels in serum-Exos were
positively correlated with LDL-C levels in the SA group. Moreover,
in the UA group, they were also positively correlated with
triglyceride, LDL-C and CRP levels, but negatively correlated with
albumin levels. Moreover, no statistically significant correlations
were observed between the serum-Exo-circRNA-006896 level and the
levels of total cholesterol, high density lipoprotein cholesterol,
glucose, glycated hemoglobin, uric acid, alanine aminotransferase,
aspartate aminotransferase, blood urea nitrogen, γ-glutamyl
transpeptidase, or creatinine in either the UA or SA group
(Table II). These findings
suggested that the increased expression of circRNA-006896 in UA
serum-Exos was positively correlated with the levels of TG, LDL-C
and CRP in UA patients.
 | Table II.Correlation between circRNA-0006896
expression and biochemical parameters in AS patients with SA or
UA. |
Table II.
Correlation between circRNA-0006896
expression and biochemical parameters in AS patients with SA or
UA.
|
| SA (n=22) | UA (n=20) |
|---|
|
|
|
|
|---|
| Biological
parameters | r | P-value | r | P-value |
|---|
| TC | 0.454 | 0.161 | 0.191 | 0.421 |
| TG | −0.006 | 0.986 | 0.653 | 0.002 |
| HDL-C | 0.126 | 0.713 | −0.505 | 0.136 |
| LDL-C | 0.432 | 0.018 | 0.547 | 0.008 |
| GLU | 0.109 | 0.749 | 0.113 | 0.755 |
| GHb | −0.225 | 0.561 | −0.421 | 0.226 |
| Uric acid | 0.276 | 0.412 | 0.662 | 0.052 |
| ALT | −0.420 | 0.199 | −0.023 | 0.915 |
| ALB | −0.579 | 0.062 | −0.927 | <0.001 |
| AST | 0.392 | 0.081 | −0.876 | 0.783 |
| BUN | 0.088 | 0.797 | 0.454 | 0.187 |
| GGT | −0.448 | 0.167 | −0.250 | 0.486 |
| sCr | 0.334 | 0.129 | 0.287 | 0.220 |
| CRP | 0.145 | 0.536 | 0.532 | 0.016 |
UA-Exos influence the
circRNA-0006896-miR-1264-DNMT1 axis in HUVECs
A previous study reported that HUVEC proliferation
and migration are important events that induce plaque
destabilization (23,24). The signaling molecules carried by
exosomes may affect the biological behavior of target cells in the
microenvironment (25). Therefore,
it was hypothesized that endothelial cell phagocytosis of
serum-Exos containing high levels of circRNA-0006896 could promote
endothelial cell proliferation and migration, leading to plaque
instability. According to the ceRNA network hypothesis, circRNAs
can abolish the inhibitory effect of miRNAs on their target genes
and cause changes in intracellular signaling pathways (26). Thus, in this study, the role of the
circRNA-0006896 ceRNA network in HUVECs following treatment with
serum-Exos was further examined.
RT-qPCR analysis suggested that miR1264 was the most
downregulated of the six circRNA-0006896-targeted miRNAs following
treatment with UA-Exos (Fig. 4A). A
previous report indicated that miR-1264 targets the DNMT1
transcript by binding to its 3′UTR, thus affecting DNMT1 expression
and enzyme activity in miR-1264--transfected cells (27,28).
Thus, it was hypothesized that circRNA-0006896 may regulate HUVEC
behavior through a circRNA-0006896-miR-1264-DNMT1 axis. The miRanda
database suggested that the 3′-UTR of circRNA-0006896 contains a
putative direct binding site for miR1264 (Fig. 4B). Dual-luciferase reporter assays
indicated that co-transfection of WT circRNA-0006896 with miR1264
mimics would lead to a significant reduction in fluorescence
intensity, compared with co-transfection with control miRNA mimics.
However, relative fluorescence intensity was not changed when the
MUT circRNA-0006896 construct was co-transfected, indicating the
specific miR1264 sponges function of circRNA-0006896 (Fig. 4C). In addition, confocal microscopy
images indicated that the amount of exosomes phagocytosed by HUVECs
did not appear different between the SA and UA-Exos groups
(Fig. 4D). FISH also confirmed that
circRNA-0006896 colocalized with miR-1264 around the nuclear
membrane (Fig. 4E).
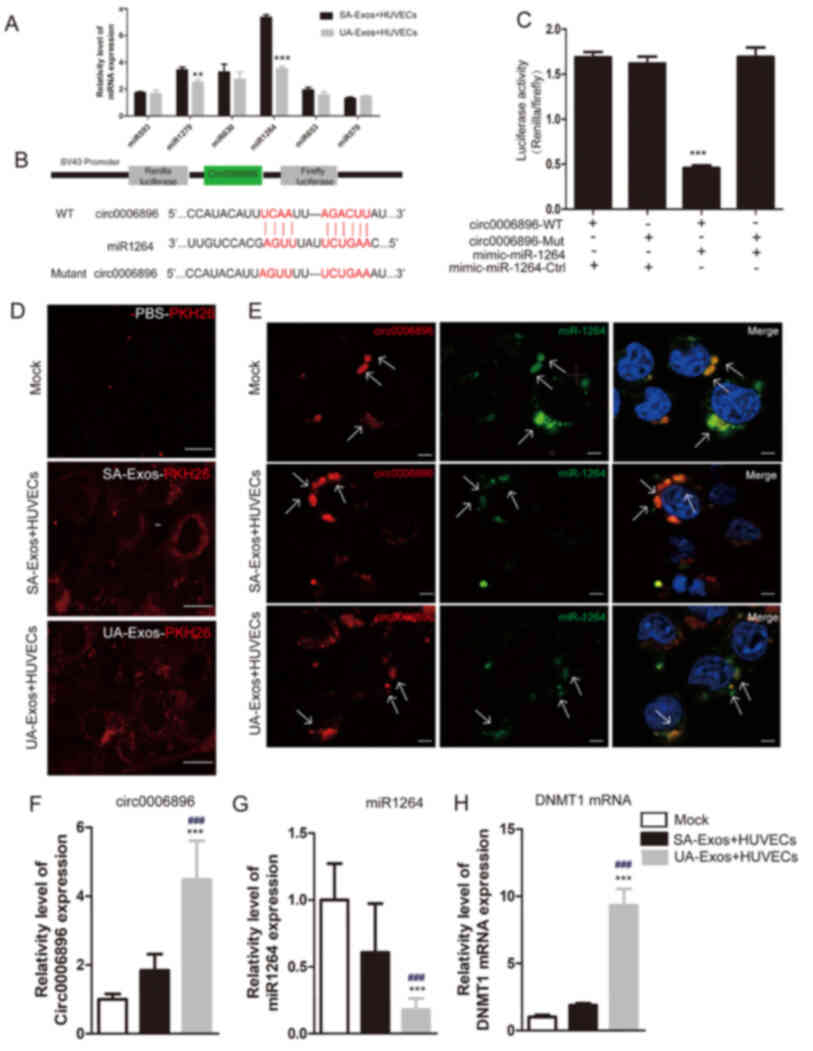 | Figure 4.circRNA-006896-miR1264-DNMT1 axis in
HUVECs. (A) RT-qPCR validation of the expression patterns of six
circRNA-006896 targeted-miRNAs in SA-Exos or UA-Exos-treated
HUVECs. (B) Predicted formation of duplexes at the miR1264 binding
sites in circRNA-0006896 is indicated. (C) A dual-luciferase
reporter gene assay was carried out to verify the binding site.
HUVECs treated with miR-1264 mimic or control miRNA were
co-transfected with psiCHECK2 constructs containing the WT or
mutant circRNA-0006896 sites. (D) Confocal microscopy analysis of
serum-Exos phagocytosis by HUVECs. (E) Colocalization of
circRNA-0006896 with miR1264 in HUVECs Fluorescence in situ
hybridization was used to evaluate the cellular localization of
circRNA-0006896 and miR-1264. Nuclei were stained with DAPI.
Relative expression levels of (F) circRNA-0006896, (G) miR-1264 and
(H) DNMT1 in the mock and serum-Exos-treated HUVECs. Scale bar, 100
µm. Data are presented as the mean ± SD. ***P<0.001,
**P<0.01, ###P<0.001 vs. SA-Exos. Serum-Exos,
serum exosomes; circRNA, circular RNA; miRNA, microRNA; HUVEC,
human umbilical vein endothelial cell; DNMT1, DNA methyltransferase
1; WT, wild-type; Mut, mutant; SA, stable plaque atherosclerosis;
UA, unstable/vulnerable plaque atherosclerosis. |
Furthermore, the expression levels of
circRNA-0006896, miR1264 and DNMT1 were measured in HUVECs
following treatment with serum-Exos from patients with UA or SA. In
the UA group, the expression levels of both circRNA-0006896 and
DNMT1 were significantly increased, compared with the SA group. In
addition, the expression of miR-1264 was significantly lower,
compared with the SA group (Fig.
4F-H). Therefore, the expression levels of circRNA-0006896,
DNMT1 and miR-1264 differed following treatment with SA or UA-Exos.
These data indicated that the high levels of circRNA-0006896
contained within UA-Exos can affect the expression of miR-1264 and
DNMT1 in HUVECs, whereas the small amounts of circRNA-0006896
contained in SA-Exos do not.
The circRNA-0006896 network in UA
exosomes increases the proliferation and migration of HUVECs
Previous studies have documented that DNMT1 is a
critical regulator of JNK, STAT, and NF-κB signaling during the
development of UA because an increase DNMT1 expression results in
CpG island hypermethylation in the promoter region of SOCS3, which
downregulates its expression (28).
Low expression of SOCS3 promotes JNK/STAT signaling, thus
increasing the proliferation and migration of HUVECs (29), which in turn leads to plaque
destabilization and promotes AS development (23,30,31).
Thus, functional assays using HUVECs treated with UA or SA-Exos
were carried out to explore the role of circRNA-0006896, miR-1264
and DNMT1 in AS development.
Western blot analysis suggested that there was an
inverse relationship between SOCS3 and DNMT1 expression. Indeed,
SOCS3 protein expression was reduced in HUVECs following treatment
with UA-Exos, compared with SA-Exos and mock groups. This was
accompanied by a significant increase in STAT3 phosphorylation
(Fig. 5A). In addition, the
proliferation of HUVECs was significantly increased in the UA-Exos
group, compared with the SA serum-Exos and mock groups at 24 and 48
h. (Fig. 5B). A wound healing assay
was conducted to investigate the effects of circRNA-006896 on HUVEC
migration. HUVEC migration increased by ~20% in the UA-Exos group,
compared with the SA-Exos and mock groups (Fig. 5C). In a Transwell assay, staining
intensity significantly increased for HUVECs after treatment with
UA-Exos for 48 h, compared with the UA and mock groups, suggesting
increased HUVEC migration (Fig.
5D). Collectively, these results suggested that high
concentrations of circRNA-0006896 in UA-Exos enhances the
proliferation and migration of HUVECs, possibly through
DNMT1/SOCS3/JNK/STAT3 signaling.
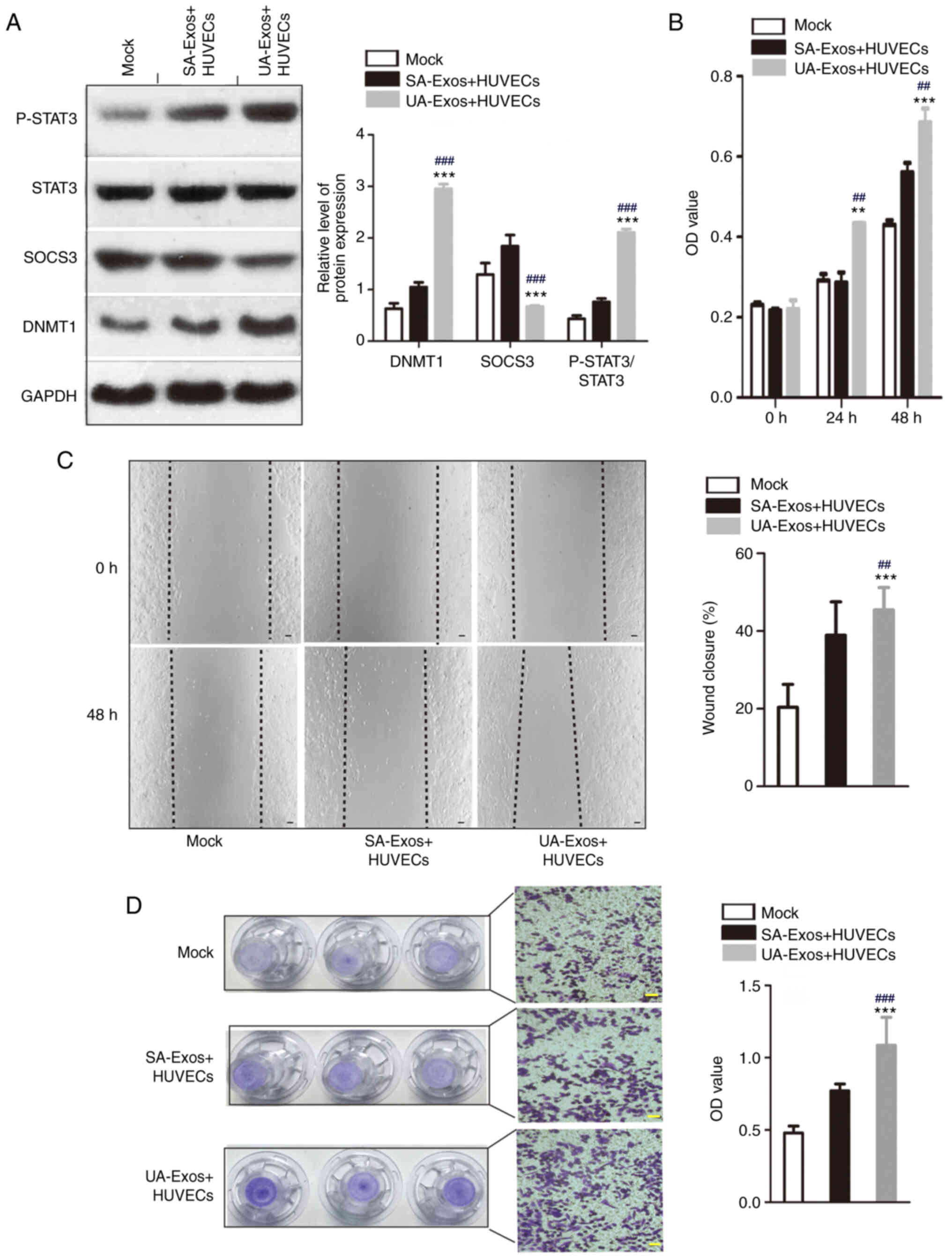 | Figure 5.Highly expressed circRNA-0006896
encapsulated in UA-serum-Exos increases the proliferation and
migration of HUVECs. (A) Protein levels of DNMT1, SOCS3, P-STAT3
and STAT3 were determined by western blot analysis. (B) MTT assay.
(C) Wound healing assay. (D) Transwell assay. Scale bar, 100 µm.
Data are presented as the mean ± SD. ***P<0.001, **P<0.01 vs.
mock group; ###P<0.001, ##P<0.01 vs.
SA-Exos. Exos, exosomes; HUVEC, human umbilical vein endothelial
cell; DNMT1, DNA methyltransferase 1; SOCS3, suppressor of cytokine
signaling 3; signal transducer and activator of transcription 3; P,
phosphorylated; SA, stable plaque atherosclerosis; UA,
unstable/vulnerable plaque atherosclerosis; OD, optical
density. |
Discussion
Previous studies have been conducted to identify
therapeutic targets for UA, most of which have been limited to
protein-coding genes, miRNAs and lncRNAs (32–35).
Recently, circRNAs have attracted attention as new diagnostic
markers for diseases, including cancer. circRNAs constitute a rich,
stable, diverse and conserved family of RNA molecules. Emerging
evidence suggests that circRNAs participate in various biological
processes, such as angiogenesis, proliferation, and differentiation
(36). Recently, Zhang et al
(37) demonstrated that the
crosstalk between circRNAs and their competing mRNAs might play
crucial roles in the development of UA by regulating cell adhesion,
migration, and activation. Moreover, exosomal circRNAs derived from
peripheral circulating serum are associated with UA formation.
Jiang et al (38) reported
that peripheral circulating serum-Exos are can induce TNF-α and
IL-6 production in macrophages through their miRNA cargo.
An increase in inflammatory cytokines has been
proven to be related to plaque vulnerability across various stages
of AS, in which vascular endothelial cells are dysfunctional and
induced by stimulatory factors. Endothelial cell functions, such as
antithrombogenicity and angiogenic sprouting capacity, are lost.
However, mesenchymal cell characteristics, such as contractility,
proliferation, and migration, are enhanced. Functionally,
endothelial cells play a vital role in the development and ultimate
rupture of unstable plaques, which cause most acute coronary artery
events (3,23). However, few studies have explored
the stimulatory effect of peripheral circulating serum-Exos on the
vascular endothelium in AS patients.
Therefore, in the present study, the circRNA
expression profiles of serum-Exos from patients with SA or UA were
used to assess the effect of exosomal circRNAs on vascular
endothelial cell activation. circRNA-0006896 was selected for
further study due to its higher expression in patients with UA,
compared with patients with SA. Furthermore, the expression levels
of circRNA-0006896 were positively correlated with the levels of
triglyceride, LDL-C and CRP, and negatively correlated with albumin
levels in patients with UA. Since increased levels of serum LDL-C
and CRP are an important clinical feature of UA formation, it was
hypothesized that serum-Exo-circRNA-0006896, which is positively
correlated with their expression, could play an important role in
the formation of unstable carotid plaques.
The circRNA-0006896-miR-1264-DNMT1 axis was selected
as a platform to study the regulatory roles of circRNA-0006896 in
serum-Exos through circRNA bioinformatic analysis. The results
indicated that following treatment with UA serum-Exos, the
expression of circRNA-0006896 in HUVECs was upregulated, which was
accompanied by downregulated expression of miR1264 and upregulated
expression of DNMT1 mRNA. Recently, DNMT1 has been identified as
the direct target of miR1264, which can bind to the 3′UTR of the
DNMT1 transcript to inhibit its expression. DNMT1 is an important
enzyme implicated in DNA methylation, particularly in the promoter
region of SOCS3. Moreover, CpG islands, one of which is located at
kb 4.261–6.673 in the SOCS3 genomic locus, regulate DNMT1 function
(27,28). miR-1264 downregulation facilitates
persistent DNMT1 expression, thereby inducing SOCS3 promoter
methylation and impacting its gene expression (27). Downregulated expression of SOCS3
results in loss of its inhibitory effect on the JNK/STAT3 pathway
(30,31). The proliferation and migration of
HUVECs were increased most significantly in the UA groups compared
to the SA and mock groups, possibly indicating its role in the
formation of unstable plaques.
In conclusion, increased expression of
circRNA-0006896 in serum-Exos from patients with UA promotes the
proliferation and migration of HUVECs by negatively regulating the
expression of miR-1264. In turn, this leads to an increase in DNMT1
expression and STAT3 phosphorylation, and a reduction in the
expression of SOCS3. These findings indicate that circRNA-0006896
is a potential therapeutic target that regulates JNK/STAT3 pathway
activation and may influence vulnerable plaque formation by forming
complex regulatory networks in patients with UA. Therefore, this
study may provide insight into novel interventions against
vulnerable plaque formation in patients with UA.
Acknowledgements
Not applicable.
Funding
The present study were supported by the Shenzhen
Sciences and Technology Project Foundation (grant no.
JCYJ201700818162010186) and the Public welfare research project in
Futian District, Shenzhen (grant. no. FTWS2018073).
Availability of data and materials
The data and materials of this study are available
from the corresponding author upon reasonable request
Authors' contributions
YW and ZHJ conceived and designed the experiments.
YC, YML, ZQL, CJ and ZWY performed the experiments. CYX, ZWQ, LH
and LSJ collected and analyzed the data. YW and CJ were the major
contributor in writing the manuscript. All authors read and
approved final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics Revie
Committee of the Eighth Affiliated Hospital of Sun Yat-Sen Unive
rsity (Shenzhen, China).
Patient consent for publication
All participants have given their written permission
for publication of obtained data.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Getz GS and Reardon CA: Atherosclerosis:
Cell biology and lipoproteins. Curr Opin Lipidol. 31:286–290. 2020.
View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Harari F, Barregard L, Östling G, Sallsten
G, Hedblad B, Forsgard N, Borné Y, Fagerberg B and Engström G:
Blood lead levels and risk of atherosclerosis in the carotid
artery: Results from a swedish cohort. Environ Health Perspect.
127:1270022019. View
Article : Google Scholar : PubMed/NCBI
|
|
3
|
Schiro A, Wilkinson FL, Weston R, Smyth
JV, Serracino-Inglott F and Alexander MY: Elevated levels of
endothelial-derived microparticles, and serum CXCL9 and SCGF-β are
associated with unstable asymptomatic carotid plaques. Sci Rep.
5:166582015. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Ferronato S, Mombello A, Posenato I,
Candiani P, Scuro A, Setacci C and Gomez-Lira M: Expression of
circulating miR-17-92 Cluster and HDAC9 gene in atherosclerotic
patients with unstable and stable carotid plaques. Genet Test Mol
Biomarkers. 21:402–405. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
5
|
Wang M, Su P, Liu Y, Zhang X, Yan J, An X,
Wang X and Gu S: Abnormal expression of circRNA_089763 in the
plasma exosomes of patients with postoperative cognitive
dysfunction after coronary artery bypass grafting. Mol Med Rep.
20:2549–2562. 2019.PubMed/NCBI
|
|
6
|
Wang Y, Xie Y, Zhang A, Wang M, Fang Z and
Zhang J: Exosomes: An emerging factor in atherosclerosis. Biomed
Pharmacother. 115:1089512019. View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Wang Y, Xie Y, Zhang A, Wang M, Fang Z and
Zhang J: Corrigendum to ‘Exosomes: An emerging factor in
atherosclerosis’. Biomed Pharmacother. 118:1091192019. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Dumache R, Ciocan V, Muresan C, Rogobete
AF and Enache A: Circulating MicroRNAs as promising biomarkers in
forensic body fluids identification. Clin Lab. 61:1129–1135. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Cortez MA, Bueso-Ramos C, Ferdin J,
Lopez-Berestein G, Sood AK and Calin GA: MicroRNAs in body
fluids-the mix of hormones and biomarkers. Nat Rev Clin Oncol.
8:467–477. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Kuner R, Brase JC, Sultmann H and Wuttig
D: microRNA biomarkers in body fluids of prostate cancer patients.
Methods. 59:132–137. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Goetzl EJ, Goetzl L, Karliner JS, Tang N
and Pulliam L: Human plasma platelet-derived exosomes: Effects of
aspirin. FASEB J. 30:2058–2063. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Lu M, Yuan S, Li S, Li L, Liu M and Wan S:
The exosome-derived biomarker in atherosclerosis and its clinical
application. J Cardiovasc Transl Res. 12:68–74. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Geng X, Jia Y, Zhang Y, Shi L, Li Q, Zang
A and Wang H: Circular RNA: Biogenesis, degradation, functions and
potential roles in mediating resistance to anticarcinogens.
Epigenomics. 12:267–283. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Wang L, Zheng Z, Feng X, Zang X, Ding W,
Wu F and Zhao Q: circRNA/lncRNA-miRNA-mRNA network in oxidized,
low-density, lipoprotein-induced foam cells. DNA Cell Biol.
38:1499–1511. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Luo J, Liu H, Luan S and Li Z: Guidance of
circular RNAs to proteins' behavior as binding partners. Cell Mol
Life Sci. 76:4233–4243. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Kuret T, Sodin-Semrl S, Mrak-Poljsak K,
Cucnik S, Lakota K and Erman A: Interleukin-1β induces
intracellular serum amyloid A1 expression in human coronary artery
endothelial cells and promotes its intercellular exchange.
Inflammation. 42:1413–1425. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Marchio P, Guerra-Ojeda S, Vila JM,
Aldasoro M, Victor VM and Mauricio MD: Targeting early
atherosclerosis: A focus on oxidative stress and inflammation. Oxid
Med Cell Longev. 2019:85638452019. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Lima Junior JC, Moura-Assis A, Cintra RM,
Quinaglia T, Velloso LA and Sposito AC: Central role of obesity in
endothelial cell dysfunction and cardiovascular risk. Rev Assoc Med
Bras (1992). 65:87–97. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Tanemura H, Maeda M, Ichikawa N, Miura Y,
Umeda Y, Hatazaki S, Toma N, Asakura F, Suzuki H, Sakaida H,
Matsushima S and Taki W: High-risk plaque for carotid artery
stenting evaluated with 3-dimensional T1-weighted gradient echo
sequence. Stroke. 44:105–110. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Yamada K, Yoshimura S, Kawasaki M, Enomoto
Y, Asano T, Hara A, Minatoguchi S and Iwama T: Embolic
complications after carotid artery stenting or carotid
endarterectomy are associated with tissue characteristics of
carotid plaques evaluated by magnetic resonance imaging.
Atherosclerosis. 215:399–404. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Method. 25:402–408. 2001.
View Article : Google Scholar
|
|
22
|
Wang Y, Shao S, Luo M, Huang S, Feng L,
Yuan N, Wu F, Dang C and Zhao X: Effects of rat bone marrow-derived
mesenchymal stem cells on breast cancer cells with differing
hormone receptor status. Oncol Lett. 14:7269–7275. 2017.PubMed/NCBI
|
|
23
|
Yang XP, Irani K, Mattagajasingh S,
Dipaula A, Khanday F, Ozaki M, Fox-Talbot K, Baldwin WM III and
Becker LC: Signal transducer and activator of transcription 3alpha
and specificity protein 1 interact to upregulate intercellular
adhesion molecule-1 in ischemic-reperfused myocardium and vascular
endothelium. Arterioscler Thromb Vasc Biol. 25:1395–1400. 2005.
View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Chen LY, Wang X, Qu XL, Pan LN, Wang ZY,
Lu YH and Hu HY: Activation of the STAT3/microRNA-21 pathway
participates in angiotensin II-induced angiogenesis. J Cell
Physiol. 234:19640–19654. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Maia J, Caja S, Strano Moraes MC, Couto N
and Costa-Silva B: Exosome-based cell-cell communication in the
tumor microenvironment. Front Cell Dev Biol. 6:182018. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Awasthi R, Singh AK, Mishra G, Maurya A,
Chellappan DK, Gupta G, Hansbro PM and Dua K: An overview of
circular RNAs. Adv Exp Med Biol. 1087:3–14. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
27
|
Boosani CS, Dhar K and Agrawal DK:
Down-regulation of hsa-miR-1264 contributes to DNMT1-mediated
silencing of SOCS3. Mol Biol Rep. 42:1365–1376. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Boosani CS, Gunasekar P, Block M, Jiang W,
Zhang Z, Radwan MM and Agrawal DK: Inhibition of DNA
methyltransferase-1 instigates the expression of DNA
methyltransferase-3a in angioplasty-induced restenosis. Can J
Physiol Pharmacol. 96:1030–1039. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li S, Geng Q, Chen H, Zhang J, Cao C,
Zhang F, Song J, Liu C and Liang W: The potential inhibitory
effects of miR19b on vulnerable plaque formation via the
suppression of STAT3 transcriptional activity. Int J Mol Med.
41:859–867. 2018.PubMed/NCBI
|
|
30
|
Santana FPR, da Silva RC, Grecco SDS,
Pinheiro AJMCR, Caperuto LC, Arantes-Costa FM, Claudio SR,
Yoshizaki K, Macchione M, Ribeiro DA, et al: Inhibition of MAPK and
STAT3-SOCS3 by sakuranetin attenuated chronic allergic airway
inflammation in mice. Mediators Inflamm. 2019:13563562019.
View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Baus D and Pfitzner E: Specific function
of STAT3, SOCS1, and SOCS3 in the regulation of proliferation and
survival of classical Hodgkin lymphoma cells. Int J Cancer.
118:1404–1413. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lopez-Pedrera C, Barbarroja N,
Patino-Trives AM, Collantes E, Aguirre MA and Perez-Sanchez C: New
biomarkers for atherothrombosis in antiphospholipid syndrome:
Genomics and epigenetics approaches. Front Immunol. 10:7642019.
View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Johnson JL: Elucidating the contributory
role of microRNA to cardiovascular diseases (a review). Vascul
Pharmacol. 114:31–48. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Berkan O, Arslan S, Lalem T, Zhang L,
Sahin NO, Aydemir EI, Korkmaz O, Egilmez HR, Cekin N and Devaux Y:
Regulation of microRNAs in coronary atherosclerotic plaque.
Epigenomics. 11:1387–1397. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Fasolo F, Di Gregoli K, Maegdefessel L and
Johnson JL: Non-coding RNAs in cardiovascular cell biology and
atherosclerosis. Cardiovasc Res. 115:1732–1756. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Pan RY, Zhao CH, Yuan JX, Zhang YJ, Jin
JL, Gu MF, Mao ZY, Sun HJ, Jia QW, Ji MY, et al: Circular RNA
profile in coronary artery disease. Am J Transl Res. 11:7115–7125.
2019.PubMed/NCBI
|
|
37
|
Zhang F, Zhang R, Zhang X, Wu Y, Li X,
Zhang S, Hou W, Ding Y, Tian J, Sun L and Kong X: Comprehensive
analysis of circRNA expression pattern and circRNA-miRNA-mRNA
network in the pathogenesis of atherosclerosis in rabbits. Aging
(Albany NY). 10:2266–2283. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Jiang K, Yang J, Guo S, Zhao G, Wu H and
Deng G: Peripheral circulating exosome-mediated delivery of miR-155
as a novel mechanism for acute lung inflammation. Mol Ther.
27:1758–1771. 2019. View Article : Google Scholar : PubMed/NCBI
|