Introduction
Isocitrate dehydrogenase (IDH) genes are mutated in
multiple types of tumor, including glioma, chondrogenic tumors,
leukemia and other bone marrow proliferative tumors. In glioma and
leukemia, IDH1 and IDH2 mutations occur in >70% of low-grade
tumors (level II and III) (1,2). IDH
mutation is the most important genetic change in glioma. The
mutation is located at the isocitric acid binding site (Arg/R132 of
IDH1 and R172 of IDH2) of a single amino acid (3,4).
IDH1/2 mutations are common in World Health Organization (WHO)
grade II and III gliomas and secondary glioblastoma (GBM; 70–80%),
while primary GBM (WHO grade IV) IDH1/2 mutations are rare (<5%)
(5,6). IDH1 mutations account for >90% of
glioma IDH1/2 mutations (7). IDH1
Arg132His (R132H) mutation can affect the proliferation of glioma
cells, which is slower than the corresponding wild-type IDH1 cells
(8,9). Clinical studies (10,11)
have shown that mutations in IDH1 were found to be associated with
younger age, secondary GBMs (grade IV tumors that arise from
biopsy-proven lower-grade predecessors), and increased overall
survival (OS) (12). Further
studies (13,14) have revealed that IDH1/2 mutations as
good prognostic markers are universally present in grade II and III
glioma and secondary glioblastoma, and serve an important role in
the occurrence, development and evolution of glioma (15). Therefore, studying the role of the
IDH1 R132H mutation in the occurrence of glioma may provide new
ideas for clinical treatment.
There is growing evidence that dysregulation of the
circadian clock serves an important role in the development of
malignant tumors, including glioma (16,17).
Circadian timing is a basic biological process that affects most
aspects of eukaryotic and prokaryotic physiology. Circadian
dysrhythmia may lead to an increased risk of cancer, as well as
affect the response of patients with cancer to treatment (18). In a circadian rhythm, the oscillator
is coordinated by a set of interlocked transcription-translation
feedback loops. Brain-Muscle Arnt-Like protein 1 (BMAL1) and
Circadian Locomotor Output Cycles Kaput (CLOCK) are the main
biological rhythm genes (19).
CLOCK and BMAL1 heterogeneous dimers combined with Period (PER) and
Cryptochrom (CRY) proteins in E-box device drive rhythmic
transcription (20–22). The PER and CRY proteins inhibit
CLOCK-BMAL1 complexes, which inhibit PER and CRY protein
degradation after release (23–25).
Upon epigenetic modification and increase post-translational
modifications, the core CLOCK proteins in the suprachiasmatic
nucleus can maintain peripheral CLOCK oscillation and rhythmic
expression (26), and the
downstream targets are similar to those in the suprachiasmatic
nucleus; the molecular biological clock of the peripheral tissues
and organs of the body is also composed of a
transcription-translation feedback loop regulated by clock genes
(27–32). Disruption of circadian rhythms may
negatively affect normal cellular function and may lead to an
increased incidence of multiple types of cancer, such as colorectal
cancer (33), breast cancer
(34), prostate cancer (35), pancreatic cancer (36), osteosarcoma (37) and others (38,39).
Therefore, it is very important to study the role of the circadian
clock in the development and progression of cancer. Mutations in
circadian clock components can increase the proliferation rate of
cells through the general dysregulation of the cell cycle, thereby
causing the cell to become cancerous (40). Several studies have revealed an
interaction between the biological clock and the cell cycle. For
example, a previous study has indicated that light-induced phase
shifts in mouse behavior lead to corresponding changes in the time
of intestinal cell proliferation (41). Another study has suggested that
clock genes are involved in the regulation of important cell cycle
checkpoints (42).
Understanding the mechanism of circadian clock
changes in tumors is of great importance for the improvement of
tumor therapy. However, to the best of our knowledge, the mechanism
of biorhythm change in glioma with IDH1 R132H mutation has not been
previously reported. In the present study, the IDH1 R132H mutant
gene was introduced into human glioma U87-MG cells to observe the
effect of IDH1 on biological rhythm genes and to analyze its
relevant mechanism, laying a theoretical foundation for the study
of the effect of biological rhythm on the biological function of
malignant tumors.
Materials and methods
Cell line and culture
The full name of the cell line used is U87-MG human
brain astroblastoma, which is a GBM of unknown origin. Cells were
cultured in Minimum Essential Medium (MEM; Shanghai Zhongqiao
Xinzhou Biotechnology Co., Ltd.) containing 10% fetal bovine serum
(FBS; HyClone; Cytiva) at 37°C with 5% CO2. The cell
line was purchased from Shanghai Zhongqiao Xinzhou Biotechnology
Co., Ltd.
Reagents and instruments
pLVX–IDH1-mCMV-ZsGreen-PGK-Puro and
pLVX–IDH1(MUT)-mCMV-ZsGreen-PGK-Puro lentiviruses were purchased
from Beijing Xibei Hongcheng Biotechnology Co., Ltd. (http://www.xbhcbio.com/). Trypsin (0.25%)-EDTA was
purchased from Invitrogen (Thermo Fisher Scientific, Inc.) and
0.45-µm PVDF membranes and a chemiluminescence kit (Immobilon
Western Chemiluminescent HRP Substrate, eCl@ss cat. no. 42029053)
were purchased from EMD Millipore. A total protein extraction kit
(Whole protein extraction kit, cat. no. KGP250), a BCA assay
protein content detection kit (BCA Protein Quantitation Assay, cat.
no. KGPBCA), an SDS-PAGE gel preparation kit (KGI SDS-PAGE Gel
Preparation kit, cat. no. KGP113) and a flow cytometry cell cycle
analysis kit (Cell Cycle Detection kit, cat. no. KGA511) were
purchased from Jiangsu Kaiji Biotechnology Co., Ltd. (http://keygentec.com.cn/index.php?cid=1). The BSA
(BSA-V, cat. no. A8020) used to dilute the antibody was purchased
from Beijing Solarbio Science & Technology Co., Ltd. Mouse
anti-IDH1 (R132H; cat. no. SAB4200548) was purchased from
Sigma-Aldrich (Merck KGaA). Rabbit anti-IDH1 (cat. no. 8137S),
rabbit anti-BMAL1 (cat. no. 14020S), rabbit anti-CLOCK (cat. no.
5157S), rabbit anti-phosphorylated (p)-Smad2 (cat. no. 18338S),
rabbit anti-p-Smad3 (cat. no. 9520S), rabbit anti-Smad2 (cat. no.
5339S), rabbit anti-Smad2-3 (cat. no. 86855S), rabbit anti-Smad3
(cat. no. 9513S), rabbit anti-Smad4 (cat. no. 46535S), mouse
anti-β-actin (cat. no. 3700S) and mouse anti-GAPDH (cat. no.
51332S) primary antibodies, as well as HRP-labelled goat
anti-rabbit IgG (cat. no. 7074S) and goat anti-mouse IgG (cat. no.
7076S) secondary antibodies, were purchased from Cell Signaling
Technology, Inc. Rabbit anti-Cyclin A (cat. no. AF0142), rabbit
anti-CyclinB1 (cat. no. DF6786), rabbit anti-CyclinD3 (cat. no.
DF6229), rabbit anti-CDK2 (cat. no. AF6237), rabbit anti-CDK4 (cat.
no. DF6102), rabbit anti-P18 (cat. no. AF0620), rabbit anti-P21
(cat. no. AF6290), rabbit anti-P27 (cat. no. AF6324), rabbit
anti-PER1 (cat. no. DF9080), rabbit anti-PER2 (cat. no. DF12304),
rabbit anti-PER3 (cat. no. DF7349), rabbit anti-Cry1 (cat. no.
DF8932) and rabbit anti-Cry2 (cat. no. DF12919) primary antibodies
were purchased from Affinity Biosciences. An IX73 inverted
fluorescence microscope was purchased from Olympus Corporation, and
an Amersham Imager 600 gel imaging analysis system was purchased
from General Electric.
The cancer genome atlas (TCGA)
database analysis
BMAL1 and CLOCK expression data of all types of
tumor analyzed in the present study were derived from TCGA database
(https://portal.gdc.cancer.gov/) and the
online analysis software GEPIA2 (http://gepia2.cancer-pku.cn/#index).
U87-MG human glioma cell culture and
lentiviral infection
Construction of recombinant lentiviral vector: 1 µg
Plasmid and pLVX/IDH1, 1 µl EcoRI, 1 µl BamHI, 2 µl
pLVX, 6 µl target gene, 1 µl 10X DNA Ligase Buffer and 1 µl T4 DNA
Ligase, were used to construct lentiviral vectors. The 239T cell
line (Beijing Xibei Hongcheng Biotechnology Co., Ltd.) was used for
virus purification. After culturing for 12 to 16 h, the medium was
discarded (500 µl HET Buffer A + 10 µl recombinant lentiviral
vector + 15 µl lentiviral packaging vector + HET Buffer B 50 µl +
ddH2O 425 µl), and replaced with 10 ml fresh complete
medium solution (DMEM + 10% FBS + P/S). The cells were placed in a
37°C, 5% CO2 incubator for 48 h, the supernatant was
collected after centrifugation at 500 × g and room temperature for
5 min and the cell debris was discarded. The supernatant was
filtered with a 45 µm PVDF filter into a 50 ml round bottom
centrifuge tube. This was centrifugated at a high speed of 6,000 ×
g at 4°C for 2 h, and purified recombinant lentivirus was obtained
after centrifugation. Lentivirus can be introduced into U87-MG
cells after 48 h of infection with MOI value (20:1) (43). U87-MG cells were inoculated in MEM
containing 10% FBS at a density of 2×105 cells/100-mm
culture dish, incubated at 37°C and then subjected to lentivirus
infection 6–8 h after becoming adherent. The titre of the treatment
and control lentiviruses was set to an MOI of 20:1, and MEM with 2%
FBS was used to prepare viral titre gradient solutions for
subsequent experiments. The experimental groups were the IDH1
wild-type (WT) and the IDH1 R132H mutant (MUT) groups, and the
corresponding lentiviral vectors were
pLVX–IDH1-mCMV-ZsGreen-PGK-Puro and
pLVX–IDH1(MUT)-mCMV-ZsGreen-PGK-Puro, respectively. Subsequent
experiments were performed 72 h after lentiviral infection.
Western blot analysis
Following lentiviral transfection, U87-MG cells in
each group were collected for extraction of total protein using the
aforementioned whole protein extraction kit. The BCA assay kit was
used to quantify the protein, and equal amounts of protein (40
µg/10 µl) were separated via 8% SDS-PAGE. Subsequently, the
proteins were transferred to PVDF membranes, which were blocked at
room temperature in 5% skimmed milk for 1 h and then incubated at
4°C overnight with the following primary antibodies diluted with 3%
BSA: Anti-IDH1 (R132H; 1:1,000), anti-IDH1 (1:1,000), anti-GAPDH
(1:2,000), anti-β-actin (1:2,000), anti-BMAL1 (1:1,000), anti-CLOCK
(1:1,000), anti-PER1/2/3 (1:1,000), anti-CRY1/2 (1:1,000),
anti-CyclinA (1:1,000), anti-CyclinB1 (1:1,000), anti-Cyclin D3
(1:1,000), anti-CDK2 (1:1,000), anti-CDK4 (1:1,000), anti-P18
(1:1,000), anti-P21 (1:1,000), anti-P27 (1:1,000), anti-p-Smad2
(1:1,000), anti-p-Smad3 (1:1,000), anti-Smad2 (1:1,000),
anti-Smad2-3 (1:1,000), anti-Smad3 (1:1,000) and anti-Smad4
(1:1,000). The membranes were incubated with secondary antibodies
(HRP-conjugated goat anti-rabbit IgG and goat anti-rat IgG; both
1:2,000) at room temperature for 1 h. The chemiluminescence reagent
was used for color development for 1 min, and the membranes were
exposed for greyscale measurement. The protein expression level was
semi-quantified using Image Pro Plus 6.0 (Media Cybernetics,
Inc.).
Colony formation experiments
After seeding 1,000 lentivirus-infected U87-MG cells
into 100-mm culture dishes, the cells were divided into the WT and
MUT groups. After 5 days of culture, the medium was changed once,
and whether the cells formed clumps or sheets was observed under a
10× light microscope. The medium was then discarded. The cells were
fixed in 4% paraformaldehyde at room temperature for 20 min.
Methylene violet solution (4 ml) was added to each dish, and the
cells were stained at room temperature for 30 min before rinsing
with pure water. After drying at room temperature, images were
captured, and the colonies, which are clumps or flakes formed by
cells, were counted using Image-Pro Plus 6.0 (Media Cybernetics,
Inc.) for statistical analysis.
Cell cycle distribution detection by
flow cytometry
A cell cycle analysis kit (Cell Cycle Detection kit,
cat. no. KGA511) was used for cell cycle detection. The cells in
each group were digested with trypsin at 37°C for 1 min, collected
by centrifugation at 1,500 × g for 5 min at normal temperature and
rinsed with PBS to form a cell suspension. The concentration was
adjusted to 1×106 cells/ml. After removing the
supernatant by centrifugation at 1,500 × g for 5 min at normal
temperature, 500 µl precooled 75% ethanol was used to fix the cells
overnight at 4°C. The fixation solution was removed by
centrifugation at 1,500 × g for 5 min at 4°C, 100 µl RNase A was
added and the cells were incubated in a water bath at 37°C for 30
min. A 400 µm mesh screen was used for filtration. Then, 400 µl PI
dye was added and the solution was gently mixed and incubated at
4°C for 1 h in the dark. Analysis was performed using a flow
cytometer (FACSCalibur, BD Biosciences; FlowJo, Version 10, FlowJo
LLC).
Statistical analysis
SPSS 21.0 (IBM Corp.) statistical software was used
for statistical analysis. The data are expressed as the mean ± SD.
Each independent experiment was repeated three times. Independent
sample t-test was used to analyze differences between two groups of
continuous data. One-way ANOVA was used for the comparison among
multiple groups followed by Tukey's post hoc test. P<0.05 was
considered to indicate a statistically significant difference.
Results
Biological rhythm genes are abnormally
expressed in glioma
BMAL1 and CLOCK expression was analyzed in TCGA
database. The expression levels of BMAL1 and CLOCK appeared to
differ among the 33 types of tumor. BMAL1 expression in acute
myeloid leukemia was higher than that in normal tissues (Fig. 1A). By contrast, BMAL1 expression was
lower in diffuse large adrenocortical carcinoma (ACC), cervical
squamous cell carcinoma, endocervical adenocarcinoma (CESC), colon
adenocarcinoma (COAD), lung adenocarcinoma, lung squamous cell
carcinoma, ovarian serous cystadenocarcinoma, prostate
adenocarcinoma, rectum adenocarcinoma, skin cutaneous melanoma,
testicular germ cell tumors (TGCT), uterine corpus endometrial
carcinoma and uterine carcinosarcoma compared with that in normal
tissues (Fig. 1A). Additionally,
BMAL1 expression in brain lower grade glioma (LGG) tissues was
lower than that in normal tissues (Fig.
1C). The expression of BMAL1 in GBM tissue was not
significantly different from that in normal tissue (Fig. 1C). CLOCK expression in TGCT tissues
was lower than that in normal tissues (Fig. 1B). By contrast, CLOCK expression was
higher in diffuse large pancreatic adenocarcinoma (PAAD), stomach
adenocarcinoma (STAD) and thymoma (THYM) tissues compared with that
in normal tissues (Fig. 1B).
Additionally, CLOCK expression in brain LGG was higher than that in
normal tissues (Fig. 1D). The
expression of CLOCK in GBM tissues was slightly higher compared
with that in normal tissues (Fig.
1D). However, how the rhythm genes are expressed in IDH1 R132H
mutated glioma is unclear. Therefore, further experiments were
performed to investigate this.
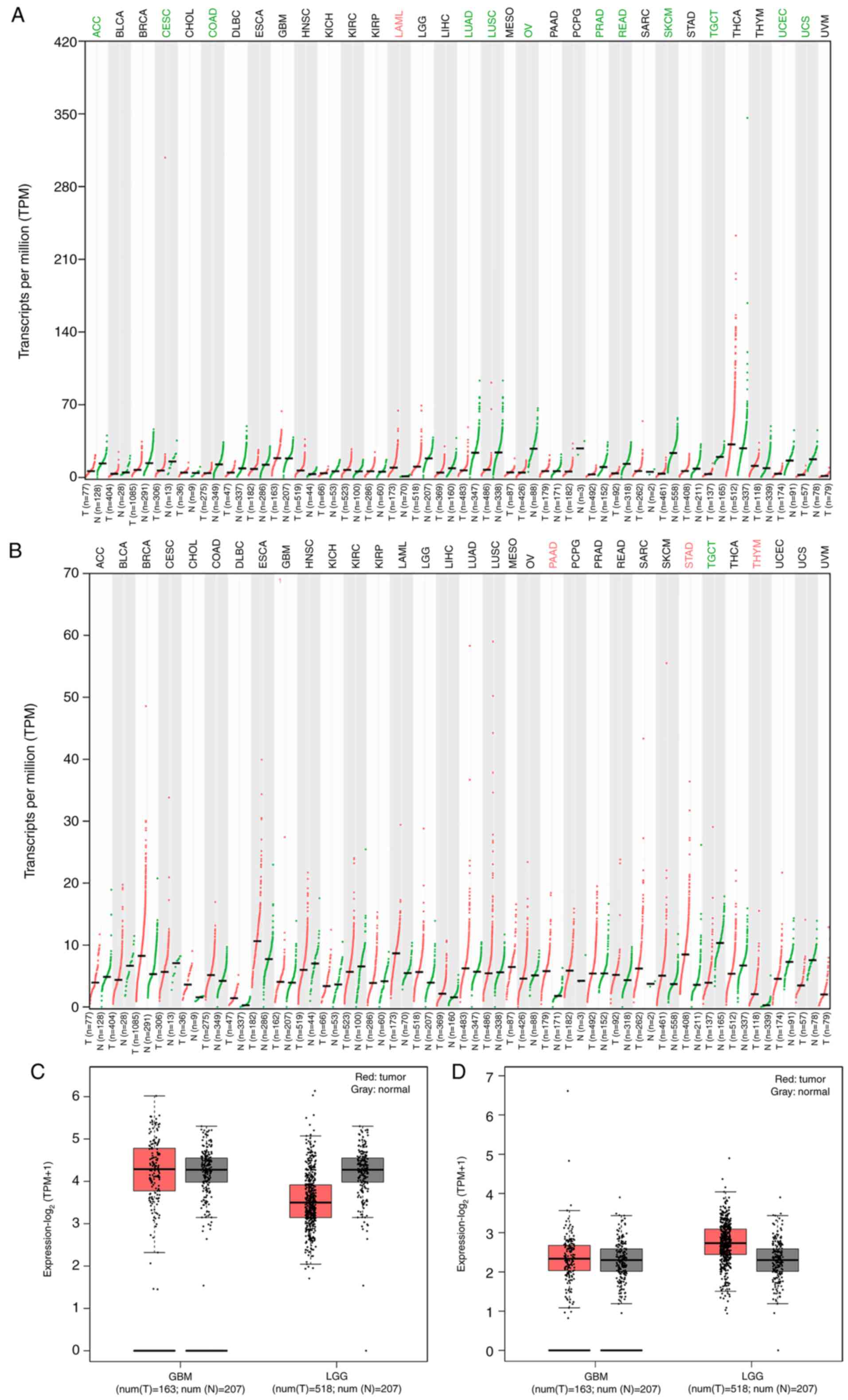 | Figure 1.Expression of BMAL1 and CLOCK in
different tumors in TCGA database. (A) BMAL1 and (B) CLOCK
expression in 33 types of tumor from The Cancer Genome Atlas
database. Red represents tumor samples, while green represents
normal tissue samples. (C) BMAL1 and (D) CLOCK expression in GBM
and LGG tumor and normal tissues. The red box represents tumor
samples, while the grey box represents normal tissue samples. T,
tumor; N, normal; TPM, transcripts per million; BMAL1, Brain-Muscle
Arnt-Like protein 1; CLOCK, Circadian Locomotor Output Cycles
Kaput; ACC, Adrenocortical carcinoma; BLCA, Bladder Urothelial
Carcinoma; BRCA, Breast invasive carcinoma; CESC, Cervical squamous
cell carcinoma and endocervical adenocarcinoma; CHOL,
Cholangiocarcinoma; COAD, Colon adenocarcinoma; DLBC, Lymphoid
Neoplasm Diffuse Large B-cell Lymphoma; ESCA, Esophageal carcinoma;
GBM, Glioblastoma multiforme; HNSC, Head and Neck squamous cell
carcinoma; KICH, Kidney Chromophobe; KIRC, Kidney renal clear cell
carcinoma; KIRP, Kidney renal papillary cell carcinoma; LAML, Acute
Myeloid Leukemia; LGG, Brain Lower Grade Glioma; LIHC, Liver
hepatocellular carcinoma; LUAD, Lung adenocarcinoma; LUSC, Lung
squamous cell carcinoma; MESO, Mesothelioma; OV, Ovarian serous
cystadenocarcinoma; PAAD, Pancreatic adenocarcinoma; PCPG,
Pheochromocytoma and Paraganglioma; PRAD, Prostate adenocarcinoma;
READ, Rectum adenocarcinoma; SARC, Sarcoma; SKCM, Skin Cutaneous
Melanoma; STAD, Stomach adenocarcinoma; TGCT, Testicular Germ Cell
Tumors; THCA, Thyroid carcinoma; THYM, Thymoma; UCEC, Uterine
Corpus Endometrial Carcinoma; UCS, Uterine Carcinosarcoma; UVM,
Uveal Melanoma. |
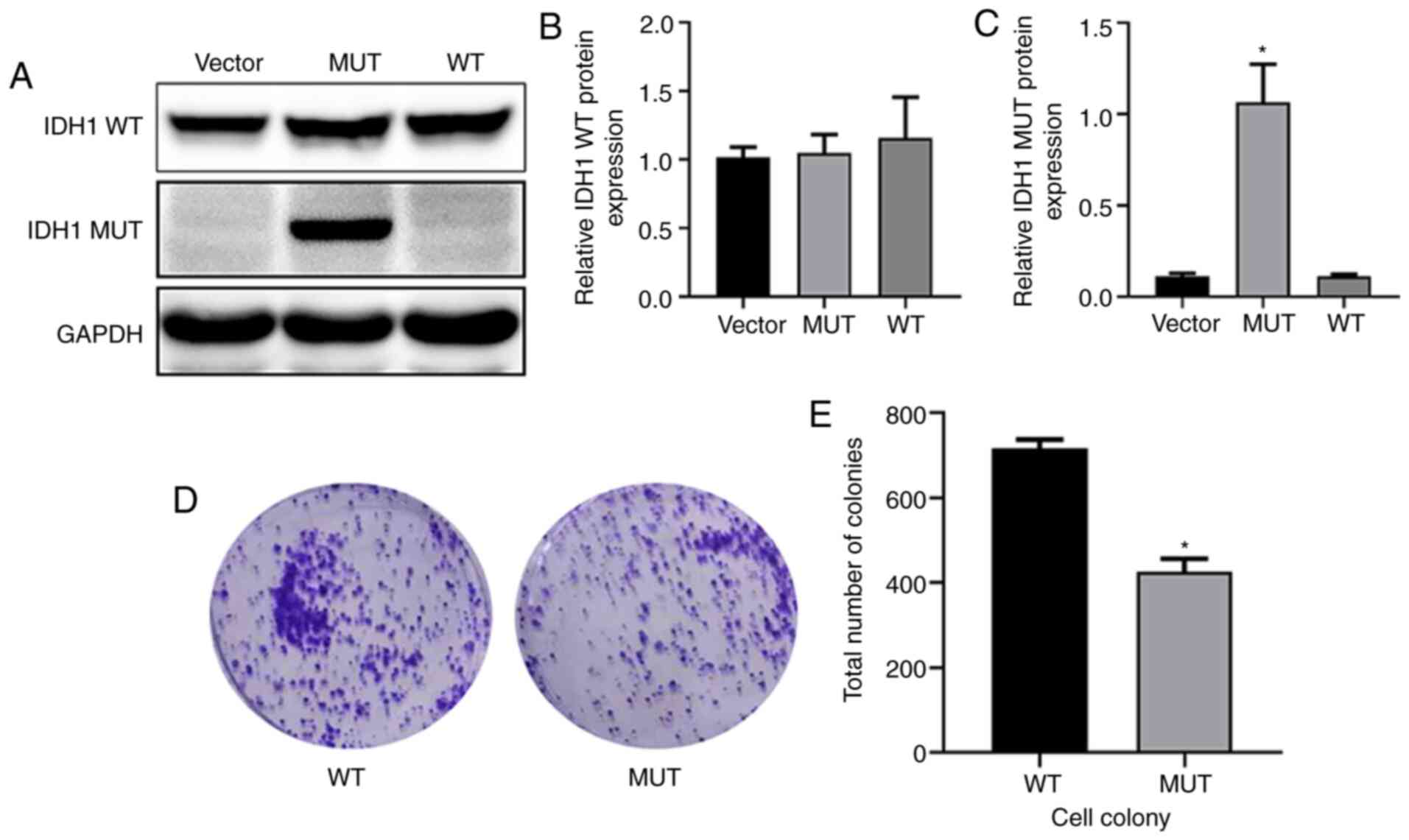
Introduction and expression analysis
of WT and MUT IDH1 genes in U87-MG cells
U87-MG cells were transfected with lentivirus.
Western blot analysis (Fig. 2A)
indicated that after 72 h of lentiviral infection, IDH1 WT protein
levels in WT U87-MG cells were slightly higher than those in the
MUT U87-MG cells (Fig. 2B). The
IDH1 MUT protein was highly expressed in the MUT group (Fig. 2C). The effect of IDH1 gene mutation
on U87-MG cell proliferation was detected through colony formation
experiments, and the results indicated that significantly fewer new
colonies were formed in the MUT group than in the WT group
(Fig. 2D and E). The current
results indicated that a cell model of IDH1 lentivirus transfection
was successfully established and that IDH1 R132H mutation
influenced the colony formation of glioma cells.
Effects of IDH1 gene mutation on the
cell cycle of U87-MG cells
Cell cycle distribution was detected by flow
cytometry. In the WT group, 34.41% of cells were in S phase, while
in the MUT group, 19.39% of cells were in S phase (Fig. 3A-C and G). The proportion of cells
in S phase in the MUT group was markedly lower than those in the WT
and Vector groups (Fig. 3A-C and
G). The proportion of cells in G1 phase in the MUT
group was 72.83%, which was higher than that in the WT group
(57.83%); additionally, there was no marked difference in the
proportion of cells in G2 phase among the groups
(Fig. 3A-C and G), suggesting that
the IDH1 R132H mutation may inhibit cells from entering S phase. To
further describe the changes in the cell cycle, the expression
levels of S phase-associated proteins (Cyclin A and CDK2),
G2/M phase-associated protein (Cyclin B1), G1
phase-associated proteins (Cyclin D3 and CDK4), inhibition of early
G1 phase P18 protein via inhibiting Cyclin D-CDK4/6
(44,45) and inhibition of late G1
phase P27 and P21 proteins via Cyclin E-CDK2 (46) were analyzed by western blot analysis
(Fig. 3D). The results revealed
that the protein expression levels of CyclinA and CDK2 were
significantly lower in the MUT group than in the WT and vector
groups (Fig. 3E and F). There were
no significant differences in CyclinB1 expression among the groups
(Fig. 3H). The expression levels of
Cyclin D3 and CDK4 were significantly higher in the MUT group
(Fig. 3I and J). The protein
expression levels of P18 protein in the MUT group were
significantly lower than in the WT group, and the expression levels
of P27 and P21 protein in the MUT group were significantly higher
than those in the WT and vector groups (Fig. 3K-M). Therefore, the flow cytometry
results were verified by detecting the protein expression levels of
relevant cyclins, indicating that the IDH1 R132H mutation may
affect the regulation of the cell cycle.
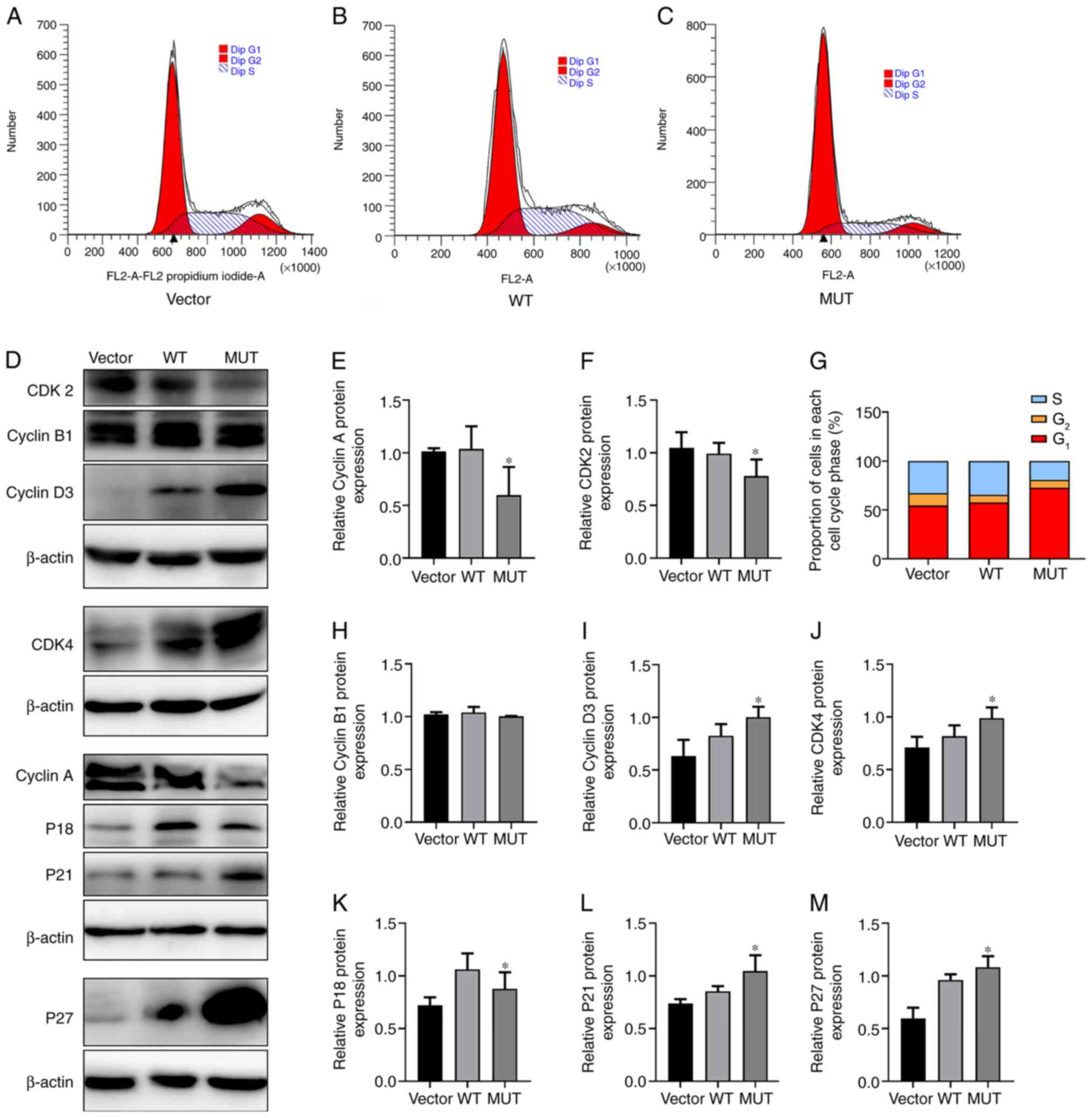 | Figure 3.Effect of IDH1 gene mutation on cell
cycle. Changes in (A) vector, (B) WT and (C) MUT U87-MG cells after
transfection with lentivirus. The cell cycle changes in each group
were detected by flow cytometry. (D) Protein expression levels of
CyclinA, CyclinB1, CyclinD3, CDK2, CDK4, P18, P21 and P27 in each
group of U87-MG cells were detected by western blot analysis.
Quantification of relative protein expression levels of (E) CyclinA
and (F) CDK2. (G) Proportion of cells in each cell cycle phase in
each group. Semi-quantification of relative protein expression
levels of (H) CyclinB1, (I) CyclinD3, (J) CDK4, (K) P18, (L) P21
and (M) P27. *P<0.05 vs. WT. MUT, mutant; WT, wild-type; IDH1,
isocitrate dehydrogenase 1. |
Effect of IDH1 gene mutation on
biological rhythm genes in U87-MG cells
Abnormal clock gene expression and circadian rhythm
disorders are closely associated with the occurrence and
development of tumors (47).
Western blot analysis revealed that the expression levels of the
biological rhythm genes BMAL1 and CLOCK in the IDH1 MUT group were
significantly lower than those in the WT group (Fig. 4A-C). Transcriptionally, the clock is
driven by positive regulators of the loop. Basic helix-loop-helix
heterodimeric transcription factors (CLOCK/BMAL1 or BMAL1/NPAS2)
regulate the expression levels of key circadian genes, including
CRY genes (Cry1 and Cry2) and PER genes (PER1, PER2, and PER3),
which are negative regulators of the circadian loop (48,49).
CRY and PER form a transcriptional repressor complex that enters
the nucleus to repress CLOCK/BMAL1 activity, thus creating a
negative feedback loop to control the clock (50). Therefore, the expression levels of
negative regulators of clock genes were also examined. The negative
regulatory factors of biological rhythm PER1, PER2, PER3, Cry1 and
Cry2 were significantly downregulated compared with the WT group
(Fig. 4D-I). These results
indicated that the IDH1 R132H mutation may change the expression
levels of biological rhythm genes and may participate in the
regulation of the cell biological rhythm.
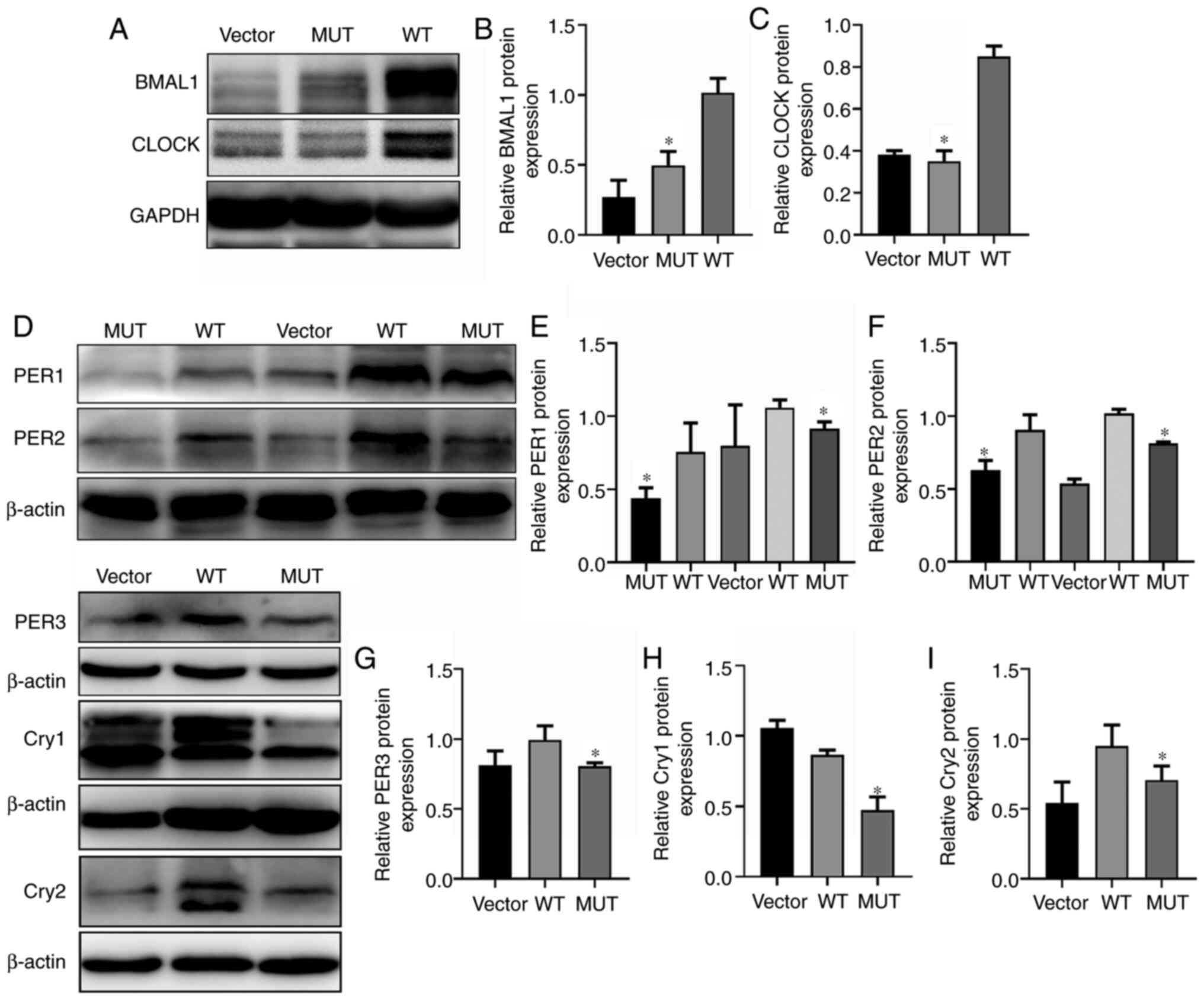 | Figure 4.Effects of IDH1 gene mutation on
biological rhythm genes. (A) Protein expression levels of BMAL1 and
CLOCK in the WT, MUT and vector groups. Quantification of relative
protein expression levels of (B) BMAL1 and (C) CLOCK. (D) Protein
expression levels of PER1, PER2, PER3, CRY1 and CRY2 in the WT, MUT
and vector groups. Quantification of relative protein expression
levels of (E) PER1, (F) PER2, (G) PER3, (H) Cry1 and (I) Cry2.
*P<0.05 vs. WT. MUT, mutant; WT, wild-type; IDH1, isocitrate
dehydrogenase 1; BMAL1, Brain-Muscle Arnt-Like protein 1; CLOCK,
Circadian Locomotor Output Cycles Kaput; PER, Period; Cry,
Cryptochrom. |
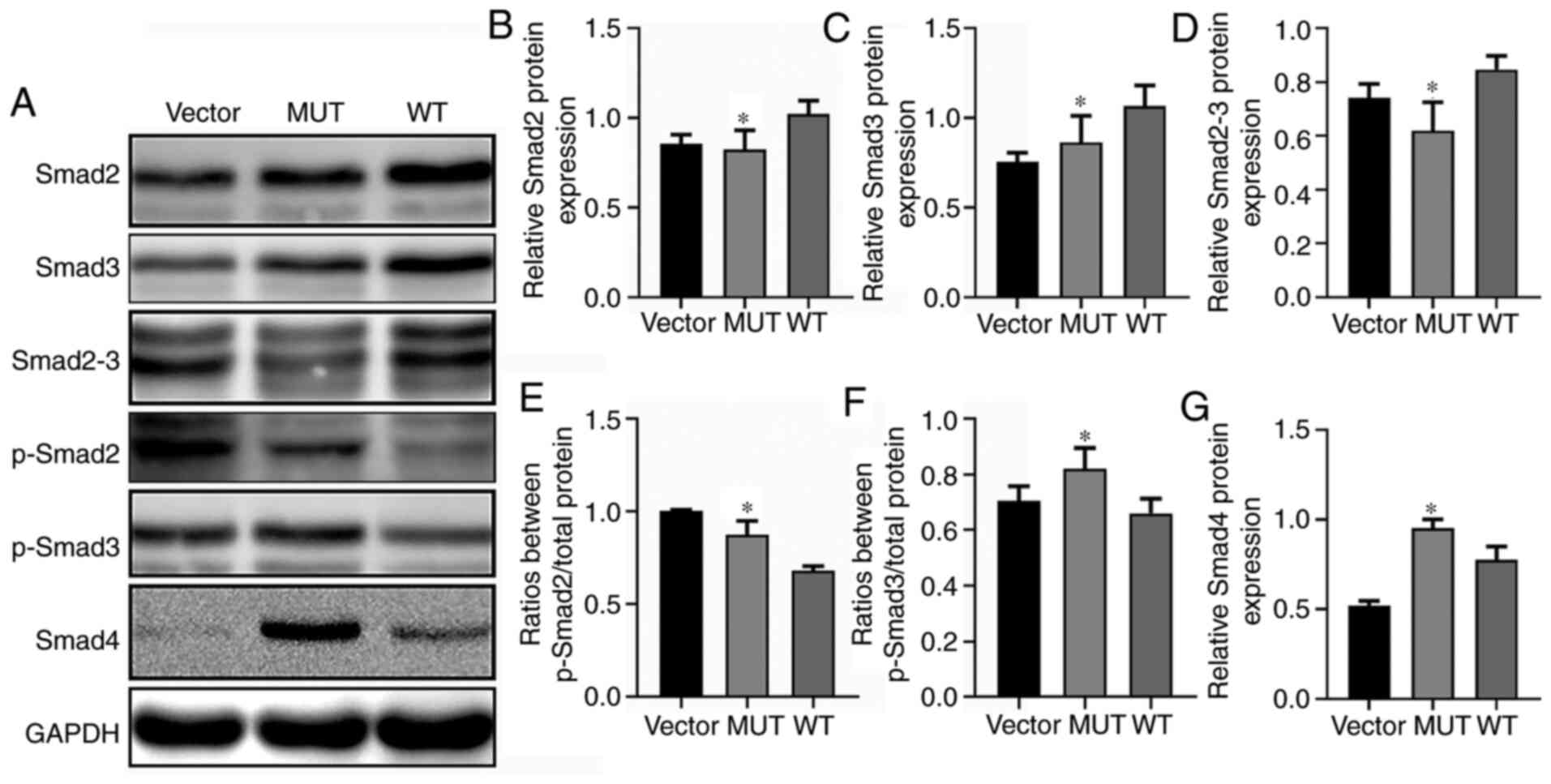
Effects of IDH1 gene mutation on Smad
signaling molecules
IDH1 regulates the TGF-β signaling feedback loop
through α-ketoglutarate (α-KG) and TGF-βR)-IDH1-Canine adenovirus 1
(Cav1) to enhance TGF-β signaling in a regulatory network between
cellular signaling and cell metabolism (51). Additionally, TGF-β is an important
regulator of the physiological clock (52,53).
Therefore, the present study further investigated any changes in
the TGF-β/Smad signaling pathway in IDH1 R132H mutated glioma
cells. Western blot assays were used to detect the effects of MUT
and WT IDH1 on the levels of p-Smad2, p-Smad3, Smad2, Smad2-3,
Smad3 and Smad4 in U87-MG cells (Fig.
5A). Compared with the WT group, the MUT group exhibited
significantly decreased protein expression levels of Smad2, Smad3
and Smad2-3 (Fig. 5B-D) and
significantly increased levels of p-Smad2 and p-Smad3 (Fig. 5E and F). Furthermore, Smad4
expression was significantly upregulated in the MUT group compared
with in the WT group (Fig. 5G).
TGF-β initiates signaling pathways by phosphorylating Smad2 and
Smad3, and phosphorylated Smad2 and Smad3 bind to Smad4 to form
complexes that are transferred to the nucleus to interact with
multiple transcription factors in order to induce cellular
responses (54,55). Thus, it was hypothesized that the
IDH1 R132H mutation may affect biological rhythm genes of glioma
cells through the TGF-β/Smad signaling pathway.
Discussion
Glioma arises from glial cells and most often occurs
in the brain (56). Glioma accounts
for ~30% of central nervous system tumors and 80% of malignant
brain tumors (57). It is
characterized with highly infiltrative growth and a poor prognosis.
The most common genetic changes in glioma include IDH1/2 mutations,
TP53 mutations and 1p/19q heterozygous deletion, which are present
in most cases (>90%) of glioma (58,59).
The most common and early genetic change in glioma is the IDH1/2
mutation. The most common IDH1 mutation results in the amino acid
substitution of R132, which is located at the active site of the
enzyme (60). IDH1 mutated glioma
cells have a reduced proliferative rate (61). Numerous diseases follow circadian
rhythms, and there is growing evidence that circadian disruption
may be a risk factor for cancer in humans (62). A previous study has reported that
the BMAL1 gene serves a role as a potential tumor suppressor gene
in pancreatic cancer by activating the P53 tumor suppressor
signaling pathway (63). Circadian
rhythm genes activate certain pathways in tumors or are activated
by certain pathways to affect the development of tumors. β-catenin
showed an increased expression in NIH-3T3 cells after BMAL1
overexpression, indicating that activation of the canonical Wnt
pathway may be the mechanism underlying the effect of the circadian
clock gene BMAL on promoting cell proliferation (64). BMAL1 suppresses cancer cell invasion
by blocking the PI3K-Akt-MMP-2 signaling pathway (65).
However, to the best of our knowledge, there are no
studies on the mechanism between IDH1 R132H mutation and biological
rhythm genes. Therefore, the present study aimed to describe the
role of biological rhythm genes in glioma cells with IDH1 R132H
mutation to determine how the IDH1 R132H mutation affects
biological rhythm genes and the proliferation of glioma cells.
The current study revealed that the IDH1 R132H
mutation in U87-MG cells decreased the number of new colonies
formed by tumor cells. The effect of the IDH1 R132H mutation on the
cell cycle was further investigated, leading to increased cells in
G1 phase and decreased cells in S phase. The effect of
the mutation on cyclins was also examined. The expression levels of
various cyclically related proteins was altered, including the S
phase-associated proteins Cyclin A and CDK2. No change in Cyclin B1
expression in G2/M phase was observed. The expression
levels of the G1 phase-associated proteins Cyclin D3 and
CDK4 were increased in the MUT group compared with in the WT group.
The protein expression levels of P18 were decreased, while those of
P21 and P27 were increased in the MUT group compared with in the WT
group. Subsequently, the effect of the IDH1 R132H mutation was
examined on biological rhythm genes. To the best of our knowledge,
the current study was the first to analyze the effect of the IDH1
R132H mutation on biological rhythm. The IDH1 R132H mutation
significantly decreased the protein expression levels of both BMAL1
and CLOCK compared with in the WT group, as well as the expression
levels of the negative regulation factors PER1, PER2, PER3, Cry1
and Cry2. A previous study has demonstrated that the disturbance of
the circadian clock has a strong influence on tumor transformation
and growth by affecting various cancer regulatory signaling
pathways, including cell cycle, apoptosis and metabolism (40). The detection of biological rhythm
genes in the present study revealed that IDH1 R132H mutation
inhibited the protein expression levels of CLOCK and BMAL1, and
further inhibited PER and CRY proteins. Therefore, it was shown
that the IDH1 R132H mutation has an effect on the expression of
biological rhythm genes and has been confirmed in in vitro
experiments.
Furthermore, how the IDH1 R132H mutation may affect
biological rhythm genes was further investigated. Previous studies
have shown that the TGF-β signaling pathway is closely associated
with the occurrence of glioma (66,67).
TGF-β is a factor that strongly inhibits the proliferation of
epithelial, astrocyte and immune cells, and is considered a tumor
suppressor (68). Some tumors
acquire mutations in components of the TGF-β signaling pathway to
evade the TGF-β cellular inhibitory response (69). On the other hand, in some malignant
tumors, including glioma, the ability of TGF-β to inhibit cell
proliferation and maintain the integrity of the TGF-β signaling
pathway is selectively lost (70).
TGF-β signaling decreases differences in IDH1 expression by
normalizing the Smad signaling pathway, and inhibition of Cav1
expression by IDH1 is regulated by α-KG epigenetic regulation.
Finally, downregulation of Cav1 expression interrupts the
degradation of TGFBR and enhances the Smad signal. IDH1 regulates
the TGF-β signaling feedback loop through α-KG and TGFBR-IDH1-Cav1
to enhance TGF-β signaling in a regulatory network between cellular
signaling and cell metabolism (51). In the present study, it was observed
that the IDH1 R132H mutation significantly decreased the protein
expression levels of Smad2, Smad3 and Smad2-3, and significantly
upregulated the levels of p-Smad2, p-Smad3 and Smad4. A previous
study has suggested that TGF-β is a multipotent cytokine that
controls tissue homeostasis and embryonic development. TGF-β binds
and activates a membrane receptor serine threonine kinase complex
to phosphorylate Smad2 and Smad3; after phosphorylation, Smad
proteins accumulate in the nucleus and form complexes with
transcription factors, such as Smad4, to regulate transcription
(54). The present study revealed
that the IDH1 R132H mutation affected the TGF-β/Smad signaling
pathway. Additionally, TGF-β is an important regulator of the
physiological clock. A previous study has demonstrated that TGF-β,
by regulating the expression of positive and negative regulators of
circadian rhythm oscillation, serves a vital role in regulating
circadian rhythm. Adenovirus-mediated TGF-β expression can
significantly induce BMAL1 and NPAS2 expression (71). It has been demonstrated that the
expression levels of the TGF-β-activated transcription factor Smad3
display similar expression patterns with BMAL1, and that Smad3
functions as an upstream molecule of BMAL1, explaining how TGF-β
induces BMAL1 expression (53).
However, TGF-β strongly inhibits the expression levels of PER1,
PER2, PER3, Rev-erbα, retinoic acid receptor-related orphan
receptor α and D-site albumin promoter-binding protein (71). Therefore, increased expression
levels of TGF-β and negative regulators of the circadian clock
prolong the arousal cycle (52).
The impact of CLOCK and BMAL1 on cancer pathogenesis is highly
context- and disease-dependent (50). For instance, CLOCK or BMAL1 provide
tumor suppressor-like functions in prostate, breast, ovarian and
pancreatic cancer, but exhibit tumor-promoting roles in colorectal
cancer and acute myeloid leukemia (50,72).
In glioma, CLOCK or BMAL1 are tumor-promoting factors that regulate
glioma cell proliferation and migration via regulating the NF-κB
signaling pathway (73), and can
support glioma stem cell function via regulation of anabolic
metabolism (74). Therefore, the
current study hypothesized that the IDH1 R132H mutation may affect
biological rhythm genes through the TGF-β/Smad signaling pathway,
thus affecting the proliferation of glioma cells.
In summary, the IDH1 gene serves crucial roles in
the occurrence and development of numerous types of tumor (75), and imbalance in the circadian clock
plays an important role in the development of malignant tumors,
including glioma (76). The present
study revealed that the IDH1 R132H mutation affected the expression
levels of cyclin and biological rhythm genes, and thus may affect
the occurrence of glioma. Based on the current results, it was
hypothesized that IDH1 mutation may affect the expression levels of
biological rhythm genes of cells through the TGF-β/Smad signaling
pathway. To the best of our knowledge, the present results are the
first to report the association between IDH1 R132H mutation and
biological rhythm genes, as well as the effect on glioma cell
proliferation and the possible underlying mechanisms. However,
further studies are required to confirm the current results.
Acknowledgements
Not applicable.
Funding
The present study was funded by the National Natural
Science Foundation of China (grant no. 81560501), the Ningxia
Natural Science Foundation (grant no. 2020AAC03351) and the Ningxia
Innovation Team of the foundation and clinical researches of
diabetes and its complications (grant no. NXKJT2019010).
Availability of data and materials
The datasets used and/or analyzed during the current
study are available from the corresponding author on reasonable
request.
Authors' contributions
XC and SH designed the experiments, guided the study
and revised the draft. YW performed the experiments and wrote the
manuscript with support from all the other authors. YG and NZ
performed the data analysis and contributed to the writing of the
manuscript. HY, JY and JM participated in some of the experiments
and in data collection. FW and HX participated in some of the data
analyses. XC, YW and YG confirmed the authenticity of the raw data.
All authors read and approved the final manuscript.
Ethics approval and consent to
participate
Not applicable.
Patient consent for publication
Not applicable.
Competing interests
The authors declare that they have no competing
interests.
References
|
1
|
Sjoblom T, Jones S, Wood LD, Parsons DW,
Lin J, Barber TD, Mandelker D, Leary RJ, Ptak J, Silliman N, et al:
The consensus coding sequences of human breast and colorectal
cancers. Science. 314:268–274. 2006. View Article : Google Scholar : PubMed/NCBI
|
|
2
|
Mardis ER, Ding L, Dooling DJ, Larson DE,
McLellan MD, Chen K, Koboldt DC, Fulton RS, Delehaunty KD, McGrath
SD, et al: Recurring mutations found by sequencing an acute myeloid
leukemia genome. N Engl J Med. 361:1058–1066. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
3
|
Cardaci S and Ciriolo MR: TCA cycle
defects and cancer: When metabolism tunes redox state. Int J Cell
Biol. 2012:1618372012. View Article : Google Scholar : PubMed/NCBI
|
|
4
|
Pastore M, Lori G, Gentilini A, Taddei ML,
Di Maira G, Campani C, Recalcati S, Invernizzi P, Marra F and Raggi
C: Multifaceted aspects of metabolic plasticity in human
cholangiocarcinoma: An overview of current perspectives. Cells.
9:5962020. View Article : Google Scholar
|
|
5
|
Bosnyák E, Michelhaugh SK, Klinger NV,
Kamson DO, Barger GR, Mittal S and Juhász C: Prognostic molecular
and imaging biomarkers in primary glioblastoma. Clin Nucl Med.
42:341–347. 2017. View Article : Google Scholar : PubMed/NCBI
|
|
6
|
Krell D, Assoku M, Galloway M, Mulholland
P, Tomlinson I and Bardella C: Screen for IDH1, IDH2, IDH3, D2HGDH
and L2HGDH mutations in glioblastoma. PLoS One. 6:e198682011.
View Article : Google Scholar : PubMed/NCBI
|
|
7
|
Yang H, Ye D, Guan KL and Xiong Y: IDH1
and IDH2 mutations in tumorigenesis: Mechanistic insights and
clinical perspectives. Clin Cancer Res. 18:5562–5571. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
8
|
Zhang Y, Lv W, Li Q, Wang Q, Ru Y, Xiong
X, Yan F, Pan T, Lin W and Li X: IDH2 compensates for IDH1 mutation
to maintain cell survival under hypoxic conditions in IDH1-mutant
tumor cells. Mol Med Rep. 20:1893–1900. 2019.PubMed/NCBI
|
|
9
|
Chittaranjan S, Chan S, Yang C, Yang KC,
Chen V, Moradian A, Firme M, Song J, Go NE, Blough MD, et al:
Mutations in CIC and IDH1 cooperatively regulate 2-hydroxyglutarate
levels and cell clonogenicity. Oncotarget. 5:7960–7979. 2014.
View Article : Google Scholar : PubMed/NCBI
|
|
10
|
Hersh DS, Peng S, Dancy JG, Galisteo R,
Eschbacher JM, Castellani RJ, Heath JE, Legesse T, Kim AJ,
Woodworth GF, et al: Differential expression of the TWEAK receptor
Fn14 in IDH1 wild-type and mutant gliomas. J Neurooncol.
138:241–250. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
11
|
Sakai Y, Yang C, Kihira S, Tsankova N,
Khan F, Hormigo A, Lai A, Cloughesy T and Nael K: MRI radiomic
features to predict IDH1 mutation status in gliomas: A machine
learning approach using gradient tree boosting. Int J Mol Sci.
21:80042020. View Article : Google Scholar
|
|
12
|
Kim W and Liau LM: IDH mutations in human
glioma. Neurosurg Clin N Am. 23:471–480. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
13
|
Ducray F, El Hallani S and Idbaih A:
Diagnostic and prognostic markers in gliomas. Curr Opin Oncol.
21:537–542. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
14
|
Bergo E, Lombardi G, Pambuku A, Della
Puppa A, Bellu L, D'Avella D and Zagonel V: Cognitive
rehabilitation in patients with gliomas and other brain tumors:
State of the Art. Biomed Res Int. 2016:30418242016. View Article : Google Scholar : PubMed/NCBI
|
|
15
|
Lu C, Ward PS, Kapoor GS, Rohle D, Turcan
S, Abdel-Wahab O, Edwards CR, Khanin R, Figueroa ME, Melnick A, et
al: IDH mutation impairs histone demethylation and results in a
block to cell differentiation. Nature. 483:474–478. 2012.
View Article : Google Scholar : PubMed/NCBI
|
|
16
|
Chen Z, Liu P, Li C, Luo Y, Chen I, Liang
W, Chen X, Feng Y, Xia H and Wang F: Deregulated expression of the
clock genes in gliomas. Technol Cancer Res Treat. 12:91–97. 2013.
View Article : Google Scholar : PubMed/NCBI
|
|
17
|
Luo Y, Wang F, Chen LA, Chen XW, Chen ZJ,
Liu PF, li FF, Li CY and Liang W: Deregulated expression of cry1
and cry2 in human gliomas. Asian Pac J Cancer Prev. 13:5725–5728.
2012. View Article : Google Scholar : PubMed/NCBI
|
|
18
|
Chang WH and Lai AG: Timing gone awry:
Distinct tumour suppressive and oncogenic roles of the circadian
clock and crosstalk with hypoxia signalling in diverse
malignancies. J Transl Med. 17:1322019. View Article : Google Scholar : PubMed/NCBI
|
|
19
|
Chan AB, Huber AL and Lamia KA:
Cryptochromes modulate E2F family transcription factors. Sci Rep.
10:40772020. View Article : Google Scholar : PubMed/NCBI
|
|
20
|
Young MW and Kay SA: Time zones: A
comparative genetics of circadian clocks. Nat Rev Genet. 2:702–715.
2001. View Article : Google Scholar : PubMed/NCBI
|
|
21
|
Lowrey PL and Takahashi JS: Genetics of
the mammalian circadian system: Photic entrainment, circadian
pacemaker mechanisms, and posttranslational regulation. Ann Rev
Genet. 34:533–562. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
22
|
Dunlap JC: Molecular bases for circadian
clocks. Cell. 96:271–290. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
23
|
Kume K, Zylka MJ, Sriram S, Shearman LP,
Weaver DR, Jin X, Maywood ES, Hastings MH and Reppert SM: mCRY1 and
mCRY2 are essential components of the negative limb of the
circadian clock feedback loop. Cell. 98:193–205. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
24
|
van der Horst GT, Muijtjens M, Kobayashi
K, Takano R, Kanno S, Takao M, de Wit J, Verkerk A, Eker AP, van
Leenen D, et al: Mammalian Cry1 and Cry2 are essential for
maintenance of circadian rhythms. Nature. 398:627–630. 1999.
View Article : Google Scholar : PubMed/NCBI
|
|
25
|
Vitaterna MH, Selby CP, Todo T, Niwa H,
Thompson C, Fruechte EM, Hitomi K, Thresher RJ, Ishikawa T,
Miyazaki J, et al: Differential regulation of mammalian period
genes and circadian rhythmicity by cryptochromes 1 and 2. Proc Natl
Acad Sci USA. 96:12114–12119. 1999. View Article : Google Scholar : PubMed/NCBI
|
|
26
|
Dibner C, Schibler U and Albrecht U: The
mammalian circadian timing system: Organization and coordination of
central and peripheral clocks. Ann Rev Physiol. 72:517–549. 2010.
View Article : Google Scholar
|
|
27
|
Harmer SL, Panda S and Kay SA: Molecular
bases of circadian rhythms. Ann Rev Cell Dev Biol. 17:215–253.
2001. View Article : Google Scholar
|
|
28
|
Oster H: The genetic basis of circadian
behavior. Genes Brain Behav. 5 (Suppl 2):S73–S79. 2006. View Article : Google Scholar
|
|
29
|
Balsalobre A, Brown SA, Marcacci L,
Tronche F, Kellendonk C, Reichardt HM, Schütz G and Schibler U:
Resetting of circadian time in peripheral tissues by glucocorticoid
signaling. Science. 289:2344–2347. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
30
|
Damiola F, Le Minh N, Preitner N, Kornmann
B, Fleury-Olela F and Schibler U: Restricted feeding uncouples
circadian oscillators in peripheral tissues from the central
pacemaker in the suprachiasmatic nucleus. Genes Dev. 14:2950–2961.
2000. View Article : Google Scholar : PubMed/NCBI
|
|
31
|
Yagita K, Tamanini F, van Der Horst GT and
Okamura H: Molecular mechanisms of the biological clock in cultured
fibroblasts. Science. 292:278–281. 2001. View Article : Google Scholar : PubMed/NCBI
|
|
32
|
Shearman LP, Sriram S, Weaver DR, Maywood
ES, Chaves I, Zheng B, Kume K, Lee CC, van der Horst GT, Hastings
MH and Reppert SM: Interacting molecular loops in the mammalian
circadian clock. Science. 288:1013–1019. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
33
|
Mazzoccoli G, Vinciguerra M, Papa G and
Piepoli A: Circadian clock circuitry in colorectal cancer. World J
Gastroenterol. 20:4197–4207. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
34
|
Blakeman V, Williams JL, Meng QJ and
Streuli CH: Circadian clocks and breast cancer. Br Cancer Res.
18:892016. View Article : Google Scholar
|
|
35
|
Cao Q, Gery S, Dashti A, Yin D, Zhou Y, Gu
J and Koeffler HP: A role for the clock gene per1 in prostate
cancer. Cancer Res. 69:7619–7625. 2009. View Article : Google Scholar : PubMed/NCBI
|
|
36
|
Oda A, Katayose Y, Yabuuchi S, Yamamoto K,
Mizuma M, Shirasou S, Onogawa T, Ohtsuka H, Yoshida H, Hayashi H,
et al: Clock gene mouse period2 overexpression inhibits growth of
human pancreatic cancer cells and has synergistic effect with
cisplatin. Anticancer Res. 29:1201–1209. 2009.PubMed/NCBI
|
|
37
|
Zhang S, Zhang J, Deng Z, Liu H, Mao W,
Jiang F, Xia Z and Li JD: Circadian clock components RORα and Bmal1
mediate the anti-proliferative effect of MLN4924 in osteosarcoma
cells. Oncotarget. 7:66087–66099. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
38
|
Papagiannakopoulos T, Bauer MR, Davidson
SM, Heimann M, Subbaraj L, Bhutkar A, Bartlebaugh J, Vander Heiden
MG and Jacks T: Circadian rhythm disruption promotes lung
tumorigenesis. Cell Metab. 24:324–331. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
39
|
Yang MY, Lin PM, Hsiao HH, Hsu JF, Lin HY,
Hsu CM, Chen IY, Su SW, Liu YC and Lin SF: Up-regulation of PER3
expression is correlated with better clinical outcome in acute
leukemia. Anticancer Res. 35:6615–6622. 2015.PubMed/NCBI
|
|
40
|
Farshadi E, van der Horst GTJ and Chaves
I: Molecular links between the circadian clock and the cell cycle.
J Mol Biol. 432:3515–3524. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
41
|
Scheving LE, Tsai TH and Scheving LA:
Chronobiology of the intestinal tract of the mouse. Am J Anatomy.
168:433–465. 1983. View Article : Google Scholar
|
|
42
|
Farshadi E, Yan J, Leclere P, Goldbeter A,
Chaves I and van der Horst GTJ: The positive circadian regulators
CLOCK and BMAL1 control G2/M cell cycle transition
through Cyclin B1. Cell Cycle. 18:16–33. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
43
|
Shan DZ, Zhang C and Cao XM: Preparation
and expression of isocitrate dehydrogenase 1 and mutant recombinant
lentivirus. Chin J Neuroanatomy. 36:200–206. 2020.
|
|
44
|
Yu H, Yuan Y, Shen H and Cheng T:
Hematopoietic stem cell exhaustion impacted by p18 INK4C and p21
Cip1/Waf1 in opposite manners. Blood. 107:1200–1206. 2006.
View Article : Google Scholar : PubMed/NCBI
|
|
45
|
Yuan Y, Shen H, Franklin DS, Scadden DT
and Cheng T: In vivo self-renewing divisions of haematopoietic stem
cells are increased in the absence of the early G1-phase
inhibitor, p18INK4C. Nat Cell Biol. 6:436–442. 2004. View Article : Google Scholar : PubMed/NCBI
|
|
46
|
Sherr CJ: Cancer cell cycles. Science.
274:1672–1677. 1996. View Article : Google Scholar : PubMed/NCBI
|
|
47
|
Mandal AS, Biswas N, Karim KM, Guha A,
Chatterjee S, Behera M and Kuotsu K: Drug delivery system based on
chronobiology-A review. J Control Release. 147:314–325. 2010.
View Article : Google Scholar : PubMed/NCBI
|
|
48
|
Zhao Q, Zheng G, Yang K, Ao YR, Su XL, Li
Y and Lv XQ: The clock gene PER1 plays an important role in
regulating the clock gene network in human oral squamous cell
carcinoma cells. Oncotarget. 7:70290–70302. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
49
|
Qu M, Duffy T, Hirota T and Kay SA:
Nuclear receptor HNF4A transrepresses CLOCK:BMAL1 and modulates
tissue-specific circadian networks. Proc Natl Acad Sci USA.
115:E12305–E12312. 2018. View Article : Google Scholar : PubMed/NCBI
|
|
50
|
Shafi AA and Knudsen KE: Cancer and the
circadian clock. Cancer Res. 79:3806–3814. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
51
|
Hou X, Zhang J, Wang Y, Xiong W and Mi J:
TGFBR-IDH1-Cav1 axis promotes TGF-beta signalling in
cancer-associated fibroblast. Oncotarget. 8:83962–83974. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
52
|
Lopez M, Meier D, Muller A, Franken P,
Fujita J and Fontana A: Tumor necrosis factor and transforming
growth factor beta regulate clock genes by controlling the
expression of the cold inducible RNA-binding protein (CIRBP). J
Biol Chem. 289:2736–2744. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
53
|
Sato F, Sato H, Jin D, Bhawal UK, Wu Y,
Noshiro M, Kawamoto T, Fujimoto K, Seino H, Morohashi S, et al:
Smad3 and Snail show circadian expression in human gingival
fibroblasts, human mesenchymal stem cell, and in mouse liver.
Biochem Biophys Res Commun. 419:441–446. 2012. View Article : Google Scholar : PubMed/NCBI
|
|
54
|
Kang Y, He W, Tulley S, Gupta GP,
Serganova I, Chen CR, Manova-Todorova K, Blasberg R, Gerald WL and
Massagué J: Breast cancer bone metastasis mediated by the Smad
tumor suppressor pathway. Proc Natl Acad Sci USA. 102:13909–13914.
2005. View Article : Google Scholar : PubMed/NCBI
|
|
55
|
Dou C, Lee J, Liu B, Liu F, Massague J,
Xuan S and Lai E: BF-1 interferes with transforming growth factor
beta signaling by associating with Smad partners. Mol Cell Biol.
20:6201–6211. 2000. View Article : Google Scholar : PubMed/NCBI
|
|
56
|
Parisot S, Wells W III, Chemouny S, Duffau
H and Paragios N: Concurrent tumor segmentation and registration
with uncertainty-based sparse non-uniform graphs. Med Image Anal.
18:647–659. 2014. View Article : Google Scholar : PubMed/NCBI
|
|
57
|
de Robles P, Fiest KM, Frolkis AD,
Pringsheim T, Atta C, St Germaine-Smith C, Day L, Lam D and Jette
N: The worldwide incidence and prevalence of primary brain tumors:
A systematic review and meta-analysis. Neurooncology. 17:776–783.
2015.
|
|
58
|
Eckel-Passow JE, Lachance DH, Molinaro AM,
Walsh KM, Decker PA, Sicotte H, Pekmezci M, Rice T, Kosel ML,
Smirnov IV, et al: Glioma Groups Based on 1p/19q, IDH, and TERT
promoter mutations in tumors. N Engl J Med. 372:2499–2508. 2015.
View Article : Google Scholar : PubMed/NCBI
|
|
59
|
Miller JJ, Shih HA, Andronesi OC and
Cahill DP: Isocitrate dehydrogenase-mutant glioma: Evolving
clinical and therapeutic implications. Cancer. 123:4535–4546. 2017.
View Article : Google Scholar : PubMed/NCBI
|
|
60
|
Ferreira MSV, Sørensen MD, Pusch S, Beier
D, Bouillon AS, Kristensen BW, Brümmendorf TH, Beier CP and Beier
F: Alternative lengthening of telomeres is the major telomere
maintenance mechanism in astrocytoma with isocitrate dehydrogenase
1 mutation. J Neurooncol. 147:1–14. 2020. View Article : Google Scholar : PubMed/NCBI
|
|
61
|
Karpel-Massler G, Nguyen TTT, Shang E and
Siegelin MD: Novel IDH1-targeted glioma therapies. CNS Drugs.
33:1155–1166. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
62
|
Salavaty A, Mohammadi N, Shahmoradi M and
Naderi Soorki M: Bioinformatic analysis of circadian expression of
oncogenes and tumor suppressor genes. Bioinform Biol Insights.
11:11779322177469912017. View Article : Google Scholar : PubMed/NCBI
|
|
63
|
Jiang W, Zhao S, Jiang X, Zhang E, Hu G,
Hu B, Zheng P, Xiao J, Lu Z, Lu Y, et al: The circadian clock gene
Bmal1 acts as a potential anti-oncogene in pancreatic cancer by
activating the p53 tumor suppressor pathway. Cancer Lett.
371:314–325. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
64
|
Lin F, Chen Y, Li X, Zhao Q and Tan Z:
Over-expression of circadian clock gene Bmal1 affects proliferation
and the canonical Wnt pathway in NIH-3T3 cells. Cell Biochem Funct.
31:166–172. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
65
|
Jung CH, Kim EM, Park JK, Hwang SG, Moon
SK, Kim WJ and Um HD: Bmal1 suppresses cancer cell invasion by
blocking the phosphoinositide 3-kinase-Akt-MMP-2 signaling pathway.
Oncol Rep. 29:2109–2113. 2013. View Article : Google Scholar : PubMed/NCBI
|
|
66
|
Guan F, Kang Z, Wang L, Wang K, Mao BB,
Peng WC, Zhang BL, Lin ZY, Zhang JT and Hu ZQ: Retinol
dehydrogenase 10 promotes metastasis of glioma cells via the
transforming growth factor-β/SMAD signaling pathway. Chin Med J
(Engl). 132:2430–2437. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
67
|
Derynck R and Zhang YE: Smad-dependent and
Smad-independent pathways in TGF-beta family signalling. Nature.
425:577–584. 2003. View Article : Google Scholar : PubMed/NCBI
|
|
68
|
Frei K, Gramatzki D, Tritschler I,
Schroeder JJ, Espinoza L, Rushing EJ and Weller M: Transforming
growth factor-β pathway activity in glioblastoma. Oncotarget.
6:5963–5977. 2015. View Article : Google Scholar : PubMed/NCBI
|
|
69
|
Huse K, Bakkebø M, Wälchli S, Oksvold MP,
Hilden VI, Forfang L, Bredahl ML, Liestøl K, Alizadeh AA, Smeland
EB and Myklebust JH: Role of Smad proteins in resistance to
BMP-induced growth inhibition in B-cell lymphoma. PLoS One.
7:e461172012. View Article : Google Scholar : PubMed/NCBI
|
|
70
|
Sadeghi Y, Tabatabaei Irani P, Rafiee L,
Tajadini M, Amouheidari A and Javanmard SH: Evaluation of rs1982073
polymorphism of transforming growth factor-β1 in glioblastoma. J
Res Med Sci. 24:402019. View Article : Google Scholar : PubMed/NCBI
|
|
71
|
Dong C, Gongora R, Sosulski ML, Luo F and
Sanchez CG: Regulation of transforming growth factor-beta1
(TGF-β1)-induced pro-fibrotic activities by circadian clock gene
BMAL1. Respir Res. 17:42016. View Article : Google Scholar : PubMed/NCBI
|
|
72
|
Puram RV, Kowalczyk MS, de Boer CG,
Schneider RK, Miller PG, McConkey M, Tothova Z, Tejero H, Heckl D,
Järås M, et al: Core circadian clock genes regulate leukemia stem
cells in AML. Cell. 165:303–316. 2016. View Article : Google Scholar : PubMed/NCBI
|
|
73
|
Li A, Lin X, Tan X, Yin B, Han W, Zhao J,
Yuan J, Qiang B and Peng X: Circadian gene Clock contributes to
cell proliferation and migration of glioma and is directly
regulated by tumor-suppressive miR-124. FEBS Lett. 587:2455–2460.
2013. View Article : Google Scholar : PubMed/NCBI
|
|
74
|
Dong Z, Zhang G, Qu M, Gimple RC, Wu Q,
Qiu Z, Prager BC, Wang X, Kim LJY, Morton AR, et al: Targeting
glioblastoma stem cells through disruption of the circadian clock.
Cancer Discov. 9:1556–1573. 2019. View Article : Google Scholar : PubMed/NCBI
|
|
75
|
Liu WS, Chan SH, Chang HT, Li GC, Tu YT,
Tseng HH, Fu TY, Chang HY, Liou HH, Ger LP and Tsai KW: Isocitrate
dehydrogenase 1-snail axis dysfunction significantly correlates
with breast cancer prognosis and regulates cell invasion ability.
Breast Cancer Res. 20:252018. View Article : Google Scholar : PubMed/NCBI
|
|
76
|
Xia HC, Niu ZF, Ma H, Cao SZ, Hao SC, Liu
ZT and Wang F: Deregulated expression of the Per1 and Per2 in human
gliomas. Can J Neurol Sci. 37:365–370. 2010. View Article : Google Scholar : PubMed/NCBI
|