Introduction
Mucinous cystic neoplasms of the pancreas (MCNs) are
a group of lesions that are usually benign but potentially
malignant. They include mucinous cystadenoma (MCA), borderline
mucinous cystadenoma (MCB) and mucinous cystadenocarcinoma (MCC).
MCNs are common in women, with a male-female incidence rate of
1:9-1:20 (1–5). Pancreatic MCC is a rare malignant
tumor, accounting for <1% of pancreatic cancer types. In 1934,
Lichtenstein (6) first reported a
pancreatic MCC case that is notable due to it being a rare tumor
and due to the complete clinical record in a period of 6 years may
throw possible light on its pathogenesis. The case was an
encapsulated cystic tumor of the tail of the pancreas, the size of
a child's head and had in part undergone carcinomatous change after
an interval of ~5 years, invading the capsule and metastasizing to
the peritoneum, omentum and liver (6). Le Borgne et al (7) analyze 398 cases of cystadenomas of the
pancreas between 1984 and 1996 in 73 institutions of the French
Surgical Association in 1999. They identified 150 MCA and 78 MCC
cases. In the 1970s, Compagno and Oertel (8) proposed that pancreatic MCA will
eventually become malignant over time and indicate that the
potential to invade is an innate intrinsic characteristic of
pancreatic tumor exocrine cells rather than anacquired
phenotype.
Non-coding RNAs, especially microRNAs (miRNAs/miRs),
have gained attention due to their participation in numerous
pathological processes, such as tumorigenesis, invasion and
metastasis of tumors. miRNAs are an endogenous and small molecular
non-coding RNA with a single strand. They can negatively regulate
the target gene by combining with its 3′-untranslated region
(3′-UTR) (9–11). Dynamic comparison of samples from
different diseases in the progression of pancreatic cyst neoplasm,
including the tumor tissue, peripheral blood and tumor cystic
fluid, can reflect the pathogenesis of the malignant transformation
of the pancreatic cells (12).
Consistent with the WHO 2000 grade standard (13), the current research group used the
expression profile chip of Agilent 16.0 to perform differential
miRNA screening of the pathological tissues, cystic fluid and serum
of patients with different diseases of MCN, including MCA, MCB and
MCC. The present study anchored the key molecule miR-224-5p in
miRNA by qPCR verification.
Although studies on miR-224-5p have received
increasing attention (14–16), most of have been limited to common
tumors. However, there area few studies on pancreatic MCC, which is
rare in the clinic. In 1963, Cullen et al (17) searched 2.4 million medical records
of Mayo Clinic and found only 17 patients who could be diagnosed
with pancreatic cystadenocarcinoma. Thus, it is extremely difficult
to conduct a study with fresh tissues of pancreatic MCC. The
pancreatic disease group of Changhai Hospital is a national key
discipline and >500 patients with pancreatic cancer are admitted
annually. Paraffin specimens of MCC, which were surgically removed
in Changhai hospital Affiliated to Naval Military Medical
University between January 2012 and December 2016 were retrieved
and only four cases were identified. In PubMed, there are few
studies related to pancreatic MCC, most of which are case reports
and the basic research related to pancreatic MCC was almost absent.
Meanwhile, there is only one cell line of pancreatic MCC (MCC1
cell) globally.
Therefore, to further understand this rare tumor,
more studies related to the functions and mechanisms of pancreatic
MCC are required. To the best of the authors' knowledge, the
present study is the first to systematically analyze the biological
function and mechanism of miR-224-5p in pancreatic MCC in
vitro and in vivo.
Materials and methods
Clinical samples
A total of four paired paraffin-embedded pancreatic
MCC tumor samples and matched adjacent normal tissue samples were
obtained from the Department of Pathology, Changhai Hospital
(Shanghai, China). All cases were histologically confirmed. All
four patients were female and the median age was 54.5 years (range,
47–76 years). Lymph node metastasis was confirmed by pathology in
three cases. In addition, the locations of the primary tumor in the
patients were the tail and body of the pancreas, the tail in one
patient and the body and tail in three patients. There was no
history of radiotherapy or chemotherapy preoperatively. Informed
consent was obtained from the subjects prior to specimen
collection.
Cell culture
Human pancreatic ductal epithelial (HPDE) cells were
obtained from the Shanghai Institute of Biochemistry and Cell
Biology of the Chinese Academy of Sciences. The cells were cultured
in DMEM (Thermo Fisher Scientific, Inc.) supplemented with 10% FBS
(HyClone; Cytiva) and 1% penicillin/streptomycin (Thermo Fisher
Scientific, Inc.). MCC1 cells were provided by Professor Claudio
Sorio from the University of Verona (Italy). They were incubated in
RPMI-1640 medium (HyClone; Cytiva) supplemented with 10% FBS, 2 mM
glutamine, 80 µg/ml gentamicin sulphate and 2.5 µg/ml amphotericin
B (Sigma-Aldrich; Merck KGaA). All cells were incubated at 37°C in
a humidified atmosphere with 5% CO2.
MTT assay
MTT assays wereperformed in 96-well plates (Corning,
Inc.). The cells (4×103 cells/100 µl) were seeded for 24, 48, 72
and 96 h at 37°C in a humidified atmosphere with 5% CO2.
After 24, 48, 72 and 96 h of culture, 10 µl MTT (5 mg/ml,
Sigma-Aldrich; Merck KGaA) solution was added to each well and
cultured for another 4 h. The culture medium was carefully removed
and 100 µl DMSO (Sigma-Aldrich; Merck KGaA) was added at room
temperature for 30 min. The absorbance value of each well was
measured at 490 nm using a microplate reader (Bio-Rad Laboratories,
Inc.).
Tumor xenografts
The animal experiments in the present study were
performed in accordance with the protocol approved by the
Institutional Animal Care and Use Committee of Shanghai Institute
for Biological Sciences, Chinese Academy of Sciences (approval no.
SIBS-2018-ZLX-2). In total, six 4-week-old female nude mice
(BALB/c, nu/nu, weight, 20–25 g) were purchased from the Shanghai
Experimental Animal Centre and maintained under pathogen-free
conditions with 60–65% humidity at 22–25°C under a 12-h light/dark
cycle. All mice were allowed free access to drinking water and
sterilized standard diet. Prior to the experiment, the nude mice
were placed in pathogen-free conditions for 1 week. The nude mice
were randomly divided into two groups with three in each group:
Lenti-miR-224-5p-mimic group (Lenti-m) and Lenti-miR-224-5p-mimic
negative control group (Lenti-c). On day 0, tumor cells (1×106
cells/mouse) suspended in 100 µl serum-free medium and mixed 1:1
(v/v) with Matrigel were injected subcutaneously into the lower
flank of each nude mouse. The tumor dimensions [length (L) and
width (W)] were measured twice a week for the tumor volume (volume
= W2× L × 0.5). After 35 days frominoculation, the nude mice were
sacrificed using cervical dislocation. Tumors were harvested,
imaged, fixed and stored in liquid nitrogen.
Wound healing assay
The wound healing test was performed in 6-well
plates. Tumor cells (3×105 cells/1,000 µl) were seeded per well and
incubated at 37°C in 5% CO2. At 80% confluence, the
scratches were created using a sterile 200 µl pipette tip. Then,
cells were washed gently to remove the floating cells and the
medium was replaced with serum free medium for 24 h. Images of the
cells that had migrated into the wound were captured under an
Olympus IX51 light microscope (Olympus Corporation; magnification,
×10). The details of the wound healing assay were conducted as
previously described (18–20).
Transwell migration and invasion
assay
Transwell migration assay was performed in 24-well
plates with 8-µm pores (Corning, Inc.), whereas invasion assay was
performed in 24-well plates with 8-µm pores coated with Matrigel
(Corning, Inc.) according to the manufacturer's protocol. The tumor
cells (1×105 cells/200 µl per well) were seeded into the upper
chamber. The Matrigel was precoated at 37°C for 30 min. In both the
migration and invasion experiments, 500 µl medium containing 10%
FBS was added into the lower chamber as a chemoattractant. After 24
h, the cells on the top surface of the Transwell chamber were
removed using cotton swabs. The cells on the bottom surface were
fixed at room temperature with 100% methanol for 30 min, and then
stained with 0.05% crystal violet for 30 min at room temperature.
In total, five visual fields were randomly selected to be imaged
with an Olympus IX51 light microscope (Olympus Corporation;
magnification, ×20). The details of the Transwell migration and
invasion assay were conducted as previously described (18–20).
Target gene prediction and luciferase
assay
The online tools TargetScanv7.2 (http://www.targetscan.org), PicTar version.2005
(https://pictar.mdc-berlin.de/cgi-bin/PicTar_vertebrate.cgi)
and miRanda version.2010 (http://www.microrna.org/microrna/getGeneForm.do) were
used to predict miR-224-5p targets. The fragment of the PTEN
3′-UTR sequence containing one putative miR-224-5p binding site was
cloned into the luciferase reporter plasmids (Shanghai GeneChem
Co., Ltd.). GV369-miR-224 mimic-transfected MCC1 cells and
GV369-miR-224 mimic-negative control-transfected MCC1 cells were
transfected with the luciferase reporter using
Lipofectamine® 2000 (Invitrogen; Thermo Fisher
Scientific, Inc.). A microplate reader was used to detect firefly
luminescence and Renilla luminescence. Results were
evaluated via normalization of the firefly luciferase activity with
Renilla luciferase activity at 48 h post transfection using
a Dual Luciferase Reporter assay (Promega Corporation). The details
of the target gene prediction and luciferase assay were conducted
as previously described (18–20).
Protein extraction and western blot
assays
Cells were lysed with RIPA buffer (Cell Signaling
Technology, Inc.) containing complete protease inhibitor cocktail
(Roche Diagnostics), phosphatase inhibitors (Roche Diagnostics), 5
mM dithiothreitol (Sigma-Aldrich; Merck KGaA) and 1 mM
phenylmethylsulfonyl fluoride (Sigma-Aldrich; Merck KGaA) and
incubated on ice for 30 min. The cell lysate was centrifuged at
12,000 × g for 10 min at 4°C. The supernatant was collected and
protein concentrations were determined using a BCA protein assay
kit (Thermo Fisher Scientific, Inc.) according to the
manufacturer's instructions. Western blot assays were performed as
previously described (18). Then,
20 µg protein was loaded onto a 10% gel, resolved using SDS-PAGE
and transferred onto PVDF membranes. Membranes were then blocked
with 5% fat-free milk for 2 h at room temperature. Subsequently,
membranes were incubated with primary antibodies against PTEN
(1:1,000; cat. no. 9552; Cell Signaling Technology, Inc.) and GAPDH
(1:5,000; cat. no. M2006M; Abmart Pharmaceutical Technology Co.,
Ltd.) at 4°C overnight, washed three times with TBS-Tween-20 (TBST;
0.05% Tween-20) and incubated with anti-mouse HRP-conjugated
secondary antibody (1:2,000; cat. no. 7054S; Cell Signaling
Technology, Inc.) at room temperature for 2 h. Following three
washes with TBST, immunoreactive bands were visualized using ECL
working fluid (Biochannel; http://www.biochannel.cn/page19.html?product_id=299).
RNA extraction and reverse
transcription-quantitative PCR (RT-qPCR)
For RT-qPCR, TRIzol® reagent (Thermo
Fisher Scientific, Inc.) was used to isolate the RNA of MCC-1 cells
at 90% confluence, which was reverse transcribed into cDNA by
Takara Prime Script RT kit (Takara Biotechnology Co., Ltd.)
according to the manufacturer's instructions. The amount of cDNA
was detected with a SYBR® Premix Ex Taq™ kit (Takara
Biotechnology Co., Ltd.) in a StepOne Real-Time PCR Detection
system (Thermo Fisher Scientific, Inc.). All expression data were
normalized to U6-encoding transcript levels. RT-qPCR was performed
as previously described (18). The
total RNA was extracted from cells as aforementioned, and then
miRNA was reverse transcribed and amplified using a Takara
PrimeScript RT Reagent kit according to the manufacturer's
instructions. The amplification reactions were performed in
triplicate in a 96-well plate using the following thermocycling
conditions for qPCR as follows: 5 min at 95°C, followed by 40
cycles of 10 sec at 95°C and 30 sec at 60°C. The Cq values were
calculated using ABI Sequence Detection System software (version
2.1; Thermo Fisher Scientific, Inc.). Each samplewas analyzed in
triplicate and levels were quantified using the 2 ΔΔCq
method (21). The primers used for
RT-qPCR are presented in Table
I.
 | Table I.Primer sequences used in reverse
transcription-quantitative PCR. |
Table I.
Primer sequences used in reverse
transcription-quantitative PCR.
| Primer | Sequence
(5′→3′) |
|---|
| miR-224-5p | F:
CTCAACTGGTGTCGTGGAGTCGGCAATTCAGTTGAGCAAGGCAA |
|
| R:
ACACTCCAGCTGGGCAAGTCACTAGTGGT |
| U6 | F:
CTCGCTTCGGCAGCACA |
|
| R:
AACGCTTCACGAATTTGCGT |
| PTEN | F:
TGGATTCGACTTAGACTTGACCT |
|
| R:
GGTGGGTTATGGTCTTCAAAAGG |
| GAPDH | F:
GGGGAGCCAAAAGGGTCATCATCT |
|
| R:
GACGCCTGCTTCACCACCTTCTTG |
Small interfering (si)RNA and cell
transfection
siRNA was purchased from Guangzhou RiboBio Co., Ltd.
The sequence of PTEN siRNA was 5′-ACCAGGACCAGAGGAAACCT-3′
and negative control siRNA (siRNA NC) was
5′-AUUGGCUACUACCGAAGAG-3′. pcDNA vector expression PTEN and
empty pcDNA vector were freely provided by Dr Renxu Chang (Shanghai
Institute of Nutrition and Health, Shanghai Institutes for
Biological Sciences). The siRNA was co-transfected in miR-224-5p
inhibitor and NC MCC1 cells (5×104) and pcDNA vector were
co-transfected in miR-224-5p mimic and negative control MCC1 cells
(5×104), using Lipofectamine® 2000 reagent (Thermo
Fisher Scientific, Inc.) according to the manufacturer's protocol;
the siRNA and Lipofectamine® 2000 was mixed at room
temperature and was added to the cells after 25 min. The ratio of
siRNA to Lipofectamine® 2000 was 1 µg:2 µl. Cells were
transfected for 48 h before performing subsequent experiments.
Vectors and lentiviral
transduction
All recombinant lentiviruses were obtained from
Shanghai Genechem Co., Ltd. The packaged lentiviruses contained
miR-224-5p mimic, miR-224-5p inhibitor and miR-224-5p mimic
control, miR-224-5p inhibitor and miR-224-5p inhibitor control. All
negative controls were empty vectors. Lentiviral vector of
miR-224-5p mimic and miR-224-5p inhibitor was GV280 and GV390,
respectively. The sequences were as follows: hsa-miR-224-5p-mimic
forward, 5′-GAGGATCCCCGGGTACCGGCCAGCTAACCATGGGCCTGCCTC-3′ and
reverse, 5′-CACACATTCCACAGGCTAGAGGAGAAAGAAGACCTCTTTTC-3′,
hsa-miR-224-5p-inhibitor forward, 5′-CAAGTCACTAGTGGTTCCGTT-3′ and
reverse, 5′-AACGGAACCACTAGTGACTTG-3′. Lentiviral transduction was
performed according to the manufacturer's protocol. The packaged
virus solution was thawed and the virus stock solution was diluted
with the infection medium value = 10 using fresh medium containing
the gene transfection enhancer polyamine. After 72 h of infection
at room temperature, the successes transduction was observed by
green fluorescent protein-positive cells observed under an inverted
fluorescence microscope (magnification, ×200; Olympus Corporation).
The lentivirus-infected cells were treated with 2 µg/ml puromycin
for 1 week and the cells resistant to puromycin were selected.
Then, the expression of miR-224-5p in the groups of Lent-c, Lent-m
and Lent-i were quantified by qPCR.
Immunohistochemistry (IHC)
IHC analysis was performed as previously described
(18). Deparaffinized and
rehydrated sections were incubated with 3%
H2O2 in methanol for 10 min at room
temperature to block endogenous peroxidase activity. The sections
were then subjected to antigen retrieval for 10 min in a pressure
cooker containing sodium citrate, and were incubated in 5% normal
goat serum (cat. no. DXT 50197Z; Invitrogen; Thermo Fisher
Scientific, Inc.) for 20 min at 37°C, permeabilized in PBS Triton
solution and incubated with a primary antibody against PTEN (1:200;
cat. no. 9552; Cell Signaling Technology, Inc.) at 4°C overnight.
The sections were then incubated with anti-mouse HRP-conjugated
secondary antibody (1:1,000; cat. no. 7054S; Cell Signaling
Technology, Inc.) at 37°C for 1 h. The sections were then
counterstained with hematoxylin for 5 min at room temperature, and
finally dehydrated and covered with a coverslip. The sections were
observed under an IX51 light microscope (Olympus Corporation;
magnification, ×40). Each experiment was performed three times. IHC
evaluation of protein expression intensity in normal adjacent
tissues and paired pancreatic MCC tissues was performed
independently by two pathologists from the Department of Pathology,
Changhai Hospital. Staining intensity was scored as previously
described (22).
Statistical analysis
Data are presented as mean ± standard deviation from
at least three independent experiments. Student's t-test was used
to compare the differences between the two groups; one-way ANOVA
followed by Tukey's post hoc test was used to compare the
differences among multiple groups. SPSS 21.0 software (IBM Corp.)
was used to conduct statistical analysis. The experimental images
were edited and processed using Photoshop 19.0 (Adobe Systems,
Inc.) and GraphPad Prism 5.0 (GraphPad Software, Inc.). P<0.05
was considered to indicate a statistically significant
difference.
Results
Effect of miR-224-5p on the
proliferation of MCC1 cells in vitro and in vivo
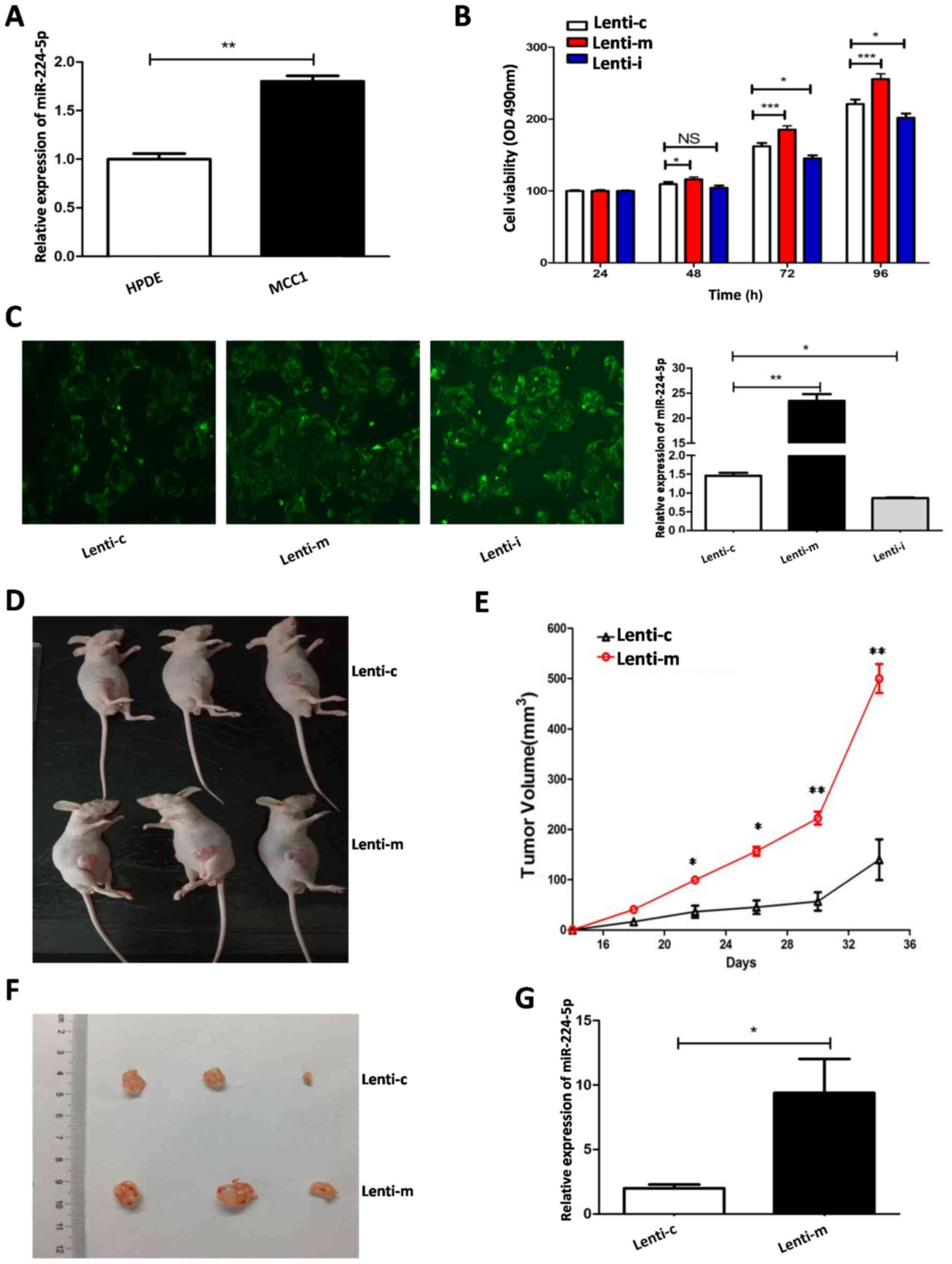
The results demonstrated that the expression of
miR-224-5p in MCC1 cells was significantly higher compared with
HPDE cells (Fig. 1A). Lentivirus
with different expression of miR-224-5p (high, low and negative
control) was used to transfect MCC1 cells. A total of three groups
of MCC1 cells: Lenti-miR-224-mimic group (Lenti-m), lenti-miR-224
inhibitor group (Lenti-i) and Lenti-miR-224 control group (Lenti-c)
were obtained. Following puromycin screening, MCC1 cells with
stably different expression of miR-224-5p were obtained (Fig. 1C). Fig.
1C (left) illustrates the cells resistant to
puromycinasselected under an inverted fluorescence microscope.
Fig. 1C (right) illustrates the
expression of miR-224-5p in the groups of Lent-c, Lent-m and Lent-i
from qPCR.
MTT was then used to measure the effect of
miR-224-5p on the proliferation of MCC1 cells. The results
demonstrated that the proliferation ability of MCC1 cells in the
Lenti-m group was significantly higher compared withthe Lenti-c
group after 48 h and the difference was more pronounced after 72 h,
while the proliferation activity of MCC1 cells in the Lenti-i group
was significantly lower compared with the Lenti-c group after 72 h
(Fig. 1B). To further observe the
effects of miR-224-5p on the promotion of the proliferation of MCC1
cells in vivo, a tumorigenesis assay in nude mice was
performed using MCC1 cells with stable miR-224-5p overexpression.
The authors of the present study were the first to report MCC
subcutaneous tumors in nude mice. The tumors were cystic with white
fluid (Fig. 1D). The tumor
dimensions (L and W) were measured twice a week for the tumor
volume (volume = W2 × L × 0.5). The tumors in the Lenti-m group
grew faster than those in the Lenti-cgroup (Fig. 1E). The tumors in the Lenti-m group
were larger than those in the stable transfer Lenti-cgroup
(Fig. 1F). RT-qPCR was used to
detect miR-224-5p expression in the tumor tissues. The results
verified that miR-224-5p expression in the Lenti-m group was higher
compared with the Lenti-c group in vivo (Fig. 1G).
Effect of miR-224-5p on the migration
and invasion of MCC1 cells
Wound healing assay and Transwell assay were used to
detect cell migration and invasion of MCC1 cells. miR-224-5p
overexpression promoted the proliferation and migration abilities
of MCC1 cells and knockdown of miR-224-5p expression inhibited the
proliferation and migration abilities of MCC1 cells in vitro
(Fig. 2A-C).
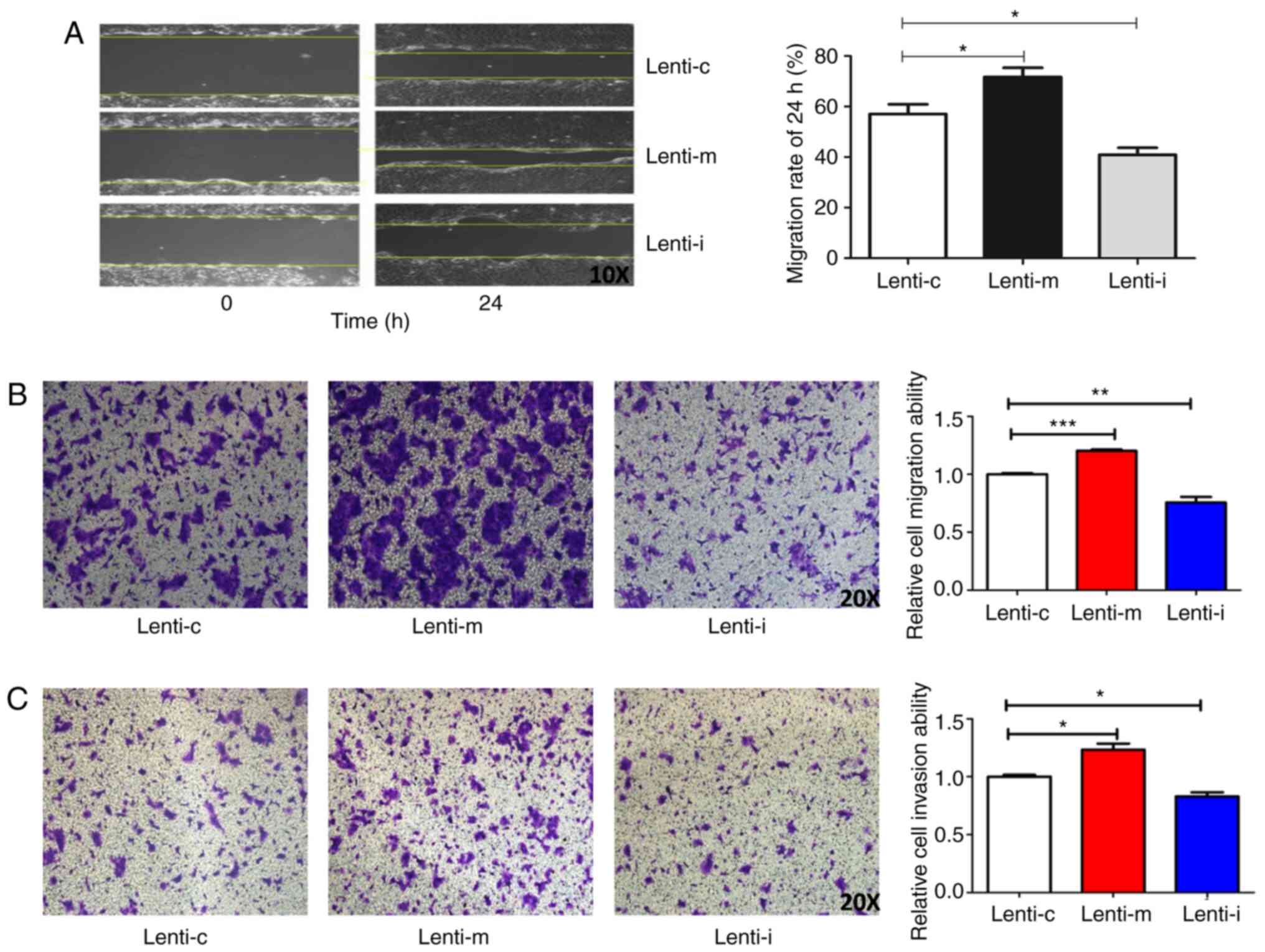 | Figure 2.Effect of miR-224-5p on the migration
and invasion of MCC1 cells. (A) Effect of miR-224-5p on the
migration of MCC1 cells by the wound healing assay (magnification,
×10). (B) Effect of miR-224-5p on the migration of MCC1 cells by
Transwell migration assay (magnification, ×10). (C) Effect of
miR-224-5p on the invasion of MCC1 cells by Transwell assay
(magnification, ×10). *P<0.05, **P<0.01, ***P<0.001. miR,
microRNA; MCC, mucinous cystadenocarcinoma; Lent-c,
lenti-miR-224-5p-mimic negative control group; Lent-m,
lenti-miR-224-5p-mimic group; Lent-I, lenti-miR-224 inhibitor
group. |
PTEN is the target gene of
miR-224-5p
The potential target genes of miR-224-5p that
regulate proliferation and invasion were screened using the
bioinformatics algorithm TargetScan Human 7.2. There was a
potential binding site of 6 bp between miR-224-5p and the
PTEN gene, which was selected for experimental validation
(Fig. 3A). To confirm whether
PTEN was a direct target of miR-224-5p, luciferase reporter
assay was performed. The results demonstrated that miR-224-5p
overexpression reduced luciferase activity in the PTEN
wild-type (WT) 3′-UTR reporter but had no effect on the PTEN
mutant (MUT) 3′-UTR reporter (Fig.
3A). To further confirm that PTEN was a target of
miR-224-5p, the mRNA and protein expression of PTEN was
examined using RT-qPCR and western blot analysis. The results
demonstrated that PTEN mRNA and protein levels were
significantly downregulated in normal MCC1 cells and xenograft
tumors transfected with miR-224-5p-overexpressing MCC1 cells,
whereas the expression of these levels was upregulated in HPDE
cells and xenograft tumors transfected with
miR-224-5p-overexpressing-control MCC1 cells (Fig. 3C and D). miR-224-5p expression was
negatively associated with PTEN in MCC1 cells in
vitro (Figs. 1A and 3C) and in vivo (Figs. 1G and 3D). These findings indicated that
PTEN was a target of miR-224-5p in MCC1 cells. The
expression of PTEN protein in the tumor and matched adjacent
normal tissues was detected by IHC. The results demonstrated that
the expression of PTEN protein in the tumor was lower
compared with matched adjacent normal tissues (Fig. 3B).
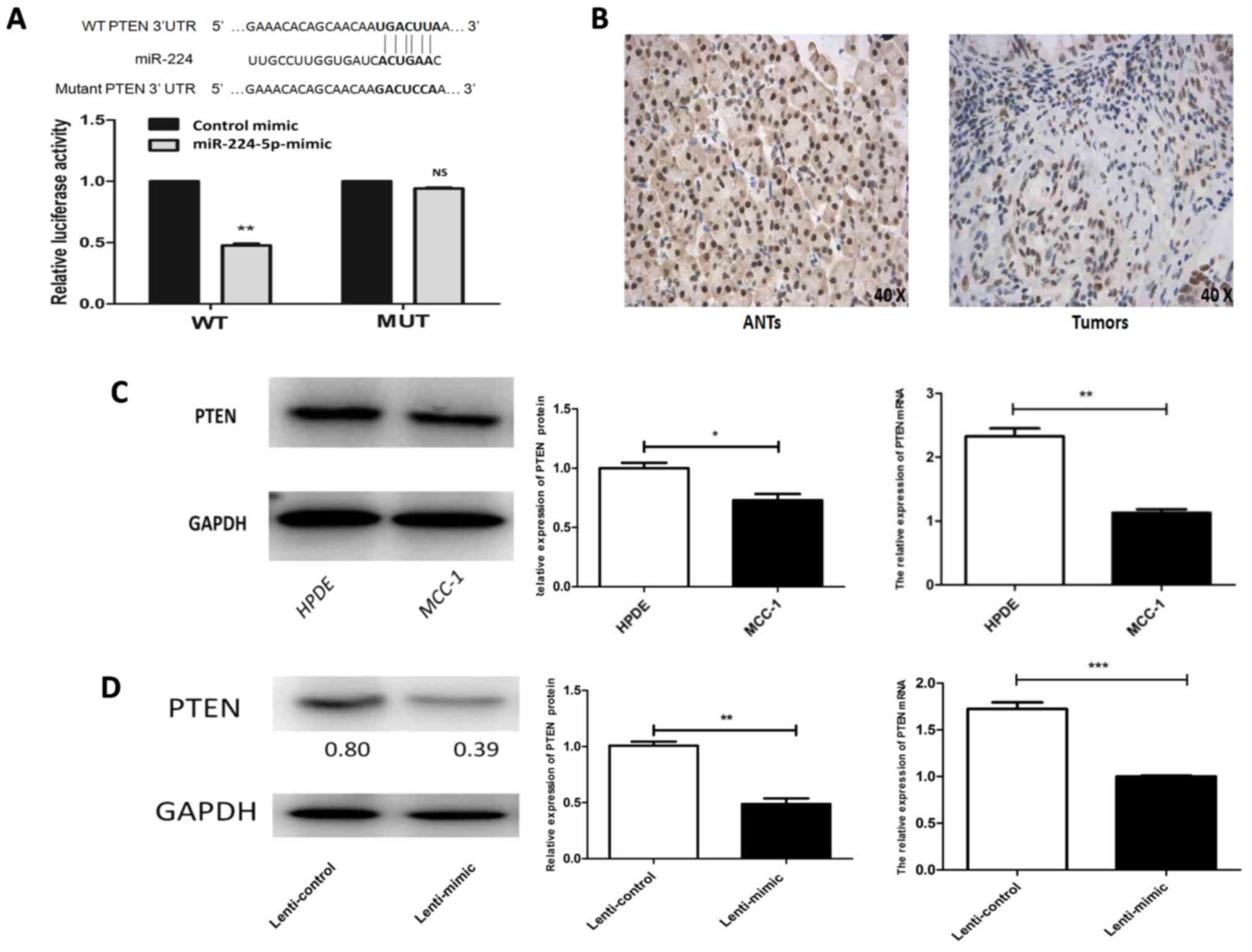 | Figure 3.PTEN is the target gene of
miR-224-5p. (A) Luciferase report on miR-224-5p and PTEN
target gene. (B) Expression of PTEN in pancreatic MCC
tissues and matched adjacent normal tissues (magnification, ×40).
(C) Expression of PTEN in MCC1 and HPDE cells. (D) Protein and mRNA
expression level of PTEN in MCC1 cells in vivo.
*P<0.05, **P<0.01, ***P<0.001. NS, P>0.05. miR,
microRNA; MCC, mucinous cystadenocarcinoma; HPDE, human pancreatic
ductal epithelial; WT, wild-type; MUT, mutation type; ANTs,
adjacent normal tissues. |
miR-224-5p regulates the
proliferation, migration and invasion of MCC1 cells by targeting
PTEN
The transfection of si-PTEN to MCC1 cells was
successful (Fig. 4A). The
miR-224-5p-inhibitor, PTEN siRNA, or negative control was
transfected into MCC1 cells to investigate the effect of
PTEN on MCC1 cells. The expression of PTEN was
detected by western blot analysis (Fig.
4B). MTT and Transwell assays were used to evaluate the effect
on proliferation, migration and invasion in MCC1 cells. The results
demonstrated that the proliferation, migration and invasion of MCC1
cells transfected with Lenti-miR-224-5p inhibitor decreased
compared to those of the MCC1 cells transfected with the negative
control. The effect of PTEN on proliferation, migration and
invasion in MCC1 cells downregulated by miR-224-5p were then
investigated. It was found that inhibiting PTEN expression
could reverse the inhibitory effect of miR-224-5p on MCC1 cells
(Fig. 4C-E) after which the
transfection of PTEN plasmid into the MCC1 cells was
successful (Fig. 5A). miR-224-5p
mimic, PTEN plasmid or NC was also transfected into MCC1
cells to investigate the effect of PTEN on MCC1 cells.
PTEN expression was detected by western blot analysis
(Fig. 5B). MTT and Transwell assays
were used to test the effect of proliferation, migration and
invasionin MCC1 cells. The results demonstrated that the
proliferation, migration and invasion of MCC1 cells transfected
with Lenti-miR-224-5p mimic increased compared to those in MCC1
cells transfected with the NC. Then, the effect of PTEN on
proliferation, migration and invasion in MCC1 cells upregulated by
miR-224-5p were investigated. It was found that increasing
PTEN expression could reverse the inhibitory effect of
miR-224-5p on MCC1 cells (Fig.
5C-E).
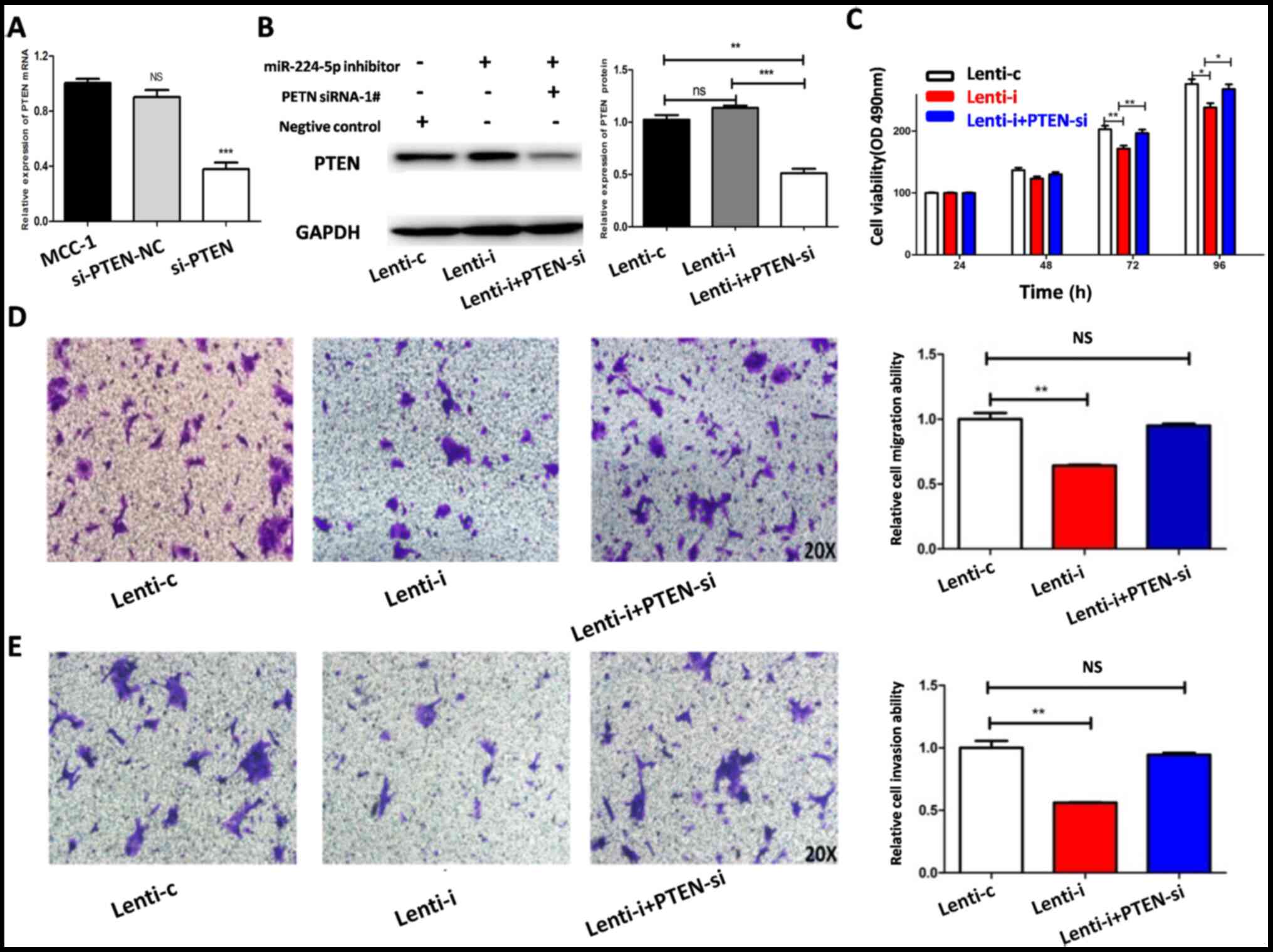 | Figure 4.Inhibiting PTEN expression in
MCC1 cells reverses the inhibition of proliferation, migration and
invasion induced by low miR-224-5p expression. (A) The
transfections of si-PTEN were verified as successful by PCR.
(B) Expression of PTEN in MCC1 cells of Lenti-c, Lenti-i and
Lenti-i+PTENsi group. Inhibition of PTEN expression
in MCC1 cells reverses the inhibition of (C) viability, (D)
migration and (E) invasion induced by low miR-224-5p expression.
*P<0.05, **P<0.01, ***P<0.001. NS, P>0.05. MCC,
mucinous cystadenocarcinoma; miR, microRNA; si, small interfering;
Lent-c, lenti-miR-224-5p-mimic negative control group; Lent-m,
lenti-miR-224-5p-mimic group; Lent-i, lenti-miR-224 inhibitor
group; Lenti-i+PTENsi, Lenti-i+PTEN siRNA-1#. |
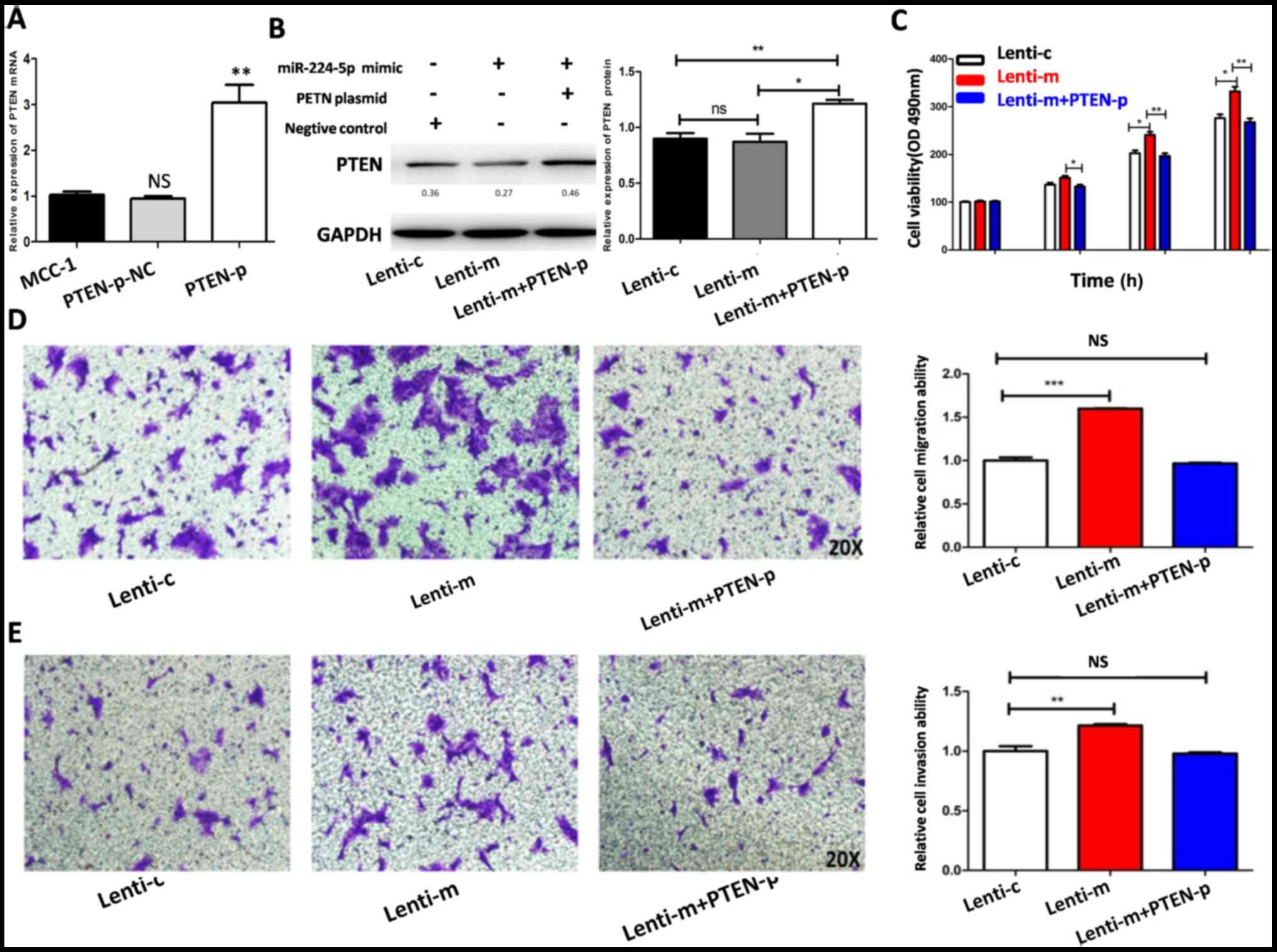 | Figure 5.Increasing PTEN expression in
MCC1 cells reverses the promotion of proliferation, migration and
invasion induced by miR-224-5p overexpression. (A) The
transfections of PTEN plasmid were verified as successful by
PCR. (B) Expression of PTEN in MCC1 cells of Lenti-c,
Lenti-m and Lenti-m+PTEN-p groups. Increasing PTEN
expression in MCC1 cells reverse the promotion of (C) viability,
(D) migration and (E) invasion induced by miR-224-5p
overexpression. *P<0.05, **P<0.01, ***P<0.001. NS,
P>0.05. MCC, mucinous cystadenocarcinoma; miR, microRNA; Lent-c,
lenti-miR-224-5p-mimic negative control group; Lent-m,
lenti-miR-224-5p-mimic group; Lent-i, lenti-miR-224 inhibitor
group; Lenti-m+PTENp, Lenti-m+PTEN plasmid. |
Discussion
Abnormal proliferation is a unique feature of tumor
cells andthe inhibition of tumor cell proliferation is
controversial (23). miRNA serves
an important role in tumorigenesis and the development of malignant
tumors and regulates the proliferation and invasion of tumor cells
by combining with target genes (24–26).
Pancreatic MCC is a rare malignant tumor, with a
limited number of studies, to the best of the authors' knowledge,
and there is only one cell line of pancreatic MCC (MCC1) worldwide.
Therefore, to further understand this rare tumor, more studies on
the functions and mechanisms of pancreatic MCC are urgently
required. We had previously established and verified the key
molecule miR-224-5p in pancreatic MCNs (27). The present study found that
miR-224-5p expression in MCC1 cells was significantly higher
compared with HPDE cells, indicating that miR-224-5p might serve an
oncogenic role in pancreatic MCC. It was observed that miR-224-5p
overexpression in MCC1 cells promoted proliferation, migration and
invasion, while low miR-224-5p expression in MCC1 cells
significantly inhibited proliferation, migration and invasion. This
is consistent with previously reported studies (28–30).
The present study also assessed the effect of miR-224-5p on MCC1
cells in nude mice. To the best of the authors' knowledge, this is
the first study to construct subcutaneous tumors in pancreatic MCC
in vivo. The tumors that were cystic and contained white
fluid were observed at 10–14 days in all nude mice. These
characteristics of tumors in mice were similar to the pathological
characteristics of patients diagnosed with pancreatic MCC in the
clinic. The present study verified that miR-224-5p overexpression
in MCC1 cells promoted proliferation in nude mice. To the best of
the authors' knowledge, the present study was the first on
pancreatic MCC in nude mice. miRNAs can regulate the biological
function of cells by binding with the 3′-UTR of the target gene
(31). Bioinformatics software,
including miRanda, TargetScan and PicTar, is commonly used to
predict miRNA target genes (32,33).
The three databases (TargetScan, PicTar and miRanda) were used to
explore the target genes of miR-224-5p. Combined with the research
reported in the literature, the PTEN gene was screened,
which is related to tumor proliferation and invasion (34). The present study verified the
targeting relationship between miR-224-5p and PTEN using a
luciferase reporter assay, which provided direct evidence of
miR-224-5p targeting the PTEN gene. Simultaneously, RT-qPCR
and western blot analysis were used to further confirm that
miR-224-5p expression was negatively associated with PTEN in
MCC1 cells in vitro and in vivo. As pancreatic MCC is
rare in the clinic, it is extremely difficult to detect miR-224-5p
expression in fresh tissues of pancreatic MCC and miR-224-5p was
degraded in the paraffin tissue of pancreatic MCC. Therefore,
PTEN protein was only detected in pancreatic MCC
paraffin-embedded samples and it was found that PTEN protein
expression in tumors was lower compared with adjacent normal
tissues. Therefore, these results directly and indirectly support
the finding that PTEN is the target gene of miR-224-5p in
MCC1 cells.
PTEN, a tumor suppressor gene, mainly
regulates the PI3K/AKT, MAPK and focal adhesion kinase signaling
pathways and serves an important role in regulating cell
proliferation, apoptosis, cell signal transduction, tumor cell
infiltration, metastasis, drug resistance and angiogenesis
(35–37). miRNAs serve a role in the
inactivation of PTEN. In breast cancer tissues and cells,
miR-182-5p is highly expressed and patients with breast cancer with
high miR-182-5p expression are associated with a low survival rate.
Knocking down miR-182-5p expression in breast cancer cells can
inhibit the proliferation and invasion of breast cancer cells and
PTEN is considered the target of miR-182-5p. The recovery of
PTEN expression can reverse the miR-182-5p-mediated
promotion of breast cancer cell proliferation and invasion
(38). In hepatocellular carcinoma,
Jiang et al (39) found that
miR-19a-3p promoted tumor metastasis and chemoresistance through
the PTEN/AKT pathway.
To clarify that miR-224-5p mediates the
proliferation, migration and invasion of pancreatic MCC by
targeting PTEN, PTEN siRNA and plasmids were used to
regulate the expression of PTEN in MCC1 cells. The present
study found that low miR-224-5p expression could inhibit the
proliferation, migration and invasion of MCC1 cells, while
PTEN inhibition could reverse the biological effect of low
miR-224-5p expression in MCC1 cells. Meanwhile, miR-224-5p
overexpression in MCC1 cells promoted proliferation, migration and
invasion, while PTEN overexpression reversed the biological
effect of miR-224-5p overexpression in MCC1 cells. These results
confirmed that miR-224-5p partly mediates the proliferation,
migration and invasion of MCC1 cells by regulating PTEN.
Tumorigenesis and the development of pancreatic MCC
involve many factors, including genes, proteins andmolecules
(40). The present study was the
first, to the best of the authors' knowledge, to systematically
evaluate the relationship between miR-224-5p and the PTEN
gene in vitro and in nude mice and clinical tissue samples.
It was confirmed that miR-224-5p can regulate the proliferation,
migration and invasion of pancreatic MCC by targeting PTEN.
These results suggested that miR-224-5p serves an oncogenic role in
MCC. The present study analyzed the relationship between miR-224-5p
and pancreatic MCC with respect to miRNA. The present study
enriched the basic and clinical knowledge on pancreatic MCC. It is
hoped that the present study will bring breakthroughs in the
targeted and precise treatment of pancreatic MCC in the future.
There were some limitationsin the present study.
First, the number of patients was too small (only four patients).
Second, there was only one pancreatic MCC cell line (MCC1)
worldwide. Third, the study only verified that miR-224-5p
overexpression in MCC1 cells promoted proliferation in nude mice
in vivo. Last, as pancreatic MCC is rare in the clinicand
miR-224-5p was degraded in paraffin tissue of pancreatic MCC, the
present study only detected PTEN protein in pancreatic MCC
paraffin-embedded samples. Some of these concerns are worthy of
further study.
Acknowledgements
The authors would like to thank Professor Claudio
Sorio (University of Verona, Italy) for the MCC1 cell line.
Funding
The present study was supported by grants from the
National Natural Science Foundation of China (grant nos. 81672892
and 82072707) and the Shanghai Anticancer Association EYAS PROJECT
(grant no. SACA-CY1C12).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XZ and LZ conceived the study. XP, CG, LS and YW
acquired the data. LS and MY analyzed the data. XZ acquired
funding. XP, CG, YW and RC performed the experiments. RC and MY
developed the methodology. XZ and MY were responsible for project
administration. XP, CG and YW interpreted the results. XP wrote the
manuscript. XZ and LZ revised the manuscript. XP, CG and YW
confirmed the authenticity of all the raw data. All authors have
reviewed and approved the final version of the manuscript.
Ethics approval and consent to
participate
The present study was approved by the Ethics
Committee of the Changhai Hospital (approval no. CHEC-2016
8167111578). The animal experiments was approved by the
Institutional Animal Care and Use Committee of Shanghai Institute
for Biological Sciences, Chinese Academy of Sciences (approval no.
SIBS-2018-ZLX-2).
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Brewer Gutierrez OI and Lennon AM:
Pancreatic Cysts: Sinister Findings or Incidentalomas? Med Clin
North Am. 103:163–172. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Dorobanţu BM, Matei E, Herlea V, Boroş M,
Tivadar B and Ciurea SH: Diagnosis, morphopathological profile and
treatment of mucinous cystadenoma of the pancreas - a single center
experience. Rom J Morphol Embryol. 59:1155–1163. 2018.PubMed/NCBI
|
|
3
|
Becker WF, Welsh RA and Pratt HS:
Cystadenoma and cystadenocarcinoma of the pancreas. Ann Surg.
161:845–863. 1965. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Katoh H, Rossi RL, Braasch JW, Munson JL,
Shimozawa E and Tanabe T: Cystadenoma and cystadenocarcinoma of the
pancreas. Hepatogastroenterology. 36:424–430. 1989.PubMed/NCBI
|
|
5
|
Doulamis IP, Mylonas KS, Kalfountzos CE,
Mou D, Haj-Ibrahim H and Nasioudis D: Pancreatic mucinous
cystadenocarcinoma: Epidemiology and outcomes. Int J Surg.
35:76–82. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Lichtenstein L:
Papillarycystadenocarcinoma of pancreas. case report, with notes on
classification of malignant cystic tumors of pancreas. Am J Cancer.
21:542–553. 1934. View Article : Google Scholar
|
|
7
|
Le Borgne J, de Calan L and Partensky C;
French Surgical Association, : Cystadenomas and cystadenocarcinomas
of the pancreas: A multiinstitutional retrospective study of 398
cases. Ann Surg. 230:152–161. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Compagno J and Oertel JE: Mucinous cystic
neoplasms of the pancreas with overt and latent malignancy
(cystadenocarcinoma and cystadenoma). A clinicopathologic study of
41 cases. Am J Clin Pathol. 69:573–580. 1978. View Article : Google Scholar : PubMed/NCBI
|
|
9
|
Lu J, Getz G, Miska EA, Alvarez-Saavedra
E, Lamb J, Peck D, Sweet-Cordero A, Ebert BL, Mak RH, Ferrando AA,
et al: MicroRNA expression profiles classify human cancers. Nature.
435:834–838. 2005. View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Van Roosbroeck K and Calin GA: Cancer
Hallmarks and MicroRNAs: The Therapeutic Connection. Adv Cancer
Res. 135:119–149. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Rawat M, Kadian K, Gupta Y, Kumar A, Chain
PSG, Kovbasnjuk O, Kumar S and Parasher G: MicroRNA in Pancreatic
Cancer: From Biology to Therapeutic Potential. Genes (Basel).
10:752–773. 2019. View Article : Google Scholar
|
|
12
|
Zhang B, Guo X, Zhang J, Liu X, Zhan X and
Li Z: MicroRNA 224 is downregulated in mucinous cystic neoplasms of
the pancreas and may regulate tumorigenesis by targeting Jagged1.
Mol Med Rep. 10:3303–3309. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Aaltonen LA and Hamilton SR: World Health
Organization, International Agency for Research on Cancer.
Pathology and Genetics of Tumours of the Digestive System. Oxford
University Press; Lyon, Oxford: 2000
|
|
14
|
Li S, Zhang J, Zhao Y, Wang F, Chen Y and
Fei X: miR-224 enhances invasion and metastasis by targeting HOXD10
in non-small cell lung cancer cells. Oncol Lett. 15:7069–7075.
2018.PubMed/NCBI
|
|
15
|
Zhou J, Hu M, Wang F, Song M, Huang Q and
Ge B: miR-224 Controls Human Colorectal Cancer Cell Line HCT116
Proliferation by Targeting Smad4. Int J Med Sci. 14:937–942. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Lu Y, Huang W, Chen H, Wei H, Luo A, Xia
G, Deng X and Zhang G: MicroRNA-224, negatively regulated by c-jun,
inhibits growth and epithelial-to-mesenchymal transition phenotype
via targeting ADAM17 in oral squamous cell carcinoma. J Cell Mol
Med. 23:4913–4920. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Cullen PK Jr, Remine WH and Dahlin DC: A
clinicopathological study of cystadenocarcinoma of the pancreas.
Surg Gynecol Obstet. 117:189–195. 1963.PubMed/NCBI
|
|
18
|
Chang R, Song L, Xu Y, Wu Y, Dai C, Wang
X, Sun X, Hou Y, Li W, Zhan X, et al: Loss of Wwox drives
metastasis in triple-negative breast cancer by JAK2/STAT3 axis. Nat
Commun. 9:34862018. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Song L, Guo J, Chang R, Peng X, Li J, Xu
X, Zhan X and Zhan L: LKB1 obliterates Snail stability and inhibits
pancreatic cancer metastasis in response to metformin treatment.
Cancer Sci. 109:1382–1392. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Zhou Y, Chang R, Ji W, Wang N, Qi M, Xu Y,
Guo J and Zhan L: Loss of Scribble Promotes Snail Translation
through Translocation of HuR and Enhances Cancer Drug Resistance. J
Biol Chem. 291:291–302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Livak KJ and Schmittgen TD: Analysis of
relative gene expression data using real-time quantitative PCR and
the 2(-Delta Delta C(T)) Method. Methods. 25:402–408. 2001.
View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Hieronymus H, Iaquinta PJ, Wongvipat J,
Gopalan A, Murali R, Mao N, Carver BS and Sawyers CL: Deletion of
3p13-14 locus spanning FOXP1 to SHQ1 cooperates with PTEN loss in
prostate oncogenesis. Nat Commun. 8:1081–1090. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Hanahan D and Weinberg RA: Hallmarks of
cancer: The next generation. Cell. 144:646–674. 2011. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
Huang C, Wang Z, Zhang K, Dong Y, Zhang A,
Lu C and Liu L: MicroRNA-107 inhibits proliferation and invasion of
laryngeal squamous cell carcinoma cells by targeting CACNA2D1 in
vitro. Anticancer Drugs. 31:260–271. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Ma X, Feng J, Lu M, Tang W, Han J, Luo X,
Zhao Q and Yang L and Yang L: microRNA-501-5p promotes cell
proliferation and migration in gastric cancer by downregulating
LPAR1. J Cell Biochem. 121:1911–1922. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Ye J, Xie W, Zuo Y, Jing G and Tong J:
MicroRNA-496 suppresses tumor cell proliferation by targeting BDNF
in osteosarcoma. Exp Ther Med. 19:1425–1431. 2020.PubMed/NCBI
|
|
27
|
Guo C, Peng X, Song L, Ying M, Wu Y, Chang
R, Li J, Feng D, Zhan L and Zhan X: Autophagy promotes malignant
migration and invasion via miR-224-5p/BCL2 in pancreatic mucinous
cystadenocarcinoma MCC1 cells. Oncol Lett. 20:2762020. View Article : Google Scholar : PubMed/NCBI
|
|
28
|
Liu H, Li P, Li B, Sun P, Zhang J, Wang B
and Jia B: RKIP suppresses gastric cancer cell proliferation and
invasion and enhances apoptosis regulated by microRNA-224. Tumour
Biol. 35:10095–10103. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
29
|
Li Q, Ding C, Chen C, Zhang Z, Xiao H, Xie
F, Lei L, Chen Y, Mao B, Jiang M, et al: miR-224 promotion of cell
migration and invasion by targeting Homeobox D 10 gene in human
hepatocellular carcinoma. J Gastroenterol Hepatol. 29:835–842.
2014. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Lin ZY, Huang YQ, Zhang YQ, Han ZD, He HC,
Ling XH, Fu X, Dai QS, Cai C, Chen JH, et al: MicroRNA-224 inhibits
progression of human prostate cancer by downregulating TRIB1. Int J
Cancer. 135:541–550. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Brown JA and Bourke E: Practical
Bioinformatics Analysis of miRNA Data Using Online Tools. Methods
Mol Biol. 1509:195–208. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Lewis BP, Shih IH, Jones-Rhoades MW,
Bartel DP and Burge CB and Burge CB: Prediction of mammalian
microRNA targets. Cell. 115:787–798. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Krek A, Grün D, Poy MN, Wolf R, Rosenberg
L, Epstein EJ, MacMenamin P, da Piedade I, Gunsalus KC, Stoffel M,
et al: Combinatorial microRNA target predictions. Nat Genet.
37:495–500. 2005. View
Article : Google Scholar : PubMed/NCBI
|
|
34
|
Zhu B and Wei Y: Antitumor activity of
celastrol by inhibition of proliferation, invasion, and migration
in cholangiocarcinoma via PTEN/PI3K/Akt pathway. Cancer Med.
9:783–796. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
35
|
Ramírez-Moya J, Wert-Lamas L and
Santisteban P: MicroRNA-146b promotes PI3K/AKT pathway
hyperactivation and thyroid cancer progression by targeting PTEN.
Oncogene. 37:3369–3383. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Di Cristofano A and Pandolfi PP: The
multiple roles of PTEN in tumor suppression. Cell. 100:387–390.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
37
|
Lin Y, Chen Q, Liu QX, Zhou D, Lu X, Deng
XF, Yang H, Zheng H and Qiu Y: High expression of DJ-1 promotes
growth and invasion via the PTEN-AKT pathway and predicts a poor
prognosis in colorectal cancer. Cancer Med. 7:809–819. 2018.
View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Zhao YS, Yang WC, Xin HW, Han JX and Ma
SG: miR-182-5p knockdown targeting PTEN inhibits cell proliferation
and invasion of breast cancer cells. Yonsei Med J. 60:148–157.
2019. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Jiang XM, Yu XN, Liu TT, Zhu HR, Shi X,
Bilegsaikhan E, Guo HY, Song GQ, Weng SQ, Huang XX, et al:
microRNA-19a-3p promotes tumor metastasis and chemoresistance
through the PTEN/Akt pathway in hepatocellular carcinoma. Biomed
Pharmacother. 105:1147–1154. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
40
|
Macgregor-Das AM and Iacobuzio-Donahue CA:
Molecular pathways in pancreatic carcinogenesis. J Surg Oncol.
107:8–14. 2013. View Article : Google Scholar : PubMed/NCBI
|