Introduction
Nasopharyngeal carcinoma (NPC), a highly malignant
tumor originating from the nasopharyngeal epithelium, is among the
most prevalent malignancies in China (1). The combined use of magnetic resonance
imaging and intensity-modulated radiotherapy, as well as advanced
radiotherapy and chemotherapy, have substantially improved the
prognosis of patients with NPC. However, numerous patients are
diagnosed with advanced NPC, and many develop distant metastases
and recurrent disease within 4 years of treatment (2). To date, the pathogenesis of NPC has
not been fully elucidated. Therefore, further research to determine
the pathogenesis of NPC and to elucidate a potential molecular
marker for early diagnosis is urgently warranted.
MicroRNAs (miRNAs/miRs) act as tumor suppressor
genes or oncogenes to modulate tumor progression, and may provide a
new approach to explore the molecular mechanisms of the
pathogenesis of NPC (3). miRNAs are
a diverse class of short (~22 nucleotides) non-coding RNAs that can
regulate gene expression by attenuating translation or promoting
mRNA degradation in a sequence-specific manner. miRNA dysfunction
partially contributes to the development of cancer (4,5).
Aberrant miRNA expression has been observed in patients with NPC.
For example, miR-125b has been reported to be significantly
upregulated in patients with NPC and has been identified as an
independent predictor of poor patient survival. Furthermore,
miR-125b functionally promoted proliferation and inhibited the
apoptosis of NPC cells by targeting the A20/NF-κB signaling pathway
(6). Exosomal miR-24-3p impedes
T-cell function by targeting fibroblast growth factor 11 and serves
as a potential prognostic biomarker of NPC (7). Metastasis-associated miR-23a from
NPC-derived exosomes promotes angiogenesis by targeting TSGA10
(8). Notably, miR-301a-3p is
upregulated and functions as a potential oncogenic RNA in some
types of cancer, including lung cancer, pancreatic cancer, prostate
cancer and glioma (9–12). Nonetheless, the role of miR-301a-3p
and its underlying mechanism in the progression of NPC remain
poorly understood.
The present study demonstrated that miR-301a-3p
upregulation promotes NPC cell proliferation, migration, invasion
and epithelial-mesenchymal transition (EMT) in vitro,
whereas downregulation has an opposite effect. In addition, it was
identified that B-cell translocation gene 1 (BTG1) was the
direct functional target of miR-301a-3p in NPC cells. Finally, it
was shown that miR-301a-3p participates in cell-cell communication
via exosomes. These findings reveal the novel mechanism of NPC
progression and provide a molecular target for NPC research.
Materials and methods
Cell culture
Two NPC cell lines, C666-1 [Epstein-Barr virus
(EBV+)] and 5–8F (EBV-), and an immortalized nasopharyngeal
epithelial cell line (NP69) were procured from the Central South
University (Hunan, China). C666-1 and 5–8F cells were maintained in
RPMI-1640 medium and supplemented with 10% fetal bovine serum (both
from Gibco; Thermo Fisher Scientific, Inc.). NP-69 cells were
cultured in keratinocyte serum-free medium supplemented with human
recombinant epidermal growth factor and bovine pituitary extract
(Sigma-Aldrich; Merck KGaA). All cells were cultured in a
humidified chamber with 5% CO2 at 37°C.
Reverse transcription-quantitative
polymerase chain reaction (RT-qPCR)
Total RNA was extracted from the cells and subjected
to reverse transcription for 60 min at 37°C using the NCode miRNA
First-Strand cDNA Synthesis kit (Thermo Fisher Scientific, Inc.).
RT-qPCR was performed using the ABI 7900HT system with SYBR-Green
PCR Master mix (Applied Biosystems; Thermo Fisher Scientific, Inc.)
using the following thermocycling parameters: 95°C for 10 min,
followed by 40 cycles of 95°C for 10 sec, 60°C for 20 sec and 72°C
for 35 sec. GAPDH and U6 snRNA were used as internal
controls. The data from each sample were normalized with the
internal control, and fold changes were calculated via relative
quantification (2−ΔΔCq) (13). The primer sequences used were as
follows: miR-301a-3p forward, 5′-CAGTGCAATAGTATTGT-3′ and
reverse, 5′-GTGCAGGGTCCGAGGT-3′; U6 forward,
5′-GCGCGTCGTGAAGCGTTC-3′ and reverse, 5′-GTGCAGGGTCCGAGGT-3′;
BTG1 forward, 5′-ACCGTTGTATTCGCATCA-3′ and reverse,
5′-CCATCCTCTCCAATTCTGTA-3′; and GAPDH forward,
5′-ACAACTTTGGTATCGTGGAAGG-3′ and reverse,
5′-GCCATCACGCCACAGTTTC-3′.
Transfection experiments
miR-301a-3p mimic (5′-CAGUGCAAUAGUAUUGUCAAAGC-3′)
and negative control (NC mimic, 5′-UCUACUCUUUCUAGGAGGUUGUGA-3′)
were purchased from Guangzhou RiboBio Co., Ltd. miR-301a-3p
inhibitor (5′-GCUUUGACAATCTATTGCACTG-3′) and negative control
(NC-in, 5′-ACCGCUAAUCAUACGAAUACAC-3′) were purchased from Guangzhou
RiboBio Co., Ltd. Plasmids (pcDNA3.1) expressing BTG1 and
lentiviruses expressing miR-301a-3p were purchased from Hanbio
Biotechnology Co., Ltd. Oligonucleotide (100 nM) transfection was
performed using Lipofectamine® 2000 (Thermo Fisher
Scientific, Inc.). After 48 h of transfection, cells were collected
for further investigation.
Cell proliferation, EdU and colony
formation assays
To determine the effect of miR-301a-3p on cell
proliferation, Cell Counting Kit-8 (CCK-8; Dojindo Molecular
Technologies, Inc.) was used. Transfected cells were plated at
2×103 cells per well in 96-well plates in triplicate. At
various time intervals, the number of cells per well was calculated
as absorbance units at a wavelength of 450 nm.
The EdU assay was performed using the EdU-594 Cell
Proliferation kit (Beyotime Institute of Biotechnology).
Transfected cells were plated at 3×105 cells per well in
6-well plates. After 12 h, cells were then washed with PBS and
fresh medium containing 10 µM EdU was added. Cells were
subsequently incubated for 2 h at 37°C in 5% CO2 and
washed with PBS to remove the free EdU probe and medium. Cells were
then fixed in 4% paraformaldehyde at room temperature for 15 min
and stained with DAPI at room temperature for 5 min. Images were
captured using a fluorescence microscope (magnification, ×200).
For the colony formation assay, 300 cells were
seeded into individual wells of a 6-well plate and cultured at 37°C
for 12 days. The colony counts per well were determined after
staining the cells with 0.1% crystal violet at room temperature for
30 min. Images were captured using a Nikon camera (Nikon
Corporation). Only unambiguous colonies (diameter >40 µm) in the
wells were analyzed using ImageJ software (version 1.49p; National
Institutes of Health).
Transwell assay
Transwell assays were performed as previously
described (10). Transfected cells
(1×105) in serum-free RPMI-1640 medium were placed into
either control inserts or Transwell chambers with or without
Matrigel. A medium with 15% fetal bovine serum was placed in the
lower chamber as a chemoattractant. After incubation at 37°C for 12
h, cells that adhered to the lower surfaces of the membrane inserts
were fixed in 4% paraformaldehyde at room temperature for 30 min,
stained with 0.1% crystal violet at room temperature for 2 h, and
counted under a microscope (magnification, ×200; Olympus
Corporation).
Western blotting
Western blotting was performed as previously
described (8). In brief, protein
lysates (10–30 µg) were separated via sodium dodecyl
sulfate-polyacrylamide gel electrophoresis and were subsequently
transferred onto polyvinylidene difluoride membranes (EMD
Millipore). The membranes were first incubated overnight with
primary antibodies specific for E-cadherin (1:10,000; cat. no.
20874-1-AP), vimentin (1:5,000; cat. no. 10366-1-AP), BTG1
(1:1,000; cat. no. 14879-1-AP), CD63 (1:1,000; cat. no.
25682-1-AP), TSG101 (1:3,000; cat. no. 14497-1-AP) and GAPDH
(1:6,000; cat. no. 60004-1-Ig) that had been diluted in a 5%
low-fat milk-TBS with 0.1% Tween-20 solution and then with a
horseradish peroxidase-conjugated secondary antibody (1:3,000; cat.
no. PR30009). The labeled protein bands were visualized using
enhanced chemiluminescence (Beyotime Institute of Biotechnology),
and band intensities were semi-quantified using ImageJ software
(version 1.49p; National Institutes of Health). All primary and
secondary antibodies were purchased from Wuhan Sanying
Biotechnology.
Target prediction
Targets of miR-301a-3p were searched for using
different computational methods, such as miRanda (http://www.microrna.org/) and TargetScan (http://www.targetscan.org/vert_71/). These
methods identified numerous candidate genes that were commonly
predicted to be possible targets of miR-301a-3p. Gene Ontology
(http://www.geneontology.org/) analysis
was then carried out. Genes classified as having tumor-suppressive
functions involved in NPC were identified; BTG1 was of particular
interest due to its negative roles in cancer cell proliferation and
invasion.
Luciferase reporter assay
The pMIR-REPORT vector (Promega Corporation) was
used to generate luciferase reporters carrying either the wild-type
(Wt) or mutated (Mut) 3′-untranslated region (UTR) of BTG1.
C666-1 and 5-8F cells (2×104) were first seeded in
24-well plates, incubated for 24 h, and then transfected with the
miR-301a-3p mimic (100 nM) and reporter vectors (30 nM) at 37°C
using Lipofectamine® 2000. After 48 h of incubation,
luciferase activity was analyzed using the Dual-Luciferase Reporter
Assay System (Promega Corporation).
Exosome separation
Exosomes were separated from the cell culture medium
using a previously described method (14). Briefly, the culture supernatants
from C666-1 cells that stably overexpress miR-301a-3p were
differentially centrifuged at 300 × g, 1,200 × g and 10,000 × g for
3 h at 4°C. The supernatant obtained was then filtered and
ultracentrifuged at 110,000 × g for 3 h at 4°C. Exosomes were
collected and resuspended in PBS.
Transmission electron microscopy (TEM)
observation
Isolated exosomes were diluted with PBS, and 10 µl
sample was absorbed to a copper mesh. Then, the solution on the
surface of the copper mesh was absorbed with filter paper after
being placed for 5 min. The copper mesh was cleaned once and dyed
for 45 sec with 2% 10 µl uranyl acetate. The image was developed
with 80–120 kV projection electron microscope (JEM-1230; JEOL).
Digital images were collected with a Gatan model 830 ORIUS SC200
CCD camera using Gatan Digital Micrograph software version 3.11
(Gatan, Inc.).
Statistical analyses
The SPSS 17.0 software (SPSS, Inc.) was used for
statistical analyses. All data are presented as the means ±
standard deviations from at least three independent experiments.
The differences between groups were analyzed using unpaired
Student's t-test. Comparisons among multiple groups were analyzed
by one-way analysis of variance (ANOVA), followed by Tukey's post
hoc test. P<0.05 was considered to indicate a statistically
significant difference.
Results
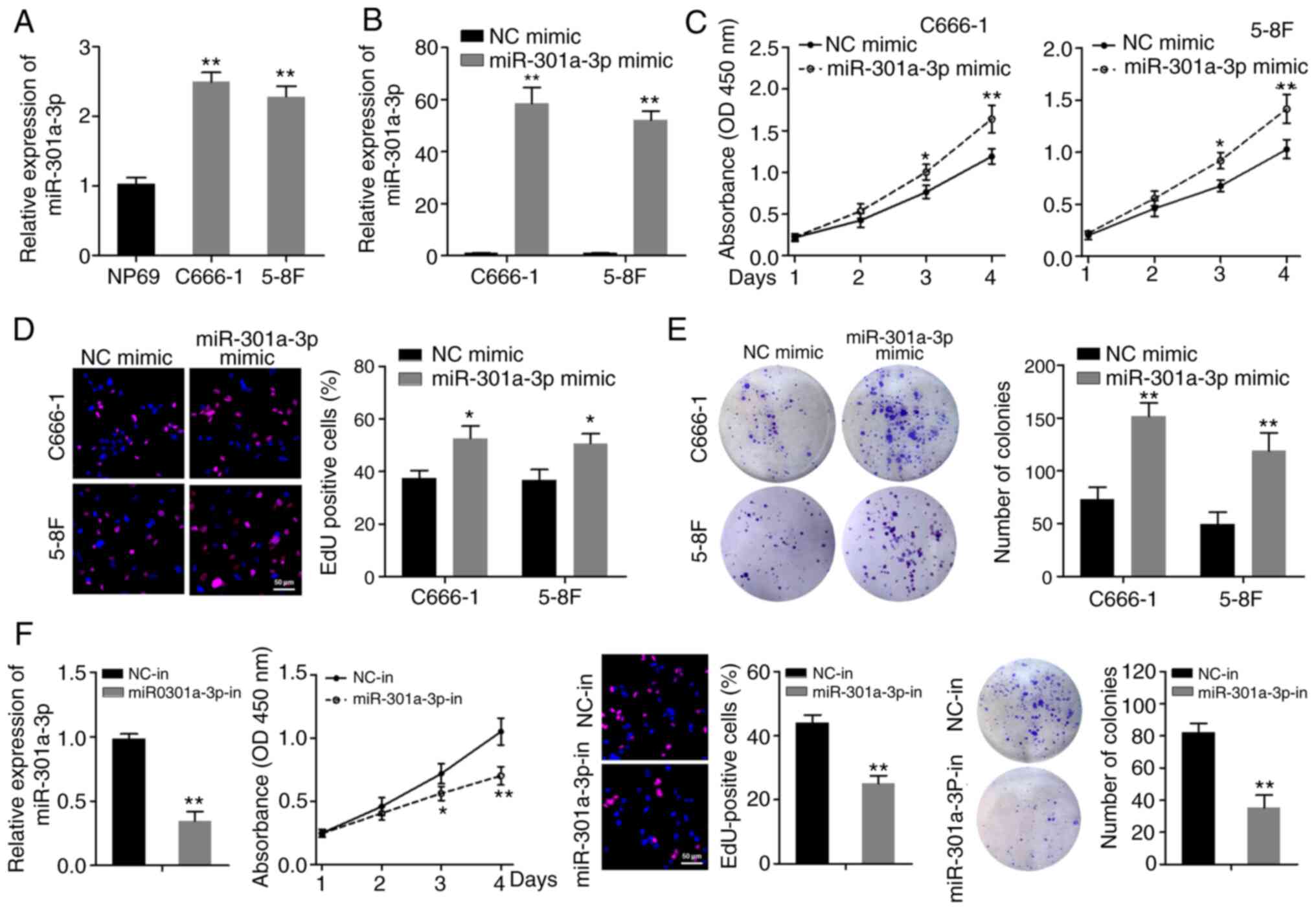
miR-301a-3p is upregulated in NPC cell
lines and promotes NPC cell proliferation in vitro
RT-qPCR was first used to examine the expression
levels of miR-301a-3p in two NPC cell lines (C666-1 and 5-8F) and
in the NP69 immortalized normal human nasopharynx epithelial cell
line. As shown in Fig. 1A,
miR-301a-3p was expressed at higher levels in NPC cell lines
compared with in NP69 cells. Subsequently, the function of
miR-301a-3p in vitro was explored via gain- and
loss-of-function assays, using miR-301a-3p mimics and a specific
inhibitor. The CCK-8 assay revealed that overexpression of
miR-301a-3p enhanced the proliferative abilities of C666-1 and 5–8F
cells (Fig. 1B and C). The EdU
assay revealed that the EdU-positive rate was significantly
increased in NPC cells when miR-301a-3p was overexpressed (Fig. 1D). Furthermore, miR-301a-3p
overexpression increased the colony-formation efficiency of C666-1
and 5–8F cells (Fig. 1E).
Accordingly, C666-1 cells subjected to miR-301a-3p knockdown
exhibited decreased cell proliferation and colony-formation
capacity (Fig. 1F). These data
indicated that miR-301a-3p can enhance the proliferation of NPC
cells.
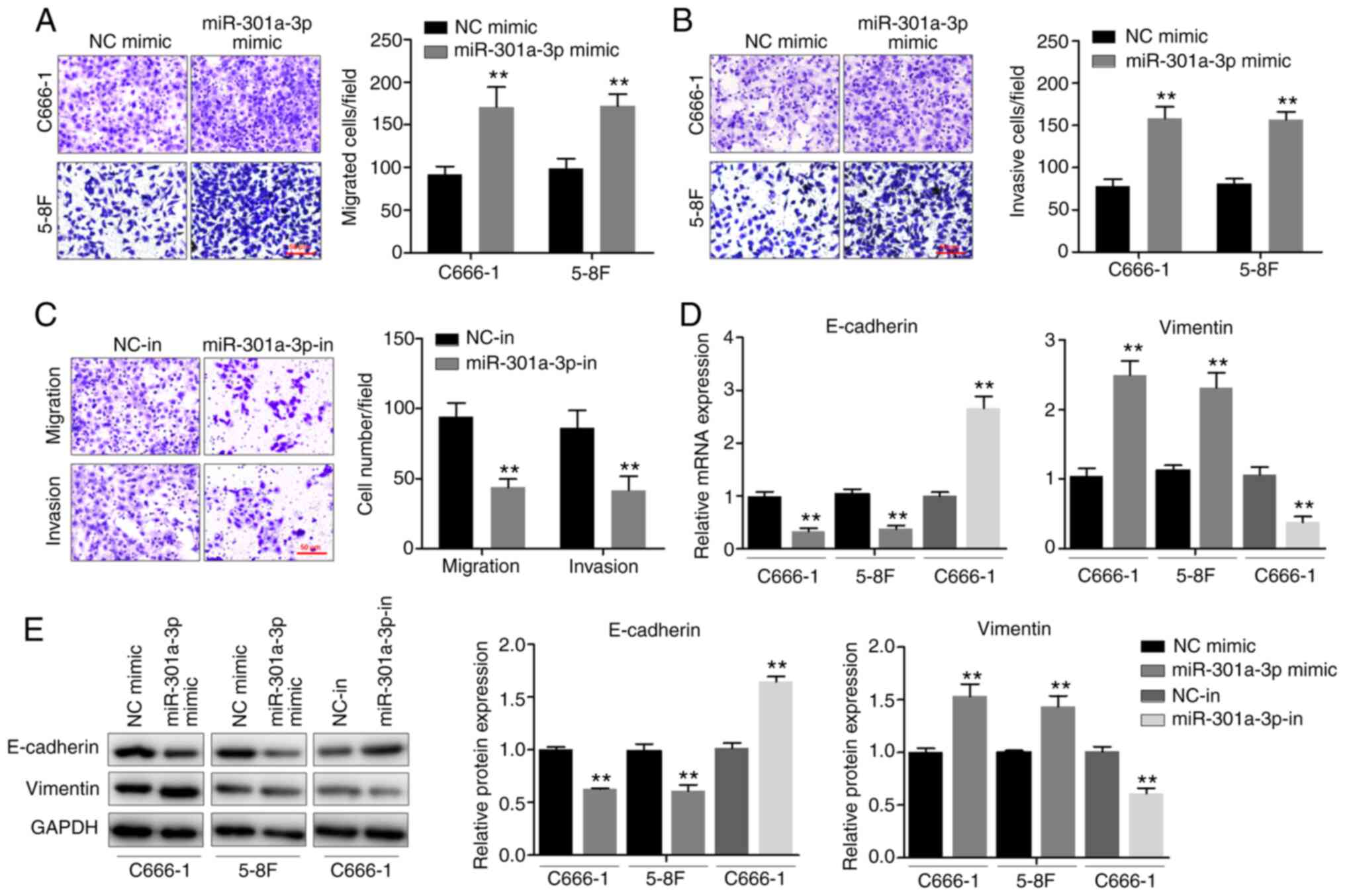
miR-301a-3p promotes NPC cell
migration, invasion and EMT in vitro
To determine the effects of miR-301a-3p on the
migration and invasion of NPC cells, the Transwell chamber assay
was performed. miR-301a-3p overexpression significantly enhanced
the invasive and migratory abilities of both C666-1 and 5–8F cells
(Fig. 2A and B). By contrast,
miR-301a-3p knockdown inhibited the invasive and migratory
abilities of C666-1 cells (Fig.
2C). Since EMT is an important process during metastasis, the
effects of miR-301a-3p on EMT was subsequently determined. The
expression levels of both epithelial and mesenchymal markers were
measured using RT-qPCR (Fig. 2D)
and western blotting (Fig. 2E). It
was observed that miR-301a-3p overexpression was associated with a
significant downregulation of E-cadherin and a notable upregulation
of vimentin, a mesenchymal marker. Furthermore, increased
E-cadherin expression and decreased vimentin expression were
observed following miR-301a-3p knockdown. Altogether, the
observations demonstrated that miR-301a-3p plays a potential role
in promoting NPC cell migration, invasion and EMT in
vitro.
BTG1 is a putative direct target gene
of miR-301a-3p
The potential regulatory targets of miR-301a-3p were
explored via bioinformatics analysis of the miRanda and TargetScan
databases. The analysis revealed BTG1 as a likely target.
One predicted binding site of miR-301a-3p was identified in the
3′-UTR region of BTG1 mRNA (Fig.
3A). RT-qPCR and western blotting assays revealed that
miR-301a-3p overexpression downregulated the expression of BTG1 in
C666-1 and 5–8F cells. In particular, miR-301a-3p overexpression
decreased BTG1 mRNA and protein expression levels, whereas
miR-301a-3p knockdown yielded an opposite effect (Fig. 3B and C). Moreover, luciferase
reporter plasmids BTG1−3′UTR-Wt and BTG1−3′UTR-Mt
were constructed, which were co-transfected in C666-1 and 5–8F
cells with either miR-301a-3p mimic or NC. miR-301a-3p mimic
suppressed the luciferase activity of the cells transfected with
BTG1−3′-UTR-Wt, but not that of cells transfected with
BTG1−3′-UTR-Mt (Fig. 3D). To
further elucidate whether the effects of miR-301a-5p were mediated
by repression of BTG1 in NPC cells, gain-of-function studies were
performed. C666-1 cells were transfected with the BTG1
plasmid without the 3′UTR region or vector control, and successful
increase in BTG1 mRNA expression was verified by RT-qPCR
(Fig. 3E). The BTG1 plasmid
and miR-301a-3p mimic were then co-transfected into C666-1 cells,
and RT-qPCR confirmed the expression of BTG1 mRNA (Fig. 3F). It was found that overexpression
of BTG1 inhibited miR-301a-3p mimic-induced cell
proliferation and invasion in CCK-8, EdU and Transwell invasion
assays (Fig. 3G-I). These
observations demonstrated that BTG1 is potentially a direct target
of miR-301a-3p.
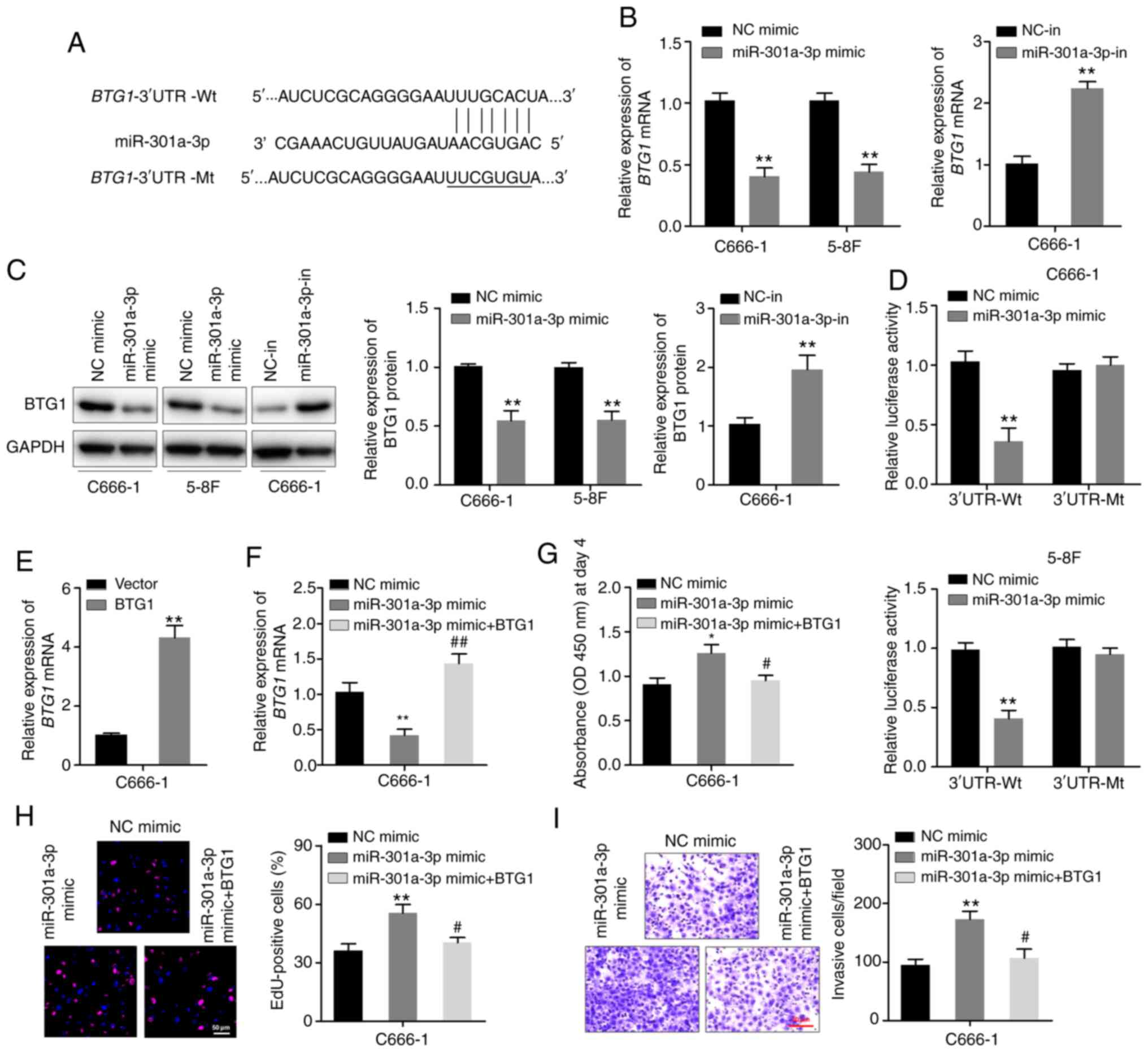 | Figure 3.miR-301a-3p binds to the 3′-UTR of
BTG1 mRNA. (A) The predicted miR-301a-3p-binding site and
mutant miR-301a-3p-binding site in the 3’-UTR of BTG1
(present in the BTG1−3′-UTR-Wt and BTG1−3′-UTR-Mt
plasmids, respectively). (B) RT-qPCR analysis of BTG1
expression of C666-1 and 5–8F cells transfected with the
miR-301a-3p mimic and that of C666-1 cells transfected with the
miR-301a-3p inhibitor. (C) Western blotting analysis of BTG1
expression in the indicated cells. (D) Luciferase activity of
BTG1−3′-UTR-Wt or BTG1−3′-UTR-Mt plasmids in C666-1
and 5–8F cells after transfection with either the miR-301a-3p mimic
or NC. (E) C666-1 cells were transfected with the BTG1
plasmid without the 3′-UTR region or vector control, and RT-qPCR
was used to determine the expression of BTG1 mRNA. (F)
C666-1 cells were co-transfected with the miR-301a-3p mimic and
BTG1 expression plasmid. BTG1 mRNA levels were
detected via RT-qPCR after 48 h. (G) Cell Counting Kit-8 assay. (H)
EdU assay. (I) Transwell invasion assay. *P<0.05, **P<0.01
vs. vector or NC mimic group; #P<0.05,
##P<0.01 vs. miR-301a-3p mimic group. miR, microRNA;
UTR, untranslated region; BTG1, B-cell translocation gene-1; NC,
negative control; in, inhibitor; RT-qPCR, reverse
transcription-quantitative PCR; Wt, wild-type; Mt, mutant. |
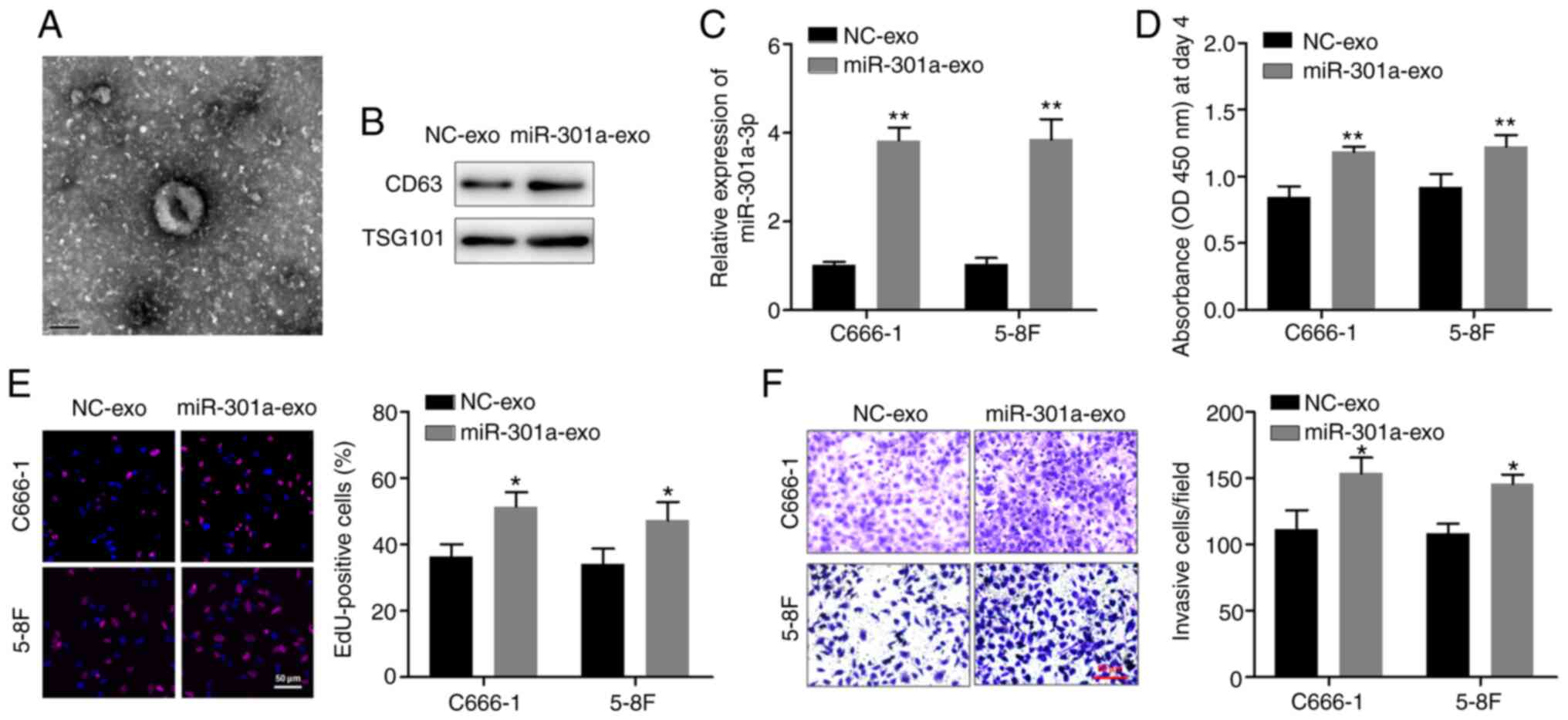
miR-301a-3p participates in cell-cell
communication via exosomes
Finally, C666-1 and 5–8F cells were co-cultured with
exosomes containing high miR-301a-3p levels, in order to determine
whether this miRNA could be delivered via exosomes and regulate the
function of recipient cells. Exosomes secreted from C666-1 cells
stably overexpressing miR-301a-3p were isolated. TEM revealed that
the exosomes had a lipid bilayer membrane structure, and western
blotting confirmed the expression of exosomal markers CD63 and
TSG101 (Fig. 4A and B). C666-1 and
5–8F cells were pretreated with the isolated exosomes, and RT-qPCR
analysis confirmed that the expression levels of miR-301a-3p in NPC
cells were significantly upregulated after treatment with exosomes
derived from miR-301a-3p-overexpressing cells (miR-301a-exo)
compared with control cells (NC-exo) (Fig. 4C). The CCK-8 and EdU assays revealed
that miR-301a-exo significantly promoted the proliferation of
C666-1 and 5–8F cells compared with NC-exo (Fig. 4D and E). Furthermore, the Transwell
invasion assay indicated that miR-301a-exo significantly enhanced
the invasion of NPC cells (Fig.
4F). These observations suggested that exosomal miR-301a-3p is
absorbed by C666-1 and 5–8F cells to participate in cell-cell
communication to further regulate the malignant characteristics of
NPC cells.
Discussion
The present study focused on the regulatory
functions of miR-301a-3p in NPC cells. It was discovered that
miR-301a-3p upregulation promotes the proliferation, migration,
invasion and EMT of NPC cells in vitro. BTG1 mRNA was
identified as the direct and functional target of miR-301a-3p in
NPC cells. Furthermore, miR-301a-3p was delivered via exosomes,
which promoted the proliferation and invasion of NPC cells. These
findings indicate that the miR-301a-3p/BTG1 pathway plays important
roles in the progression of NPC.
The aberrant expression of miR-301a-3p is associated
with cancer initiation and development. miR-301a-3p has been
reported to be upregulated in several types of cancer. For
instance, Li et al (9)
reported for the first time that miR-301a deficiency enhances
CD8+ T-cell accumulation in lung cancer cells by
negatively regulating Runx3. Furthermore, Hu et al (10) showed that miR-301a promotes the
invasion and metastasis of pancreatic cancer cells via the
JAK/STAT3 signaling pathway by targeting SOCS5. Li et al
(11) found that
hyperglycemia-induced miR-301a promotes cell proliferation in
prostate cancer cells by repressing p21 and Smad4. Nevertheless,
the functions of miR-301a-3p in NPC cells remain unknown. In the
present study, miR-301a-3p was overexpressed or knocked down in
C666-1 and 5–8F cells to evaluate the biological role of
miR-301a-3p in NPC cells. It was confirmed that miR-301a-3p
overexpression promotes the proliferation, migration, invasion and
EMT of NPC cells in vitro, whereas miR-301a-3p knockdown
inhibits the proliferation and invasion of these cells. These
observations confirm that miR-301a-3p has an oncogenic function in
NPC cells and support its potential use as a target of cancer
therapy.
Existing experimental data suggest that miR-301a-3p
directly regulates the expression of numerous target mRNAs,
including Runx3, SOCS5, p21, Smad4, TCEAL7, FOXL1, ESR1 and
PTEN (9–12,15–17).
Therefore, it is essential to identify specific
miR-301a-3p-mediated signaling pathways in NPC cells. TargetScan
and miRanda database search identified the 3′-UTR region of
BTG1 mRNA as the binding site of miR-301a-3p. Subsequently,
BTG1 was identified as the critical target gene during
experiments designed to unravel the biological role of miR-301a-3p
in NPC. Furthermore, it was found that miR-301a-3p overexpression
diminishes the mRNA and protein expression levels of BTG1 in
vitro. The luciferase reporter assays in the present study also
revealed that miR-301a-3p can directly bind to the 3′-UTR of
BTG1 mRNA.
BTG1, the key gene within the BTG/TOB family,
is implicated in various biological phenomena, including cancer,
and functions as a tumor suppressor gene (18). In a study on NPC, Sun et al
(19) reported that BTG1 expression
is downregulated in NPC cells and is possibly associated with tumor
metastasis. Based on these findings, gain-of-function assays were
conducted and it was determined that BTG1 overexpression partly
reverses the tumor-promoting influence of miR-301a-3p. In other
words, miR-301a-3p overexpression enhances the proliferation and
invasion of human NPC cells by inhibiting a BTG1-mediated signaling
pathway.
Finally, it was demonstrated that NPC cells transmit
miR-301a-3p to surrounding cancer cells via exosomes and increased
miR-301a-exo levels promote cell proliferation and invasion.
Exosomes, secreted by various cell types, can be absorbed by the
surrounding cells and exert their biological functions in these
receptor cells (20). Exosomes with
nucleic acids, including miRNAs, can contribute to cancer
progression (21). A study
demonstrated that hypoxic tumor-derived exosomal miR-301a promotes
pancreatic cancer metastasis by mediating M2 macrophage
polarization via the PTEN/PI3Kγ pathway (22). Exosomal miR-301a secreted from
hypoxic glioma cells activates the Wnt/β-catenin signaling pathway
and promotes radiation resistance by targeting TCEAL7, and serum
exosomal miR-301a concentration serves as a novel diagnostic
biomarker of glioma and a prognostic factor of advanced-stage
disease (12,23). These findings suggest that exosomal
miR-301a contributes to NPC tumorigenesis and progression. To the
best of our knowledge, no study has evaluated the role of exosomal
miR-301a-3p in NPC cells. Therefore, the present study analyzed
miR-301a-3p levels in exosomes isolated from the cell culture
supernatants of normal and NPC cells and observed high miR-301a-3p
levels in NPC cell-derived exosomes. Next, it was determined
whether miR-301a-3p exerts its functions via exosome delivery by
treating C666-1 and 5–8F cells with the exosomes extracted from
miR-301a-3p-overexpressing cells. Incubation with these highly
expressed exosomes increased the proliferation and invasion of
C666-1 and 5–8F cells. These results indicate that miR-301a-3p
promotes the malignant parameters of NPC cells via exosome
transfer.
In conclusion, the present data indicate that
miR-301a-3p promotes the proliferation and invasion of human NPC
cells by downregulating BTG1 and that this miRNA participates in
cell-cell communication via exosomes. These findings provide a new
direction for further investigating the pathogenesis of NPC.
However, expression patterns of miR-301a-3p and survival outcomes
of patients with NPC were not analyzed in the present study. Thus,
these topics remain to be verified in future research.
Acknowledgements
Not applicable.
Funding
No funding was received.
Availability of materials and data
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
QC and HJ made substantial contributions to the
study conception and design. QC, QL and LX performed data
acquisition, analyses and interpretation. QC drafted the manuscript
and critically revised it for important intellectual content. QC
and HJ confirm the authenticity of all the raw data. All authors
read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Chen W, Zheng R, Baade PD, Zhang S, Zeng
H, Bray F, Jemal A, Yu XQ and He J: Cancer statistics in China,
2015. CA Cancer J Clin. 66:115–132. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Liang TS, Zheng YJ, Wang J, Zhao JY, Yang
DK and Liu ZS: MicroRNA-506 inhibits tumor growth and metastasis in
nasopharyngeal carcinoma through the inactivation of the
Wnt/β-catenin signaling pathway by down-regulating LHX2. J Exp Clin
Cancer Res. 38:972019. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Kong YW, Ferland-McCollough D, Jackson TJ
and Bushell M: microRNAs in cancer management. Lancet Oncol.
13:e249–e258. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Filipowicz W, Bhattacharyya SN and
Sonenberg N: Mechanisms of post-transcriptional regulation by
microRNAs: Are the answers in sight? Nat Rev Genet. 9:102–114.
2008. View
Article : Google Scholar : PubMed/NCBI
|
|
5
|
Li B, Xu WW, Han L, Chan KT, Tsao SW, Lee
NP, Law S, Xu LY, Li EM, Chan KW, et al: MicroRNA-377 suppresses
initiation and progression of esophageal cancer by inhibiting CD133
and VEGF. Oncogene. 36:3986–4000. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Zheng Z, Qu JQ, Yi HM, Ye X, Huang W, Xiao
T, Li JY, Wang YY, Feng J, Zhu JF, et al: MiR-125b regulates
proliferation and apoptosis of nasopharyngeal carcinoma by
targeting A20/NF-κB signaling pathway. Cell Death Dis. 8:e28552017.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Ye SB, Zhang H, Cai TT, Liu YN, Ni JJ, He
J, Peng JY, Chen QY, Mo HY, Jun-Cui, et al: Exosomal miR-24-3p
impedes T-cell function by targeting FGF11 and serves as a
potential prognostic biomarker for nasopharyngeal carcinoma. J
Pathol. 240:329–340. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Bao L, You B, Shi S, Shan Y, Zhang Q, Yue
H, Zhang J, Zhang W, Shi Y, Liu Y, et al: Metastasis-associated
miR-23a from nasopharyngeal carcinoma-derived exosomes mediates
angiogenesis by repressing a novel target gene TSGA10. Oncogene.
37:2873–2889. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Li X, Zhong M, Wang J, Wang L, Lin Z, Cao
Z, Huang Z, Zhang F, Li Y, Liu M, et al: miR-301a promotes lung
tumorigenesis by suppressing Runx3. Mol Cancer. 18:992019.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hu H, Zhang Q, Chen W, Wu T, Liu S, Li X,
Luo B, Zhang T, Yan G, Lu H, et al: MicroRNA-301a promotes
pancreatic cancer invasion and metastasis through the JAK/STAT3
signaling pathway by targeting SOCS5. Carcinogenesis. 41:502–514.
2020. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Li X, Li J, Cai Y, Peng S, Wang J, Xiao Z,
Wang Y, Tao Y, Li J, Leng Q, et al: Hyperglycaemia-induced miR-301a
promotes cell proliferation by repressing p21 and Smad4 in prostate
cancer. Cancer Lett. 418:211–220. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
12
|
Yue X, Lan F and Xia T: Hypoxic glioma
cell-secreted exosomal miR-301a activates Wnt/β-catenin signaling
and promotes radiation resistance by targeting TCEAL7. Mol Ther.
27:1939–1949. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Lu J, Liu QH, Wang F, Tan JJ, Deng YQ,
Peng XH, Liu X, Zhang B, Xu X and Li XP: Exosomal miR-9 inhibits
angiogenesis by targeting MDK and regulating PDK/AKT pathway in
nasopharyngeal carcinoma. J Exp Clin Cancer Res. 37:1472018.
View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Wang YG, Wang T, Shi M and Zhai B: Long
noncoding RNA EPB41L4A-AS2 inhibits hepatocellular carcinoma
development by sponging miR-301a-5p and targeting FOXL1. J Exp Clin
Cancer Res. 38:1532019. View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lettlova S, Brynychova V, Blecha J, Vrana
D, Vondrusova M, Soucek P and Truksa J: MiR-301a-3p suppresses
estrogen signaling by directly inhibiting ESR1 in ERα positive
breast cancer. Cell Physiol Biochem. 46:2601–2615. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Xia X, Zhang K, Luo G, Cen G, Cao J, Huang
K and Qiu Z: Downregulation of miR-301a-3p sensitizes pancreatic
cancer cells to gemcitabine treatment via PTEN. Am J Transl Res.
9:1886–1895. 2017.PubMed/NCBI
|
|
18
|
Yuniati L, Scheijen B, van der Meer LT and
van Leeuwen FN: Tumor suppressors BTG1 and BTG2: Beyond growth
control. J Cell Physiol. 234:5379–5389. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Sun GG, Wang YD, Cheng YJ and Hu WN: The
expression of BTG1 is downregulated in nasopharyngeal carcinoma and
possibly associated with tumour metastasis. Mol Biol Rep.
41:5979–5988. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Kalluri R and LeBleu VS: The biology,
function, and biomedical applications of exosomes. Science.
367:eaau69772020. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Kalluri R: The biology and function of
exosomes in cancer. J Clin Invest. 126:1208–1215. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Wang X, Luo G, Zhang K, Cao J, Huang C,
Jiang T, Liu B, Su L and Qiu Z: Hypoxic tumor-derived exosomal
miR-301a mediates M2 macrophage polarization via PTEN/PI3Kγ to
promote pancreatic cancer metastasis. Cancer Res. 78:4586–4598.
2018. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Lan F, Qing Q, Pan Q, Hu M, Yu H and Yue
X: Serum exosomal miR-301a as a potential diagnostic and prognostic
biomarker for human glioma. Cell Oncol (Dordr). 41:25–33. 2018.
View Article : Google Scholar : PubMed/NCBI
|